Abstract
The Eph receptor tyrosine kinases regulate a variety of physiological and pathological processes not only during development but also in adult organs, and therefore they represent a promising class of drug targets. The EphA4 receptor plays important roles in the inhibition of the regeneration of injured axons, synaptic plasticity, platelet aggregation, and likely in certain types of cancer. Here we report the first crystal structure of the EphA4 ligand-binding domain, which adopts the same jellyroll β-sandwich architecture as shown previously for EphB2 and EphB4. The similarity with EphB receptors is high in the core β-stranded regions, whereas large variations exist in the loops, particularly the D-E and J-K loops, which form the high affinity ephrin binding channel. We also used isothermal titration calorimetry, NMR spectroscopy, and computational docking to characterize the binding to EphA4 of two small molecules, 4- and 5-(2,5 dimethyl-pyrrol-1-yl)-2-hydroxybenzoic acid which antagonize ephrin-induced effects in EphA4-expressing cells. We show that the two molecules bind to the EphA4 ligand-binding domain with Kd values of 20.4 and 26.4 μm, respectively. NMR heteronuclear single quantum coherence titrations revealed that upon binding, both molecules significantly perturb EphA4 residues Ile31-Met32 in the D-E loop, Gln43 in the E β-strand, and Ile131-Gly132 in the J-K loop. Molecular docking shows that they can occupy a cavity in the high affinity ephrin binding channel of EphA4 in a similar manner, by interacting mainly with the EphA4 residues in the E strand and D-E and J-K loops. However, many of the interactions observed in Eph receptor-ephrin complexes are absent, which is consistent with the small size of the two molecules and may account for their relatively weak binding affinity. Thus, our studies provide the first published structure of the ligand-binding domain of an EphA receptor of the A subclass. Furthermore, the results demonstrate that the high affinity ephrin binding channel of the Eph receptors is amenable to targeting with small molecule antagonists and suggest avenues for further optimization.
The erythropoietin-producing hepatocellular (Eph)3 carcinoma receptors constitute the largest family of receptor tyrosine kinases, with 16 individual receptors throughout the animal kingdom, which are activated by nine ephrins (1-6). Eph receptors and their ligands are both anchored onto the plasma membrane and are subdivided into two subclasses (A and B) based on their sequence conservation and binding preferences. Usually, EphA receptors (EphA1-A10) interact with glycosylphosphatidylinositol-anchored ephrin-A ligands (ephrin-A1-A6), whereas EphB receptors (EphB1-B6) interact with transmembrane ephrin-B ligands (ephrin-B1-B3) that have a short cytoplasmic portion carrying both Src homology domain 2 and PDZ domain-binding motifs (7, 8).
The Eph receptors have a modular structure, consisting of a unique N-terminal ephrin-binding domain followed by a cysteine-rich linker and two fibronectin type III repeats in the extracellular region. The intracellular region is composed of a conserved tyrosine kinase domain, a C-terminal sterile α-domain, and a PDZ-binding motif. The N-terminal 180-residue globular domain of the Eph receptors has been shown to be sufficient for high affinity ephrin binding (9-11). EphA subclass receptors remarkably differ from EphB receptors because they lack a 4-residue insert in the H-I loop of the ligand-binding domain. Previously, the structures of the EphB2 and EphB4 ligand-binding domains have been determined in both the free state and in complex with ephrins or peptide antagonists (10, 11, 12-15). These studies have shown that the ligand-binding domains of EphB2 and EphB4 adopt the same jellyroll β-sandwich architecture composed of 11 antiparallel β-strands connected by loops of various lengths. In particular, the D-E and J-K loops have been revealed to play a critical role by forming the high affinity Eph-ephrin binding channel.
Interactions between Eph receptors and ephrins initiate bidirectional signals that direct pattern formation and morphogenetic processes, such as axon growth, cell assembly and migration, and angiogenesis (1-8). The roles of Eph receptors and ephrins in bone remodeling, immune function, blood clotting, and stem cells are also starting to be characterized. In general, although interactions between the Eph receptors and ephrins of the same subclass are quite promiscuous, interactions between subclasses are relatively rare. However, EphA4 is a receptor capable of interacting with ephrins of both subclasses to generate a diverse spectrum of biological activities (16-18).
