Abstract
Hepatitis C virus often causes persistent infection and hepatocellular carcinoma. Studies have demonstrated the roles of viral nonstructural protein 5A (NS5A) in the induction of chromosome aneuploidy, but the molecular mechanisms are not clear. In this study, hydrodynamics-based in vivo transfection was applied to a mouse system. Mouse hepatocytes that successfully expressed NS5A protein were isolated by laser capture microdissection. Gene expression profiles of the NS5A-expressing hepatocytes were examined by an Affymetrix oligonucleotide microarray system. Aspm (abnormal spindle-like, microcephaly associated), which encodes the mitotic spindle protein ASPM, was identified to be differentially expressed in the absence and the presence of NS5A. The down-regulation of Aspm mRNA and ASPM protein was confirmed by real time polymerase chain reaction and Western blot analysis, respectively, both in mouse model systems and in viral subgenomic replicon and in vitro transfection culturing systems. In addition, cultured cells that constitutively expressed NS5A protein showed G2/M cell cycle block and chromosome aneuploidy. Overexpression of ASPM relieved the G2/M cell cycle block. Furthermore, NS5A protein repressed the promoter activity of Aspm gene in a dose-dependent manner. The regulatory effect was abolished when amino acid substitutions P2209L, T2214A, and T2217G known to interrupt the NS5A-PKR interaction were introduced into the NS5A protein. This indicates that the down-regulation of Aspm expression is via the PKR-p38 signaling pathway. These results suggest that NS5A protein down-regulates the expression of the mitotic spindle protein ASPM and induces aberrant mitotic cell cycle associated with chromosome instability and hepatocellular carcinoma.
Hepatitis C virus (HCV)3 is the main causative agent for transfusion-associated and sporadic non-A, non-B hepatitis throughout the world (1). HCV often causes a prolonged and persistent infection (2) and plays an important role in the pathogenesis of virus-associated hepatocellular carcinoma (HCC) (3-5). The current hypothesis is that HCV-associated HCC may be resulted from immune-mediated inflammatory damage and cirrhosis. However, viral proteins involved in the progression of liver diseases are not clear.
HCV is the only member of the genus hepacivirus, within the family Flaviviridae. It has a single-stranded positive sense RNA genome of ∼9.6 kb with a single open reading frame encoding four structural (core, E1, E2, and p7) and six nonstructural proteins (NS2, NS3, NS4A, NS4B, NS5A, and NS5B) (6).
The NS5A protein of genotype 1b contains 447 amino acid residues and has multiple functions (7). It forms heterodimer with PKR (the double-stranded RNA-activated protein kinase R) and blocks the interferon signaling pathway in HCV resistance to interferon therapy (8, 9). Through interacting with PKR, NS5A inhibits PKR activity and the p38 mitogen-activated protein kinase (MAPK) signaling pathway (10). Dysregulation of MAPK signaling perturbs the control of host cell cycle (11-13). It has been suggested that NS5A is involved in the control of cell growth and cell viability and plays a role in the induction of chromosome instability; the NS5A-induced chromosome instability is associated with aberrant mitotic regulation (14, 15). In addition, NS5A interacts with the adaptor protein Grb2 and inhibits the extracellular signal-regulated kinase (ERK) MAPK (16, 17). NS5A may also take a part in the regulation of stress-activated protein kinase/c-Jun N-terminal kinase (SAPK/JNK) pathway (18). These observations provide insights into the molecular mechanisms of HCV-associated hepatocarcinogenesis. Nevertheless, molecular mechanisms of the NS5A protein involved in the dysregulation of mitotic cell cycle are not clear.
ASPM (human abnormal spindle-like, microcephaly-associated) protein consists of 3,477 amino acid residues; it appears to be the orthologue product of the Drosophila abnormal spindle gene Asp and the mouse Aspm gene (19). Recessive mutations in the Aspm gene are the most common genetic cause of primary microcephaly in humans (20-22). Even though mouse ASPM protein consists of 3,122 amino acid residues, which is 355 residues shorter than the human ASPM protein, both human and mouse ASPM proteins possess an N-terminal microtubule-binding domain, an actin-binding calponin homology domain, and a calmodulin-binding domain consisting of a large number of isoleucine-glutamine repeats (19, 23). Human ASPM localizes to the centrosome in interphase and to the spindle poles during mitosis (24). Similarly, ASPM protein was found to localize to the spindle poles in embryonic mouse brain (25). These indicate that ASPM might function, as its orthologue ASP does, in regulating the organization of centrosomal processes and mitotic spindle during cell cycle progression (26). Knockdown of ASPM inhibits cellular proliferation in mice (25).
In this study, we showed a down-regulation of Aspm mRNA in HCV NS5A-expressing mouse hepatocytes. The NS5A protein inhibited Aspm expression via the down-regulation of the PKR-p38 MAPK signaling pathway. In addition, overexpression of ASPM relieved the G2/M cell cycle block imposed by NS5A. These results certify that ASPM is a downstream target of NS5A and suggest a novel role of ASPM in the pathogenesis of HCV.
EXPERIMENTAL PROCEDURES
Plasmids
pAdTrack(-CMV) and pTTRB-T7HisNS5A—Plasmid pAd-Track(-CMV) was derived from plasmid pAdTrack-CMV by replacing its 2.9-kb EcoRI/SalI fragment with the 2.3-kb EcoRI/SalI fragment of pShuttle (Stratagene). This resulted in a removal of one of the two CMV promoters present in pAdTrack-CMV. For generation of plasmid pTTRB-T7His-NS5A, plasmid pET15b-NS5A that encodes a His-tagged full-length NS5A protein of HCV (27) and PAP-TTRBsv that contains the liver-specific promoter and enhancer of transthyretin (kindly provided by R. H. Costa, Department of Biochemistry and Molecular Genetics, University of Illinois at Chicago) were digested with HindIII and EcoRI restriction endonucleases, respectively. The linearized DNAs of both plasmids were treated with the Klenow fragment of DNA polymerase I prior to a further digestion with BglII restriction endonuclease. The resultant T7HisNS5A DNA fragment was then cloned into the modified PAP-TTRBsv to generate pTTRB-T7HisNS5A.
pAdTrack-TTRB-T7HisNS5A, pAdTrack-CMV-T7HisNS5A, and pAdTrack-CMV-T7HisNS5A-mPTT—Plasmid pAdTrack-TTRB-T7HisNS5A contains a NS5A cDNA fragment under the control of the liver-specific transthyretin promoter, and in the opposite direction, it contains the cDNA of green fluorescence protein (GFP) under the control of CMV promoter. For generation of plasmid pAdTrack-TTRB-T7HisNS5A, plasmid pTTRB-T7HisNS5A was digested with PacI and PmeI restriction endonucleases. The resultant TTRB-T7HisNS5A DNA fragment was treated with the T4 DNA polymerase and then cloned into the EcoRV site of pAdTrack(-CMV). Plasmid pAdTrack-CMV-T7HisNS5A contains the cDNA fragments of NS5A and GFP under the control of two independent CMV promoters. For generation of plasmid pAdTrack-CMV-T7His-NS5A, plasmid pAdTrack-TTRB-T7HisNS5A was digested with BglII and NotI restriction endonucleases. The resultant T7HisNS5A DNA fragment was cloned into the BglII-NotI sites of pAdTrack-CMV. Plasmid pAdTrack-CMV-T7His-NS5A-mPTT contains amino acid substitutions P2209L, T2214A and T2217G in the PKR-binding region (amino acid residues 2209-2274) of the HCV genotype 1b NS5A and was derived from the wild type plasmid pAdTrack-CMV-T7HisNS5A. Site-directed mutagenesis was performed followed the procedures as described by the manufacturer (QuikChange II XL site-directed mutagenesis kit; Stratagene). The primer set used was 5′-CTGTCTGCACTTTCTCTGAAGGCAGCATGCACTGGCCGTCATGAC-3′ (underlining corresponds to the amino acid mutations at Pro2209, Thr2214, and Thr2217) and 5′-GTCATGACGGCCAGTGCATGCTGCCTTCAGAGAAAGTGCAGACAG-3′.
