Abstract
Mature thrombin-activable fibrinolysis inhibitor (TAFIa) is a highly unstable metallocarboxypeptidase that stabilizes blood clots by clipping C-terminal lysine residues from partially degraded fibrin. In accordance with its in vitro antifibrinolytic activity, animal studies have reported that inhibition of mature TAFI aids in the prevention of thrombosis. The level of TAFI activity is stringently regulated through (i) controlled proteolytic truncation of the zymogen (TAFI), generating the mature enzyme, TAFIa, and (ii) the short half-life of TAFIa. TAFI itself exhibits an intrinsic enzymatic activity, which is likely required to provide a baseline level of antifibrinolytic activity. The novel crystal structure presented here reveals that the active site of TAFI is accessible, providing the structural explanation for the its intrinsic activity. It also supports the notion that an “instability region” exists, in agreement with site-directed mutagenesis studies. Sulfate ions, bound to this region, point toward a potential heparin-binding site and could explain how heparin stabilizes TAFIa.
Thrombin-activable fibrinolysis inhibitor (TAFI)6 (1), also known as plasma carboxypeptidase B (CPB), carboxypeptidase B2, carboxypeptidase R, or procarboxypeptidase U (2-4), is a member of the M14A subfamily of metallocarboxypeptidases (MEROPS data base; Ref. 5). It has been described as a fibrinolysis inhibitor that circulates in plasma as a plasminogen-binding protein. TAFI is involved in the fibrinolytic pathway and therefore seems to be highly regulated (6). It is a 60-kDa glyco-protein secreted by the liver with an isoelectric point of 5.0 (2). The glycans are mainly located on the pro-domain. They account for around 20% of the human TAFI mass and are assumed to play a role in stability and solubility (7). The pro-domain consists of a 92-residue N-terminal peptide, which can be removed by proteolysis, generating a 304-residue catalytic region called TAFIa (1-3).
The removal of this peptide takes place by proteolytic cleavage of a specific Arg-Ala peptide bond at the loop between the pro-domain and the catalytic moiety. These two residues are Arg-92-Ala-93, numbered according to the N terminus of TAFI. The scission and dissociation of the two moieties causes the pI to shift from ∼5 to 8 for mature TAFIa, rendering the protein more basic and less soluble than TAFI (7). Several proteases, including trypsin, thrombin, and plasmin, are able to cleave the Arg-92-Ala-93 bond. In vivo, the thrombin/thrombomodulin complex is most likely responsible for generation of TAFIa, although plasmin may also be able to function in this manner (8). TAFIa down-regulates fibrinolysis by removing C-terminal lysine residues from fibrin that has already been partially degraded by plasmin. Those lysine residues act as ligands for the lysine-binding sites of plasminogen and tissue-type plasminogen activator (9). Removal of the lysine residues attenuates the fibrin cofactor function in the activation of plasminogen by tissue-type plasminogen activator, resulting in the prevention of accelerated plasmin formation and subsequent down-regulation of fibrinolysis (10). Consequently, inhibition of TAFIa may potentiate thrombolysis and thrombolytic therapies (11). TAFIa has also been implicated in the regulation of the inflammatory response by inactivating complement-derived inflammatory peptides (12). This enzyme is therefore considered an important biomedical target (13). TAFI shows around 40% sequence identity to several metallocarboxypeptidases of the M14A subfamily (Fig. 1), sharing with them the most important residues implicated in catalysis, zinc binding, and substrate binding (2, 14). It displays carboxypeptidase B (CPB)-like specificity, although, as mentioned above, it is designed to trim a polymeric natural protein substrate (fibrin) and not peptides like most of the CPB-like enzymes.
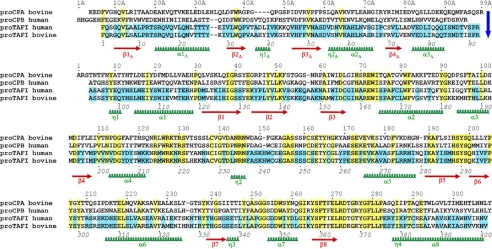
FIGURE 1.
Sequence alignment of human and bovine TAFI with secondary structure elements corresponding to bovine TAFI indicated below. The blue arrow indicates the limit between the pro-domain and the catalytic domain. The sequences of bovine proCPA1 and human proCPB are included at the first two rows of the alignment as a reference. Identical residues in bovine and human TAFI are indicated over a blue background, and those identical in all four sequences are indicated over a yellow background. Residue numbers in the pro-domain are followed by an A, whereas the mature enzyme moiety follows the traditional numbering of bovine CPA1. Thus, although the sequence of bovine TAFI is continuous from its N terminus to its C terminus, its pro-domain starts at position Phe-7A and ends up at position Arg-98A, whereas the mature enzyme moiety starts at position Ala-4. A complete explanation of this numbering system is given by García-Sáez et al. (28). The continuous numbering shown below the aligned sequences does not consider insertions or deletions derived from structural alignments with other proMCPs and permits an easier reference to most available references on TAFI.