EphA4 has important functions in the developing and adult nervous system and is expressed in brain regions characterized by extensive synaptic remodeling (19, 20). In the adult, EphA4 is particularly enriched in the hippocampus and cortex, two brain structures important for learning and memory processes. Although EphA4 interacts with ephrin-A ligands to mediate a variety of critical biological processes, such as inhibiting integrin downstream signaling pathways (19) and modulating sensory and motor projections (21), this receptor is also able to bind all three ephrin-B ligands. For example, EphA4 interacts with ephrin-B1 expressed in human platelets to stabilize blood clot formation through an integrin-dependent mechanism (22). By interacting with ephrin-B2 and/or ephrin-B3, EphA4 regulates neuronal circuits important for coordinated movement and may inhibit the regeneration of injured spinal cord axons (23-25).
The critical roles of EphA4 in various physiological and pathological processes validate this receptor as a promising target for the development of small molecule drugs to treat human diseases, such as spinal cord injury, abnormal blood clotting, and certain types of cancer (22-29). Despite intensive efforts, only several small molecule inhibitors of Eph receptors have been reported previously, which target the ATP-binding site in the receptor cytoplasmic kinase domain (30-33). However, these molecules also inhibit the activities of other families of kinases (30, 31). On the other hand, although the high affinity ephrin binding pocket of the Eph receptors appears to be an attractive target for design of small molecules capable of inhibiting the Eph receptor signaling by blocking ephrin binding, only now two small molecules have been identified by a high throughput screening, which are able to antagonize ephrin-induced effects in EphA4-expressing cells (see accompanying article, Ref. 54). Hence, it is of significant interest to gain structural insight into the binding interactions between the two small molecules and the EphA4 ligand-binding domain, with the ultimate goal to develop small molecule antagonists capable of inhibiting Eph-ephrin binding with high affinity and specificity.
So far, no structure has been published for the ligand-binding domain of any EphA subclass member. In this study, we determined the crystal structure of the EphA4 ligand-binding domain and characterized its binding to two antagonistic small molecules, namely 4- and 5-(2,5 dimethyl-pyrrol-1-yl)-2-hydroxybenzoic acid by using isothermal titration calorimetry, CD, NMR spectroscopy, and computational docking.
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification of the EphA4 Ligand-binding Domain—The DNA fragment encoding the human EphA4 ligand-binding domain (residues 28-208) was amplified from a HeLa cell cDNA library by using two primers containing BamHI and XhoI restriction sites, 5′-GGATCCAATGAAGTTACCTTATTGGATTCC-3′ (forward) and 5′-CTCGAGTCAGCGGACTGTGAGTGGACAC-3′ (reverse). The PCR fragment was cloned into a modified pET32a vector (Novagen), and the vector was transformed into Escherichia coli Rosetta-gami (DE3) cells (Novagen), as described previously (34), allowing more efficient formation of disulfide bonds and expression of eukaryotic proteins containing codons rarely used in E. coli. To enhance the solubility of the EphA4 ligand-binding domain for NMR studies, in this construct we also included a C-terminal tail (residues 175-181), which was found to be totally unstructured in all structures determined so far. The free Cys176 in this extra tail was mutated to Ala by use of the site-directed mutagenesis kit (Stratagene) to avoid the formation of non-native disulfide bridges.