pCRII-Topo-mAspm-TLS(-475/-59) and pGL3-mAspm-TLS(-475/-59)—For the construction of plasmid pCRII-Topo-mAspm-TLS(-475/-59), DNA fragment mAspm-TLS(-475/-59) that contains the mouse Aspm gene from nucleotides -475 to -59 upstream of the translation start site was amplified from the genomic DNA of mouse NIH3T3 cells with the primer set TLS-59 (5′-GAAGCCAACGACCAGGACAAGG-3′) and TLS-475 (5′-CTCAGCTATTCAGGACCGCATG-3′) and cloned into pCRII-TOPO (Invitrogen). Isolation of the genomic DNA was performed using the Gentra Systems PUREGENE DNA purification kit (Qiagen). For generation of plasmid pGL3-mAspm-TLS(-475/-59), plasmid pCRII-Topo-mAspm-TLS(-475/-59) was digested with XhoI and HindIII restriction endonucleases, and the resultant mAspm-TLS(-475/-59) fragment was cloned into the XhoI-HindIII sites of pGL3-basic (Promega).
pcDNA-hASPM(1-5141)V5HisTopo, pCRII-Topo-hASPM-(4121-10431Not), and pcDNA-hASPM-V5HisTopo—For the construction of plasmid pcDNA-hASPM(1-5141)V5HisTopo, DNA fragment hASPM(1-5141) that contains the coding sequences of the human Aspm from nucleotides 1 to 5141 was amplified from cDNA of human 293T cells with the primer set hASPM1F (5′-CACCATGGCGAACCGGCGAGT-3′) and hASPM5141R (5′-GAACGGTAACATTGCTGGAT-3′) and cloned into pcDNA3.1D/V5-His-TOPO (Invitrogen). For the construction of plasmid pCRII-Topo-hASPM(4121-10431Not), DNA fragment hASPM(4121-10431Not) that contains the coding sequences of the human Aspm from nucleotides 4121 to 10431 was amplified from cDNA of human 293T cells with the primer set hASPM4121F (5′-CAATCATCCTGCAATCTAGG-3′) and hASPM10431NotR (5′-GCGGCCGCCATAAGGAATGCCAAGCGTATCC-3′) and cloned into pCRII-TOPO. Plasmid pcDNA-hASPM-V5HisTopo that encodes the full-length human ASPM protein with V5His-tag was generated from plasmids pcDNA-hASPM(1-5141)V5HisTopo and pCRII-Topo-hASPM-(4121-10431Not) following partial digestion of the plasmids with restriction endonucleases, and ligation of the resultant 5.9-kb ScaI-NotI DNA fragment from pCRII-Topo-hASPM(4121-10431Not) with the 10-kb ScaI-NotI partial digestion product of pcDNA-hASPM(1-5141)V5HisTopo. Sequence analysis revealed a difference of two nucleotides (7064-AG-7065 for Gln2355) in the human Aspm gene of plasmid pcDNA-hASPM-V5HisTopo as compared with that previously identified (NM_018136; 7064-GA-7065 for Arg2355).
Cell Lines, DNA Transfection, and Western Blot Analysis
Huh7 (a human hepatoma cell line), HepG2 (a human hepatocellular carcinoma cell line), A549 (a human lung carcinoma cell line), COS7 (an African Green Monkey kidney fibroblast cell line), 293T (a human embryonic kidney cell line), and NIH3T3 cells (a mouse fibroblast cell line) were maintained at 37 °C with 5% CO2 in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% heat-inactivated fetal bovine serum plus 100 units of penicillin and 100 μg of streptomycin/ml.
Unless indicated, DNA transfection was performed with the Lipofectin reagent (Invitrogen) as described by the manufacturer. The cells were harvested 48 h post-transfection and subjected to Western blot analysis as described previously (28).
To establish stable cell lines that constitutively express NS5A, NIH3T3 cells cultured on 10-cm dishes were transfected with 1.5 μg of pcDNA3.1(+) and 15 μg of pAdTrack-CMV-T7HisNS5A. At 48 h post-transfection, the cells were split at a 1:10 ratio and cultured in the presence of 1.2 mg of G418/ml of medium. Drug-resistant colonies that appeared 10-14 days post-transfection were clonally isolated and screened for the expression of HCV NS5A protein by Western blot analysis with NS5A monoclonal antibody (Chemicon). NIH3T3-NS5A stable cell lines that constitutively express HCV NS5A were maintained in medium containing 1.2 mg of G418/ml of medium. H7-HCVR cells were derived from Huh7 cells by inclusion of a bicistronic HCV subgenomic replicon carrying neo gene as shown in supplemental Fig. S1. H7-HCVR cells were maintained in medium containing 1 mg of G418/ml of medium.
Expression of Transgenes in Mice by Hydrodynamics-based in Vivo Transfection
Hydrodynamics-based in vivo transfection was performed as previously described (29) in male BALB/c mice with plasmid pAdTrack-TTRB-T7HisNS5A and the control plasmid, pAdTrack(-CMV). Two days post-injection, the mice were sacrificed. Fresh tissues including liver, heart, kidney, and lung were collected for the preparation of cryosection and examination of the expression of HCV NS5A protein and its regulatory host genes.
Preparation of Tissue Section and Laser Capture Microdissection (LCM)
To prepare for cryosection, the mouse tissues were embedded in optimal cutting temperature solution on dried ice. After cutting the frozen blocks into 14-μm-thick sections by microtome (Leica), the tissue sections were attached to slides and deep frozen immediately on dried ice. To further prepare for LCM, the cryosections were fixed in 70% ethanol for 1 min before proceeding to dehydration. The dehydration processes include a 1-min submergence in 95% ethanol, a 30-s immersion in 100% ethanol followed by another 1-min immersion in 100% ethanol. Lastly, to complete the dehydration process, the cryosections were treated by 30-s and 5-min submergence in xylene. The above were done with gentle shaking at each step. After being air-dried, cryosections of the mouse liver tissue that successfully expressed GFP and NS5A proteins were isolated by LCM with a fluorescence detector (PixCell IIe; Arcturus) setting at laser spot size of 7.5 μm, pulse power of 50 milliwatt, pulse width of 3.5 ms, and threshold voltage of 270 mV.