Following the removal of the pro-domain, the activity of TAFIa exhibits a short half-life, both in vivo and in vitro, with reported half-lives at 37 °C from 8 to 15 min (15). Due to its physiological role and the need for a very tight regulation of the blood coagulation cascade, it is likely that TAFIa has evolved to acquire its intrinsic instability, which ensures rapid inactivation at the site of action. The irreversible activity decay has been shown to be accompanied by structural changes of the enzyme (16-18). TAFIa can also be inactivated in vivo by further proteolytic degradation, indicating that other parts of the molecule become flexible and accessible. It was therefore suggested that the instability of the activity of TAFIa is due to intrinsic structural lability of the enzyme (19). This is quite unusual among metallocarboxypeptidases, which are generally very stable enzymes in the mature form (14).
The three-dimensional structure of TAFI described here is an advance in the long pursued structural knowledge. It will help to understand the instability of the mature enzyme and predict its ligands and ligand-binding sites. Significantly, these results reinforce a recent report of one of our groups (20) describing the significant intrinsic catalytic activity of TAFI and its potential capability to act in fibrinolysis, a function until recently exclusively assigned to its mature form, TAFIa.
EXPERIMENTAL PROCEDURES
Protein Purification and Crystallization—Bovine TAFI was purified using the same protocol as used for human TAFI (2, 21) and crystallized by hanging drop vapor diffusion at 18 °C using 8 mg/ml protein in 20 mm Tris-HCl, 120 mm NaCl at pH 7.4. Preliminary crystallization trials were conducted using the commercial Crystal Screen and Additive Screen I, II, and III (Hampton Research, Aliso Viejo, CA). The initial crystals were optimized using the micro-seeding technique. The best crystals, which were of bipyramidal shape, were obtained by using 100 mm Tris-HCl, pH 8.0, 1.5 m ammonium sulfate, 2% benzamidine as a precipitant with 10 μl of sodium malonate added to the reservoir the next day after setting up the drop. Crystals of dimensions ∼0.3 × 0.25 × 0.3 mm3 grew in about 10 days. Crystallization plate setup using aged protein (of about 1 week) gave better quality crystals than those from freshly purified protein. Elucidation of optimal cryoconditions involved many trials with conventional cryoprotectants, none of which worked. Interestingly, a quick rinse in unrefined mustard oil (mustard seed oil) was more successful to bring the diffraction from 7 to 3.5 Å. Reannealing with 15% ethylene glycol and 2% glycerol resulted in diffraction beyond 2.5 Å. Crystals were immediately flash-cooled in liquid nitrogen. Data were collected from a single loop-mounted crystal held in a stream of cooled nitrogen gas at 100 K (Oxford Cryosystems). The crystals were fragile and suffered from radiation damage when exposed for longer times to X-rays. The data set up to 2.5 Å resolution (Table 1) were collected at station ID14-EH3 of the European Synchrotron Radiation Facility (ESRF, Grenoble, France) with an ADSC-q4 (Area Detector Systems Corp.) CCD detector. The data were processed using XDS and XSCALE (22).
TABLE 1.
Data collection and refinement statistics
Numbers in parentheses are for the highest-resolution shell. One crystal was used for the data set. No reflection cutoffs were applied.
| Data collection | |
| Space group | P42212 |
| Cell dimensions | |
| a, b, c (Å) | 146.5, 146.5, 231.7 |
| α, β, γ (°) | 90, 90, 90 |
| Resolution (Å) | 50.0-2.5 (2.6-2.5) |
| RmergedF(%) | 5.6 (23.5) |
| I/σI | 29.2 (4.8) |
| Completeness (%) | 99.2 (95.4) |
| Redundancy | 32.2 (30.4) |
| Refinement | |
| Resolution (Å) | 50-2.5 |
| No. Reflections (work/free) | 82547 (4345) |
| Rwork/Rfree (%) | 20.5 (24.3) |
| No. atoms | |
| Protein | 9622 |
| Ligand/ion | 15 NAG, 2 FUC, 1 MAN, 3 MAL, 22 SO4 |
| Water | 1019 |
| B-factors | |
| Molecule I | 47.2 |
| Molecule II | 51.7 |
| Molecule III | 120.9 |
| Ligand/ion | 97.4 |
| Water | 63.2 |
| r.m.s. deviations | |
| Bond lengths (Å) | 0.006 |
| Bond angles (°) 0.9 |
Structure Solution and Refinement—The crystal structure was determined by molecular replacement using the software package Phaser (23). The catalytic domain of the refined structure of porcine proCPB (Protein Data Bank (PDB) code 1NSA; Ref. 24) was used as the search model. The space group was P42212 with three TAFI molecules in the asymmetric unit, the solvent percentage is 66.7%, and Vm = 3.8Å2/Da. The molecular replacement procedure was non-trivial. Initial attempts found only two molecules that did not build a continuous crystal lattice. By allowing a large number of clashes, a third molecule was found that overlapped with the first molecule found. Only by asking Phaser to find four molecules was a third independent molecule found that completed the crystal packing. The