Abstract
The hypoxia-inducible factor-1α (HIF-1α) is a master regulator of the cellular response to decreased oxygen levels. This transcription factor is highly unstable at normal oxygen concentrations and is rapidly stabilized by hypoxia. At normoxia two specific proline residues (Pro402 and Pro563) of mHIF-1α are hydroxylated and recognized by the von Hippel-Lindau E3 ubiquitin ligase (pVHL) complex, which upon binding mediates degradation of the protein. Previous studies have demonstrated that these two proline residues are critical for high affinity binding to pVHL. We have performed a detailed analysis of a mutant form of HIF-1α, where both these proline residues have been mutated, and we have uncovered a novel degradation pathway, to which the HIF-1α mutant protein is not resistant. Our results show that the HIF-1α double proline mutant undergoes ubiquitination and proteasome-dependent degradation, and retains the ability to be stabilized in response to hypoxia and CoCl2 treatment. However in contrast to the wild-type protein, stabilization of the mutant was only observed within short periods of hypoxia exposure (1-2 h). Degradation assays in the presence of the expressed prolyl hydroxylases (PHDs) 1-3 showed that, unlike the wild-type protein, the HIF-1α mutant was resistant to these hydroxylases. However, experiments knocking-down expression of pVHL by RNA interference showed that the HIF-1α mutant is degraded and ubiquitinated by a pVHL-mediated mechanism. In conclusion, we show the first evidence of a novel mechanism of degradation of HIF-1α at normoxia that involves pVHL but is not mediated by PHDs 1-3 or by degradation boxes surrounding Pro402 and Pro563.
Prokaryotic and eukaryotic cells have the ability to sense and adapt to changes in oxygen concentration. In metazoans, oxygen homeostasis is regulated by the hypoxia-inducible factor-1 (HIF-1)2 that activates gene expression in response to hypoxia. HIF-1 target genes encode proteins critical for anaerobic metabolism (e.g. glycolytic enzymes and glucose transporters), angiogenesis (e.g. vascular endothelial growth factor), and erythropoiesis (e.g. erythropoietin). HIF-1 heterodimers consist of a constitutively expressed subunit, Arnt, and an oxygen-regulated factor, HIF-1α. Both HIF-1α protein stability and transcriptional activity are regulated by oxygen levels. At normoxia, two specific proline residues, Pro402 and Pro564 (Pro564 in human HIF-1α, Pro563 in mouse HIF-1α), have been shown to be hydroxylated by a new family of prolyl hydroxylases (PHDs) (1-6). These modified residues are recognized by the von Hippel-Lindau tumor suppressor protein (pVHL) as part of a complex with E3 ubiquitin ligase activity that leads to ubiquitination and proteasome-dependent degradation of HIF-1α (7-12). The transcriptional activity of HIF-1α is also regulated by another hydroxylase, the factor inhibiting HIF-1α (FIH-1) that at normoxia hydroxylates an asparagine residue leading to inhibition of CBP recruitment to the HIF-1α C-terminal transactivation domain (C-TAD) (13, 14). Molecular oxygen is used as a cosubstrate by the different hydroxylases that regulate HIF-1α activity, and therefore these enzymes have been proposed to function as cellular oxygen sensors (15).
In the present work, we show that mutation to alanine of both the Pro402 and Pro563 residues does not result in inhibition of HIF-1α degradation. Thus, this HIF-1α mutant was still degraded at normoxia, and showed hypoxia-induced stabilization. Furthermore, the HIF-1α mutant underwent ubiquitination and was stabilized in response to inhibition of the proteasome proteolytic activity. Although we observed that this mutant was resistant to PHD-dependent degradation, the protein could be stabilized following siRNA-mediated knock-down of pVHL expression levels. These results indicate that, in addition to the previously identified proline residues, additional structures of HIF-1α are involved in the regulation of protein degradation at normoxia.
EXPERIMENTAL PROCEDURES
Plasmid Constructs—pFLAG-PHD1 and pFLAG-PHD3 were cloned from cDNAs generated by RT-PCR using RNA from HeLa cells. The PCR products, flanked by EcoRI/SmaI restriction sites, were inserted into pFLAG-CMV2 (Eastman Kodak Co). pFLAG-PHD2 was cloned by amplification of the PHD2-encoding region from the PHD2-GFP plasmid (16) using PCR. The PCR product carrying EcoRI and BamHI ends was inserted into pFLAG-CMV2. pFLAG-mHIF-1α(391Δ628) (Δoxygen-dependent degradation (ODD) domain) was generated by cloning a PCR fragment spanning amino acids 629-822 carrying a AflII in the 5′-end and KpnI in the 3′-end into pFLAG-mHIF-1α (17) previously digested with AflII (amino acid 390) and KpnI. pFLAG-mHIF-1α.puro and pFLAG-mHIF-1α(P402A/P563A).puro were generated by blunt insertion of a BstXI/BamHI fragment from pGEM-T-FLAG-mHIF-1α (17) or the corresponding mutant into a blunted EcoRI site in pEFIRESpuro (a kind gift from Dr. Murray Whitelaw). The frame orientation was analyzed by restriction digestion. Constructs generated by PCR were completely sequenced using the DYEnamic sequencing kit (Amersham Biosciences). All amino acid mutations were introduced using the QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions, and subsequently screened by sequencing. The hemagglutinin (HA)-tagged ubiquitin expression plasmid, pMT123, was obtained from Dr. D. Bohmann. The pMyc-Arnt was a kind gift from Dr. Katarina Gradin. pT81/HRE-luc, pFLAG-mHIF-1α, and pFLAG-mHIF-1α(P402A/P563A) are described elsewhere (11, 17-18).
Cell Culture and Transient Transfection Experiments—Human embryonic kidney (HEK) 293A and H cells were maintained in Dulbecco's modified Eagle's/F12 (1:1) medium containing glutaMAX I supplemented with 10% fetal calf serum, 50 international units/ml penicillin, and 50 mg/ml streptomycin sulfate. SKRC-7, SKRC-17, and HepG2 cells were grown in RPMI 1640 medium containing l-glutamine supplemented as for HEK 293 cells. All media and growth factors were purchased from Invitrogen. Transient transfections of HEK 293A cells were performed using Lipofectamine (Invitrogen) while transfection of HepG2, and HEK 293H cells was achieved using FuGene 6 (Roche Applied Sciences) according to the manufacturers' instructions. For reporter gene assays, HEK 293A cells were seeded in 6-well plates and allowed to grow for 24 h prior to transfection with 500 ng of pT81/HRE-luc and 100 ng of pFLAG-mHIF-1α or pFLAG-mHIF-1α(P402A/P563A). After transfection, cells were allowed to recover for 18 h and subsequently exposed for 24 h to normoxia (21% O2), hypoxia (1% O2), or CoCl2 (200 μm). Cells were then harvested, and extracts were prepared and analyzed for luciferase activity. The total protein concentration of whole cell extracts was determined by a colorimetric method (Bio-Rad). A pCMV vector was used to keep the DNA concentration constant. For degradation assays, HEK 293A cells were transfected with 1.2 μg of pFLAG-mHIF-1α, or pFLAG-mHIF-1α(P402A/P563A), and increasing amounts of pFLAG-PHD1, pFLAG-PHD2, or pFLAG-PHD3, as detailed in the figure legends. After a recovery period of 24 h, cells were kept at normoxia, or treated with 200 μm CoCl2 for an additional 12-h period. When indicated in figure legends, transfected cells were concomitantly treated with 10 μm of a proteasome inhibitor (MG132, Sigma), during the last 12-h period. Whole cell extracts were prepared using modified RIPA buffer. 25 μg of whole cell extract proteins were separated by SDS-PAGE and analyzed by immunoblotting. For immunoprecipitation experiments, HEK 293A cells were transfected with 50 ng of pFLAG, 500 ng of pFLAG-mHIF-1α, 500 ng of pFLAG-mHIF-1α(P402A/P563A), or 500 ng of pFLAG-mHIF-1α(392Δ622) (ΔODD), together with 900 ng of pVHL, and 100 ng of pHA-ubiquitin. 24 h after transfection, cells were treated with 10 μm MG132 and harvested 12 h later.