The cells were cultured in Luria-Bertani medium at 37 °C until the absorbance at 600 nm reached ∼0.7. Then 0.4 mm isopropyl 1-thio-d-galactopyranoside was then added to induce EphA4 expression at 20 °C overnight. The harvested cells were sonicated in the lysis buffer containing 150 mm sodium chloride, 20 mm sodium phosphate, pH 7.2, to release soluble His-tagged proteins, which were subsequently purified by affinity chromatography using nickel-nitrilotriacetic acid-agarose (Qiagen). In-gel cleavage of the EphA4 fusion protein was performed at room temperature by incubating the fusion protein attached to nickel-nitrilotriacetic acid-agarose with thrombin overnight. The released EphA4 protein was further purified on an AKTA FPLC machine (Amersham Biosciences) using a gel filtration column (HiLoad 16/60 Superdex 200) equilibrated with a buffer containing 150 mm NaCl, 50 mm Tris-HCl, pH 7.5, followed by ion-exchange chromatography on an anion-exchange column (Mono Q 5/50). The eluted fraction containing the EphA4 ligand-binding domain was collected and buffer-exchanged to a buffer containing 150 mm NaCl, 25 mm Tris-HCl, and 5 mm CaCl2, pH 7.8, for storage.
The generation of the isotope-labeled proteins for NMR studies followed a similar procedure except that the bacteria were grown in M9 medium with the addition of (15NH4)2SO4 for 15N labeling and (15NH4)2SO4/[13C]glucose for 15N-/13C-double labeling (34, 35). The purity of the protein samples was verified by the SDS-polyacrylamide gel, and the molecular weight of the recombinant EphA4 ligand-binding domain was verified by a Voyager STR matrix-assisted laser desorption ionization time-of-flight-mass spectrometer (Applied Biosystems). The concentration of protein samples was determined by use of a previously described spectroscopic method in the presence of denaturant (36).
Crystallization, Data Collection, and Structure Determination—The EphA4 ligand-binding domain was prepared at a concentration of 12 mg/ml and crystallized by setting up 2-μl hanging drops at room temperature in a well containing the reservoir solution (20% PEG 4000, 15% isopropyl alcohol, and 0.1 m Hepes, pH 7.5). Rock-like crystals formed after 4 days, and dehydration of the crystals was subsequently performed by moving the coverslips to a new well containing dehydration buffer (20% PEG 4000, 15% isopropyl alcohol, 10% glycol, and 0.1 m Hepes, pH 7.5).
The x-ray diffraction images for a single crystal were collected by using an in-house Rigaku/MSC FR-E x-ray generator with an R-AXIS IV++ imaging plate detector at the Biopolis shared equipment facility. The crystal was protected by the cryoprotectant (20% PEG 4000, 15% isopropyl alcohol, 25% glycol, and 0.1 m Hepes, pH 7.5). The data were indexed and scaled using the program d*Trek (37, 38). After an all-space-group search, the crystal was identified as belonging to the space group P22121 with a = 53.75, b = 71.12, and c = 127.00 with two molecules per asymmetric unit. The Matthews coefficient was 2.91 with 57.68% solvent constant.
The initial model of the EphA4 ligand-binding domain was generated by the program Phaser from the Phenix suite (39) using the EphB2 structure (Protein Data Bank code 1NUK) as a search model through the molecular replacement method. This model was completed by manual fitting using the program COOT (40) and refined using the program Phenix for many rounds (41). During model building and refinement, 9.11% of the data were reserved for cross-validation to monitor the refinement progress. The final R-factor was 0.2335 (Rfree = 0.2869) at 2.8-Å resolution. The final structure was analyzed by PROCHECK (42), and details of the data collection and refinement statistics are shown in Table 1. The atomic coordinates were deposited in the Protein Data Bank with the Protein Data Bank code 3CKH. All the figures were prepared using the PyMOL molecular graphics system (W. L. DeLano, DeLano Scientific LLC, San Carlos, CA).
TABLE 1.
Crystallographic data and refinement statistics for the EphA4 ligand-binding domain structure
| Data collection | |
| Wavelength (Å) | 1.5418 |
| Resolution limit (Å) | 63.52 to 2.80 (2.90 to 2.80)a |
| Space group | P22121 |
| Cell parameters | |
| a (Å) | 53.75 |
| b (Å) | 71.12 |
| c (Å) | 127.00 |
| Observed reflections | 93,170 |
| Unique reflections | 12,572 |
| Completeness | 99.7% (99.7%)a |
| Redundancy | 7.41 (7.52)a |
| Linear R-factor | 0.087 (0.395)a |
| Overall I/(I) | 11.6 (3.5)a |
| Refinement | |
| Resolution range (Å) | 19.70 to 2.80 (2.90 to 2.80)a |
| Rworkb | 0.233 (0.305)a |
| No. of reflections | 11,229 |
| Rfreec | 0.286 (0.371)a |
| No. of reflections | 1126 |
| r.m.s.d. bond lengths | 0.007 Å |
| r.m.s.d. bond angles | 1.17° |
| Ramachandran plot | |
| Favored | 83.0% |
| Allowed | 16.7% |
| Generously allowed | 0.3% |
| Disallowed | 0% |
Values in parentheses are for highest resolution shell.