search for the first molecule, which was repeated in all possible related space groups, gave a very clear signal: a single solution for the rotation and translation functions and a log-likelihood gain of 367 versus 187 in space group P4122, the related space group with the next best result. Also, for the second molecule, a clear solution was found with a log-likelihood gain of 1160 versus 458 for the second best solution. Even for the third independent molecule, the signal was clear: a log-likelihood gain of 2047 (all molecules combined) versus 1586 for the next best solution. Apparently, the signal of the first molecule was so strong that only after fitting two overlapping molecules at this position was the signal sufficiently attenuated for the signal of the third independent molecule to come through. In retrospect, one probably could have reduced the threshold for potential solutions to be tested and refined in Phaser. Crystallographic refinement was carried out with the programs CNX (25) and Refmac (26). Cycles of manual inspection and rebuilding were done using Quanta (Accelrys Inc., San Diego, CA). Although the first two molecules showed very good electron density, even for the pro-domain, which was not included in the search model, the electron density of the third molecule was very poor. Nevertheless, electron density consistently persisted when parts were omitted, and therefore, most of the third molecule was finally built as well, guided by the well defined structure of the first two molecules. Fig. 2 shows the final electron density map around the active site for the well defined molecule I and the less well defined molecule III. The electron density for molecule II is similar to the electron density for molecule I. Although the average temperature factors of the first two molecules are 47.2 and 51.7 Å2, the average temperature factor of the third molecule was 120.9 Å2. These values are very high but consistent with the overall B-factor of 57.3 Å2 derived from the diffraction data using a Wilson plot. Glycosylation sites were observed at Asn-57A, Asn-69A, and Asn-92A of the pro-domain and a total of 15 N-acetylglucosamine, two fucose, and one mannose molecules were fitted. No glycosylation was observed for Asn-129, which was reported to be glycosylated in human TAFI (7). Interestingly, this residue is completely buried in the TAFI structure. In addition, a large number of sulfate ions, 22 in total, were fitted, as well as three malonate molecules bound in the three independent active sites. In the Ramachandran plot as calculated with the program Molprobity (27), 1045 residues (89.9%) have ϕψ angles in preferred regions, 76 (6.5%) in allowed regions, and 42 (3.6%) in disallowed regions. The final statistics are listed in Table 1. The structure factors and final coordinates have been deposited with the PDB under code 3DGV.
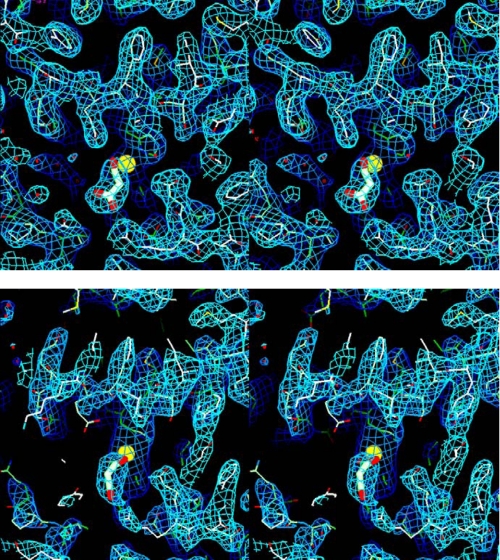
FIGURE 2.
Stereo figures of the electron density of the active site of bovine TAFI with the bound malonate as a stick model and the catalytic zinc as a yellow sphere. Shown is the final 2Fo - Fc map, contoured at 1σ. Top, active site of molecule I. The electron density for molecule II is equally well defined. Bottom, active site of molecule III. This molecule has extremely high temperature factors, and in particular, many side chains are much less well defined as in molecules I and II.
RESULTS
Crystallization, Structure Determination, and Overall Structure—Previously, systematic attempts from our groups to crystallize human TAFI and TAFIa failed due to the low recovery levels of the enzyme purified from human plasma. Attempts to produce crystals using TAFI purified from pooled human plasma were unsuccessful, possibly due to individual glycosylation differences resulting in a heterogeneous population of TAFI molecules. Consequently, we refocused our strategy toward the crystallization and x-ray structure determination of bovine TAFI, for which we were able to purify large amounts of homogeneously glycosylated material from the blood of a single animal. Bovine TAFI has 77% sequence identity and 86% sequence similarity to human TAFI (Fig. 1). In addition, as discussed below, sequence comparison suggests that the bovine form is more stable than the human, which would also facilitate crystallization. The generated bovine TAFI crystals diffracted to 2.5 Å and contained three protein molecules in the asymmetric unit (Fig. 3). These molecules were named I, II, and III in the order they were discovered in the molecular replacement procedure (see “Experimental Procedures”). As shown in Fig. 4, the three independent molecules are very similar in the three-dimensional structure. The r.m.s. deviations are 0.25 Å for all 401 equivalent Cα atoms between molecules I and II and 0.62 Å for all 379 equivalent Cα atoms between molecules I and III as well as II and III.
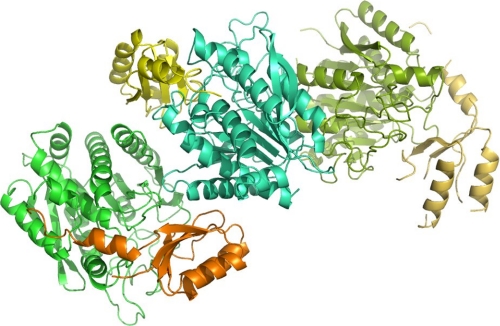
FIGURE 3.
