Abstract
Eph receptors are found in a wide variety of cells in developing and mature tissues and represent the largest family of receptor tyrosine kinases, regulating cell shape, movements, and attachment. The receptor tyrosine kinases conduct biochemical signals across plasma membrane via lateral dimerization in which their transmembrane domains play an important role. Structural-dynamic properties of the homodimeric transmembrane domain of the EphA1 receptor were investigated with the aid of solution NMR in lipid bicelles and molecular dynamics in explicit lipid bilayer. EphA1 transmembrane segments associate in a right-handed parallel α-helical bundle, region (544-569)2, through the N-terminal glycine zipper motif A550X3G554X3G558. Under acidic conditions, the N terminus of the transmembrane helix is stabilized by an N-capping box formed by the uncharged carboxyl group of Glu547, whereas its deprotonation results in a rearrangement of hydrogen bonds, fractional unfolding of the helix, and a realignment of the helix-helix packing with appearance of additional minor dimer conformation utilizing seemingly the C-terminal GG4-like dimerization motif A560X3G564. This can be interpreted as the ability of the EphA1 receptor to adjust its response to ligand binding according to extracellular pH. The dependence of the pKa value of Glu547 and the dimer conformational equilibrium on the lipid head charge suggests that both local environment and membrane surface potential can modulate dimerization and activation of the receptor. This makes the EphA1 receptor unique among the Eph family, implying its possible physiological role as an “extracellular pH sensor,” and can have relevant physiological implications.
Erythropoietin-producing hepatocellular (Eph)3 receptor and corresponding membrane-bound Eph receptor-interacting proteins (ephrins) transduce signal in a cell-cell contact-dependent fashion, thereby coordinating growth, differentiation, and patterning of almost every organ and tissue during vertebrate and invertebrate embryogenesis (1, 2). In adult organism, Eph-ephrin interactions can also trigger a wide array of cellular responses, including cell boundary formation, motility, adhesion, and repulsion, especially for neuronal and endothelial cells, whereas deregulated reemergence of Eph function appears to contribute to mechanism of tissue injury and of tumor invasion and metastasis. Intriguingly the Eph-ephrin interactions may have a role in synaptic plasticity, learning, memory formation, and mental disease (3, 4). The Eph receptors represent the largest family of receptor tyrosine kinases and are divided into subclasses A and B based on the sequence homology of their extracellular parts, the structure, and the binding affinity (5). Ephrin-A ligands share a membrane-tethered glycosylphosphatidylinositol anchor, whereas ephrin-B ligands have a transmembrane domain and a short cytoplasmic tail. The Eph receptors and ephrins are not only numerous, but their relationship is also complex (6). Receptor-ligand binding is highly promiscuous within each subclass, and there are several examples of interactions between members of different subclasses (7). In addition to their role as conventional ligands for the related receptor tyrosine kinase counterparts (frequently referred to as “forward signaling”), the ephrins possess a receptor-like ability to transduce signals into the cell that presents these molecules on its surface (so-called “reverse signaling”) (2).
Like other receptor tyrosine kinases, both types of Eph receptors contain a single transmembrane (TM) helical domain with an N-terminal glycosylated extracellular region comprised of a ligand-binding domain with immunoglobulin-like motifs, a cysteine-rich region with an epidermal growth factor-like motif, and two fibronectin type III repeats. The cytoplasmic region includes a juxtamembrane segment, the kinase domain, and a sterile α-motif (2). Upon cell-cell contact, the N-terminal globular domains of two Eph receptors recognize two ephrins in the opposing membrane and bind with high affinity and specificity to form a heterotetrameric ephrin-Eph receptor signaling complex. Although the activation of signal transduction cascade of most receptor tyrosine kinases is controlled by a ligand-induced receptor dimerization or by reorientation of monomers in preformed receptor dimers (8-10), additional formation of higher order Eph-ephrin signaling clusters is required as the signals resulting from higher order clusters seem to differ from those resulting from a single dimer (6). Such oligomerization is restricted within specialized plasma membrane microdomains enriched by glycosphingolipids and cholesterol, known as “lipid rafts,” which can impart additional specificity to the ephrin-Eph signaling (11). Several weak ephrin-ephrin and receptor-receptor contacts could promote the association of the complex into an interconnected network. Regions that mediate low affinity intermolecular binding have been identified in the extracellular domain and in the juxtamembrane cytoplasmic segment of ephrins as well as in the ephrin-binding domain, cysteine-rich region, and cytoplasmic sterile α-motif domain of Eph receptors (2, 12).
Association of TM helices plays an important role both in the assembly and in the function of membrane proteins. Nevertheless despite the fact that structural characterization of membrane proteins is of particular interest in contemporary structural biology, the examples of TM domains of single spanning membrane proteins whose noncovalent dimeric spatial structure has been determined experimentally are exceptional (GpA (13), BNip3 (14), and ErbB2 (15)) compared with the hundreds of structures available for membrane proteins to date partly because of difficulties associated with expression and solubilization of transmembrane proteins. This disproportion also reflects that many biologically relevant TM helix interactions are weak, providing a mechanism for modulation of protein function through reversible association. Accumulating evidences reveal that the proper lateral dimerization of TM domains of receptor tyrosine kinase members is required for biochemical signal transduction across plasma membrane (16, 17). The EphA1 receptor tyrosine kinase initially isolated from a hepatoma cell line (18) is widely expressed in human tissues and found to be up-regulated in breast, colon, lung, kidney, gastric, and head cancers (7). Recently our studies have demonstrated that fluorescently labeled TM fragments of the EphA1 receptor self-associate in liposomes, forming helical dimers crossing the lipid bilayer (19). Consistent with this notion and with the fact that Eph-ephrin signaling requires lateral dimerization or aggregation in cell membrane (2, 6, 12, 20), we present the high resolution spatial structure of the homodimeric TM domain of the human EphA1 receptor obtained by heteronuclear NMR in lipid bicelles combined with molecular dynamics (MD) relaxation in explicit membrane. The found dimerization specificity and dependence of structural-dynamic properties of the EphA1 TM domain on ionization state of the membrane embedded glutamate carboxyl group tempted us to speculate how extracellular conditions and local cell membrane environment can modulate biological activity of the receptor, providing a better understanding of the molecular mechanism of the Eph-ephrin signaling.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification—The DNA sequence encoding human EphA1 fragment 536-573 (EphA1tm) was synthesized from six oligonucleotides by PCR. The TrxA-EphA1tm fusion protein was constructed by fusing the coding sequence for thioredoxin of Escherichia coli with the N-terminal His tag extension to the coding sequence for EphA1tm in the pGEMEX1 vector (Promega). To facilitate purification, the His tag and enterokinase cleavage site were placed between the coding sequences for thioredoxin and EphA1tm. The fusion protein was expressed in E. coli BL21(DE3)pLysS and grown in 2-liter flasks at 37 °C in M9 minimal medium containing (15NH4)2SO4 or (15NH4)2 SO4/[U-13C]glucose (both from Cambridge Isotope Laboratories, Inc.) for the production of uniformly 15N- or 15N/13C-labeled protein samples. After induction with 50 μm isopropyl β-d-thiogalactopyranoside and an additional 40 h of growth at 13 °C, the cells were harvested and stored at -20 °C.
Cell pellets (1-liter equivalents) were resuspended in 50 ml of lysis buffer (50 mm Tris, pH 8.0, 150 mm NaCl, 10 mm 2-mercaptoethanol, 10 mm imidazole, 15 mm Triton X-100, 0.2 mm phenylmethylsulfonyl fluoride) and lysed by ultrasonication. Centrifugally clarified lysate was applied to Chelating Sepharose Fast Flow beads (Amersham Biosciences) pretreated with NiSO4, washed with 70 mm imidazole, and eluted with 200 mm imidazole. After overnight incubation with recombinant human enterokinase light chain, the cleaved EphA1tm was passed through Chelating Sepharose Fast Flow, loaded onto an SP-Sepharose Fast Flow column (Amersham Biosciences), and eluted with a gradient of NaCl. An additional purification step was applied using reverse phase chromatography to remove a trace of thioredoxin, resulting in >97% purity of the EphA1tm sample. Yields of ∼7 mg of protein/liter of the cell culture could be obtained by this procedure. Protein identity and purity were confirmed by gel electrophoreses, mass spectrometry, and NMR spectroscopy in a 1:1 methanol/chloroform mixture containing 5-10% water.