Rwork = Σ |Fobs - Fcalc|/ΣFobs, where Fcalc and Fobs are the calculated and observed structure factor amplitudes, respectively.
Rfree = as for Rwork, but for 9.11% of the total reflections chosen at random and omitted from refinement.
Oligomerization Status Characterized by FPLC, Dynamic Light Scattering, and Analytic Ultracentrifugation—The oligomerization status of the EphA4 ligand-binding domain was assessed by FPLC gel filtration, dynamic light scattering, as well as analytic ultracentrifugation in solution. Briefly, as described previously (34), the FPLC gel filtration experiments were conducted using a fast protein liquid chromatography AKTA instrument (Amersham Biosciences) with a gel filtration column (HiLoad 16/60 Superdex 200). The column was calibrated with a low molecular weight protein kit (Amersham Biosciences) including four proteins as follows: ribonuclease A (15.6 kDa), chymotrypsinogen A (22.8 kDa), ovalbumin (48.9 kDa), and albumin (65.4 kDa). Dynamic light scattering experiments were performed at 20 °C on a DynaPro-MS/X instrument (Protein Solutions Inc.), and the apparent molecular mass values were calculated from 10 readings using the Protein Dynamics analysis software (43). Sedimentation velocity experiments were done at 20 °C using a Beckman Coulter XL-I analytical ultracentrifuge as described previously (37).
Binding Characterization by Isothermal Titration Calorimetry and Circular Dichroism—Isothermal titration calorimetry experiments were performed using a Microcal VP isothermal titration calorimetry machine as described previously (44). Titrations were conducted in 10 mm phosphate buffer, pH 6.3, at 25 °C. The two small molecule antagonists were purchased from Matrix Scientific, with 4-(2,5-dimethyl-pyrrol-1-yl)-2-hydroxybenzoic acid designated as compound 1 and 5-(2,5 dimethyl-pyrrol-1-yl)-2-hydroxybenzoic acid designated as compound 2. The powders of the two compounds were weighed and then dissolved in 10 mm phosphate buffer with the final pH values adjusted to 6.3. The EphA4 receptor at a concentration of 70 μm was placed in a 1.8-ml sample cell, and the compounds at a concentration of 2 mm were loaded into a 300-μl syringe. The samples were degassed for 15 min to remove bubbles before the titrations were initiated. Control experiments with the same parameter settings were also performed for the two compounds without EphA4, to subtract the effects because of sample dilution. To obtain thermodynamic binding parameters, the titration data after subtracting the values obtained from the control experiments were fit to a single binding site model using the built-in software ORIGIN version 5.0 (Microcal Software Inc.). The detailed setup and the results are documented in Table 2.
TABLE 2.
Thermodynamic parameters for the binding interactions between EphA4 and two small molecules by isothermal titration calorimetry
Compound 1 is 4-(2,5-dimethyl-pyrrol-1-yl)-2-hydroxylbenzonic acid, and compound 2 is 5-(2,5-dimethyl-pyrrol-1-yl)-2-hydroxylbenzonic acid.