Packing of the three TAFI monomers in the asymmetric unit of our crystals. The two monomers on the left (orange/green and yellow/cyan) have extensive contacts and are packed as a dimer. Dimer contacts between these two monomers stabilize some regions (see “Results”), which are extremely flexible in the third monomer (top, right).
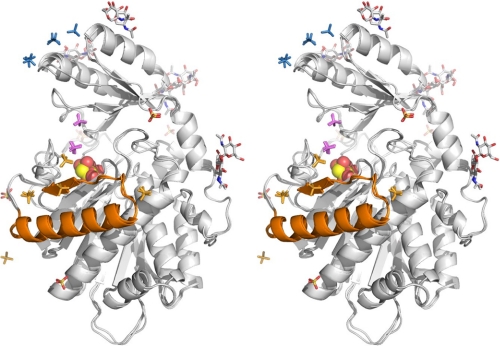
FIGURE 4.
Stereo figure of the superposition of the three independent TAFI molecules with the attached sugars, sulfate, and malonate ions. The conformation of the three molecules is almost identical. The malonate molecule, bound in the active site is shown as a Corey, Pauling, Koltun model. The putative instability region is shown in gold. The bound sulfates are concentrated in three regions: a; the instability region, colored in gold; b, across the active site cleft toward the pro-domain, colored in magenta; and c, at the top of the pro-domain, colored in blue.
Bovine TAFI contains 401 amino acid residues, with residues Phe-7A-Arg-98A (see Fig. 1 for an explanation of the numbering scheme) and Ala-4 -Val-308 corresponding to the N-terminal pro-domain and the catalytic domain, respectively. The pro-domain consists of an N-terminal globular segment encompassing ∼80 residues (running from Phe-7A to Gln-89A) followed by a partially helical connecting region that links it to the carboxypeptidase moiety and contains the target peptide bond for proteolytic activation; this is located in a loosely arranged loop where Arg-98A is the most exposed residue. The catalytic domain has a compact globular shape and displays the typical α/β-hydrolase fold topology consisting of a central mixed eight-stranded twisted β-sheet surrounded by eight α-helices.
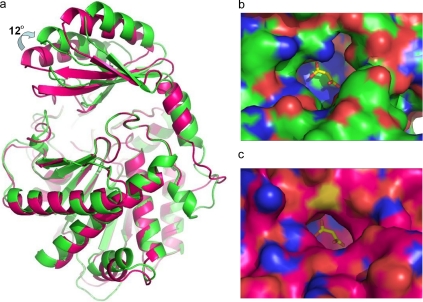
Topologically, the structure of bovine TAFI is similar to the structure of porcine proCPB (PDB code 1NSA; 24), being that the r.m.s. difference is 1.15 Å for all 300 equivalent Cα atoms between the catalytic domains of molecule I and 1NSA and 0.30 Å for the 180 “core” Cα atoms that deviate less than 3σ. For the pro-domain, the r.m.s. differences are 2.52 Å for all 90 equivalent Cα atoms and 0.42 Å for the 52 core Cα atoms. Interestingly, the position of the pro-domain in bovine TAFI is rotated 12° with respect to the orientation of the pro-domain in porcine proCPB (Fig. 5a). As a result, the pro-domain is farther away from the catalytic domain, leading to a more open structure in which the active site is not completely shielded, and an open channel that connects it to the solvent can be visualized (Fig. 5b); this channel is ∼8 Å wide in its narrowest point, whereas it is ∼7 Å wide and globally much narrower in the case of porcine proCPB (Fig. 5c). This entrance channel is also wider than those observed for the carboxypeptidase A1 and A2 proenzymes (28) despite the presence of an inserted 310 helix in the structure of TAFI (i.e. Fig. 1, residues Ala-43-Ala-46), which is typical of the CPB-like forms (29).
FIGURE 5.
Comparison of bovine TAFI and porcine proCPB. a, superposition of the crystal structures of bovine TAFI (green) and porcine proCPB (purple). The entrance to the active site of bovine TAFI is wider than in porcine CPB due to the rotation of the pro-domain. b, entrance to the active site in TAFI. c, entrance to the active site in proCPB. The bound malonate ion is shown in yellow and red. As a reference, a superimposed malonate ion from bovine TAFI is also shown in the structure of proCPB.
Comparison with Human TAFI—Sequence alignment of human and bovine TAFI (Fig. 1) and three-dimensional fitting of the structure derived in this study revealed three clusters of low sequence homology. The first cluster, residues Lys-92-His-96 and Lys-101-Met-103, is located in the loop between helices α2 and α3 and between helix α3 and strand β4. The second cluster, residues Leu-132-Lys-135, is located in the long loop between helices α4 and α5. These clusters are all in solvent-exposed loops, and it is not clear whether they have functional significance. The last cluster involves residues Asn-232-Ile-236 and is right in the middle of the instability region (see below). Mutations to Ile in two residues of this cluster at positions 233 and 236 (Table 2) are in human TAFI associated with increased stability. Since bovine TAFI displays Ile at both positions, it is expected to be more stable than human TAFI; this may in part explain why the bovine form is able to crystallize, whereas the human counterpart is not.