RNA Amplification and Microarray Analysis
For microarray analysis, 1,000 hepatocytes were captured by LCM with a fluorescence detector. RNA isolated from the hepatocytes using the PicoPure RNA isolation kit was subjected to two rounds of amplification with RiboAmp RNA amplification kit containing T7 RNA polymerase followed the procedures as described by the manufacturer (Arcturus) with modifications. In brief, the Agilent Lab-on-a-Chip System with the BioAnalyzer 2100 (Agilent) was used to examine the quality of the ribosomal RNA in the RNA preparation of hepatocytes to ensure the accuracy and reproducibility in the analysis of gene expression profiles. In addition, following the first amplification, the resultant cDNAs were added to the in vitro transcription reaction of Affymetrix microarray system to generate biotinylated cRNAs in the second round of transcript amplification. Gene expression profiles were performed with an Affymetrix high density oligonucleotide microarray (Mouse Genome 430 2.0 Array) using the Affymetrix microarray system following the protocol as described by the manufacturer. GCOS and RMA statistics were applied for data analysis.
RNA Isolation, Reverse Transcription, and Real Time PCR
Total RNA was isolated from culture cells by using TRIzol® reagent (Invitrogen). Reverse transcription was performed with the RNA templates, avian myeloblastosis virus reverse transcriptase (Roche Applied Science), and oligo(dT) primer. The products were subjected to real time PCR with primer sets of specific genes and SYBR Green PCR Master Mix (Bio-Rad). The primer set used for mouse Aspm was 5′-GCTTCATCACCTGCTCACCTAC-3′ and 5′-GTAGATACCGCTCCGCTTTCAG-3′, whereas the primer sets used in parallel as internal controls were 5′-CTATTGGCAACGAGCGGTTCC-3′ and 5′-GCACTGTGTTGGCATAGAGGTC-3′ for Actb and 5′-TGTGTCCGTCGTGGATCTGAC-3′ and 5′-GATGCCTGCTTCACCACCTTC-3′ for Gapdh. The results were analyzed with the iCycler iQ real time PCR detection system (Bio-Rad).
Synchronization of Culture Cells and Cell Cycle Analysis
To generate synchronized cell population arrested at the G1/S boundary, the cells were cultured in the medium without fetal bovine serum for 24 h. After release from the serum starvation, the cells were harvested by trypsinization at various time points. Following two washes with phosphate-buffered saline containing 2 mm EDTA, the cells were fixed in 75% (v/v) ethanol and stained with propidium iodide (40 mg/ml) in the presence of RNase A (50 mg/ml) for 30 min at room temperature. Samples of 10,000 cells were then analyzed on a Becton Dickinson FACScan flow cytometer (BD Biosciences), and the data were analyzed with the CellQuest software (BD Biosciences).
Antibodies
Mouse monoclonal antibodies against NS5A were purchased from Chemicon. Mouse monoclonal antibodies against cyclin B1 and rabbit polyclonal antibodies against p-p38 and p38 were purchased from Cell Signaling. Mouse monoclonal antibodies against Sp1 and rabbit polyclonal antibodies against GAPDH were purchased from Santa Cruz. Mouse monoclonal antibodies against His tag were purchased from Upstate Biotechnology Inc. Rabbit monoclonal antibodies against p-PKR (Thr(P)446) and PKR were purchased from Abcam. Rabbit polyclonal antibodies against GFP were kindly provided by S. L. Doong (Institute of Microbiology, National Taiwan University College of Medicine). For generation of an antiserum specific for both human and mouse ASPM, a highly conserved antigenic peptide (487PKRRPILSATVTKRK501) between human and mouse (supplemental Fig. S2) was synthesized, conjugated to Multiple Antigenic Peptides (Kelowna International Scientific Inc.), and subjected to immunization of rabbits as previously described (28).
Luciferase Reporter Assay
For analysis of the promoter activity of Aspm, NIH3T3 cells were co-transfected with the effector plasmid pAdTrack-CMV-T7HisNS5A (or pAdTrack-CMV-T7HisNS5A-mPTT), the Firefly luciferase reporter plasmid pGL3-mAspm-TLS(-475/-59), and the Renilla luciferase-expressing control plasmid phRL-TK (Promega). At 48 h post-transfection, the cells were harvested by trypsinization. Following two washes with phosphate-buffered saline, the cells were suspended with 100 μl of phosphate-buffered saline. One half of the cells were subjected to Western blot analysis, and the other half of the cells were subjected to the analysis of firefly luciferase activity and Renilla luciferase activity according to the protocol provided by the manufacturer (Dual-Glo™ luciferase assay system; Promega). The firefly luciferase activity was normalized against the Renilla luciferase activity.
RESULTS
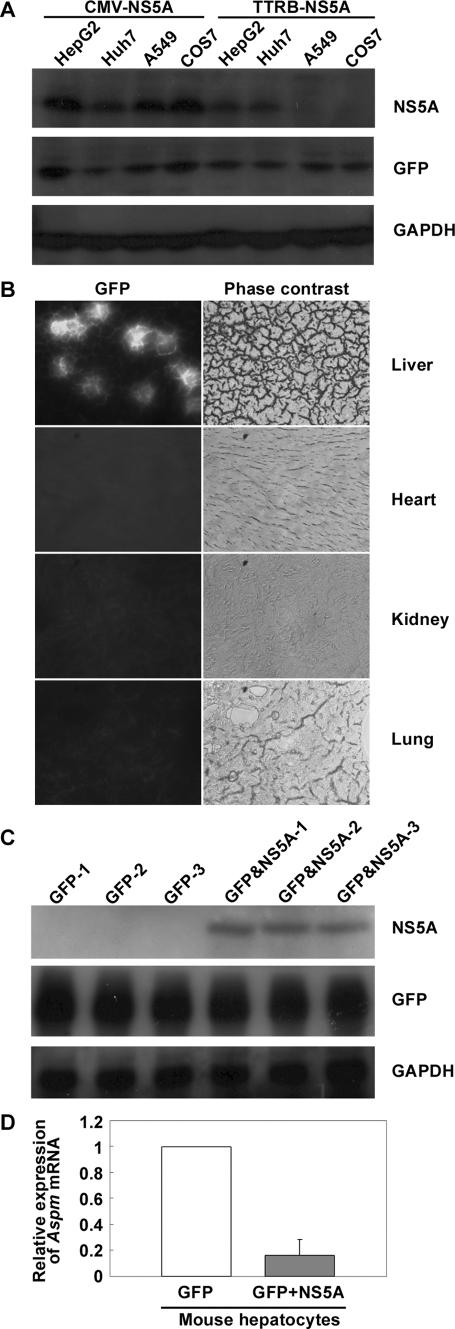
Gene Expression Profiles in Mouse Hepatocytes Expressing HCV NS5A Protein—To study the pathogenesis of HCV NS5A protein, plasmid pAdTrack-TTRB-T7HisNS5A that contains the HCV NS5A gene under the control of liver specific transthyretin promoter and GFP gene under the control of CMV promoter was constructed. Expression of the viral NS5A protein and GFP was initially examined in cultured cells of various tissue origins. The results demonstrated that the NS5A protein co-expressed with GFP only in the hepatocyte-derived HepG2 and Huh7 cells but not in the nonhepatic A549 and COS7 cells (Fig. 1A), indicating the tissue-specific expression of the viral NS5A protein driven by the liver-specific promoter. Hydrodynamics-based in vivo transfection was then applied to a mouse system. The hydrodynamics-based method mimics the postnatal HCV infection unlike prenatal existence of the viral protein in transgenic mouse model. Two days following the rapid injection of the plasmid pAdTrack-TTRB-T7HisNS5A into mouse tail vein, GFP expression was detected specifically in the liver; no detection was observed in heart, kidney, and lung tissues (Fig. 1B). The GFP-expressing hepatocytes were isolated by LCM with a fluorescence detector. Co-expression of the viral NS5A protein in the GFP-expressing hepatocytes was further confirmed by Western blot analysis (Fig. 1C). In addition, plasmid pAdTrack(-CMV) that expresses GFP only was used in parallel in hydrodynamic injection as a control. To learn the differential expression profiles of host genes in the presence and the absence of the viral NS5A protein, RNA was isolated from the captured hepatocytes that successfully co-expressed GFP and NS5A proteins and from the control hepatocytes that expressed GFP only. The RNA samples were amplified and biotin-labeled as described under “Experimental Procedures.” Analysis of gene expression profiles with Affymetrix oligonucleotide microarray system containing 34,000 genes revealed 63 genes that were up-regulated for more than 2-fold and 34 genes that were down-regulated for more than 2-fold in the NS5A and GFP co-expressing hepatocytes as compared with the control (data not shown). Among these differentially expressed genes, 10 genes are involved in multiple cellular signaling pathways including cell growth, inflammation, and apoptosis. The spindle gene Aspm is one of the genes whose down-regulation in the NS5A-expressing hepatocytes was further confirmed by real time PCR analysis (Fig. 1D).