Whole Cell Extracts—Cell extracts were prepared using modified RIPA buffer (150 mm NaCl, 50 mm Tris-Cl pH 8.7, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 2 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, 0.2 mm N-ethylmaleimide) containing a protease inhibitor mixture (Roche Applied Science), vortexed for 1 min followed by clarification at 14,000 rpm for 30 min at 4 °C.
Stable Transfections—HEK 293H cells were transfected with pFLAG-mHIF-1α.puro, pFLAG-mHIF-1α(P402A/P563A).puro, or pEFIRESpuro using Lipofectamine. 24 h after transfection, cells were expanded, and selection was initiated adding 1 μg/ml of puromycin (Sigma). Surviving cells were selected for monoclonal expansion and assayed by Western blot under normoxic or CoCl2 (200 μm) treatment as indicated in figure legends. For time course assays, cells were exposed to hypoxia, CoCl2 (200 μm), dimethyloxallyl glycine (DMOG, 1 mm), 2,2′-dipyridyl (200 μm), deferoxamine mesylate (200 μm) (all chemical from Sigma), or kept at normoxia, for constant end-point experiments. Whole cell extracts were prepared using modified RIPA buffer. 25 μg of whole cell extract proteins were separated by SDS-PAGE and analyzed by immunoblotting.
Ubiquitination Assays—HEK 293A cells were cultured for 24 h in 10-cm diameter dishes prior to transfection of 4.5 μg of pFLAG, pFLAG-mHIF-1α or pFLAG-mHIF-1α(P402A/P563A), and 0.25 μg of pHA-ubiquitin (pMT123). After 24 h of transfection, cells were kept at normoxia or treated with CoCl2 (200 μm) for 12 h in the presence or absence of MG132 (10 μm). Whole cell extracts were prepared using modified RIPA buffer as described previously. Protein G-Sepharose beads were blocked with 1% bovine serum albumin in TBS (150 mm NaCl, 50 mm Tris-Cl, pH 7.4) before immunized with anti-FLAG antibodies (Sigma). 300 μg of whole cell extract protein was immunoprecipitated onto the immunized beads at 4 °C for 16 h. After washing three times with RIPA/TBS (1:1), proteins were eluted at room temperature for 90 min with 2.5 μg/μl of FLAG-peptide (Sigma). Eluates and 25 μg of whole cell extract proteins were separated by SDS-PAGE and analyzed by immunoblotting.
RNA Interference Assays—Stable cell lines expressing FLAG-mHIF-1α or FLAG-mHIF-1α(P402A/P563A) were transfected with 200 nm negative control siRNA (Qiagen, AllStars negative control siRNA) or VHL siRNA (Qiagen, SI02664550; Dharmacon, J-003936-11) using Oligofectamine (Invitrogen) according to the manufacturer's instructions. 60 h after transfection, cells were treated with CoCl2 (200 μm) or kept at normoxia for an additional 12-h period. For protein analysis, whole cell extracts were prepared using modified RIPA buffer, and immunoblotting assays were performed using 50 μg of whole cell extract protein. For ubiquitination assays, stably transfected HEK 293H were cultured for 24 h in 10-cm diameter dishes prior to transfection with 200 nm negative control or VHL siRNAs. Cells were allowed to recover for 24 h prior to transfection of 1.2 μg of pHA-ubiquitin. 36 h after transfection with pHA-ubiquitin cells were treated for 8 h with 15 μm of MG132 and harvested. Whole cell extracts were prepared by sonication with a buffer containing 200 mm KCl and 0.05% Nonidet P-40. For precipitation of ubiquitinated-HIF-1α proteins, 5 μl of anti-FLAG antibody was incubated with 25 μl of protein G-Sepharose beads after blocking with 1% bovine serum albumin and followed by incubation with 1.0 mg of whole cell extract overnight at 4 °C. Subsequent washing was carried out in a buffer containing 300 mm KCl and 0.1% Nonidet P-40. Precipitated complexes were eluted from the beads using 2.5 μg/μl of FLAG-peptide in a buffer containing 100 mm KCl and 0.1% Nonidet P-40. Eluates were analyzed by SDS-PAGE and immunoblotting.
RT-PCR Experiments—Total RNAs were isolated using PureLink™ (Invitrogen) under different conditions as depicted in the figure legends. First-strand cDNA was generated from 1 μg of total RNA, using random primers by SuperScript™ II Reverse Transcriptase (Invitrogen). PCR was performed using Platinum® Pfx DNA Polymerase (Invitrogen), and 1 μl of cDNA as template. All protocols were preformed according to the manufacturer's instructions. Specific primers for the different cDNAs were as follows: (5′-3′): FLAG_F (GGACTACAAAGACGATGACGAC); HIF-1α_R (CGTCATCTGTTAGCACCATC); VHL_F (TCTTCTGCAATCGCAGTC); VHL_R (CAAAGGTGACCTCGGTAG); PGK-1_F (TGTGGCTTCTGGCATACCTG); PGK-1_R (AGGAGCTCCAAACTGGCACC); actin_F (GACAGGATGCAGAAGGAGAT); actin_R (TTGCTGATCCACATCTGCTG).
Immunoprecipitation Assays—Cells were lysed using sonication in a buffer with 180 mm KCl and 0.05% Nonidet P-40. For precipitation of HIF-1α protein/pVHL complexes, 5 μl of anti-FLAG antibody was incubated with 25 μl of protein G-Sepharose beads after blocking with 1% bovine serum albumin, and followed by incubation with 1.5 mg of whole cell extracts overnight at 4 °C. Subsequent washing was carried out in a buffer containing 250 mm KCl and 0.1% Nonidet P-40. Precipitated complexes were eluted from the beads using 2.5 μg/μl of FLAG-peptide in a buffer containing 100 mm KCl and 0.1% Nonidet P-40. Eluates were analyzed by SDS-PAGE and immunoblotting.
Immunoblotting Assays—After separation by SDS-PAGE, samples were blotted onto nitrocellulose filters, and blocking was performed using 5% nonfat milk in TBS (nfm/TBS). Filters were then incubated with primary antibodies: anti-FLAG (1:500) in 5% nfm/TBS (Sigma), anti-VHL (1:500) in 5% nfm/TBS (Abcam, ab 28434), anti-hemagglutinin (1:500) in 5% nfm/TBS (Santa Cruz Biotechnology), anti-Myc (1:500) in 1% nfm/TBS (Clontech), anti-HIF-1α (1:500) in 1% nfm/TBS (Abcam, ab 6489), anti-Ubiquitin (1:50) in 5% nfm/TBS (Dako), and anti-actin (1:2500) in 5% nfm/TBS (Abcam). Secondary antibodies were used at a 1:2000 dilution in 1% nfm/TBS of anti-mouse or anti-rabbit IgG-horseradish peroxidase conjugates (Amersham Biosciences). Extensive washings were performed after both incubations with primary or secondary antibodies using TBS containing 0.05% Tween-20. Proteins were visualized using enhanced chemiluminescence (Amersham Biosciences).