TABLE 2.
Selected human TAFI variants with longer half-lives as wild-type
| Mutationa | t½ | WT t½b | Mutant/WT | Ref.c |
|---|---|---|---|---|
| T233I | 15 min | 8 min | 1.9 | A |
| T233I | 9.4 min | 6.3 min | 1.5 | B |
| T233I-T236I | 15 min | 6.3 min | 2.4 | B |
| T233I-S213C | 41 min | 6.3 min | 6.5 | B |
| T233I-S213C-T236I | 70 min | 6.3 min | 11.1 | B |
| T233I | 18 min | 12 min | 1.5 | C |
| L261Q | 16 min | 12 min | 1.3 | C |
| K55N-H242Q | 31 min | 12 min | 2.6 | C |
| I139T-H242P | 31 min | 12 min | 2.6 | C |
| 168F-H242A | 55 min | 12 min | 4.6 | C |
| H240Y | 47 min | 12 min | 3.9 | C |
| K55N-H201R-S213C-H242P | 4.4 h | 0.2 h | 22 | C |
| S213C-S234N-H242Q | 2.9 h | 0.2 h | 15 | C |
| H240Y-H242Q | 3.0 h | 0.2 h | 15 | C |
| S213C-H240Y | 4.3 h | 0.2 h | 22 | C |
| T233I-H240Y-H242Q | 3.5 h | 0.2 h | 18 | C |
| S213C-H240Y-H242Q | 26 h | 0.2 h | 130 | C |
| S213C-T233I-T236I-H240Y-H242Q | 19 h | 0.1 h | 181 | D |
Numbering adapted to the scheme of Figure 1.
Since the measured t½ is dependent on the exact assay conditions, we have listed for each mutant the wild-type t½ measured under exactly the same conditions. The ratio mutant t½ to WT t½ should be more or less independent of the details of the assay.
Glycosylation Sites—Our electron density maps provided clear evidence for attached carbohydrates at Asn-28A, Asn-57A, Asn-69A, and Asn-92A, all in the pro-domain, although they do not allow a full description of the glycan structures. Equivalent carbohydrate attachment sites have been previously identified in human TAFI by one of our groups (7). A fifth carbohydrate site at Asn-129, identified in the same report in the catalytic domain of human TAFI, could not be confirmed in our bovine TAFI crystal structure, where Asn-129 is buried and cannot be glycosylated.
The Active Site—The active site catalytic machinery follows the same arrangement described for all A/B type M14 metallocarboxypeptidases; the catalytic zinc ion is pentahedrally coordinated by His-69 Nδ1, His-196 Nδ1, and both Oε1 and Oε2of Glu-72 and O3 of the bound malonate that occupies the position normally occupied by a solvent water molecule polarized by the side chain of the general base, Glu-270. Further residues traditionally identified as responsible for substrate binding and catalysis (Arg-71, Arg-145, Arg-127, Ser-197, Tyr-198, Ser-199, Tyr-248, and Glu-270) are found in equivalent positions. The CPB-like specificity toward basic residues of TAFIa is expected to be mainly determined by the acidic residue Asp-255 typically present in B forms at this position in the crystal structure (24, 28). The active site of TAFI is in the active conformation, similar to e.g. proCPB, where the conformation of the catalytic domain within the proenzyme is identical to the conformation of the catalytic domain in the activated form (24). Interestingly, the active site of proCPB is blocked by a salt bridge between Asp-41A in the pro-domain and Arg-145. In contrast, this interaction is absent in TAFI because the residue equivalent to the aspartate in proCPB is here a valine, unable to make a salt link. Considering that the TAFI pro-domain is ∼1.2 Å farther away from the catalytic domain as compared with proCPB due to the 12° rotation described above, it may be feasible that TAFI (zymogen) is able to accommodate substrates. Biochemical evidences (20, 30) indicate that, in contrast to other B forms, TAFI possesses a significant intrinsic proteolytic activity (20). This experimental observation can now be rationalized in view of the structural determinants described here. TAFI displays a wide channel, significantly larger and more open than in the zymogen forms of other carboxypeptidases and lacks the Asp-41A-Arg-145 salt bridge that blocks the active site, both features nicely explaining its intrinsic activity.
A malonate molecule from the crystallization buffer was found bound to the active site in the crystal structure (Figs. 2, 4, and 5). One of the carboxylate groups of the malonate interacts with the catalytic zinc; the other interacts with the side chains of Arg-127 and Arg-145. The same binding sites are used by the peptide substrates (14), further illustrating that the active site is unblocked in our TAFI structure.