FIGURE 1.
Down-regulation of Aspm gene in mouse hepatocytes expressing HCV NS5A protein. A, liver-specific expression of NS5A protein driven by the transthyretin promoter. HepG2, Huh7, A549, and COS7 cells were transfected with 15 μg of plasmid pAdTrack-CMV-T7HisNS5A or pAdTrack-TTRB-T7HisNS5A as indicated. Forty-eight hours post-transfection, the cells were harvested, and cell lysates were subjected to Western blot analysis with antibodies specific to the viral NS5A protein, GFP, and the control GAPDH. B, expression of GFP protein in the tissue sections of hydrodynamic-injected mouse. Plasmid pAdTrack-TTRB-T7HisNA5A that encodes GFP and NS5A protein was injected into mouse tail vein by hydrodynamics-based method. Two days post-injection, mouse tissue sections from various origins as indicated were prepared. Expression of GFP in the tissue sections was monitored by a fluorescence detector. C, co-expression of HCV NS5A protein in the GFP-expressing mouse hepatocytes. Hepatocytes from three mice injected with the control plasmid pAdTrack(-CMV) that expresses GFP protein only (GFP-1, -2, and -3) and hepatocytes from three mice injected with plasmid pAdTrack-TTRB-T7HisNS5A that co-express GFP and NS5A protein (GFP&NS5A-1, -2, and -3) were harvested for Western blot analysis with antibodies as indicated. D, real time PCR analysis for expression of Aspm gene in mouse hepatocytes. One thousand mouse hepatocytes were captured with a fluorescence detector from each of the group of mouse injected with plasmid pAdTrack-TTRB-T7HisNS5A (GFP+NS5A), and the group was injected with control plasmid pAdTrack(-CMV) (GFP) as indicated. RNA isolation, amplification, and real time PCR analysis of the Aspm gene followed the procedures as described under “Experimental Procedures.” The results shown represent averages from three independent experiments.
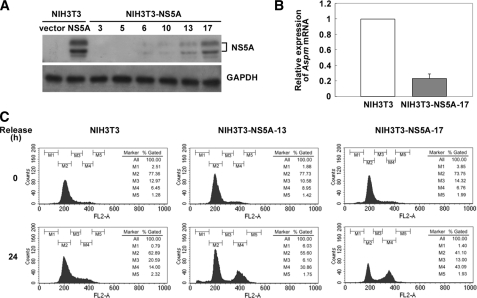
Growth Arrest of the Stable Cell Lines Constitutively Expressing NS5A—HCV NS5A protein has been demonstrated to induce chromosome aneuploidy (15). To understand whether the down-regulation of Aspm is involved in the NS5A-induced mitotic dysregulation, stable cell lines that constitutively express the NS5A protein were established following a transfection of plasmid pAdTrack-CMV-T7HisNS5A into NIH3T3 cells and G418 selection. Among the G418-resistant clones examined, the NIH3T3-NS5A-17 stable line that showed the highest expression level of the NS5A protein (Fig. 2A) and a remarkable reduction of the growth rate (data not shown) was chosen for further analysis. RT real time PCR confirmed the down-regulation of Aspm gene in NIH3T3-NS5A-17 stable line (Fig. 2B). The effect of NS5A on the mitotic dysregulation was examined in the NIH3T3-NS5A-17 cells. The cells were synchronized at the G1/S boundary by serum starvation and subjected to flow analysis 24 h after release from the starvation. As shown in Fig. 2C, the NS5A-expressing NIH3T3-NS5A-17 cells had an increased cell population of G2/M phase (43%) as compared with that of the parental NIH3T3 cells (14%). The NIH3T3-NS5A-13 stable line that expressed a lower level of NS5A protein as compared with the NIH3T3-NS5A-17 cells showed a G2/M cell cycle block similar to NIH3T3-NS5A-17 cells but to a lower extent (31%). These results further correlate the expression level of NS5A with the extent of the G2/M cell cycle block.
FIGURE 2.
The G2/M arrest in NS5A-expressing cells. A, expression of the HCV NS5A protein in NIH3T3-NS5A stable clones. The cell lysates were prepared from NIH3T3-NS5A stable clones (numbered 3, 5, 6, 10, 13, and 17) and subjected to Western blot analysis with antibodies specific to the NS5A protein. Protein lysates prepared from NIH3T3 cells transfected with plasmid pAdTrack-CMV (vector) and plasmid pAdTrack-CMV-T7HisNS5A (NS5A) as indicated were used as negative and positive controls, respectively. B, RT real time PCR analysis for the expression of Aspm gene in NIH3T3-NS5A-17. Total RNA was isolated from NIH3T3-NS5A-17 cells by TRIzol® reagent and subjected to RT real time PCR analysis of the Aspm gene. The results shown represent averages from three independent experiments. C, G2/M arrest of the NIH3T3-NS5A-13 and NIH3T3-NS5A-17 stable cell lines. NIH3T3-NS5A-13, NIH3T3-NS5A-17, and the parental NIH3T3 cells were arrested at the G1/S boundary by serum starvation. At 0 and 24 h after release from the serum starvation, 10,000 cells were gated for cell cycle analysis. M1, sub-G1 phase; M2, G1 phase; M3, S phase; M4, G2/M phase; M5, multiple nucleus phase.