RESULTS
Mutation of Both Pro402 and Pro563 Does Not Render HIF-1α Protein Stable at Normoxia—Hydroxylation of specific proline residues has been shown to mediate recognition of HIF-1α by pVHL. pVHL belongs to a multiprotein complex with E3 ubiquitin ligase activity that mediates ubiquitination of HIF-1α leading to its proteasome-dependent degradation. Previous studies have identified two independent degradation boxes surrounding Pro402 and Pro564 (Pro563 on mouse HIF-1α) on human HIF-1α. Mutation of these two residues was suggested to render the HIF-1α protein stable at normoxia (3, 20). Here we have investigated the protein levels of HIF-1α mutants expressed in HEK 293 and HepG2 cells to evaluate how the critical proline residues affect protein stability. Cells were treated with CoCl2, an iron antagonist that is known to act as an hypoxia-mimicking agent because Fe(II) is a critical cofactor required for the reactions performed by the different hydroxylases that regulate HIF-1α activity (1, 2). As expected (11), wild-type HIF-1α expressed in both HEK 293 (Fig. 1A) and HepG2 (Fig. 1B) cells was degraded at normoxia and stabilized after treatment of cells with CoCl2. In analogy to the wild-type protein, levels of expressed HIF-1α(P402A/P563A) were lower in cells kept at normoxia than in cells treated with CoCl2, indicating that the mutant underwent degradation in non-treated cells. In contrast with the HIF-1α mutant, expression of Arnt in HEK 293 cells showed not to be regulated by CoCl2 treatment (Fig. 1C).
FIGURE 1.
Mutation of the identified degradation boxes fails to stabilize HIF-1α. A, HIF-1α(P402A/P563A) is degraded at normoxia in HEK 293 cells. Cells were transfected with 400 ng (+), 600 ng (++), and 800 ng (+++) of plasmids encoding mHIF-1α, or mHIF-1α(P402A/P563A) (PP-A) and exposed to normoxia (-) or CoCl2 (+) for 12 h. Whole cell extracts were separated by SDS-PAGE and analyzed by immunoblotting with anti-FLAG (α-FLAG) and anti-actin (α-actin) antibodies. B, expression of mHIF-1α(P402A/P563A) in HepG2 cells is regulated by CoCl2. HepG2 cells were transfected with 500 ng (+), and 1000 ng (++) of pFLAG-mHIF-1α or pFLAG-mHIF-1α(P402A/P563A), allowed to grow for 24 h at normoxia, and then kept at normoxia (-), or treated with CoCl2 (+) for 12 h. FLAG-tagged proteins were immunoprecipitated from whole cell extracts (500 μg) and analyzed by SDS-PAGE and immunoblotting using an anti-FLAG antibody (α-FLAG). C, Arnt protein expressed in HEK 293 cells. Cells were transfected with 200 ng (+), 400 ng (++), and 600 ng (+++) of pMyc-Arnt and kept at normoxia (-), or treated with CoCl2 (+). D, mutation of Pro402 and Pro563 does not abrogate hypoxia-inducible transactivation of mHIF-1α. HEK 293 cells were transfected with a HRE-driven luciferase reporter plasmid and mHIF-1α or mHIF-1α(P402A/P563A) expression plasmids, and exposed to normoxia (N), hypoxia (H), or CoCl2 (Co) for 24 h. Data are presented as luciferase activity relative to cells transfected with pCMX (CMX) and cultured at normoxia. Values represent mean ± S.E. of three independent experiments performed in duplicate.
To evaluate the transactivation potency mediated by the HIF-1α double proline mutant we performed HRE-driven luciferase reporter gene assays in HEK 293 cells. These experiments (Fig. 1D) showed that mutation of Pro402 and Pro563 in mHIF-1α does not affect the responsiveness of the transcriptional activator to hypoxia and CoCl2. Although we cannot exclude the activity of the C-TAD, we did not observe any decrease on transactivation induction in response to hypoxia or CoCl2 with the expressed HIF-1α mutant when compared with the wild-type protein. Taken together, these results indicate that mutation of the two critical proline residues, which disrupts the interaction of pVHL with the known degradation boxes of HIF-1α (3, 20) is not sufficient to stabilize the full-length protein at normoxia.
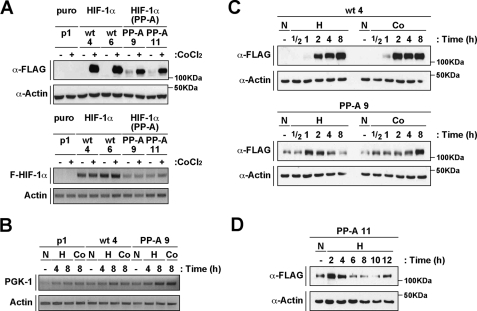
Generation and Characterization of Stable Cell Lines Expressing HIF-1α or HIF-1α(P402A/P563A)—To further study the mechanism and kinetics of degradation and stabilization of the HIF-1α double proline mutant, we generated stable cell lines expressing either wild-type or mutant HIF-1α alleles. Several HEK 293H cell lines were selected by analysis of protein expression after 12 h of treatment with CoCl2. In Fig. 2A, upper panel, we compare the expression levels of wild-type and mutant HIF-1α proteins in four independent lines (wt 4, wt 6, PP-A 9, and PP-A 11). In contrast to stable cell lines expressing wild-type HIF-1α, cells expressing the HIF-1α mutant presented detectable levels of protein at normoxia. In most of performed experiments we observed that the HIF-1α mutant protein could be detected at normoxia, albeit at low levels. These indicate that, although mutation of the two critical proline residues does not lead to complete stabilization of the protein, it may yield low but yet detectable levels of protection of HIF-1α against proteasome-mediated degradation. Wild-type or mutant HIF-1α RNAs were analyzed by semiquantitative RT-PCR. As presented in Fig. 2A, lower panel, HIF-1α(P402A/P563A) mRNA levels were lower than the corresponding wild-type HIF-1α transcript. This observation offers an explanation for the lower protein accumulation detected in CoCl2-treated cells expressing the HIF-1α double proline mutant (Fig. 2A, upper panel). In conclusion, our results show that the HIF-1α double proline mutant expressed in stable cell lines still undergoes degradation at normoxia, and CoCl2-dependent stabilization.
FIGURE 2.
HIF-1α(P402A/P563A) is transiently stabilized in response to hypoxia. A, HIF-1α double proline mutant expressed in stable cell lines is degraded at normoxia. A, upper panel, analysis of mHIF-1α protein levels in stable cell lines expressing wild-type (wt 4 and 6) or HIF-1α(P402A/P563A) (PP-A 9 and 11). Whole cell extracts were separated by SDS-PAGE and analyzed by immunoblotting using anti-FLAG (α-FLAG), or anti-actin (α-Actin) antibodies. Cells were treated with CoCl2 as indicated (+). A pEFIRESpuro-transfected cell line is also shown (p1). A, lower panel, RNA levels of mHIF-1α wild-type or double proline mutant analyzed by semiquantitative RT-PCR. Levels of transcripts were evaluated using primers to FLAG-HIF-1α, and actin. B, RNA levels of PGK-1 are up-regulated in cells expressing HIF-1α or HIF-1α(P402A/P563A). Levels of transcripts were evaluated by semiquantitative RT-PCT using primers to PGK-1, and actin. C, a transient stabilization of HIF-1α(P402A/P563A) is observed at early time points of hypoxia treatment. Stable cell lines (wt 4 and PP-A 9) were treated with CoCl2 (Co) or hypoxia (H) during the indicated times or kept at normoxia (N). Whole cell extracts were separated by SDS-PAGE and analyzed by immunoblotting using anti-FLAG (α-FLAG), or anti-actin (α-Actin) antibodies. D, a different stable cell line expressing HIF-1α double proline mutant confirms the transient regulation of the protein at hypoxia. The stably transfected cell line PP-A 11 was treated with hypoxia (H) as indicated, and HIF-1α expression was analyzed as in C.