NMR Spectroscopy and Structure Determination—NMR experiments were performed on 600-MHz (1H) Varian Unity spectrometer equipped with a pulsed field gradient unit and a triple resonance probe. NMR spectra were acquired at 40 °C using 1 mm samples of EphA1tm incorporated into 1:4 dimyristoylphosphatidylcholine (DMPC)/dihexanoylphosphatidylcholine (DHPC) lipid bicelles (with a lipid/protein molar ratio of 35) dissolved in a buffer solution (at pH 4.3, 4.9, and 6.3) containing 10 mm deuterated sodium acetate, 0.15 μm sodium azide, 1 mm EDTA, and either 5 or 99.9% D2O unless otherwise specified. For the NMR sample preparation the DMPC and DHPC lipids with deuterated hydrophobic tails were used. The deuterated 1,2-di-[2H27]myristoyl-sn-glycero-3-phosphocholine (d54 -DMPC) and 1,2-di-[2 H11]myristoyl-sn-glycero-3-phosphocholine (d22-DHPC) were synthesized from sn-glycero-3-phosphocholine by acylation with anhydride of d27-myristic acid and d11-hexanoic acid, respectively, as described previously (21). Three EphA1tm samples were prepared: uniformly 15N/13C-labeled, 15N-labeled, and a 1:1 mixture of uniformly 15N/13C-labeled and unlabeled proteins (“heterodimer” sample). The samples were initially subjected to several freeze/thaw cycles for the achievement of uniform protein distribution among the lipid bicelles. To verify the validity of NMR experimental conditions, circular dichroism studies of EphA1tm solubilized with the DMPC/DHPC bicelles and DMPC unilamellar liposomes at pH 4.9 were performed (see supplemental experimental procedures and supplemental Fig. S1A). To assess the size of the bicelles with the protein and demonstrate their stability in the pH range used, dynamic light-scattering measurements were performed at pH 4.3, 4.9, and 6.3 (see supplemental experimental procedures). There was no difference of the bicelle size in the entire range of pH; therefore only the results for pH 4.3 and 6.3 are presented in supplemental Fig. S1B.
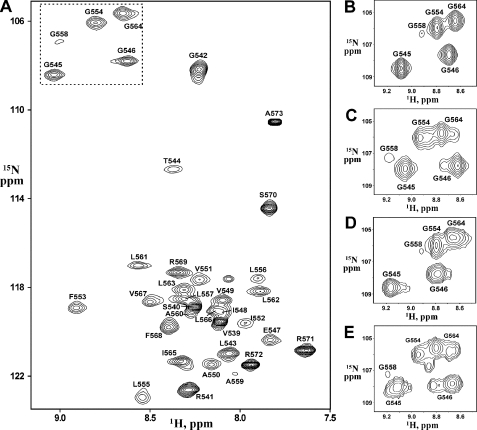
The backbone and side chain 1H, 13C, and 15N resonances of EphA1tm were assigned at pH 4.3, 4.9, and 6.3 using standard triple resonance techniques (22, 23). Two- and three-dimensional 1H-15N (Fig. 1A) and 1H-13C heteronuclear single quantum coherence (HSQC), 15N-edited total correlation spectroscopy (40-ms mixing time), and HNCA, HN(CO)CA, HNCACB, and CBCA(CO)NH spectra in H2O provided backbone and partial side chain assignments, whereas HCCH total correlation spectroscopy (15.6- and 23.4-ms mixing times) and 1H nuclear Overhauser effect (NOE) spectroscopy (NOESY; 60-ms mixing time) experiments in D2O facilitated side chain assignments. Resonance assignments were performed with the CARA software.
FIGURE 1.
NMR spectra of the EphA1tm dimer at different pH. A, The 1H-15N HSQC spectrum of 1 mm 15N-labeled EphA1tm in DMPC/DHPC bicelles, 10 mm deuterated sodium acetate at 40 °C and pH 4.9. The 1H-15N backbone resonance assignments are shown. B and C, fragments of the 1H-15N HSQC spectra of EphA1tm embedded into the zwitterionic DMPC/DHPC bicelles at pH 4.3 and 6.3, respectively. D and E, fragments of the 1H-15N HSQC spectra of EphA1tm embedded into the negatively charged DMPC/DHPC/DMPG bicelles at pH 4.5 and 6.7, respectively. B-E include the cross-peaks of Gly residues situated in the EphA1tm TM region. The corresponding region of the spectrum shown in A is highlighted by a dashed line.
Side chain pH titration of Glu547 was monitored by the chemical shift changes of neighboring amide NH groups using a set of 1H-15N HSQC spectra collected on the 15N-labeled sample of the EphA1tm dimer solubilized in 1:4 DMPC/DHPC or in 1:8:1 DMPC/DHPC/dimyristoylphosphatidylglycerol (DMPG) (Avanti Polar Lipids) bicelle suspension at a pH range from 4.2 to 6.7. The pH was adjusted by adding small amounts of HCl and NaOH and measured with a Radiometer glass electrode. The pKa value of titrating carboxyl group of Glu547 was obtained by nonlinear least-square fits of the pH dependence of the chemical shifts to a modified Hill equation (24) using the Mathematica 5.1 software (Wolfram Research).
The values of heteronuclear 15N{1H} steady-state NOE, 15N longitudinal (T1), and transverse (T2) relaxation times (supplemental Fig. S5) were obtained for the 15N-labeled sample at pH 4.3, 4.9, and 6.3 as described previously (25). The local effective rotation correlation times (τR) for the individual 15N nuclei were calculated from the T1/T2 ratio using the DASHA software (26). The effective molecular mass of the EphA1tm dimer embedded into the DMPC/DHPC bicelle was estimated according to the empirical dependence (27) from overall rotation correlation time averaged over 15NH groups with 1H{15N} NOE >0.6 and without noticeable cross-peak broadening in 1H-15N HSQC spectra.
NMR spatial structures of the EphA1tm dimer were calculated using the CYANA program (28). Intramonomeric NOE distance restraints (supplemental Fig. S2) were identified with CARA through the analysis of three-dimensional 15N- and 13C-edited NOESY (60- and 80-ms mixing times) spectra (23) obtained for 15N- and 15N/13C-labeled samples in H2O and D2O (at pH 4.3, 4.9, and 6.3), respectively. The two-dimensional 1H NOESY (80-ms mixing time) spectrum acquired for the unlabeled sample was used as an additional source of structural information concerning aromatic ring protons. Intermonomeric distance restraints (supplemental Fig. S3) were derived from three-dimensional 15N-edited NOESY and three-dimensional 13C F1-filtered/F3-edited NOESY spectra (29) acquired with a 80-ms mixing time for the 15N-labeled and heterodimer samples in H2O and D2O (at pH 4.3, 4.9, and 6.3), respectively. Stereospecific assignments and torsion angle restraints for ϕ, ψ, and χ1 were obtained by the analysis of local conformation in CYANA using sequential NOE data and the available 3JHNα and 3JNβ coupling constants evaluated qualitatively from three-dimensional 1H-15N HNHA and 1H-15N HNHB experiments (23). Backbone dihedral angle restraints were also estimated based on the assigned chemical shifts using the TALOS program (30). Slowly hydrogen-deuterium exchanging amide groups were identified by reconstituting lyophilized 15N-labeled sample in D2O at pH 4.9 and recording a series of 1H-15N HSQC spectra (supplemental Fig. S2). Taking into account the 15N relaxation data at different pH values (Fig. 2A), the slowly exchanging amide protons were assigned as donors of backbone hydrogen bonds with related hydrogen acceptor partners on the basis of preliminary structure calculations. In addition, based on the observed chemical shift perturbations, local NOE contact rearrangement and backbone dynamics perturbation upon pH titration, the characteristic N-capping hydrogen bonds (31-33) between residues Thr544, Gly546, and Glu547 (see “Results and Discussion”) were identified. Corresponding hydrogen bond restraints were used in subsequent calculations for d(O,N), d(O,HN), and d(C,HN) in accordance with angle and distance criteria for different types of hydrogen bonds (34). The EphA1tm dimer was modeled as the dimer of two EphA1tm fragments linked by 20 pseudoresidues to give the monomers enough mutual arrangement. The standard CYANA simulated annealing protocol was applied to 100 random structures using the angle and distance restraints symmetrically doubled for each dimer subunit, and the resulting 12 NMR structures of the EphA1tm dimer with the lowest target function were selected for pH 4.3 and 6.3. Protein-lipid NOE contacts (supplemental Fig. S4) were identified from three-dimensional 15N-edited NOESY and three-dimensional 13C F1-filtered/F3-edited NOESY spectra acquired with an 80-ms mixing time for the 15N- and 15N/13C-labeled samples in H2O and D2O, respectively.