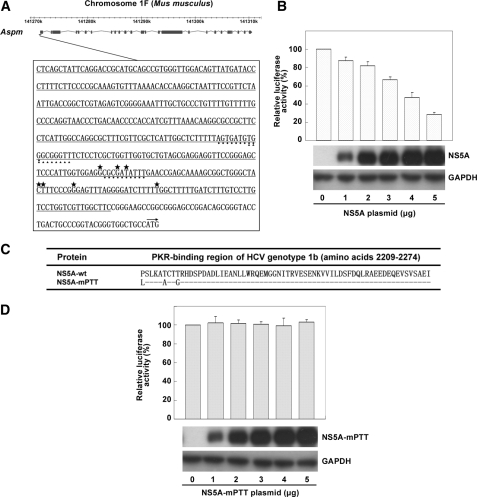
HCV NS5A Regulates the Promoter Activity of Aspm Gene—To examine whether the down-regulation of Aspm gene in NS5A-expressing cells was controlled at the transcriptional level, the promoter activity of Aspm gene was examined. Because the major transcriptional start site of the Aspm gene has not yet been determined, we decided to generate a reporter plasmid that contains a 417-bp DNA fragment of the mouse Aspm gene from nucleotides -475 to -59 upstream of the translation start site based on the information derived from the Data Base of Transcriptional Start Sites and the analytic programs of cpgplot and cpgreport (Fig. 3A). Luciferase reporter assay was performed following a co-transfection of cultured cells with various amounts of the NS5A effector plasmid, the firefly luciferase reporter plasmid, and the Renilla luciferase-expressing control plasmid. The results demonstrated a dose-dependent inhibitory effect of the NS5A protein on the promoter activity of Aspm gene (Fig. 3B). Potential regulatory sequences present in the promoter region of Aspm were analyzed by the TFSEARCH program. This analysis revealed cis-elements conserved in human and mouse for interacting with transcriptional factors Sp1 and GATA1 (Fig. 3A and data not shown). It has been known that Sp1 and GATA1 are generally regulated by MAPK signaling pathways (30-33).
FIGURE 3.
Transcriptional regulation of Aspm gene mediated by the HCV NS5A protein. A, illustration of the chromosome location of the mouse Aspm gene and its putative promoter sequences. The illustration of the chromosome location of the mouse Aspm gene was modified from MGI Mouse Genome Browser. The nucleotide sequences shown represent the predicted promoter region and 5′-untranslated region of the Aspm gene. The nucleotide sequences underlined indicated the 417-bp DNA fragment used to construct the luciferase reporter plasmid pGL3-mAspm-TLS(-475/-59). Potential transcription start sites (*), GATA1 binding cis-elements (•), Sp1 binding cis-element (▴), and the translation start site (→) were marked. B, the inhibitory effect of NS5A protein on the promoter activity of the Aspm gene. NIH3T3 cells cultured on 6-cm dishes were co-transfected with various amounts of the effector plasmid pAdTrack-CMV-T7HisNS5A, 0.5 μg of firefly luciferase reporter plasmid pGL3-mAspm-TLS(-475/-59), and 0.05 μg of Renilla luciferase-expressing control plasmid phRL-TK. At 48 h post-transfection, the cells were harvested for luciferase activity assay and Western blot analysis with antibodies specific to the viral NS5A protein and the control GAPDH. The results shown represent the averages from three independent experiments. C, amino acid sequences of the PKR-binding region of the wild type NS5A and NS5A-mPTT mutant proteins. D, PKR-binding region is important for the NS5A protein to down-regulate the promoter activity of the Aspm gene. Co-transfection followed by luciferase activity assay and Western blot analysis were performed as described in the legend for B except that the effector plasmid used was pAdTrack-CMV-T7HisNS5A-mPTT that encodes the NS5A mutant protein, NS5A-mPTT. The results shown represent averages from three independent experiments.
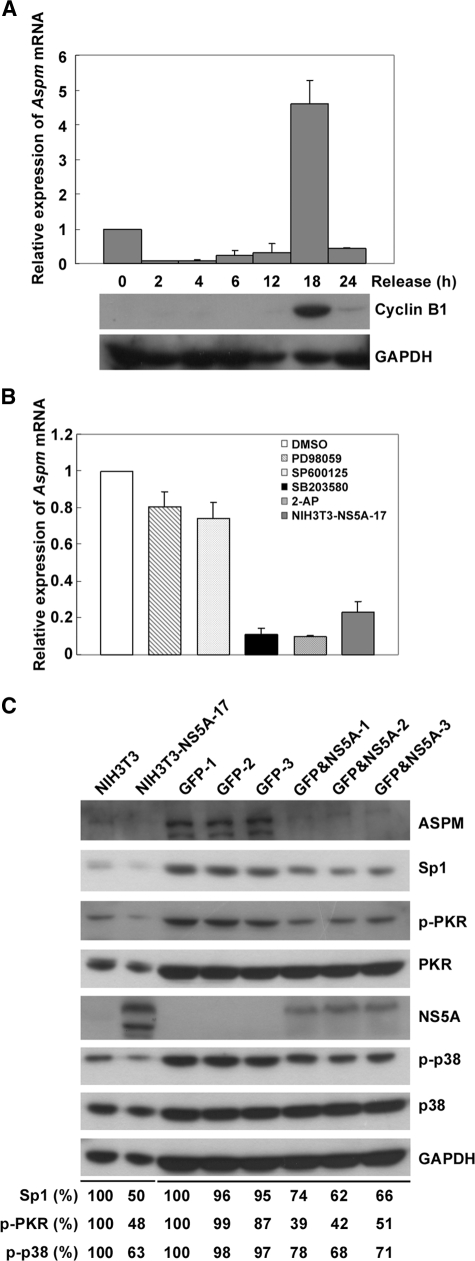
HCV NS5A Reduces the Expression of Aspm Gene via the Inhibition of p38 Phosphorylation and the PKR-p38 Signaling Pathway—To learn the expression profile of the Aspm gene at different cell cycle stages, NIH3T3 cells synchronized at G1/S boundary were subjected to RT real time PCR and Western blot analysis at various time points after release from serum starvation. As shown in Fig. 4A, Aspm was mainly expressed at 18 h after release from serum starvation at which cyclin B1 showed a maximal expression. This result indicated that Aspm mainly expressed at the G2/M phase of cell cycle. To clarify which signal transduction pathway is involved in the NS5A-mediated down-regulation of Aspm, MAPK inhibitors that target ERK (PD98059), JNK (SP600125), and p38 (SB203580) pathways were applied 18 h after the culture cells were released from G1/S arrest. As shown in Fig. 4B, the expression level of Aspm mRNA reduced to ∼10% in the presence of p38 inhibitor SB203580, 80% in the presence of ERK inhibitor PD98059 and 73% in the presence of JNK inhibitor SP600125. These results indicate that the p38 signaling pathway is the major upstream regulator for Aspm. It is previously known that p38 is a downstream target of PKR. It is also known that binding of NS5A to PKR inhibits PKR dimerization and results in an inhibition of PKR autophosphorylation, MKK6 phosphorylation, and p38 signaling pathway activation (34, 35). To test whether PKR-p38 signaling pathway is involved in the down-regulation of Aspm, PKR inhibitor 2-aminopurine was used. At 18 h after release of culture cells from serum starvation, expression level of Aspm mRNA was analyzed. The effect that 2-aminopurine induced ∼90% reduction of Aspm gene expression is similar to that of p38 inhibitor SB203580 as well as that of the HCV NS5A protein in the NIH3T3-NS5A-17 stable cells (Fig. 4B). Furthermore, the phosphorylation level of PKR and p38 and the expression of transcription factor Sp1 were reduced in both NIH3T3-NS5A-17 stable cells and in mouse hepatocytes expressing the NS5A protein (Fig. 4C). These results strongly suggest that NS5A inhibits the expression of Aspm via inhibiting PKR-p38 signaling pathway. Interestingly, a NS5A mutant with amino acid substitutions at Pro2209, Thr2214, and Thr2217 (Fig. 3C) that have known to be critical for NS5A to interact with PKR (8, 9) abolished the inhibitory effect of NS5A on the promoter activity of Aspm gene (Fig. 3D).