We next analyzed mRNA expression of the HIF-1α target gene phosphoglycerate kinase 1 (PGK-1) (21) in the different cell lines by semiquantitative RT-PCR. As shown in Fig. 2B, PGK-1 gene expression was up-regulated in response to hypoxia and CoCl2 in cells transfected with an empty expression vector, showing that endogenous HIF factors upregulate PGK-1 mRNA in these cells. Induction of the PGK-1 transcript in response to hypoxia and CoCl2 was also observed in cell lines expressing wild-type or double proline mutant HIF-1α. However, higher levels of mRNA expression were detected in the mutant expressing cell line when compared with wild-type HIF-1α cell line (Fig. 2B). These results demonstrate that inducibility of the PGK-1 mRNA is maintained in the cell line expressing the HIF-1α double proline mutant.
HIF-1α Double Proline Mutant Is Transiently Stabilized in Response to Hypoxia—We next wanted to investigate how hypoxia affects HIF-1α double proline mutant stabilization. In our first experiments we kept the cells at hypoxia or treated them with CoCl2 for 4, 8, and 12 h (data not shown). In stable cell lines expressing wild-type HIF-1α stabilization of the protein was observed in response to both treatments (data not shown). In contrast, the HIF-1α double proline mutant was stabilized in cells treated with CoCl2, but did not accumulate in response to 4 to 12 h of hypoxia treatment (data not shown). This unexpected result led us to investigate the impact of hypoxia at early time-points. Exposure of cells to 1 or 2 h of hypoxia proved to stabilize the HIF-1α double proline mutant while at later time points the protein expression levels returned to normoxic levels (Fig. 2C). To confirm hypoxia regulation of the HIF-1α double proline mutant, we analyzed another stable cell line expressing the mutant. As shown in Fig. 2D, we also observed stabilization of the protein after 2 h of treatment with return to normoxic levels with longer periods of hypoxia treatment. Thus, in contrast to the wild-type protein, the HIF-1α double proline mutant is only very transiently stabilized by hypoxia treatment.
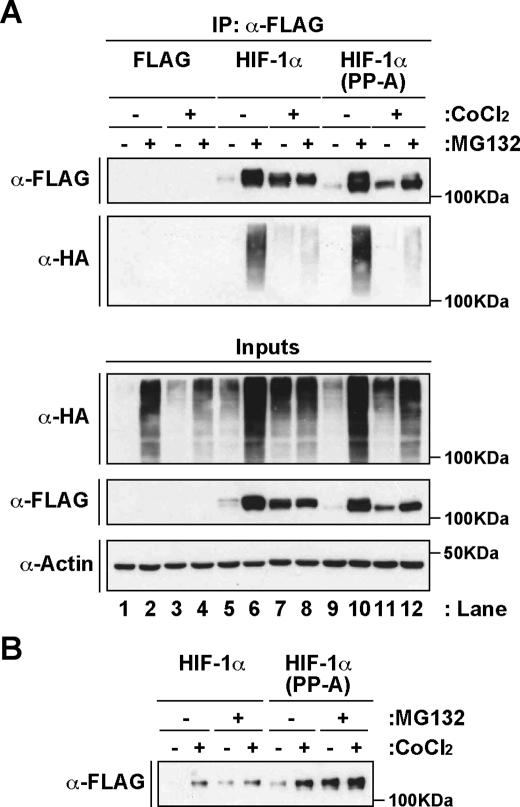
HIF-1α(P402A/P564A) Is Ubiquitinated and Degraded in a Proteasome-dependent Fashion—We next investigated if the double mutation of Pro402 and Pro563 affected the ubiquitination status of the protein. We transfected HEK 293 cells with plasmids encoding FLAG-tagged wild-type or mutant HIF-1α together with an expression plasmid for hemagglutinin (HA)-tagged ubiquitin and treated the cells with or without the proteasome inhibitor MG132, as indicated in Fig. 3A. FLAG-tagged proteins were precipitated from whole cell extracts using an anti-FLAG antibody, and ubiquitination levels were analyzed by Western blot assays using an anti-HA antibody. As shown in Fig. 3A, no ubiquitination was observed in cells expressing the FLAG-tag alone and treated with MG132 (lanes 2 and 4). In contrast, both HIF-1α wild-type (lane 6) and HIF-1α(P402A/P563A) (lane 10) presented high levels of ubiquitination in cells treated with the proteasome inhibitor and kept at normoxia. Concomitantly, FLAG-tagged proteins proved to accumulate in response to the inhibition of proteasome proteolytic activity (Fig. 3A, upper panel). As expected (3), treatment with CoCl2 significantly reduced the levels of ubiquitination in MG132-treated cells in both wild-type (lane 8) and mutant protein (lane 12). Expression of HIF-1α double proline mutant in HepG2 cells was also stabilized following treatment with either CoCl2 or MG132, or both (Fig. 3B). In conclusion, these results demonstrate that mutation of Pro402 and Pro563 to alanine does not prevent HIF-1α from being ubiquitinated and degraded at normoxia by a proteasome-dependent mechanism.
FIGURE 3.
HIF-1α(P402A/P563A) undergoes ubiquitination and proteasome-dependent degradation. A, mutation of Pro402 and Pro563 to alanine does not prevent ubiquitination of HIF-1α. HEK 293 cells were cotransfected with pHA-ubiquitin and pFLAG, pFLAG-mHIF-1α, or pFLAG-mHIF-1α(P402A/P562A) (PP-A), and treated with CoCl2 and the proteasome inhibitor MG132 as indicated (+). Whole cell extracts (inputs) and immunoprecipitated FLAG-tagged proteins (IP) were analyzed by SDS-PAGE and immunoblotting using anti-FLAG (α-FLAG), anti-hemagglutinin (α-HA), or anti-actin (α-Actin) antibodies. B, HIF-1α double proline mutant expressed in HepG2 is stabilized in response to MG132 treatment. HepG2 cells were transfected with 1 μg of HIF-1α and mutant expression plasmids and treated for 12 h with CoCl2 and MG132 as indicated (+). FLAG-tagged proteins were immunoprecipitated from whole cell extracts (500 μg) and analyzed by SDS-PAGE and immunoblotting using an anti-FLAG antibody (α-FLAG).