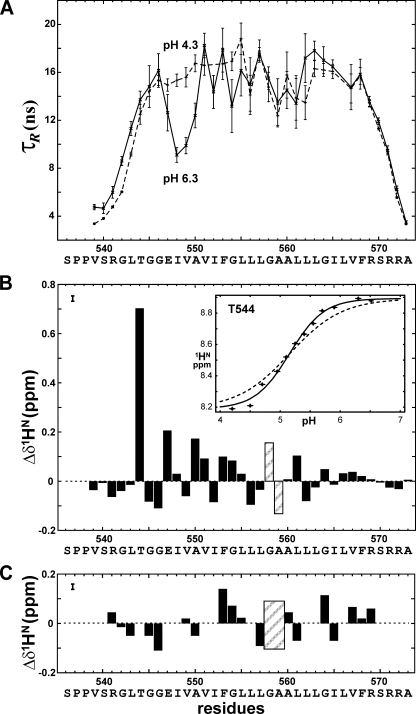
FIGURE 2.
Internal dynamics and chemical shift perturbations upon deprotonation of Glu547 carboxyl group of EphA1tm. A, local effective rotation correlation times (τR) for the EphA1tm dimer embedded in the DMPC/DHPC bicelles at pH 4.3 (dashed line) and pH 6.3 (solid line). Uncertainties are shown by bars. B, the HN chemical shift differences for the major form of the EphA1tm dimer embedded in the DMPC/DHPC bicelles at pH 6.3 and pH 4.3. The chemical shift uncertainty is shown by a bar in the left top corner. In the inset panel the pH dependence of HN chemical shift of Thr544 is presented. The experimental and theoretical sigmoid-like curves with Hill coefficients equal to 1.5 and 1 are shown by dashed and solid black lines, respectively. C, the HN chemical shift differences between major and minor components of doubling cross-peaks in the 1H-15N HSQC spectra acquired for the EphA1tm dimer embedded in the DMPC/DHPC bicelles at pH 6.3. Areas of the Gly558 and Ala559 residues subjected to strong HN resonance broadening in the NMR spectra over the entire pH range used are shaded.
Restrained energy relaxation of the representative NMR structures of the EphA1tm dimer at pH 4.3 and 6.3 was performed by molecular dynamics in hydrated explicit DMPC bilayer using the GROMACS 3.3.1 package (35). Construction of the protein/lipid system and MD protocol are described in the supplemental experimental procedures. Two 5-ns MD runs for the representative models of the EphA1tm dimer with protonated (uncharged) or negatively charged carboxyl side chain group of Glu547 in both dimer subunits corresponding to pH 4.3 and 6.3, respectively, were carried out with the NMR-derived intra- and intermonomeric distance restraints. To check the stability of the resulting systems, 3-ns continuations of the MD runs were performed without distance restraints. Equilibrium parts of MD trajectories (last 2 ns) were analyzed using home-made software and utilities supplied with the GROMACS package.
The TM regions of Eph receptors were predicted with TMpred (36). Hydrophobic properties of α-helices were calculated using the molecular hydrophobicity potential approach (see supplemental experimental procedures for details) (37). The contact area between the dimer subunits was calculated using the DSSP program (38) as a difference between the accessible surface areas of EphA1tm residues in the monomer and dimer. The EphA1tm structures were visualized with MOL-MOL (39) and PyMOL (40).
RESULTS AND DISCUSSION
Validation of EphA1tm NMR Sample—The 38-residue EphA1 fragment 536-573 (named EphA1tm) with the sequence SPPVSRGLTGGEIVAVIFGLLLGAALLLGILVFRSRRA including predicted hydrophobic TM segment (underlined) flanked by polar N- and C-terminal regions was prepared as described above. Although different membrane-mimicking systems have been used for investigations of membrane-bound proteins, currently discoidal mixed bicelles composed of a small circular bilayer of long-chain lipids surrounded by a rim of short-chain lipids have proved more suitable for structural studies by high resolution NMR (for a review, see Ref. 41). Indeed our recent experience of resolving the dimeric structure of the TM domain of human proteins, including a representative of receptor tyrosine kinase, allows stating that bicelles are a fairly adequate model of lipid membrane (14, 15). Therefore, the 15N and 13C isotope-labeled EphA1tm samples were solubilized in an aqueous suspension of bicelles consisting of DMPC and DHPC lipids with deuterated hydrophobic tails using DMPC/DHPC and lipid/protein molar ratios of 0.25 and 35, respectively. The prepared sample solutions were clear, and the preliminary NMR spectra (Fig. 1A) seen were suitable for further structural NMR studies with conventional 15N/13C heteronuclear NMR technique. Circular dichroism spectra, recorded to validate the NMR experimental conditions, proved virtually identical for EphA1tm incorporated into the DMPC/DHPC bicelles and the DMPC unilamellar liposomes (phospholipid bilayer), revealing an α-helical structure of the fragment in the membrane mimetics (supplemental Fig. S1). The measured 15N{1H} NOE, 15N T1 and T2 values (supplemental Fig. S5), and calculated local effective rotation correlation times (τR) (Fig. 2A) are indicative of a relatively stable TM segment 544-569 flanked by highly flexible N- and C-terminal regions. Overall rotation correlation time estimated from T1/T2 ratio on the TM region is ∼16 ns, which corresponds well to a particle mass (∼43 kDa) of the EphA1tm dimer surrounded by ∼70 lipid molecules composing a bicelle with effective hydrodynamic radius of 24 ± 2Å measured by dynamic light scattering (supplemental Fig. S1). In addition, protein-lipid NOE contacts with polar lipid heads observed in the regions 542-549 and 564-571 revealed that the region spans the hydrophobic phase of the bicelle (supplemental Fig. S4), therefore mimicking the embedding of the EphA1 TM domain into a bilayer lipid membrane reasonably well.
Conformational Exchange and Diversity of the EphA1tm Dimer—The NMR experiments were carried out in a pH range from 4.3 to 6.3 to overlap the pKa value (see below) of Glu547 carboxyl group and to check the influence of its ionization state on the structural and dynamic properties of EphA1tm. The preliminary analysis of NMR data showed that, being incorporated into lipid bicelles, the TM part of EphA1tm is involved in a micro- to millisecond conformational exchange (intermediate on the NMR time scale) revealed by a cross-peak broadening in the NMR spectra acquired over the pH range used (Fig. 1). The appearance of a disproportional doubling of some amide cross-peaks in the 1H-15N HSQC spectra at high pH values indicates the presence of another slow conformational exchange process induced by deprotonation of the carboxyl group of Glu547. Besides upon pH titration, the 15N{1H} NOE, 15N T1 and T2, and local τR values (Fig. 2A and supplemental Fig. S5) exhibit noticeable variations on the region 547-550, assuming a local pH-dependent perturbation of backbone dynamics of these residues in pico- to nanosecond time scale. An additional slow local conformational exchange in the solvent-exposed N terminus of EphA1tm is observed because of cis-trans isomerization of the Ser536-Pro537 and Pro537-Pro538 peptide bonds as confirmed by characteristic NOE contacts (42). Thus, there are several types of internal motions of EphA1tm embedded into lipid bicelles, each of them having a different time scale and different degree of pH dependence.