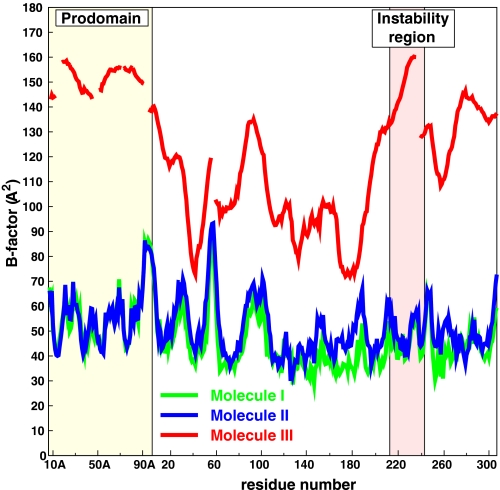
The Instability Region—TAFIa is unstable with a half-life of about 10 min at 37 °C (15), and it is thought that this instability plays an important physiological function. Extensive site-directed mutagenesis has been carried out to clarify this issue, and several of the most relevant results are summarized in Table 2. One of the conclusions of these studies points out to the instability of TAFIa primarily being due to a conformational change and not to proteolytic degradation (17). Natural variants and mutagenesis studies of human TAFI revealed a number of residues that influence the stability of TAFIa (31-34) and are concentrated in the region Ser-213-His-242, which encompasses helix α6 and strand β7 (Fig. 1, and in Fig. 4, the regions colored orange). In this respect, it is noteworthy that the naturally occurring T233I variant in bovine TAFI almost doubles the half-life of human TAFIa (31-34). In molecules I and II, in the asymmetric unit of our structure of bovine TAFI, the equivalent region is involved in dimer contacts with the pro-domain of a neighboring molecule and is well defined in the electron density maps. However, in molecule III, where this region is not tethered by crystal contacts, it is the region of the catalytic domain with the highest temperature factors in excess of 130 Å2 (Fig. 6). The first half of what would be strand β7 in the other molecules is completely disordered. Interestingly, this high temperature factor/disordered region contains 7 of the 12 amino acids in Table 2, which are shown to influence the stability of TAFIa. Combining five stabilizing mutations in this region, Ceresa et al. (34) obtained a dramatic 180-fold increase in the half-life of TAFIa. We therefore postulate that TAFIa inactivation starts in this region, which evolves from a partially disordered to a fully disordered state followed by the unfolding of the remaining parts of the protein. We thus propose to call this region the instability region of TAFI. Further support for this instability region is given below in the section on the heparin-binding site.
FIGURE 6.
Temperature factor distribution of bovine TAFI. Although the temperature factors of molecules I and II are very similar and in the range of 30-90 Å2, molecule III has extremely high temperature factors up to 160 Å2! Nevertheless, significant electron density, especially for the backbone, is still visible for this molecule (Fig. 2).
A Potential Heparin-binding Site?—The 22 bound sulfate ions contained in our crystal structure (Fig. 4) were analyzed in detail in the search for a putative heparin-binding site. Mao et al. (8) report that glycosaminoglycans (heparin) have two effects on TAFI: (i) an increase in generation of TAFIa by plasmin and (ii) a 2.3-fold increase in the half-life of TAFIa at 25 °C, from 74 to 170 min. Based on a sequence analysis, the same authors discovered that the sequence Trp-120-Ser-131 was homologous to the heparin-binding sequence in GAG proteins and postulated this site to be the heparin-binding site of TAFI. However, in our structure, only one sulfate in molecule III was bound to the NZ of Lys-122 at this site and none in molecules I and II.
In contrast, five sulfate ions were found to be bound to the instability region (Ser-213-His-242; region a; Fig. 4, gold sulfates) in molecule I, and three of them were also found at the same position in molecule II. The first sulfate is bound to His-234 and Arg-235 at the C terminus of helix α6 as well as to the side chain of Lys-276 of a neighboring molecule. This site is next to position 233, where the Ile variant is twice as stable as the Thr variant. The second sulfate is bound to Arg-237 and close to the side chains of Glu-49A at the pro-region. Residues Arg-224 and His-240 at about 5 Å distance provide additional charge compensation. Coincidentally, mutations of His-240 were identified by random mutagenesis to influence the half-life of TAFIa. The third sulfate is bound to the side chains of His-216, Ser-220, and Arg-224. Arg-237 and Lys-276 of a neighboring TAFI molecule bind the fourth sulfate. Finally, the fifth sulfate is bound to the side chain of Lys-214 but is closer to a neighboring TAFI molecule.
Another chain of bound sulfates is running from the instability region toward the active site cleft (region b; Fig. 4, magenta sulfates). However, a heparin chain bound in this position is likely to hinder substrate binding and therefore contradict biochemical data. Finally, five sulfate ions are bound at three sites near the very basic top of the pro-domain (region c; Fig. 4, blue sulfates); although this region is a potential heparin-binding site, it cannot explain the stability increase of TAFIa where the pro-domain is absent. Since heparin binding to region b in the active site cleft is likely to hinder catalytic activity and heparin binding to region c in the pro-domain is unable to influence the stability of TAFIa (pro-domain-free TAFI), we conclude that region a is the most likely heparin-binding site. Since region a coincides with the instability region, this finding could well explain the observed increase in TAFIa stability after heparin binding.