FIGURE 4.
HCV NS5A inhibits Aspm gene expression via p38 dephosphorylation. A, the expression profiles of the Aspm mRNA at various stages of the cell cycle. NIH3T3 cells arrested at G1/S boundary were released by adding 10% fetal bovine serum in the culture medium. At various time points, the expression of Aspm mRNA and cyclin B1 were analyzed by RT real time PCR and Western blot analysis, respectively. GAPDH was analyzed in parallel as a loading control. The results shown represent the averages from three independent experiments. B, relative expression level of Aspm in the presence of MAPK and PKR inhibitors. NIH3T3 cells synchronized at the G1/S boundary were incubated with 20 μm PD98059 (ERK inhibitor, Tocris Cookson Inc.), SP600125 (JNK inhibitor, Calbiochem), SB203580 (p38 inhibitor, Calbiochem), or 500 μm PKR inhibitor 2-aminopurine (Sigma-Aldrich) as indicated for 1 h. At 18 h after release from starvation, the cells were harvested for RT real time PCR analysis. In addition, cells treated with 1% Me2SO (SIGMA-ALDRICH) were analyzed in parallel as a negative control. The expression of Aspm gene in NIH3T3-NS5A-17 was also examined as a comparison. The results shown represent the averages from three independent experiments. C, the expression levels of PKR, phosphorylated PKR (p-PKR), p38, phosphorylated p38 (p-p38), and Sp1 in NIH3T3 cells and mouse hepatocytes expressing HCV NS5A protein. The cell lysates prepared from NIH3T3 cells, NIH3T3-NS5A-17, and hepatocytes from individual mouse injected with plasmid pAdTrack-TTRB-T7HisNS5A expressing GFP and NS5A (GFP&NS5A-1, -2, and -3) or plasmid pAdTrack(-CMV) expressing GFP alone (GFP-1, -2, and -3) as described in the legend for Fig. 1 were subjected to Western blot analysis with antibodies specific to ASPM, Sp1, p-PKR, PKR, NS5A, p-p38, p38, and GAPDH as indicated. Relative expression levels of Sp1, p-PKR, and p-p38 shown were calculated by normalization of the intensities of the Sp1, p-PKR, and p-p38 signals from cells expressing NS5A protein to those of the control cells.
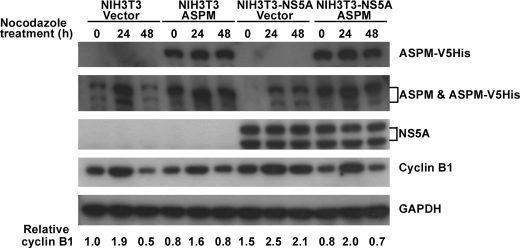
Overexpression of ASPM Relieves the G2/M Cell Cycle Block Imposed by the NS5A Protein—The degradation of cyclin B1 has been known to be required for the transition from metaphase to anaphase (36). We have earlier shown that Aspm co-expressed with the G2/M mitotic marker cyclin B1 at 18 h after the cultured cells were released from serum starvation (Fig. 4A). To examine whether the down-regulation of Aspm is involved in the NS5A-induced aberrant cell growth, the expression levels of cyclin B1 in the parental NIH3T3 cells and the NIH3T3-NS5A-17 cells that constitutively express NS5A were analyzed. NIH3T3 and NIH3T3-NS5A-17 cells were cultured in the presence of 0.2 μg/ml nocodazole for various time periods before harvested for Western blot analysis. The expression level of ASPM increased at 24 h after nocodazole treatment when cyclin B1 expression was maximal. The level of cyclin B1 started to decrease afterward. However, cyclin B1 was accumulated and maintained at similar levels in the NIH3T3-NS5A-17 cells up to 48 h (Fig. 5). These results indicated that NS5A induced a delay of mitotic exit. Association between the G2/M cell cycle block and the NS5A-mediated down-regulation of Aspm was further examined. Plasmid encoding the full-length ASPM was transfected into parental NIH3T3 and NIH3T3-NS5A-17 cells. As shown in Fig. 5, overexpression of ASPM in NIH3T3-NS5A-17 cells relieved the G2/M cell cycle block imposed by the NS5A protein as indicated by a decreased expression level of cyclin B1. Taken together, these results indicate that Aspm is a key target gene of the HCV NS5A protein in the induction of aberrant cell cycle progression.
FIGURE 5.
Overexpression of ASPM rescues the NS5A-induced mitotic exit delay. NIH3T3 and NIH3T3-NS5A-17 cells were infected with recombinant vaccinia virus (vTF7-3) harboring the T7 RNA polymerase gene, followed by DNA transfection with the Lipofectin reagent (Invitrogen) as previously described (40) with plasmid pcDNA-hASPM-V5HisTopo (ASPM) encoding the human full-length ASPM protein with V5His tag or control plasmid pcDNA-V5HisTopo (Vector). Forty-eight hours post-transfection, the cells were treated with 0.2 μg/ml nocodazole and harvested at various time points as indicated. Western blot analysis were performed with antibodies against the His tag of the ectopically expressed human ASPM (ASPM-V5His), with antibodies against ASPM to detect both the mouse endogenous ASPM and the ectopically expressed human ASPM with V5His tag (ASPM and ASPM-V5His) and with antibodies against NS5A, cyclin B1, and GAPDH as indicated. The two forms of the mouse endogenous ASPM protein are predicted as 364 and 212 kDa. The ectopically expressed human ASPM-V5His protein is predicted to be 414 kDa. Relative expression levels of cyclin B1 are shown.
DISCUSSION
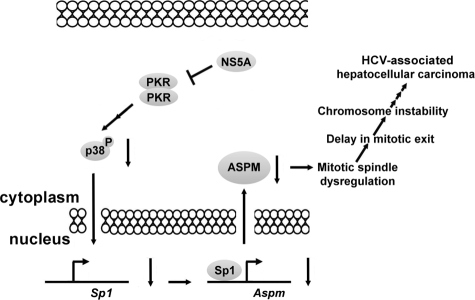
In this study, we demonstrated that the expression of Aspm was down-regulated in the mouse model system (Fig. 1D) and in cultured cells expressing HCV NS5A protein (Fig. 2B). The down-regulation of Aspm was also observed in HCV type 1b subgenomic replicon (supplemental Fig. S1) and in type 1a replicon (37). Our further studies demonstrated that Aspm gene is a downstream target of NS5A involved in HCV-associated pathogenesis. The NS5A protein down-regulated the expression of Aspm gene by repressing the PKR-p38 signaling pathway. A NS5A protein to which the PKR-interacting domain (8, 9) has been mutated failed to regulate the promoter activity of Aspm gene (Fig. 3D). The expression of transcription factor Sp1 was significantly reduced in responding to the down-regulation of the expression of the spindle protein ASPM in all of the three NS5A-expressing systems examined (Fig. 4C and supplemental Fig. S1). Overexpression of the mitotic spindle protein ASPM rescued the delay of cell cycle G2/M exit observed in the NS5A-expressing cells. We propose that, upon HCV infection, NS5A represses the activity of PKR and its downstream molecules, MKK6 and p38 by forming heterodimer with PKR. Down-regulation of p38 phosphorylation may result in the reduction of the expression of Sp1 that is involved in the transcriptional regulation of the Aspm gene. The down-regulation of the Aspm gene affects spindle formation and cell cycle progression. Mitotic dysregulation occurs. As a long term consequence, HCV infection causes chromosomal instability and HCC (Fig. 6). NS5A protein forms a heterodimer with PKR and blocks the translation inhibition of cellular and viral proteins (8, 9). In the present study, we demonstrated that inhibition of PKR-p38-ASPM pathway induced cell cycle G2/M arrest. The effects of NS5A protein on both cellular proliferation and anti-proliferation suggest that NS5A protein plays a key role in the HCV-induced HCC. Mitotic dysregulation was examined in the late passage NIH3T3-NS5A-17 cells by flow cytometry. As shown in supplemental Fig. S3, an increased cell population with chromosome number more than four copies (supplemental Fig. S3, M4 phase) was detected in the late passage NIH3T3-NS5A-17 cells (9.4%) as compared with that of the parental NIH3T3 cells (4.2%). The characteristics of multiple nuclei with abnormal size were also observed by immunofluorescence assay in the late passage NIH3T3-NS5A-17 cells (supplemental Fig. S3). These results indicate that cells expressing NS5A protein are prone to chromosome aneuploidy.