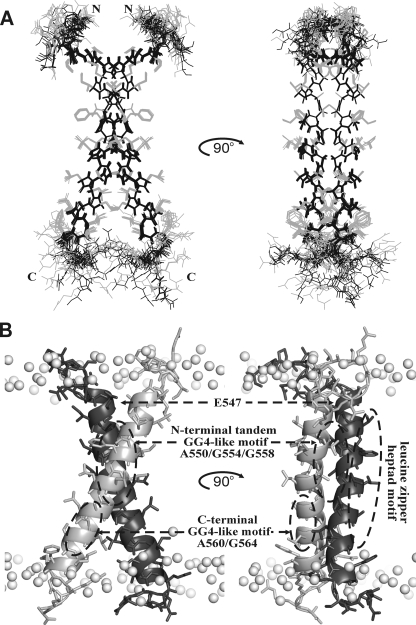
Major Conformation of EphA1tm in Lipid Bicelles—The unique patterns of intra- and intermonomeric NOE contacts (supplemental Figs. S2 and S3) identified at pH values of 4.3 and 6.3 directly demonstrated that in the major conformation the TM fragment forms a dimer with parallel α-helical subunits symmetrical on the NMR time scale (at least in the millisecond range). Therefore, the NMR-derived dihedral angle restraints and both intra- and intermonomeric distance restraints obtained at pH 4.3 or 6.3 were assigned for each dimer subunit that resulted in a dimer symmetrical over the ensemble of calculated structures (Fig. 3A and supplemental Fig. S6). The representative NMR structures of the EphA1tm dimer for both pH cases were subjected to energy relaxation via MD in explicit hydrated lipid bilayer of DMPC with the experimentally derived distance restraints that allowed adapting the structures to the explicit membrane and refining the helix packing in the dimer (supplemental Figs. S7 and S8, A and B). A survey of the structural statistics for the final ensembles of the 12 NMR-derived structures of the EphA1tm dimer and for the representative dimer structures relaxed in the explicit membrane is provided in Table 1. After restrained energy relaxation, the membrane-spanning α-helices of EphA1tm cross at an angle θ ∼-44° with the distance d ∼ 6.2 Å between helix axes, forming a right-handed parallel dimer tilted slightly from the membrane normal (Fig. 3B). The flexible N and C termini of EphA1tm are unstructured and exposed to water. It is important to note that before and after the energy relaxation both helix crossing angle and distance between helix axes are increased somewhat with pH (Table 1 and Fig. 4, A and B). The 3-ns continuation of MD without NMR restraints did not cause considerable change of the overall structure of the dimer, indicating its relative stability in the membrane.
FIGURE 3.
Spatial structure of the EphA1tm dimer. A, ensemble of 12 NMR-derived structures of the EphA1tm dimer at pH 4.3 after alignment of the backbone atoms of residues (544-569)2 of both dimer subunits. Side chain and backbone heavy atom bonds of residues (540-573)2 are shown in gray and black, respectively. B, ribbon diagrams of the EphA1tm dimer relaxed in the explicit DMPC bilayer. Gray balls show phosphorus atoms of lipid heads. The spatial locations of the Glu547 residues and the three characteristic dimerization motifs of EphA1tm are marked by dashed ovals.
TABLE 1.
Structural statistics for the ensemble of 12 best NMR structures of the EphA 1 tm dimer and results of restrained MD relaxation
| pH 4.3 | pH 6.3 | |
|---|---|---|
| NMR distance and dihedral restraints | ||
| Total unambiguous NOE restraints | 998 | 914 |
| Intraresidue | 568 | 508 |
| Inter-residue | 386 | 378 |
| Sequential (|i - j| = 1) | 212 | 214 |
| Medium range (1 < |i - j| ≤ 4) | 176 | 164 |
| Long range (|i - j| > 4) | 0 | 0 |
| Intermonomeric | 44 | 28 |
| Hydrogen bond restraints (upper/lower) | 134/134 | 120/120 |
| Total torsion angle restraints | 102 | 102 |
| Backbone ϕ | 40 | 40 |
| Backbone ψ | 34 | 34 |
| Side chain χ1 | 28 | 28 |
| Structure calculation statistics | ||
| CYANA target function (Å2) | 1.39 ± 0.13 | 0.97 ± 0.33 |
| Restraint violations | ||
| Distance (>0.2 Å) | 0 | 0 |
| Dihedral (>5°) | 0 | 0 |
| Average pairwise r.m.s.d.a (Å), stable α-helical region (551-569)2 | ||
| Backbone atoms | 0.50 ± 0.14 | 0.42 ± 0.15 |
| All heavy atoms | 0.88 ± 0.14 | 0.64 ± 0.19 |
| Ramachandran analysisb | ||
| Residues in most favored regions (%) | 80.8 | 82.3 |
| Residues in additional allowed regions (%) | 14.6 | 16.8 |
| Residues in generously allowed regions (%) | 3.7c | 0.6c |
| Residues in disallowed regions (%) | 0.9c | 0.3c |
| Helix-helix packing contact surface area (Å2), TMl region (544-569)2 | 590 ± 10 | 490 ± 10 |
| Angle θ (°) between the TM helix axes | −39 ± 2 | −48 ± 2 |
| Distance d (Å) between the TM helix axes | 7.1 ± 0.1 | 7.3 ± 0.1 |
| Restrained MD relaxation (last 2 ns) | ||
| Contact surface area (Å2), TM region (544-569)2 | 640 ± 20 | 530 ± 30 |
| Angle θ (°) between the TM helix axes | −42 ± 2 | −46 ± 2 |
| Distance d (Å) between the TM helix axes | 5.9 ± 0.2 | 6.5 ± 0.3 |
Root mean square deviation.
Ramachandran statistics were determined using CYANA (28).
Residues from unfolded and flexible regions.
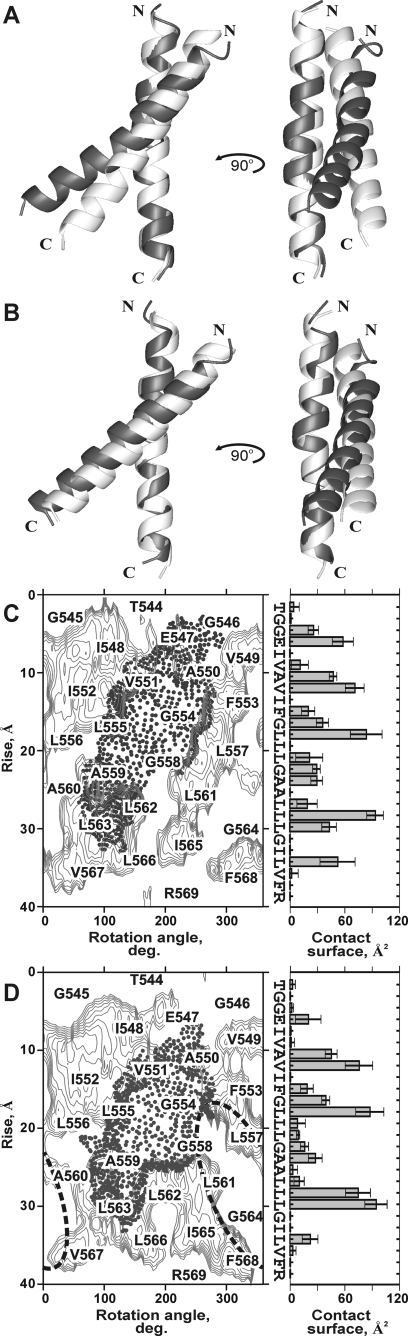
FIGURE 4.
Helix packing perturbations in the EphA1tm dimer caused by deprotonation of Glu547 carboxyl group. A and B, ribbon diagrams of the NMR-derived spatial structures of the EphA1tm dimer before (A) and after (B) MD relaxation in explicit lipid bilayer. The dimer structures (superimposed on one subunit) obtained for EphA1tm embedded in the DMPC/DHPC bicelles at pH 4.3 and 6.3 are shown in light and dark gray, respectively. C and D, the EphA1tm helix packing interface after MD relaxation in explicit lipid bilayer. Hydrophobicity maps (on the left) for EphA1tm helix surface with contour isolines encircling hydrophobic regions with high values of molecular hydrophobicity potential are covered by areas of dark points indicating the N-terminal dimerization interface realized in the major right-handed dimer conformation at pH 4.3 (C) and pH 6.3 (D). A possible C-terminal dimerization interface implying a left-handed crossing of the EphA1tm TM helices is highlighted by dashed curved lines (D). The helix packing contact areas per EphA1tm residue averaged over the equilibrium part (last 2 ns) of restrained MD relaxation of the dimer structure are presented at the right of the maps. The spreads of the contact area are shown by bars. deg., degree.