DISCUSSION
The crystal structure presented here firmly establishes that the fold of bovine TAFI and, because of the very high sequence similarity, also of human TAFI, is similar to the fold of proCPB, except for the position of the pro-domain shielding the access to the catalytic site. Two of the three independent molecules in the asymmetric unit, I and II, form a dimer and are well defined in the electron density maps since their flexible parts are fixed by dimer contacts. By contrast, molecule III is not stabilized by equivalent contacts and shows significant disorder, with average temperature factors in the range between 75 and 160 Å2. We kept this molecule in our crystallographic model because its electron density consistently persisted, even after refinement with molecule III deleted. The disorder is influenced by the crystal packing but may also be partially caused by the instability of TAFIa. It is interesting to notice that the instability region, as defined by mutagenesis studies (31-34), fully coincides with the region with the highest temperature factors in the catalytic domain of molecule III. The pro-domain of this molecule has even higher temperature factors (Fig. 6), which appear to be caused by a hinge-bending movement.
Several bound sulfate ions were identified in the TAFI crystal structure, indicating the existence of a heparin-binding site. Only one sulfate ion was bound to the residues Trp-120-Ser-131 previously proposed as a heparin-binding site (8). In addition, this region is mainly buried as shown, for instance, by Arg-127. This residue is located in the active-site pocket and is believed to play an important role in catalysis. Consequently, residues Trp-120-Ser-131 are not likely to represent the TAFI heparin-binding site. In contrast, five sulfate ions were bound to the instability region, and heparin binding here may stabilize this region, leading to the 2.3-fold increase in t½ of TAFIa as described previously (8). One should bear in mind, however, that the evidence presented here is indirect and that additional evidence (mutagenesis studies, cocrystal structure) are necessary to establish the heparin-binding site with certainty.
We observed four glycosylated sites in the pro-domain, which have also been detected in human TAFI using mass spectroscopic analyses (7). The fifth site, also identified by mass spectrometry in the mature catalytic domain of the human form, is not glycosylated in the bovine structure. The corresponding residue, Asn-129, is conserved but buried, suggesting that the folding of this region is different in the two despite their similar protein sequences.
The finding of a malonate molecule bound in a substrate-like manner in the potential active site of TAFI, the lack of a salt bridge locking the active site, and its open structure toward the solvent are interesting properties that might merit further investigation. They are in agreement with a recent report that indicates that human TAFI displays a surprisingly high intrinsic activity toward standard substrates of B-like enzymes and large peptides and a significant fibrinolytic activity (20, 30). Since TAFIa has a half-life in blood of only minutes, it seems quite reasonable to consider the possibility that TAFI, which is much more stable and soluble (7), plays a complementary (or even a direct) role in the antifibrinolytic function assigned to the enzyme. Now, a structural basis is provided for this hypothesis. Therefore, the structure here derived might not only help in better drug design strategies but also influence some of the foundations for the design and refinement of drugs and therapeutic strategies based on the potential binding and functional abilities of TAFI. We hope that this work might stimulate further efforts toward these or related research lines.
Supplementary Material
The atomic coordinates and structure factors (code 3DGV) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
This work was supported by Grants LSHG 2006-18830-CAMP from the EC-European Commission, Directorate F, Grant BIO2007-68046 from Ministry of Education and Science of Spain (for Universitat Autònoma de Barcelona), and a grant from the Danish Natural Science Research Council (for Aarhus University). H. A. S. and K. U. W. are employed by Sanofi-Aventis, a publicly held pharmaceutical company. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
This article was selected as a paper of the week.
Footnotes
The abbreviations used are: TAFI, thrombin-activable fibrinolysis inhibitor, uncleaved (zymogen) form; TAFIa, cleaved thrombin activatable fibrinolysis inhibitor; CPA, carboxypeptidase A; CPB, carboxypeptidase B; r.m.s., root mean square.
References