HIF-1α(P402A/P563A) Is Resistant to PHD-mediated Degradation—HIF-1α Pro402 and Pro563 have been shown to be hydroxylated by a new family of prolyl hydroxylases that share homology with Caenorhabditis elegans EGL9 (1, 2). These enzymes belong to a family of Fe(II), 2-oxoglutarate-dependent dioxygenases that in mammalian cells include at least 3 members, PHDs 1-3 (1, 2). In the present study, we investigated if the HIF-1α double proline mutant was sensitive to PHD-mediated degradation. We expressed HIF-1α or HIF-1α(P402A/P563A) at levels that led to stabilization of the protein at normoxia, presumably due to saturation of the endogenous degradation machinery (11). Under these conditions, we analyzed the effect of expressing increasing amounts of PHDs 1-3 on HIF-1α levels. Our results showed that while expression of PHD1 (Fig. 4A), PHD2 (Fig. 4B), or PHD3 (Fig. 4C) mediated degradation of wild-type HIF-1α, the double proline mutant was resistant to the action of these hydroxylases, indicating that it is not an enzymatic substrate of these PHDs. Similar results were observed in either CoCl2-treated (Fig. 4) or hypoxia-exposed (data not shown) cells. The results regarding degradation of the wild-type HIF-1α protein by the distinct prolyl hydroxylases indicated that PHD2 mediates a much more efficient destruction of the protein than PHD1 or -3, consistent with the notion that PHD2 is the major contributor for HIF-1α degradation at normoxia (22). Interestingly, in our study, treatment of cells with CoCl2 did not prevent PHD2-dependent degradation of HIF-1α suggesting that although the activity of the enzyme can be inhibited by CoCl2 it is not completely abolished and that its overexpression may overcome this inhibition. These results are in agreement with a recent report showing that PHD2 can be active in degradation of HIF-1α even under hypoxic conditions (23). Interestingly, our observations showed that, in addition to the previously described regulation of PHD3 transcription by hypoxia (24), protein levels of the enzyme are also regulated by CoCl2 (Fig. 4C) and hypoxia (data not shown). Further studies are required to explain this new observation. In conclusion, the results obtained by coexpressing the three known prolyl hydroxylases together with HIF-1α proteins showed that although degradation of wild-type HIF-1α can be promoted by any of these hydroxylases, their effect is strictly dependent on the two critical proline residues, as it is not observed on HIF-1α(P402A/P563A).
FIGURE 4.
Degradation of HIF-1α(P402A/P563A) is not achieved by overexpression of PHDs 1-3. A-C, PHD1, PHD2, and PHD3 fail to induce degradation of HIF-1α(P402A/P563A). HEK 293 cells were transfected with pFLAG-mHIF-1α, or pFLAG-mHIF-1α(P402A/P563A) (PP-A) and 150 ng (+), or 300 ng (++) of plasmids encoding FLAG-tagged PHD1 (A), FLAG-tagged-PHD2 (B), or FLAG-tagged-PHD3 (C). Whole cell extracts were separated by SDS-PAGE and analyzed by immunoblotting using anti-FLAG (α-FLAG), and anti-actin (α-Actin) antibodies. Cells were treated with CoCl2 as indicated (+).
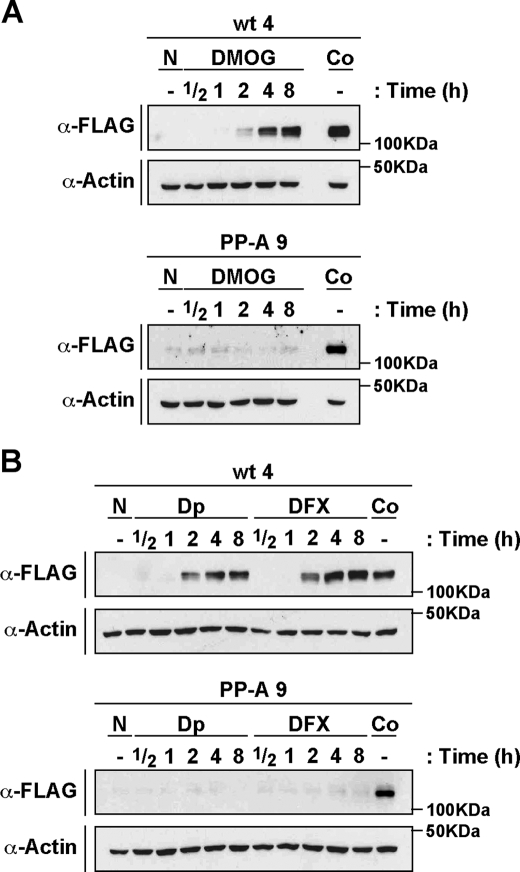
Hydroxylases Inhibitors, Other Than CoCl2, Are Unable to Stabilize the HIF-1α Double Proline Mutant—Our results show that CoCl2, an iron antagonist known to inhibit hydroxylase activity, is able to stabilize HIF-1α(P402A/P563A). However, overexpression of PHDs had no impact on HIF-1α mutant protein levels (Fig. 4). These apparently conflicting observations led us to speculate that the observed CoCl2-dependent stabilization of the HIF-1α double proline mutant is achieved by a PHD-independent mechanism and therefore other hydroxylase inhibitors may not be able to affect protein levels of the mutant. To test this hypothesis, we treated stable cell lines expressing wild-type and mutant HIF-1α with the specific hydroxylase inhibitor, DMOG, or with iron chelators such as 2,2′-dipyridyl and desferoxamine, known to inactivate PHD activity (1, 2). As shown in Fig. 5, all three compounds were able to up-regulate wild-type HIF-1α protein levels in a time-dependent manner. In contrast to these observations, the protein levels of HIF-1α double proline mutant were not changed by any of the hydroxylase inhibitors. These results led us to conclude that the PHDs are not involved in degradation of the HIF-1α mutant. On the contrary, another yet unknown pathway inhibited by Co(II) appears to participate in this mechanism.
FIGURE 5.
DMOG and iron chelators, such as 2,2′-dipyridyl and deferoxamina, are unable to up-regulate the HIF-1α double proline mutant. A and B, stable cells expressing mHIF-1α (wt 4), or mHIF-1α(P402A/P563A) (PP-A 9) were treated with DMOG, 2,2′-dipyridyl (Dp), or deferoxamine (DFX) for the indicated times, or kept at normoxia (N) or treated with CoCl2 (Co) for constant end-point experiments. Whole cell extracts were separated by SDS-PAGE and analyzed by immunoblotting using anti-FLAG (α-FLAG), or anti-actin (α-Actin) antibodies.
Degradation and Ubiquitination of HIF-1α Double Proline Mutant Is Mediated by pVHL—At normoxia, the proline hydroxylation event is followed by interaction of HIF-1α with the pVHL/E3 ubiquitin ligase complex which, in turn, promotes ubiquitination, and degradation of the protein. In renal cell carcinoma cell lines expressing nonfunctional pVHL mutants, the HIF-1α protein is known to be stable at normoxia and therefore is not responsive to hypoxia (25). Here we investigated if up-regulation of HIF-1α in response to Co(II) is pVHL-mediated by studying the impact of CoCl2 treatment on endogenous HIF-1α using a pVHL nonfunctional cell line, the renal cell carcinoma SKRC-7 (26). Cells were treated with CoCl2 for 8 h, and protein expression was analyzed by Western blot. As shown in Fig. 6A stability of HIF-1α protein expressed in this cell line is not changed by CoCl2 treatment, indicating that functional pVHL is required for Co(II)-dependent regulation.