In the major conformation, EphA1tm self-associates through a tandem variant of the GG4-like motif (17) A550X3G554X3G558 (the so-called “glycine zipper” (43)) composed of residues with small side chains allowing close approach of the helices (Fig. 3B). The weakly polar contact area of this motif located in the N-terminal part of the TM helix is shielded from lipid tails by the side chains of Val549, Val551, Phe554, Leu555, Leu557, Ala559, Leu562, and Leu563 (Fig. 4, C and D, and supplemental Fig. S7). The weak polar surface in the C-terminal part of the dimerization motif A550X3G554X3G558 is extended by the surface of the Ala559 and Ala560 residues that appears to provide a certain conformational flexibility for interacting TM helices. Indeed the remarkable NMR signal broadening observed over the entire pH range used is indicative of micro- to millisecond motions (intermediate on the NMR time scale) in the central part of the TM helix around Gly558, Ala559, and Ala560 residues. Most likely, the motions provide minor rearrangements of the helix packing within the EphA1tm dimer accompanied by some bending of the helix (see below).
Besides the N-terminal glycine zipper motif A550X3G554X3G558 used, two potential dimerization sites, the C-terminal GG4-like motif A560X3G564 and a leucine zipper heptad motif (44) IV549X5LL556X5LL563, are located on the lipid-exposed surface of the EphA1tm dimer (Figs. 3B and 4, C and D). These findings imply a possibility of the EphA1 TM domain to be involved in homodimerization with another mode and/or in additional helix-helix interactions with other partners (e.g. in higher order oligomers). This is consistent with the notions that both ephrins and Eph receptors are loosely preclustered in lipid rafts, forming low affinity ephrin-ephrin and Eph-Eph dimers, and that ephrin docking may cause an Eph receptor rearrangement, triggering aggregation into larger Eph-ephrin clusters (2, 6, 11, 12, 20).
Perturbations in Local Structure and Dynamics of the Dimeric EphA1tm Major Conformation upon Deprotonation of Glu547 Carboxyl Group—At the N-terminal part of the helix packing interface of the dimer there are two Glu547 residues facing each other (Figs. 3B and 4C). Within the range of pH used in the experiments, Glu547 is the only ionizable group in EphA1tm. In the zwitterionic DMPC/DHPC bicelles the pKa value of Glu547 is equal to 5.1 ± 0.1 (Fig. 2B and supplemental Fig. S9), exceeding the values typical for N-cap glutamate exposed to aqueous solution (32) by ∼0.6 pKa units. This is hardly surprising because Glu547 is located in the EphA1tm transmembrane part, which is buried from the bulk water. More interestingly, the Hill coefficient calculated from the titration curves is about 1.5, whereas values below 1 should be expected based on the mutual location of the Glu547 residues adjacent to each other in the dimer. The positive cooperativity of the Glu547 pH titration can be explained by conformational rearrangements induced by the process. Indeed the structure calculations of the EphA1tm dimer at pH 4.3 and 6.3 revealed considerable local rearrangements near the N terminus of the TM helix and slight overall changes of helix packing in the dimer (Table 1 and Fig. 4). The changes of local structure were accompanied by destabilization of the TM helix between the first and the second turns (residues 547-550) as evidenced by the decrease of the local effective correlation times (τR) upon the Glu547 deprotonation (Fig. 2A).
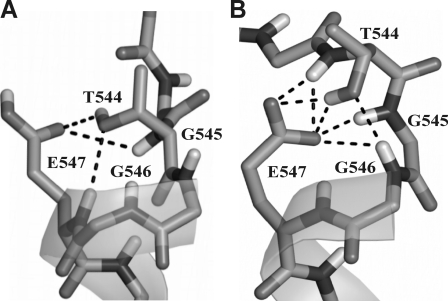
Ten HN resonances along the entire EphA1tm helix (mostly in its N-terminal part) shift by at least 0.1 ppm in the 1H-15N HSQC spectra upon Glu547 deprotonation; the downfield shift for Thr544 is exceptionally high, 0.7 ppm (Fig. 2B). The extent of the amide proton downfield pH titration shift might be used to estimate the population of transient hydrogen bond with the carboxyl group (45). Based on the observed chemical shift change, the population of the Thr544 amide and Glu547 carboxyl group hydrogen bond is approximately 40%. The number must be used with care, however, because the shift may depend on the hydrogen bond geometry as well as on the population. Thus, taking into account the overall dimer structure and the local NOE contacts, it can be concluded that the backbone HN of Thr544 forms a hydrogen bond with the side chain CδOO of Glu547; this is a widespread N-capping box interaction (31-33) stabilizing the N terminus of the helix. In addition to this interaction, structure calculations revealed that the side chain hydroxyl group of Thr544 accepts a classical N-cap hydrogen bond from the amide group of Glu547 that switches to the amide group of Gly546 upon deprotonation of Glu547 (Fig. 5). Thus, these hydrogen bonds fix the helix entry over the entire pH range used; this is consistent with the 15N relaxation data (Fig. 2A and supplemental Fig. S5), revealing that this region is relatively rigid on the pico- to nanosecond time scale. According to the results of the MD relaxation of the dimer structure, deprotonation of the carboxyl group is further accompanied by transient hydrogen bonding between the side chain CδOO of Glu547 and the amide groups of Gly545, Gly546, and Glu547 (Fig. 5) with consequential backbone rearrangements, resulting in a breaking of hydrogen bonds on the helical region 548-550. Because only a single averaged resonance is seen in the NMR spectra for each of these amide protons as well as for the Glu547 side chain, these hydrogen bonds are formed and broken in a rapid, dynamic equilibrium with lifetimes of the order of milliseconds or shorter. These rearrangements explain the increased backbone mobility evidenced by 15N relaxation data obtained at pH 6.3 according to which the high amplitude internal motions within the region 547-550 occur on the pico- to nanosecond time scale. On the millisecond time scale, the pattern of NOE contacts at pH 4.3 and 6.3 reveals α- and 310-helical structure of this region, respectively. Besides that, as revealed by the water exchange peaks in the 15N-edited NOESY spectra, the helix distortion is accompanied by an increase of water penetration inside the dimerization interface of EphA1tm from Ile548 up to Ala550 when pH was changed from 4.3 to 6.3. Thus, we observe a complex dynamic picture of the local “melting” of the N-terminal part of the EphA1tm helix (actually α-to-310-to-coil transitions) because of electrostatic repulsion of the two negatively charged carboxyl groups of the Glu547 residues spatially adjacent in the dimer.
FIGURE 5.
N-cap configurations of the EphA1tm helix depending on the ionization state of Glu547 carboxyl group. Possible transient hydrogen bond connections in the N terminus of the TM helix observed during MD relaxation of the NMR-derived structures of the EphA1tm dimer embedded in the DMPC/DHPC bicelles at pH 4.3 (A) and 6.3 (B) are shown by dashed lines.
Besides according to the obtained NMR structures, the Glu547 deprotonation caused slight bending of the TM helices of the EphA1tm dimer in the region of the dimerization motif (Fig. 4, A and B). The helix bending is also confirmed by the stepwise pattern of the HN proton chemical shift changes observed during pH titration (Fig. 2B and supplemental Fig. S10A). The changes have periodicity of 3-4 residues and are propagated far away from the carboxyl group. They can be interpreted as a periodical slight shortening and elongation of the backbone hydrogen bonds (helix bending) (46) superimposed upon the chemical shift perturbations caused by Glu547 titration.