FIGURE 6.
A possible mechanism of the HCV NS5A protein involved in the hepatocarcinogenesis via PKR-p38-ASPM pathway.
Increased expression of Aspm gene has been detected in ovarian, uterine, and brain cancers, which leads to the suggestion that ASPM may be essential for the proliferation of certain cancer cells (23, 38). However, it is not known whether ASPM is essential for the formation of hepatocellular carcinoma in the late stage of HCV infection. Using the approach of hydrodynamics-based in vivo transfection in this study, we have learned the effects of NS5A protein in mouse liver cells at the initial stage of HCV infection. In addition, possible association between the NS5A-induced down-regulation of ASPM and the development of hepatocellular carcinoma has been examined in the mouse model as well as culture systems. Our current data suggest a role for ASPM in the development of hepatocellular carcinoma different from that of the others such as ovarian, uterine, and brain cancers. Following subcutaneous injection of nude mice with the NIH3T3-NS5A cells constitutively expressing NS5A protein, we did not observe tumor formation. However, seven of eight mice injected with NIH3T3-NS5A cells of late passages formed tumors (data not shown). Tumor formation induced by NS5A-expressing cells was also observed in a previous study (39). These results indicate that the NS5A-expressing cells at the late passages, but not the early passages, may be well transformed for tumor growth in nude mice. These systems will be used in future studies to elucidate whether NIH3T3-NS5A cells overexpressing ASPM can prevent or delay tumor formation.
In general, double-stranded RNA intermediates are generated during the infection of RNA viruses. This activates PKR and phosphorylates the translation initiation factor eIF2α. The eIF2α-GDP cannot be recycled to eIF2α-GTP. Translation inhibition of both the host and viral mRNAs results in the anti-proliferation of the virus-infected cells. Nevertheless, the NS5A protein of HCV genotype 1b has a PKR-binding domain through which NS5A forms a heterodimer with PKR. Formation of NS5A-PKR heterodimer blocks PKR activity and allows recycling of eIF2α-GDP to eIF2α-GTP. Translation and cell cycle progression then proceed.
In this study, our data indicate that ASPM is a downstream target of NS5A associated with HCV-induced HCC. In the early stage of HCV infection, NS5A protein inhibits the expression of ASPM, resulting in mitotic dysregulation. However, during the late HCV chronic infection, hepatocytes are transformed and proliferated through as yet unknown mechanisms mediated by unidentified host factors and viral proteins such as NS5A, NS3 protease, and core protein. Detailed mechanisms of how down-regulation of ASPM may be involved in the development of HCC need to be further studied. Nevertheless, ASPM could be a new molecular target for drug development against HCV-associated HCC.
Supplementary Material
Acknowledgments
We thank J. H. Lin (Academia Sinica) and P. H. Lou (National Taiwan University) for statistics and analysis of microarray data. We thank Z. F. Chang (National Taiwan University) and T. L. Tseng (National Taiwan University) for helpful comments on this paper. We are grateful to S. L. Doong (National Taiwan University) for providing the GFP antibodies, to Y. H. Kou (National Taiwan University) for providing H7-HCVR replicon cells, to R. H. Costa (University of Illinois at Chicago) for providing plasmid PAP-TTRBsv, and to S. Sugano (University of Tokyo) for providing the information regarding the transcriptional start sites of Aspm.
This work was supported in part by Research Grants NSC 93-3112-B-002-025-Y, NSC 95-2752-B-002-009-PAE, NSC 96-2752-B-002-009-PAE and NSC 96-3112-B-002-028 from the National Science Council of the Republic of China. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1-S3.
Footnotes
The abbreviations used are: HCV, hepatitis C virus; NS5A, nonstructural protein 5A; MAPK, mitogen-activated protein kinase; ERK, extracellular signal-regulated kinase; SAPK/JNK, stress-activated protein kinase/c-Jun N-terminal kinase; CMV, cytomegalovirus; TTRB, liver-specific transthyretin promoter; GFP, green fluorescence protein; TLS, translation start site; LCM, laser capture microdissection; RT, reverse transcription; HCC, hepatocellular carcinoma.