FIGURE 6.
pVHL mediates degradation of HIF-1α(P402A/P563A). A, Co(II) does not upregulate HIF-1α protein levels in a nonfunctional pVHL-expressing cell line. SKRC-17 (not expressing HIF-1α) and SKRC-7 (expressing HIF-1α) cell lines were treated with CoCl2 for 8 h, and whole cell extracts were prepared. Endogenous HIF-1α protein levels were analyzed (150 μg of extracts) by immunoblotting using anti-HIF-1α (α-HIF-1α), or anti-actin (α-Act) antibodies. As a positive control, 25 μg of extracts from HEK 293A (A) cells transfected with pFLAG-mHIF-1α and treated with CoCl2 were used. B, knocking-down pVHL expression by RNA interference leads to accumulation of HIF-1α(P402A/P563A). Stable cells expressing wild-type (wt 4) or the mHIF-1α double proline mutant (PP-A 9) were transfected with negative control (C) or VHL (V) siRNA. B, upper panel. Whole cell extracts were separated by SDS-PAGE and analyzed by immunoblotting using anti-FLAG (α-FLAG), anti-VHL (α-VHL), or anti-actin (α-Actin) antibodies. Cells were treated with CoCl2 as indicated (+). B, lower panel, RNA level analysis following siRNA experiments targeting VHL expression. Semiquantitative RT-PCR was performed using primers to FLAG-HIF-1α, VHL, and actin. C, pVHL mediates HIF-1α(P402A/P563A) ubiquitination. Stable cells expressing p1 (puromycin control), wild-type HIF-1α (wt 4), or HIF-1α mutant (PP-A 9) were transfected with siRNA as in B, and exposed to MG132. C, upper panel, whole cell extracts (Inputs) and immunoprecipitated FLAG-tagged proteins (IP) were analyzed using anti-FLAG (α-FLAG), anti-ubiquitin (α-Ub), or anti-actin (α-Actin) antibodies. C, lower panel, analysis of VHL mRNA levels following RNAi experiments. Semiquantitative RT-PCR was performed using primers to VHL and actin.
To study the role of pVHL in the degradation and ubiquitination of HIF-1α double proline mutant, we used RNA interference directed against pVHL. Stable cell lines expressing wild-type or mutant HIF-1α were transfected with siRNAs targeting endogenous pVHL mRNA expression. After siRNA transfection we simultaneously analyzed protein, or ubiquitination levels following CoCl2, or MG132 treatment, respectively (Fig. 6, B and C). As shown in Fig. 6B, upper panel, pVHL protein expression was reduced in both cell lines. These results are in agreement with the levels of VHL transcript detected by semiquantitative RT-PCR (Fig. 6B, lower panel). Reduction of pVHL expression led to an increase of both wild-type and mutant HIF-1α protein accumulation at normoxia, indicating that they are both degraded by a pVHL-mediated mechanism (Fig. 6B). Similar results were obtained with an additional, distinct siRNA sequence targeting pVHL expression (data not shown) as described under “Experimental Procedures.” RNA levels of wild-type or mutant HIF-1α were unchanged by the presence of a siRNA sequence targeting VHL, as shown in Fig. 6B, lower panel. Interestingly, reduced pVHL expression also contributed to an increase of wild-type and mutant HIF-1α protein levels in CoCl2-treated cells (Fig. 6B) showing that pVHL is able to interact and mediate degradation of HIF-1α even in cells treated with an inhibitor of the prolyl hydroxylase-dependent (23) and -independent mechanisms. Ubiquitination levels of wild-type or mutant HIF-1α after transfection with a VHL-specific siRNA were analyzed has shown in Fig. 6C. A reduction in ubiquitination levels was observed in cells transfected with VHL siRNA in both wild-type (Fig. 6C, upper panel, lanes 3 and 4) and mutant (lanes 5 and 6) HIF-1α-expressing cells, indicating that both proteins are ubiquitinated by a pVHL-dependent mechanism. Analysis of VHL mRNA expression showed that transfection with the specific siRNA led to decreased pVHL mRNA levels (Fig. 6C, lower panel). In conclusion, these data show that mutation of Pro402 and Pro563 to alanine does not rescue HIF-1α from degradation by a pVHL-mediated mechanism and that pVHL regulates the ubiquitination levels of HIF-1α(P402A/P563A). It is therefore plausible that pVHL works as an HIF-1α E3 ubiquitin ligase via proline hydroxylation-dependent and -independent mechanisms.
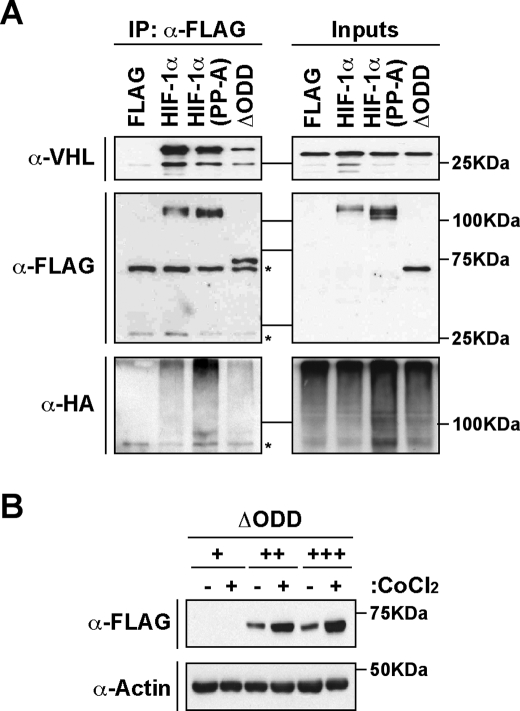
Deletion of the Oxygen-dependent Degradation (ODD) Domain of HIF-1α, Encompassing Pro402 and Pro563, Does Not Abrogate Interaction of pVHL with HIF-1α—To show that interaction of pVHL with HIF-1α can occur independently of the two critical proline residues, we investigated the binding of pVHL to different HIF-1α mutants using immunoprecipitation assays. FLAG-tagged wild-type and mutant HIF-1α together with pVHL were expressed in HEK 293 cells. An anti-FLAG antibody was used to precipitate HIF-1α proteins from whole cell extracts, and interaction with pVHL was analyzed by Western blot. These experiments were performed in the presence of coexpressed ubiquitin and upon treatment with MG132, conditions that were shown to contribute to an efficient binding of pVHL to HIF-1α (data not shown). As shown in Fig. 7A, pVHL was able to interact with similar affinity with either wild-type HIF-1α or the HIF-1α double proline mutant. Another HIF-1α mutant lacking the oxygen-dependent degradation domain (HIF-1α ΔODD) (7) that encompasses the two critical proline residues also showed positive binding to pVHL. In these experiments, ubiquitination was observed for all the expressed HIF-1α proteins including the HIF-1α ΔODD mutant (Fig. 7A). These results indicate that additional pVHL-interaction interfaces are present within the HIF-1α protein and mediate interaction with pVHL independently of the Pro402 and Pro563 residues. In excellent agreement with the results obtained in the immunoprecipitation assays, HIF-1α ΔODD that bound pVHL was degraded at normoxia and stabilized by CoCl2 treatment (Fig. 7B). Taken together, these observations show that additional pVHL-interacting interfaces are present on the HIF-1α protein and are correlated with degradation of this protein at normoxia.
FIGURE 7.
Binding of pVHL to HIF-1α occurs independently of Pro402 and Pro563. A, pVHL binds and promotes ubiquitination of HIF independently of the ODD domain. HEK 293 cells were transfected with pFLAG, pFLAG-mHIF-1α, pFLAG-mHIF-1α(P402A/P563A), or pFLAG-mHIF-1α(391Δ628) (ΔODD), together with pVHL and pHA-ubiquitin. Whole cell extracts (Inputs) and immunoprecipitated FLAG-tagged proteins (IP) were prepared and analyzed by immunoblotting using anti-FLAG (α-FLAG), anti-hemagglutinin (α-HA), or anti-VHL (α-VHL) antibodies. (Denoted unspecific bands are marked with asterisks.) B, pVHL binding correlates with degradation of HIF-1α apart from the ODD domain. HEK 293 cells were transfected with 200 ng (+), 400 ng (++), and 600 ng (+++) of plasmids encoding pFLAG-mHIF-1α(392Δ622) (ΔODD) and exposed to normoxia (-) or CoCl2 (+) for 12 h. Whole cell extracts were separated by SDS-PAGE and analyzed by immunoblotting with anti-FLAG (α-FLAG), or anti-actin (α-actin) antibodies.