Minor Form of the EphA1tm Dimer Induced by Deprotonation of Glu547—Besides the perturbations in local structure and dynamics of the major conformation of the EphA1tm dimer, deprotonation of Glu547 results in appearance of minor component of some cross-peaks in the 1H-15N HSQC spectra, reaching 15-20% of the major peak at pH 6.3 (Fig. 1C). It can be assumed that the Glu547 deprotonation results in a shift of the dimer-monomer equilibrium or induces partial transition of the dimer into a different conformation. The fact that the occupancy of the minor state is practically independent of the lipid/protein ratio in the range from 2 to 1.5 EphA1tm per bicelle supports the latter assumption. Intriguingly the chemical shift perturbations upon the cross-peak doubling (Fig. 2C and supplemental Fig. S10B) do not only affect the N-terminal glycine zipper motif A550X3G554X3G558 used but are also pronounced in the vicinity of the C-terminal GG4-like dimerization motif A560X3G564 not involved in formation of the major form of the dimer. These findings tempt us to assume that the deprotonation of the Glu547 induces partial dimerization through the C-terminal dimerization motif and that we are observing slow transitions between two alternatively packed dimers. Unfortunately relatively small occupancy and, consequentially, low NMR signal intensity did not allow obtaining the structural information about the minor state of the dimer. However, the second helix packing interface can be indirectly mapped based on the chemical shift differences between major and minor states of the dimer. A possible C-terminal dimerization interface includes Leu557, Ala560, Gly564, and Val567 residues and implies a left-handed crossing of the TM helices at an angle near 30° (Fig. 4D). Unlike the major dimer forming via the N-terminal glycine zipper motif (Figs. 3 and 4), in this case the negatively charged side chains of the Glu547 residues are separated and placed outward from the dimer interface, explaining the observed moderate pH dependence for chemical shifts of the minor component (Fig. 1, B and C, and supplemental Fig. S9B). Importantly according to the molecular hydrophobicity potential distribution, the weak polar surfaces of the N- and C-terminal dimerization motifs are spatially adjacent (Fig. 4D and supplemental Fig. S7A). The restrained MD relaxation of the NMR structures of the major dimer form revealed that the deprotonation of Glu547 leads to some rearrangements of the side chains of Leu557, Leu561, and Leu562 residues, which are located near the central part of the N-terminal helix packing interface (Fig. 4, C and D). These rearrangements open a weakly polar cleft connecting the N- and C-terminal dimerization motifs and could provide a pathway for a transition between the two states of the dimer. Hence it can be suggested that the weakly polar surface covered by residues Gly558, Ala559, and Ala560 in the central part of the TM helix is a pivotal point, making possible the transition from one state of the dimer to the other, that can be associated with the receptor activation-deactivation. Interestingly certain decreases of the local τR values can be observed in the same region (Fig. 2A) in agreement with the results of 3-ns unrestrained MD relaxations in explicit membrane, indicative of local perturbations of the helical structure without loss of overall structure of the dimer (supplemental Fig. S8C). Naturally correct disclosure of the actual reason of the observed disproportional cross-peak doubling requires additional experimental proofs, and we are working on it. Anyhow based on the data available so far it can be concluded that, being incorporated in the lipid bicelle, the EphA1tm dimer is capable of undergoing various transitions on the micro- to millisecond time scale between the conformational states separated by several energy barriers, which depend on the ionization state of the Glu547 residues.
The fact that EphA1tm forms dimers with only a limited amount of conformations implies that helix-helix interaction between the EphA1 TM domains can impose a certain restriction on the allowable conformational transitions undergone by the full-length receptor. Thus, our data suggest that the TM domains of Eph receptors do not merely play a passive role in signal transduction but can provide a leverage for underlying conformational transition and extra specificity for the receptor dimerization and activation. The existence of an additional configuration of the EphA1tm dimer is an argument in favor of the so-called rotation-coupled activation mechanism (9, 10) proposed for other families of receptor tyrosine kinases. The mechanism implies active involvement of the TM domains in the dimerization and activation of the receptor via proper helix-helix packing and rearranging. Respectively the membrane-embedded ionogenic residue Glu547 of the EphA1 receptor is capable of playing a unique role, controlling conformational flexibility of the receptor dimer and supporting different spatial orientations of the dimer subunits.
Influence of Membrane Surface Charge on the Conformational Diversity of the EphA1tm Dimer—It is well known that function of membrane proteins depends notably on the lipid environment (for a review, see Ref. 17). There are indications that the Eph-ephrin signaling events are initiated within specialized lipid microdomains of plasma membrane (2, 11). To assess the influence of the membrane charge on the structural-dynamic properties of the EphA1tm dimer, we performed pH titration experiments in the negatively charged DHPC/DMPC/DMPG (1:8:1) bicelles. The pKa of 2.9 of DMPG (47) is sufficiently far from the pKa of the Glu547 carboxyl group. The presence of 10% negatively charged DMPG lipids resulted in the increase of the Glu547 pKa value from 5.1 up to 5.7, whereas the Hill coefficient remained unchanged (supplemental Fig. S9C). This result is not unexpected because negative membrane charge usually tends to increase pKa values of the embedded carboxyl groups (48). Besides that, the presence of the negatively charged lipids leads to an alteration in the observed slow conformational exchange and slight increase of the minor form fraction of the EpA1tm dimer with the appearance of an additional (second) minor component of the cross-peaks in the 1H-15N HSQC spectra that is clearly observed for the Gly residues from the TM part (Fig. 1, D and E). The fraction of the second minor component is comparable with the first one, reaching ∼20% of the major peak at pH 6.7.
Because the deprotonation of Glu547 results in formation of a region of high local electronegativity in the helix packing interface of the major EphA1tm dimer conformation, the negatively charged polar heads of DMPG can create additional repulsion between the dimer subunits, shifting dimer-monomer equilibrium or/and promoting switching to another dimer configuration. Thus, depending on local pH and membrane composition, EphA1tm is involved in different types of association and internal motions with various amplitudes and time scales.
Possible Physiological Role of the EphA1 Receptor as an “Extracellular pH Sensor”—We have demonstrated that, depending on the ionization state of the carboxyl group of Glu547, EphA1tm undergoes conformational transitions. In our model system, the pKa value of the carboxyl group of Glu547 is in the pH range seldom attainable in physiological conditions. However, it has been shown that addition of even a small amount of negatively charged lipid into the model system substantially increases the Glu547 pKa value. The membrane surface potential, lipid composition of the cell membrane, ion concentration, and possibly extracellular domain conformation should influence the Glu547 pKa value. Therefore, certain properties of the EphA1 receptor and its biological response to the ligand binding should be sensitive to mild variations of extracellular and local membrane conditions. Hence Glu547 may serve as a sensor of the environment surrounding the EphA1 receptor on the cell surface, and its deprotonation may partly disrupt the coupling between the extracellular and cytoplasmic domains of the receptor because of transient local melting of helical structure and thus modulate signal transduction. Presence of the membrane-embedded ionogenic residue that can be protonated/deprotonated in the physiologically relevant pH range makes EphA1 a unique representative of the Eph receptor family (supplemental Fig. S11).
Intensive investigation of the expression of Eph/ephrin in various adult and embryonic tissues and of their role in different physiological processes has begun only recently (49). Although the complexity of cell-cell communications mediated by Eph/ephrin signaling is staggering, this may reflect a pivotal physiological role of these receptors and ligands in development and homeostasis of different human tissues. Ephrins and Eph receptors orchestrate a complex balance between cell repulsion and adhesion by serving directly as adhesion molecules and/or initiating repulsive signaling (50). Intriguingly EphA1 has been reported to be predominantly expressed in epithelial tissues (51, 52), including the skin where the surface pH is known to be as low as 4-6 (so-called “acid mantle” of the skin) (53). These low values are only maintained on the skin surface; they rapidly increase to normal values at the depth of about 100 μm (54). It is therefore tempting to assume that the unique ability of EphA1 to conduct ligand-induced signal at low extracellular pH values, at which its TM helix is rigid, is responsible for regulation of tight cell-cell interaction in the epidermis. Because the steady coupling between the extracellular and cytoplasmic domains is needed for the ligand-induced kinase activity, this assumption implies that the main conformation of the EphA1tm dimer (we observed at low pH in our model system) corresponds to the active state of the receptor, but this hypothesis has yet to be thoroughly tested. It should be noted that the rotation-coupled activation mechanism (9, 10) suggests that the minor conformation of the EphA1tm dimer (we observed at high pH) is presumably associated with the inactive state of the receptor and would occur for the full-length receptor at acidic pH also, in particular for the pre-existing receptor dimer before ligand-induced rearranging. Thus, the observed diversity of the dimer structures of the EphA1 TM domain can reflect the coexistence of both active and inactive dimeric conformations of the receptor.