- 1.Bajzar, L., Manuel, R., and Nesheim, M. E. (1995) J. Biol. Chem. 270 14477-14484 [DOI] [PubMed] [Google Scholar]
- 2.Eaton, D. L., Malloy, B. E., Tsai, S. P., Henzel, W., and Drayna, D. (1991) J. Biol. Chem. 266 21833-21838 [PubMed] [Google Scholar]
- 3.Hendriks, D., Wang, W., Scharpe, S., Lommaert, M. P., and van Sande, M. (1990) Biochim. Biophys. Acta 1034 86-92 [DOI] [PubMed] [Google Scholar]
- 4.Wang, W., Hendriks, D. F., and Scharpe, S. S. (1994) J. Biol. Chem. 269 15937-15944 [PubMed] [Google Scholar]
- 5.Rawlings, N. D., Morton, F. R., and Barrett, A. J. (2006) Nucleic Acids Res. 34 D270-272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bouma, B. N., Marx, P. F., Mosnier, L. O., and Meijers, J. C. (2001) Thromb. Res. 101 329-354 [DOI] [PubMed] [Google Scholar]
- 7.Valnickova, Z., Christensen, T., Skottrup, P., Thogersen, I. B., Hojrup, P., and Enghild, J. J. (2006) Biochemistry 45 1525-1535 [DOI] [PubMed] [Google Scholar]
- 8.Mao, S. S., Cooper, C. M., Wood, T., Shafer, J. A., and Gardell, S. J. (1999) J. Biol. Chem. 274 35046-35052 [DOI] [PubMed] [Google Scholar]
- 9.Suenson, E., Lutzen, O., and Thorsen, S. (1984) Eur. J. Biochem. 140 513-522 [DOI] [PubMed] [Google Scholar]
- 10.Redlitz, A., Nicolini, F. A., Malycky, J. L., Topol, E. J., and Plow, E. F. (1996) Circulation 93 1328-1330 [DOI] [PubMed] [Google Scholar]
- 11.Zirlik, A. (2004) Thromb. Haemostasis 91 420-422 [PubMed] [Google Scholar]
- 12.Campbell, W., Okada, N., and Okada, H. (2001) Immunol. Rev. 180 162-167 [DOI] [PubMed] [Google Scholar]
- 13.Willemse, J. L., and Hendriks, D. F. (2007) Front Biosci. 12 1973-1987 [DOI] [PubMed] [Google Scholar]
- 14.Arolas, J. L., Vendrell, J., Aviles, F. X., and Fricker, L. D. (2007) Curr. Pharm. Des. 13 349-366 [DOI] [PubMed] [Google Scholar]
- 15.Boffa, M. B., Wang, W., Bajzar, L., and Nesheim, M. E. (1998) J. Biol. Chem. 273 2127-2135 [DOI] [PubMed] [Google Scholar]
- 16.Boffa, M. B., Bell, R., Stevens, W. K., and Nesheim, M. E. (2000) J. Biol. Chem. 275 12868-12878 [DOI] [PubMed] [Google Scholar]
- 17.Marx, P. F., Hackeng, T. M., Dawson, P. E., Griffin, J. H., Meijers, J. C., and Bouma, B. N. (2000) J. Biol. Chem. 275 12410-12415 [DOI] [PubMed] [Google Scholar]
- 18.Nesheim, M. (2003) Chest 124 33S-39S [DOI] [PubMed] [Google Scholar]
- 19.Marx, P. F., Bouma, B. N., and Meijers, J. C. (2002) Biochemistry 41 1211-1216 [DOI] [PubMed] [Google Scholar]
- 20.Valnickova, Z., Thøgersen, I. B., Potempa, J., and Enghild, J. J. (2007) J. Biol. Chem. 282 3066-3076 [DOI] [PubMed] [Google Scholar]
- 21.Valnickova, Z., Thogersen, I. B., Christensen, S., Chu, C. T., Pizzo, S. V., and Enghild, J. J. (1996) J. Biol. Chem. 271 12937-12943 [DOI] [PubMed] [Google Scholar]
- 22.Kabsch, W. (1993) J. Appl. Crystallogr. 26 795-800 [Google Scholar]
- 23.Storoni, L. C., McCoy, A. J., and Read, R. J. (2004) Acta Crystallogr. Sect. D Biol. Crystallogr. 60 432-438 [DOI] [PubMed] [Google Scholar]
- 24.Coll, M., Guasch, A., Aviles, F. X., and Huber, R. (1991) EMBO J. 10 1-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T., and Warren, G. L. (1998) Acta Crystallogr. Sect. D Biol. Crystallogr. 54 905-921 [DOI] [PubMed] [Google Scholar]
- 26.Murshudov, G. N., Vagin, A. A., and Dodson, E. J. (1997) Acta Crystallogr. Sect. D Biol. Crystallogr. 53 240-255 [DOI] [PubMed] [Google Scholar]
- 27.Davis, I. W., Leaver-Fay, A. L., Chen, V. B., Block, J. N., Kapral, G. J., Wang, X., Murray, L. W., Arendall, W. B., III, Snoeyink, J., Richardson, J. S., and Richardson, D. C. (2007) (2007) Nucleic Acids Res., 35 W375-W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garcia-Saez, I., Reverter, D., Vendrell, J., Aviles, F. X., and Coll, M. (1997) EMBO J. 16 6906-6913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vendrell, J., Querol, E., and Aviles, F. X. (2000) Biochim. Biophys. Acta 1477 2884-2898 [DOI] [PubMed] [Google Scholar]
- 30.Willemse, J. L., Polla, M., and Hendriks, D. F. (2006) Anal. Biochem. 356 157-159 [DOI] [PubMed] [Google Scholar]
- 31.Schneider, M., Boffa, M., Stewart, R., Rahman, M., Koschinsky, M., and Nesheim, M. (2002) J. Biol. Chem. 277 1021-1030 [DOI] [PubMed] [Google Scholar]
- 32.Ceresa, E., Van de Borne, K., Peeters, M., Lijnen, H. R., Declerck, P. J., and Gils, A. (2006) J. Biol. Chem. 281 15878-15883 [DOI] [PubMed] [Google Scholar]
- 33.Knecht, W., Willemse, J., Stenhamre, H., Andersson, M., Berntsson, P., Furebring, C., Harrysson, A., Hager, A. C., Wissing, B. M., Hendriks, D., and Cronet, P. (2006) FEBS J. 273 778-792 [DOI] [PubMed] [Google Scholar]
- 34.Ceresa, E., De Maeyer, M., Jonckheer, A., Peeters, M., Engelborghs, Y., Declerck, P. J., and Gils, A. (2007) J. Thromb. Haemostasis 5 2105-2112 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.