DISCUSSION
Here we show that mutation of HIF-1α Pro402 and Pro563 to alanine generates a protein that is still degraded at normoxia and is transiently stabilized in response to hypoxia. Furthermore, we present evidence that degradation of this protein is mediated by a pVHL-dependent mechanism and does not involve the participation of any of the previously characterized PHDs.
Previous studies using proteins expressed in transiently transfected cells have suggested that mutation of prolines Pro402 and Pro564 of hHIF-1α renders the protein stable at normoxia (3). However, in transient transfections, protein expression may easily reach stabilization levels at normoxia due to saturation of the endogenous degradation machinery (11). In this context, we assessed the ability of the HIF-1α double proline mutant to be degraded at normoxia by titrating the expression levels of the proteins in transient transfections and by evaluating protein stability in stable cell lines. In both these experimental approaches, HIF-1α(P402A/P563A) showed degradation in cells kept at normoxia and was stabilized by inhibition of proteasome activity and treatment with CoCl2. A recently published report showed that, in contrast to the authors' expectations, expression of the HIF-1α double proline mutant in keratinocytes could not reconstitute the phenotype observed with pVHL inactivation (28). The results presented in our study showing that the HIF-1α double proline mutant still undergoes degradation at normoxia offer an explanation to the phenotype observed by Kim et al. (28) in keratinocytes.
Our study showed that some aspects of the mechanism underlying HIF-1α double proline mutant degradation are shared between the mutant and the wild-type proteins. Both proteins are stabilized in response to inhibition of proteasome activity, ubiquitinated, degraded by a pVHL-mediated mechanism and stabilized in response to CoCl2 treatment. The observation that mutation of Pro402 and Pro564 in hHIF-1α generated a protein that could still be stabilized in response to MG132, has been previously indicated (28). However, without further investigation of the mechanism, these authors proposed a pVHL-independent degradation process. Contrastingly, in our study the participation of pVHL in degradation and ubiquitination of HIF-1α(P402A/P563A) was clearly indicated by RNA interference experiments. Furthermore, the observed interaction of pVHL with HIF-1α(P402A/P563A) and with a HIF-1α mutant lacking the domain encompassing the critical proline residues indicates the existence of additional HIF-1α/pVHL-interaction interfaces. The role of pVHL in degradation of HIF-1α is visibly demonstrated in studies with animal models where phenotypes associated with VHL inactivation in tissues can be rescued by inactivation of HIF-1α (29-31) showing that pVHL plays a major role in the degradation of HIF-1α in these tissues. In other tissues, the pVHL phenotype is reversed by inactivation of HIF-2α or Arnt (31) depending on which target genes are responsible for the observed phenotype and on HIF-2α function in the tissue. Taken together, these observations suggest that pVHL is the major player as an E3 ubiquitin ligase in degradation of HIF protein and if other mechanisms do exist, they do not overcome pVHL function.
The mechanism of HIF-1α stabilization in response to CoCl2 is not fully understood. Co(II) is an iron antagonist and therefore it has been proposed to inhibit prolyl hydroxylases by competing with Fe(II) required for the activity of these enzymes (1, 2). However, inhibition of prolyl hydroxylases by Co(II) has been shown to be rather ineffective (32), and other mechanisms have been suggested. One study proposed binding of Co(II) directly to N-terminal domains of HIF-1α by a pVHL-independent mechanism (33), while another report suggested direct binding of Co(II) to the ODD domain (34), resulting in inhibition of pVHL-mediated degradation. Moreover, a third mechanism involving the generation of reactive oxygen species in response to CoCl2 treatment (35) has been proposed. It is tempting to speculate that, in addition to inhibition of prolyl hydroxylase activity, CoCl2 may inhibit another protein that requires divalent metal ions to be functional.
Analysis of the degradation mechanism of HIF-1α(P402A/P563A) indicated some noticeable differences between the mutant and the wild-type protein. Although both proteins are stabilized in response to CoCl2, the HIF-1α mutant showed a transient accumulation of the protein to hypoxia and was resistant to PHD1-3-mediated degradation. Our experiments using overexpressed PHDs and PHD inhibitors other than CoCl2 showed that the HIF-1α mutant is insensitive to the hydroxylase activity and does therefore not appear to be a substrate of these enzymes. The observation that CoCl2 and hypoxia could lead to stabilization of the HIF-1α double proline mutant suggests that the activity of a protein inhibited by either CoCl2 or hypoxia is required for the binding of pVHL to the HIF-1α mutant. It has been shown that expression levels of PHD2 and PHD3 increase at hypoxia (23, 24, 36), and this event has been correlated with decreased stability of HIF-1α observed at longer periods of hypoxia exposure. The transient accumulation of the HIF-1α mutant observed at hypoxia leads us to speculate that a similar negative feedback mechanism may regulate protein(s) mediating HIF-1α mutant degradation. Newly identified factors (27, 37) have been proposed to participate in the mechanism of pVHL-mediated degradation of HIF-1α. Although these proteins have been suggested to not affect the expression of the HIF-1α double proline mutant, further studies are required before their involvement can be completely excluded from the presently described mechanism. Another recent publication proposes that VHL can bind to SUMOylated HIF-1α double proline mutant (19); however, this mechanism was suggested to be only relevant at hypoxia.
In conclusion, our studies show that regions of HIF-1α other than those spanning Pro402 and Pro563 may contribute to normoxia-dependent degradation of the protein and that this mechanism requires the participation of pVHL. It remains to be elucidated what are the molecular targets for CoCl2- and transient hypoxia-dependent stabilization of HIF-1α(P402A/P563A) against degradation. Obviously, these molecular targets do not comprehend PHDs 1-3, and it will be challenging to examine this mechanism of regulation of pVHL activity in closer detail. Finally, this alternative mechanism of pVHL-mediated degradation of HIF-1α may be relevant under conditions of inhibition of PHD 1-3 activity by pharmacological means, a therapeutic approach that is presently gaining attention.
Acknowledgments
We thank Jorge Ruas and Lorenz Poellinger for critical review of the manuscript.
This study was supported by grants from the Swedish Medical Research Council, the Swedish Cancer Society, the European Union, and Karolinska Institutet. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: HIF-1, hypoxia-inducible factor-1; PHD, prolyl-hydroxylase; pVHL, von Hippel-Lindau tumor suppressor protein; TAD, transactivation domain; C-TAD, C-terminal TAD; N-TAD, N-terminal TAD; HEK, human embryonic kidney; siRNA, small interference RNA; RIPA, radioimmune precipitation buffer; HA, hemagglutinin; ODD, oxygen-dependent degradation.