Besides being widely expressed in small quantities (7, 52) in both adult and embryonic tissues, EphA1 can be overactivated during the different physiological processes accompanied by local acidification, such as synaptic cleft acidification during neural signal transduction (55), local hypoxia-induced angiogenesis (56-58), or acidosis associated with inflammatory processes (59, 60) and cancer development and invasion (61, 62). Indeed direct evidences have been provided for the involvement of the EphA1 receptor signaling at least in angiogenesis (63), inflammation (64, 65), and tumorigenesis (7, 18, 52).
At the same time, it seems likely that the EphA1 receptor is physiologically active in both slightly acidic and neutral pH environments. In the latter case the fractional unfolding of the EphA1 TM domain would result in an attenuation of the ligand-induced signal transduction, potentially altering the balance of cellular responses to ephrin binding. Moreover taking into account the possibility of various structural arrangements of the TM domain, it would appear that the EphA1 signaling mechanism is more diverse at neutral pH. Because the recruitment of Eph receptor into signaling complex may be independent of ephrin contacts (6, 66) and a limited population of active receptor dimers is supposed to exist even in the absence of ligand binding (8), the interaction between the EphA1 TM domains may also be involved in regulation of the ligand-independent receptor dimerization and activation. As was shown for receptor tyrosine kinases, the removal of their extracellular domain, either partial or total, results in constitutive activation of the receptor kinase (67). Therefore, at pH above the pKa value of Glu547 where the extracellular ligand-binding domain becomes somewhat decoupled from the TM domain and as a result from the cytoplasmic kinase domain, proper TM helix packing can enhance ligand-independent EphA1 receptor dimerization allowing active mutual orientation of the kinase domains and thus inducing a population of constitutively active receptors. Furthermore if the extracellular domain-dependent but kinase-independent functions of the Eph receptors take place (1, 6), the pH modulation of the EphA1 receptor signaling might be more complex (50, 63). Hence cell response to the Eph receptor signal transduction can vary substantially depending on pH and other cytoplasmic and cellular membrane parameters.
Thus, the membrane-embedded ionogenic residue Glu547 can play a role of extracellular pH sensor, suggesting the unique ability of the EphA1 receptor to up-regulate the response to ligand binding at decreased pH and allowing cells to interact between themselves differently depending on the external pH. Apparently specific “settings” of the sensor (pKa of the Glu547) will depend on extracellular conditions, local cell membrane environment, and possibly extracellular domain conformation and thus can vary between different tissues.
Concluding Remarks—Although the specific interactions of single TM-spanning domains were recognized to assist the ligand-binding extracellular domains during activation of some receptor tyrosine kinases (9, 10, 16, 17), the role of the TM domains in the Eph receptor function still remained unclear. We have demonstrated dimerization of the TM fragment of the EphA1 receptor and characterized its spatial structure and internal dynamics in a membrane-mimicking environment. Our study provides the dimeric structure of only one of the several states of the EphA1 TM domain with helix packing via the N-terminal tandem GG4-like motif A550X3G554X3G558. Although the Eph TM segments reveal relatively low amino acid sequence homology (supplemental Fig. S11), at least one classical dimerization GG4-like motif can be identified in each Eph TM region. Therefore, TM domains of the Eph receptors can play an important role in the receptor association and conformational switching during intercellular signal transduction as is typical for receptor tyrosine kinases.
On the other hand, the presence of a membrane-embedded ionogenic residue is a unique property of the EphA1 receptor that distinguishes the receptor among representatives of the Eph family. The observed dependence of the structural-dynamic properties of the TM domain upon the ionization state of the Glu547 residue implies that the conformational flexibility and activation of the EphA1 receptor can be regulated by such external and local factors as pH and lipid composition of the membrane. This can represent a new level of regulation of the EphA1 receptor activity integrated into a complex system of feedbacks involved in Eph-ephrin signaling. Investigation of the structural and dynamic properties of the EphA1 TM domain helps to understand the underlying mechanisms of the Ephephrin signal transduction and provides a basis to control the receptor kinase activity especially in pathological states of an organism. In particular, the obtained structural data would facilitate the design of Eph signaling inhibitors that can specifically recognize the Eph TM domains and interfere with their lateral association in cell membrane, providing a novel form of therapy for many types of human cancers associated with elevated Eph receptor expression (7).
Supplementary Material
The atomic coordinates and structure factors (codes 2k1k and 2k1l) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
The chemical shift assignments have been deposited in the Biological Magnetic Resonance Data Bank under accession code BMRB 15728.
This work was supported by the Russian Foundation for Basic Research (Grant 06-04-49740 and in part by Grants 07-04-01514 and 08-04-01372), the Russian Federation Federal Agency for Science and Innovations (State Contract 02.512.12.0019), and the Russian Funds Investment Group. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental experimental procedures, Figs. S1-S11, and references.
Footnotes
The abbreviations used are: Eph, erythropoietin-producing hepatocellular; TM, transmembrane; MD, molecular dynamics; NOE, nuclear Overhauser effect; NOESY, NOE spectroscopy; DMPC, dimyristoylphosphatidylcholine; DHPC, dihexanoylphosphatidylcholine; DMPG, dimyristoylphosphatidylglycerol; ephrin, Eph receptor-interacting protein; HSQC, heteronuclear single quantum coherence.
References
- 1.Kullander, K., and Klein, R. (2002) Nat. Rev. Mol. Cell Biol. 3 475-486 [DOI] [PubMed] [Google Scholar]
- 2.Pasquale, E. B. (2005) Nat. Rev. Mol. Cell Biol. 6 462-475 [DOI] [PubMed] [Google Scholar]
- 3.Gerlai, R. (2001) Nat. Rev. Neurosci. 2 205-209 [DOI] [PubMed] [Google Scholar]
- 4.Murai, K. K., and Pasquale, E. B. (2002) Neuron 33 159-162 [DOI] [PubMed] [Google Scholar]
- 5.Eph Nomenclature Committee (1997) Cell 90 403-404 [DOI] [PubMed] [Google Scholar]
- 6.Vearing, C. J., and Lackmann, M. (2005) Growth Factors 23 67-76 [DOI] [PubMed] [Google Scholar]
- 7.Surawska, H., Ma, P. C., and Salgia, R. (2004) Cytokine Growth Factor Rev. 15 419-433 [DOI] [PubMed] [Google Scholar]
- 8.Schlessinger, J. (2000) Cell 103 211-225 [DOI] [PubMed] [Google Scholar]
- 9.Moriki, T., Maruyama, H., and Maruyama, I. N. (2001) J. Mol. Biol. 311 1011-1026 [DOI] [PubMed] [Google Scholar]