References
- 1.Hopf, U., Moller, B., Kuther, D., Stemerowicz, R., Lobeck, H., Ludtke-Handjery, A., Walter, E., Blum, H. E., Roggendorf, M., and Deinhardt, F. (1990) J. Hepatol. 10 69-76 [DOI] [PubMed] [Google Scholar]
- 2.Takao, Y., Yamada, A., Yutani, S., Ono, T., Nagao, Y., Ando, E., Ide, T., Itoh, K., and Sata, M. (2007) Med. Microbiol. Immunol. 196 157-164 [DOI] [PubMed] [Google Scholar]
- 3.De Mitri, M. S., Morsica, G., Cassini, R., Bagaglio, S., Zoli, M., Alberti, A., and Bernardi, M. (2002) J. Hepatol. 36 116-122 [DOI] [PubMed] [Google Scholar]
- 4.Nishise, Y., Saito, T., Sugahara, K., Ito, J. I., Saito, K., Togashi, H., Nagano-Fujii, M., Hotta, H., and Kawata, S. (2007) J. Infect. Dis. 196 1006-1009 [DOI] [PubMed] [Google Scholar]
- 5.Tagger, A., Donato, F., Ribero, M. L., Chiesa, R., Portera, G., Gelatti, U., Albertini, A., Fasola, M., Boffetta, P., and Nardi, G. (1999) Int. J. Cancer 81 695-699 [DOI] [PubMed] [Google Scholar]
- 6.Choo, Q. L., Richman, K. H., Han, J. H., Berger, K., Lee, C., Dong, C., Gallegos, C., Coit, D., Medina-Selby, R., Barr, P. J., Weiner, A. J., Bradley, D. W., Kuo, G., and Houghton, M. (1991) Proc. Natl. Acad. Sci. U. S. A. 88 2451-2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaneko, T., Tanji, Y., Satoh, S., Hijikata, M., Asabe, S., Kimura, K., and Shimotohno, K. (1994) Biochem. Biophys. Res. Commun. 205 320-326 [DOI] [PubMed] [Google Scholar]
- 8.Enomoto, N., Sakuma, I., Asahina, Y., Kurosaki, M., Murakami, T., Yamamoto, C., Izumi, N., Marumo, F., and Sato, C. (1995) J. Clin. Investig. 96 224-230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gale, M. J., Jr., Korth, M. J., and Katze, M. G. (1998) Clin. Diagn. Virol. 10 157-162 [DOI] [PubMed] [Google Scholar]
- 10.He, Y., Tan, S. L., Tareen, S. U., Vijaysri, S., Langland, J. O., Jacobs, B. L., and Katze, M. G. (2001) J. Virol. 75 5090-5098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cuenda, A., and Rousseau, S. (2007) Biochim. Biophys. Acta 1773 1358-1375 [DOI] [PubMed] [Google Scholar]
- 12.Lai, C. F., Chaudhary, L., Fausto, A., Halstead, L. R., Ory, D. S., Avioli, L. V., and Cheng, S. L. (2001) J. Biol. Chem. 276 14443-14450 [DOI] [PubMed] [Google Scholar]
- 13.Rennefahrt, U., Illert, B., Greiner, A., Rapp, U. R., and Troppmair, J. (2004) Cancer Lett. 215 113-124 [DOI] [PubMed] [Google Scholar]
- 14.Arima, N., Kao, C. Y., Licht, T., Padmanabhan, R., and Sasaguri, Y. (2001) J. Biol. Chem. 276 12675-12684 [DOI] [PubMed] [Google Scholar]
- 15.Baek, K. H., Park, H. Y., Kang, C. M., Kim, S. J., Jeong, S. J., Hong, E. K., Park, J. W., Sung, Y. C., Suzuki, T., Kim, C. M., and Lee, C. W. (2006) J. Mol. Biol. 359 22-34 [DOI] [PubMed] [Google Scholar]
- 16.He, Y., Nakao, H., Tan, S. L., Polyak, S. J., Neddermann, P., Vijaysri, S., Jacobs, B. L., and Katze, M. G. (2002) J. Virol. 76 9207-9217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tan, S. L., Nakao, H., He, Y., Vijaysri, S., Neddermann, P., Jacobs, B. L., Mayer, B. J., and Katze, M. G. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 5533-5538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park, K. J., Choi, S. H., Choi, D. H., Park, J. M., Yie, S. W., Lee, S. Y., and Hwang, S. B. (2003) J. Biol. Chem. 278 30711-30718 [DOI] [PubMed] [Google Scholar]
- 19.Bond, J., Roberts, E., Mochida, G. H., Hampshire, D. J., Scott, S., Askham, J. M., Springell, K., Mahadevan, M., Crow, Y. J., Markham, A. F., Walsh, C. A., and Woods, C. G. (2002) Nat. Genet. 32 316-320 [DOI] [PubMed] [Google Scholar]
- 20.Bond, J., Scott, S., Hampshire, D. J., Springell, K., Corry, P., Abramowicz, M. J., Mochida, G. H., Hennekam, R. C., Maher, E. R., Fryns, J. P., Alswaid, A., Jafri, H., Rashid, Y., Mubaidin, A., Walsh, C. A., Roberts, E., and Woods, C. G. (2003) Am. J. Hum. Genet. 73 1170-1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar, A., Markandaya, M., and Girimaji, S. C. (2002) J. Biosci. 27 629-632 [DOI] [PubMed] [Google Scholar]
- 22.Mekel-Bobrov, N., Gilbert, S. L., Evans, P. D., Vallender, E. J., Anderson, J. R., Hudson, R. R., Tishkoff, S. A., and Lahn, B. T. (2005) Science 309 1720-1722 [DOI] [PubMed] [Google Scholar]
- 23.Kouprina, N., Pavlicek, A., Collins, N. K., Nakano, M., Noskov, V. N., Ohzeki, J., Mochida, G. H., Risinger, J. I., Goldsmith, P., Gunsior, M., Solomon, G., Gersch, W., Kim, J. H., Barrett, J. C., Walsh, C. A., Jurka, J., Masumoto, H., and Larionov, V. (2005) Hum. Mol. Genet. 14 2155-2165 [DOI] [PubMed] [Google Scholar]
- 24.Zhong, X., Liu, L., Zhao, A., Pfeifer, G. P., and Xu, X. (2005) Cell Cycle 4 1227-1229 [DOI] [PubMed] [Google Scholar]
- 25.Fish, J. L., Kosodo, Y., Enard, W., Paabo, S., and Huttner, W. B. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 10438-10443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wakefield, J. G., Bonaccorsi, S., and Gatti, M. (2001) J. Cell Biol. 153 637-648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheng, J. C., Chang, M. F., and Chang, S. C. (1999) J. Virol. 73 7044-7049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chang, M. F., Sun, C. Y., Chen, C. J., and Chang, S. C. (1993) J. Virol. 67 2529-2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kou, Y. H., Chang, M. F., Wang, Y. M., Hung, T. M., and Chang, S. C. (2007) J. Virol. 81 7999-8008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu, Y. L., Chiang, Y. J., Chen, Y. C., Papetti, M., Juo, C. G., Skoultchi, A. I., and Yen, J. J. (2005) J. Biol. Chem. 280 29533-29542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sevinsky, J. R., Whalen, A. M., and Ahn, N. G. (2004) Mol. Cell. Biol. 24 4534-4545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuo, L., Chang, H. C., Leu, T. H., Maa, M. C., and Hung, W. C. (2006) J. Cell. Physiol. 207 729-734 [DOI] [PubMed] [Google Scholar]
- 33.Towatari, M., Ciro, M., Ottolenghi, S., Tsuzuki, S., and Enver, T. (2004) Hematol. J. 5 262-272 [DOI] [PubMed] [Google Scholar]
- 34.Gale, M., Jr., Blakely, C. M., Kwieciszewski, B., Tan, S. L., Dossett, M., Tang, N. M., Korth, M. J., Polyak, S. J., Gretch, D. R., and Katze, M. G. (1998) Mol. Cell. Biol. 18 5208-5218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Silva, A. M., Whitmore, M., Xu, Z., Jiang, Z., Li, X., and Williams, B. R. (2004) J. Biol. Chem. 279 37670-37676 [DOI] [PubMed] [Google Scholar]
- 36.Clute, P., and Pines, J. (1999) Nat Cell Biol. 1 82-87 [DOI] [PubMed] [Google Scholar]
- 37.Tang, W., Lazaro, C. A., Campbell, J. S., Parks, W. T., Katze, M. G., and Fausto, N. (2007) Am. J. Pathol. 171 1831-1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Horvath, S., Zhang, B., Carlson, M., Lu, K. V., Zhu, S., Felciano, R. M., Laurance, M. F., Zhao, W., Qi, S., Chen, Z., Lee, Y., Scheck, A. C., Liau, L. M., Wu, H., Geschwind, D. H., Febbo, P. G., Kornblum, H. I., Cloughesy, T. F., Nelson, S. F., and Mischel, P. S. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 17402-17407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ghosh, A. K., Steele, R., Meyer, K., Ray, R., and Ray, R. B. (1999) J. Gen. Virol. 80 1179-1183 [DOI] [PubMed] [Google Scholar]
- 40.Hsieh, P. K., Chang, S. C., Huang, C. C., Lee, T. T., Hsiao, C. W., Kou, Y. H., Chen, I. Y., Chang, C. K., Huang, T. H., and Chang, M. F. (2005) J. Virol. 79 13848-13855 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.