References
- 1.Bruick, R. K., and McKnight, S. L. (2001) Science 294 1337-1340 [DOI] [PubMed] [Google Scholar]
- 2.Epstein, A. C., Gleadle, J. M., McNeill, L. A., Hewitson, K. S., O'Rourke, J., Mole, D. R., Mukherji, M., Metzen, E., Wilson, M. I., Dhanda, A., Tian, Y. M., Masson, N., Hamilton, D. L., Jaakkola, P., Barstead, R., Hodgkin, J., Maxwell, P. H., Pugh, C. W., Schofield, C. J., and Ratcliffe, P. J. (2001) Cell 107 43-54 [DOI] [PubMed] [Google Scholar]
- 3.Masson, N., Willam, C., Maxwell, P. H., Pugh, C. W., and Ratcliffe, P. J. (2001) EMBO J. 20 5197-5206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ivan, M., Kondo, K., Yang, H., Kim, W., Valiando, J., Ohh, M., Salic, A., Asara, J. M., Lane, W. S., and Kaelin, W. G., Jr. (2001) Science 292 464-468 [DOI] [PubMed] [Google Scholar]
- 5.Jaakkola, P., Mole, D. R., Tian, Y. M., Wilson, M. I., Gielbert, J., Gaskell, S. J., Kriegsheim, A., Hebestreit, H. F., Mukherji, M., Schofield, C. J., Maxwell, P. H., Pugh, C. W., and Ratcliffe, P. J. (2001) Science 292 468-472 [DOI] [PubMed] [Google Scholar]
- 6.Yu, F., White, S. B., Zhao, Q., and Lee, F. S. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 9630-9635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang, L. E., Gu, J., Schau, M., and Bunn, H. F. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 7987-7992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kallio, P. J., Wilson, W. J., O'Brien, S., Makino, Y., and Poellinger, L. (1999) J. Biol. Chem. 274 6519-6525 [DOI] [PubMed] [Google Scholar]
- 9.Kamura, T., Sato, S., Iwai, K., Czyzyk-Krzeska, M., Conaway, R. C., and Conaway, J. W. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 10430-10435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cockman, M. E., Masson, N., Mole, D. R., Jaakkola, P., Chang, G. W., Clifford, S. C., Maher, E. R., Pugh, C. W., Ratcliffe, P. J., and Maxwell, P. H. (2000) J. Biol. Chem. 275 25733-25741 [DOI] [PubMed] [Google Scholar]
- 11.Tanimoto, K., Makino, Y., Pereira, T., and Poellinger, L. (2000) EMBO J. 19 4298-4309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ohh, M., Park, C. W., Ivan, M., Hoffman, M. A., Kim, T. Y., Huang, L. E., Pavletich, N., Chau, V., and Kaelin, W. G. (2000) Nat. Cell Biol. 2 423-427 [DOI] [PubMed] [Google Scholar]
- 13.Lando, D., Peet, D. J., Whelan, D. A., Gorman, J. J., and Whitelaw, M. L. (2002) Science 295 858-861 [DOI] [PubMed] [Google Scholar]
- 14.Lando, D., Peet, D. J., Gorman, J. J., Whelan, D. A., Whitelaw, M. L., and Bruick, R. K. (2002) Genes Dev. 16 1466-1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berra, E., Ginouves, A., and Pouyssegur, J. (2006) EMBO Rep. 7 41-45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Metzen, E., Berchner-Pfannschmidt, U., Stengel, P., Marxsen, J. H., Stolze, I., Klinger, M., Huang, W. Q., Wotzlaw, C., Hellwig-Burgel, T., Jelkmann, W., Acker, H., and Fandrey, J. (2003) J. Cell Sci. 116 1319-1326 [DOI] [PubMed] [Google Scholar]
- 17.Ruas, J. L., Poellinger, L., and Pereira, T. (2002) J. Biol. Chem. 277 38723-38730 [DOI] [PubMed] [Google Scholar]
- 18.Pereira, T., Zheng, X., Ruas, J. L., Tanimoto, K., and Poellinger, L. (2003) J. Biol. Chem. 278 6816-6823 [DOI] [PubMed] [Google Scholar]
- 19.Cheng, J., Kang, X., Zhang, S., and Yeh, E. T. H. (2007) Cell 131 584-595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu, C. J., Wang, L. Y., Chodosh, L. A., Keith, B., and Simon, M. C. (2003) Mol. Cell. Biol. 23 9361-9374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iyer, N. V., Kotch, L. E., Agani, F., Leung, S. W., Laughner, E., Wenger, R. H., Gassmann, M., Gearhart, J. D., Lawler, A. M., Yu, A. Y., and Semenza, G. L. (1998) Genes Dev. 12 149-162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berra, E., Benizri, E., Ginouves, A., Volmat, V., Roux, D., and Pouyssegur, J. (2003) EMBO J. 22 4082-4090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stiehl, D. P., Wirthner, R., Koditz, J., Spielmann, P., Camenisch, G., and Wenger, R. H. (2006) J. Biol. Chem. 281 23482-23491 [DOI] [PubMed] [Google Scholar]
- 24.Marxsen, J. H., Stengel, P., Doege, K., Heikkinen, P., Jokilehto, T., Wagner, T., Jelkmann, W., Jaakkola, P., and Metzen, E. (2004) Biochem. J. 381 761-767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maynard, M. A., and Ohh, M. (2004) Am. J. Nephrol. 24 1-13 [DOI] [PubMed] [Google Scholar]
- 26.Grabmaier, K., MC, de Weijert, M. C. A., Verhaegh, G. W., Schalken, J. A., and Oosterwijk, E. (2004) Oncogene 23 5624-5631 [DOI] [PubMed] [Google Scholar]
- 27.Baek, J. H., Liu, Y. V., McDonald, K. R., Wesley, J. B., Hubbi, M. E., Byun, H., and Semenza, G. L. (2007) J. Biol. Chem. 282 23572-23580 [DOI] [PubMed] [Google Scholar]
- 28.Kim, W. Y., Safran, M., Buckley, M. R., Ebert, B. L., Glickman, J., Bosenberg, M., Regan, M., and Kaelin, W. G., Jr. (2006) EMBO J. 25 4650-4662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Biju, M. P., Neumann, A. K., Bensinger, S. J., Johnson, R. S., Turka, L. A., and Haase, V. H. (2004) Mol. Cell. Biol. 24 9038-9047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pfander, D., Kobayashi, T., Knight, M. C., Zelzer, E., Chan, D. A., Olsen, B. R., Giaccia, A. J., Johnson, R. S., Haase, V. H., and Schipani, E. (2004) Development 131 2497-2508 [DOI] [PubMed] [Google Scholar]
- 31.Haase, V. H. (2005) Sem. Cell Dev. Biol. 16 564-574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hirsila, M., Koivunen, P., Xu, L., Seeley, T., Kivirikko, K. I., and Myllyharju, J. (2005) FASEB J. 19 1308-1310 [DOI] [PubMed] [Google Scholar]
- 33.Kanaya, K., and Kamitani, T. (2003) Biochem. Biophys. Res. Commun. 306 750-755 [DOI] [PubMed] [Google Scholar]
- 34.Yuan, Y., Hilliard, G., Ferguson, T., and Millhorn, D. E. (2003) J. Biol. Chem. 278 15911-15916 [DOI] [PubMed] [Google Scholar]
- 35.Triantafyllou, A., Liakos, P., Tsakalof, A., Georgatsou, E., Simos, G., and Bonanou, S. (2006) Free Radic. Res. 40 847-856 [DOI] [PubMed] [Google Scholar]
- 36.D'Angelo, G., Duplan, E., Boyer, N., Vigne, P., and Frelin, C. (2003) J. Biol. Chem. 278 38183-38187 [DOI] [PubMed] [Google Scholar]
- 37.Baek, J. H., Mahon, P. C., Oh, J., Kelly, B., Krishnamachary, B., Pearson, M., Chan, D. A., Giaccia, A. J., and Semenza, G. L. (2005) Mol. Cell 17 503-512 [DOI] [PubMed] [Google Scholar]