- 10.Fleishman, S. J., Schlesinger, J., and Ben-Tal, N. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 15937-15940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gauthier, L. R., and Robbins, S. M. (2003) Life Sci. 74 207-216 [DOI] [PubMed] [Google Scholar]
- 12.Himanen, J. P., Saha, N., and Nikolov, D. B. (2007) Curr. Opin. Cell Biol. 19 534-542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mackenzie, K. R., Prestegard, J. H., and Engelman, D. M. (1997) Science 276 131-133 [DOI] [PubMed] [Google Scholar]
- 14.Bocharov, E. V., Pustovalova, Y. E., Pavlov, K. V., Volynsky, P. E., Goncharuk, M. V., Ermolyuk, Y. S., Karpunin, D. V., Schulga, A. A., Kirpichnikov, M. P., Efremov, R. G., Maslennikov, I. V., and Arseniev, A. S. (2007) J. Biol. Chem. 282 16256-16266 [DOI] [PubMed] [Google Scholar]
- 15.Bocharov, E. V., Mineev, K. S., Volynsky, P. E., Ermolyuk, Y. S., Tkach, E. N., Sobol, A. G., Chupin, V. V., Kirpichnikov, M. P., Efremov, R. G., and Arseniev, A. S. (2008) J. Biol. Chem. 283 6950-6956 [DOI] [PubMed] [Google Scholar]
- 16.Li, E., and Hristova, K. (2006) Biochemistry 45 6241-6251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mackenzie, K. R. (2006) Chem. Rev. 106 1931-1977 [DOI] [PubMed] [Google Scholar]
- 18.Hirai, H., Maru, Y., Hagiwara, K., Nishida, J., and Takaku, F. (1987) Science 238 1717-1720 [DOI] [PubMed] [Google Scholar]
- 19.Artemenko, E. O., Egorova, N. S., Arseniev, A. S., and Feofanov, A. V. (2008) Biochim. Biophys. Acta. 1778 2361-2367 [DOI] [PubMed] [Google Scholar]
- 20.Pabbisetty, K. B., Yue, X., Li, C., Himanen, J.-P., Zhou, R., Nikolov, D. B., and Hu, L. (2007) Protein Sci. 16 355-361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chakrabarti, P., and Khorana, G. (1975) Biochemistry 14 5021-5033 [DOI] [PubMed] [Google Scholar]
- 22.Sattler, M., Schleucher, J., and Griesinger, C. (1999) Prog. Nucl. Magn. Reson. Spectrosc. 34 93-158 [Google Scholar]
- 23.Cavanagh, J., Fairbrother, W. J., Palmer, A. G., and Skelton, N. J. (2006) Protein NMR Spectroscopy: Principles and Practice, 2nd Ed., Academic Press, San Diego, CA
- 24.Markley, K. L. (1975) Acc. Chem. Res. 8 70-80 [Google Scholar]
- 25.Bocharov, E. V., Korzhnev, D. M., Blommers, M. J. J., Arvinte, T., Orekhov, V. Y., Billeter, M., and Arseniev, A. S. (2002) J. Biol. Chem. 277 46273-46279 [DOI] [PubMed] [Google Scholar]
- 26.Orekhov, V. Y., Nolde, D. E., Golovanov, A. P., Korzhnev, D. M., and Arseniev, A. S. (1995) Appl. Magn. Reson. 9 581-588 [Google Scholar]
- 27.Daragan, V. A., and Mayo, K. H. (1997) Prog. Nucl. Magn. Reson. Spectrosc. 31 63-105 [Google Scholar]
- 28.Güntert, P. (2003) Prog. Nucl. Magn. Reson. Spectrosc. 43 105-125 [Google Scholar]
- 29.Zwahlen, C., Legault, P., Vincent, S. J. F., Greenblatt, J., Konrat, R., and Kay, L. E. (1997) J. Am. Chem. Soc. 119 6711-6721 [Google Scholar]
- 30.Cornilescu, G., Delaglio, F., and Bax, A. (1999) J. Biomol. NMR 13 289-302 [DOI] [PubMed] [Google Scholar]
- 31.Doig, A. J. (2002) Biophys. Chem. 101-102 281-293 [DOI] [PubMed] [Google Scholar]
- 32.Iqbalsyah, T. M., and Doig, A. J. (2004) Protein Sci. 13 32-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fonseca, N. A., Camacho, R., and Magalhaes, A. L. (2008) Proteins 70 188-196 [DOI] [PubMed] [Google Scholar]
- 34.Baker, E. N., and Hubbard, R. E. (1984) Prog. Biophys. Mol. Biol. 44 97-179 [DOI] [PubMed] [Google Scholar]
- 35.Lindahl, E., Hess, B., and van der Spoel, D. (2001) J. Mol. Model. 7 306-317 [Google Scholar]
- 36.Hofmann, K., and Stoffel, W. (1993) Biol. Chem. Hoppe-Seyler 374 166. [DOI] [PubMed] [Google Scholar]
- 37.Efremov, R. G., and Vergoten, G. (1995) J. Phys. Chem. 99 10658-10666 [Google Scholar]
- 38.Kabsch, W., and Sander, C. (1985) Biopolymers 22 2577-2637 [DOI] [PubMed] [Google Scholar]
- 39.Koradi, R., Billeter, M., and Wüthrich, K. (1996) J. Mol. Graph. 14 51-55 [DOI] [PubMed] [Google Scholar]
- 40.DeLano, W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific, Palo Alto, CA
- 41.Prosser, R. S., Evanics, F., Kitevski, J. L., and Al-Abdul-Wahid, M. S. (2006) Biochemistry 45 8453-8465 [DOI] [PubMed] [Google Scholar]
- 42.Arseniev, A. S., Kondakov, V. I., Maiorov, V. N., and Bystrov, V. F. (1984) FEBS Lett. 165 57-62 [Google Scholar]
- 43.Kim, S., Jeon, T.-J., Oberai, A., Yang, D., Schmidt, J. J., and Bowie, J. U. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 14279-14283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Langosch, D., and Heringa, J. (1998) Proteins 31 150-160 [DOI] [PubMed] [Google Scholar]
- 45.Szyperski, T., Antuch, W., Schick, M., Betz, A., Stone, S. R., and Wüthrich, K. (1994) Biochemistry 33 9303-9310 [DOI] [PubMed] [Google Scholar]
- 46.Wagner, G., Pardi, A., and Wüthrich, K. (1983) J. Am. Chem. Soc. 105 5948-5949 [Google Scholar]
- 47.Struppe, J., Whiles, J. A., and Vold, R. R. (2000) Biophys. J. 78 281-289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith, S. O., Smith, C. S., and Bormann, B. J. (1996) Nat. Struct. Biol. 3 252-258 [DOI] [PubMed] [Google Scholar]
- 49.Lackmann, M., and Boyd, A. W. (2008) Sci. Signal. 1 re2. [DOI] [PubMed] [Google Scholar]
- 50.Halloran, M. C., and Wolman, M. A. (2006) Curr. Opin. Cell Biol. 18 533-540 [DOI] [PubMed] [Google Scholar]
- 51.Coulthard, M. G., Lickliter, J. D., Subanesan, N., Chen, K., Webb, G. C., Lowry, A. J., Koblar, S., Bottema, C. D., and Boyd, A. W. (2001) Growth Factors 18 303-317 [DOI] [PubMed] [Google Scholar]
- 52.Hafner, C., Becker, B., Landthaler, M., and Vogt, T. (2006) Mod. Pathol. 19 1369-1377 [DOI] [PubMed] [Google Scholar]
- 53.Schmid-Wendtner, M. H., and Korting, H. C. (2006) Skin Pharmacol. Physiol. 19 296-302 [DOI] [PubMed] [Google Scholar]
- 54.Wagner, H., Kostka, K. H., Lehr, C. M., and Schaefer, U. F. (2003) Eur. J. Pharm. Biopharm. 55 57-65 [DOI] [PubMed] [Google Scholar]
- 55.Palmer, M. J., Hull, C., Vigh, J., and von Gersdorff, H. (2003) J. Neurosci. 23 11332-11341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liao, D., and Johnson, R. S. (2007) Cancer Metastasis Rev. 26 281-290 [DOI] [PubMed] [Google Scholar]
- 57.Xu, K., and Lamanna, J. C. (2006) J. Appl. Physiol. 100 725-730 [DOI] [PubMed] [Google Scholar]
- 58.Vihanto, M. M., Plock, J., Erni, D., Frey, B. M., Frey, F. J., and Huynh-Do, U. (2005) FASEB J. 19 1689-1691 [DOI] [PubMed] [Google Scholar]
- 59.Lardner, A. (2001) J. Leukoc. Biol. 69 522-530 [PubMed] [Google Scholar]
- 60.Kellum, J. A., Song, M., and Li, J. (2004) Crit. Care 8 331-336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stubbs, M., McSheehy, P. M., Griffiths, J. R., and Bashford, C. L. (2000) Mol. Med. Today 6 15-19 [DOI] [PubMed] [Google Scholar]
- 62.Raghunand, N., Gatenby, R. A., and Gillies, R. J. (2003) Br. J. Radiol. 1 S11-S22 [DOI] [PubMed] [Google Scholar]
- 63.Masuda, J., Usui, R., and Maru, Y. (2008) J. Biol. Chem. 283 13148-13155 [DOI] [PubMed] [Google Scholar]
- 64.Aasheim, H. C., Delabie, J., and Finne, E. F. (2005) Blood 105 2869-2876 [DOI] [PubMed] [Google Scholar]
- 65.Ivanov, A. I., and Romanovsky, A. A. (2006) IUBMB Life 58 389-394 [DOI] [PubMed] [Google Scholar]
- 66.Wimmer-Kleikamp, S. H., Janes, P. W., Squire, A., Bastiaens, P. I., and Lackmann, M. (2004) J. Cell Biol. 164 661-666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhu, H. J., Iaria, J., Orchard, S., Walker, F., and Burgess, A. W. (2003) Growth Factors 21 15-30 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.