| Syringe | Cell | Injection volume | Ka | Kd | Stoichiometry (n) | ΔS | ΔH |
|---|---|---|---|---|---|---|---|
| μl | m−1 | μm | n | cal/mol·K | kcal/mol | ||
| Compound 1 (2 mm) | EphA4 (70 μm) | 5 | 4.893 × 104 ± 5071 | 20.44 | 1.000 ± 0 | 18.11 | −1.001 ± 0.027 |
| Compound 2 (2 mm) | EphA4 (70 μm) | 5 | 3.785 × 104 ± 7575 | 26.42 | 1.000 ± 0 | 20.15 | −0.237 ± 0.013 |
The samples were prepared for circular dichroism experiments by buffer exchanging the EphA4 ligand-binding domain into a 10 mm phosphate buffer, pH 6.3, at a protein concentration of 20 μm. The far-UV circular dichroism experiments were performed using a Jasco J-810 spectropolarimeter, and data from five independent scans were averaged (44). The spectra of the EphA4 receptor in the absence or in the presence of the two compounds at a molar ratio of 1:6 (EphA4:compounds) were collected at room temperature. The contribution of the two compounds and the buffer was removed by subtracting the CD spectra of the two compounds diluted at the identical concentrations and in the same buffer.
Binding Characterization by NMR—Samples were prepared for NMR experiments in 10 mm phosphate buffer, pH 6.3, with the addition of 10% D2O for NMR spin-lock. All NMR data were collected at 25 °C on an 800-MHz Bruker Avance spectrometer equipped with a shielded cryoprobe as described previously (34, 35, 44, 45). For the preliminary sequential assignment, a pair of triple-resonance NMR spectra, HNCACB and CBCA(CO)NH, were acquired on a double-labeled EphA4 sample at a concentration of 500 μm. The obtained sequential assignments were further confirmed by analyzing other three-dimensional spectra including (H)CC(CO)NH, H(CCO)NH, and 15N-edited HSQC-total correlation spectroscopy, HSQC-nuclear Overhauser effect spectroscopy, and 13C-edited HCCH-total correlation spectroscopy and nuclear Overhauser effect spectroscopy. All NMR data were processed with NMRPipe (46) and analyzed with NMRView (47).
For NMR characterization of the binding of the EphA4 ligand-binding domain with two small molecules, two-dimensional 1H-15N HSQC spectra were acquired at a protein concentration of 100 μm in the absence or in the presence of the two molecules at different molar ratios, including 1:1; 1:2, 1:4, 1:6, and 1:8 (EphA4:compounds). By superimposing the HSQC spectra, the shifted HSQC peaks could be identified and further assigned to the corresponding EphA4 residues (44). The degree of perturbation was reflected by an integrated index calculated by the formula ((Δ1H)2 + (Δ15N)2/5)1/2. We also investigated the interactions by monitoring the line broadening and shifting of the resonance peaks of the two compounds in their one-dimensional NMR spectra upon the progressive addition of the EphA4 protein.
Molecular Docking—The models of the EphA4 ligand-binding domains in complex with two antagonistic molecules were constructed by use of the HADDOCK software (48, 49) in combination with CNS (50), which makes use of chemical shift perturbation data to derive the docking while allowing various degrees of flexibility. The docking procedure was performed by three steps as follows: first, randomization and rigid body energy minimization; second, semi-flexible simulated annealing; and third, flexible explicit solvent refinement.
To conduct the docking, several invisible residues over the loop regions were added to the EphA4 crystal structures by COOT (40), and the obtained structures were then subjected to several rounds of energy minimization by PHENIX (41). Subsequently, hydrogen atoms were added to the structures by use of the CNS protocol. On the other hand, the geometric coordinates and parameters for the two small molecules were generated and energy-minimized by the on-line PRODRG server (51).
All EphA4 residues with a chemical shift perturbation greater than the threshold value of 0.08 (2.5 times of the standard deviation) were set to be “active” residues (52), whereas neighbors of active residues were defined as “passive” residues according to HADDOCK definition. These active residues included Gln43 on the E β-strand, Ile31-Met32 and Ile39 on the D-E loop, and Asp123 and Ile131-Gly132 on the J-K loop. Furthermore, all residues with heteronuclear NOE intensities of less than 0.7 were found to be located on the N and C termini, or on the loops, and thus set to be “fully flexible” during the molecular docking. One thousand structures were generated during the rigid body docking, and the best 50 structures were selected for semi-flexible simulated annealing, followed by water refinement. Three structures with the lowest energies were selected for detailed analysis and display.
RESULTS
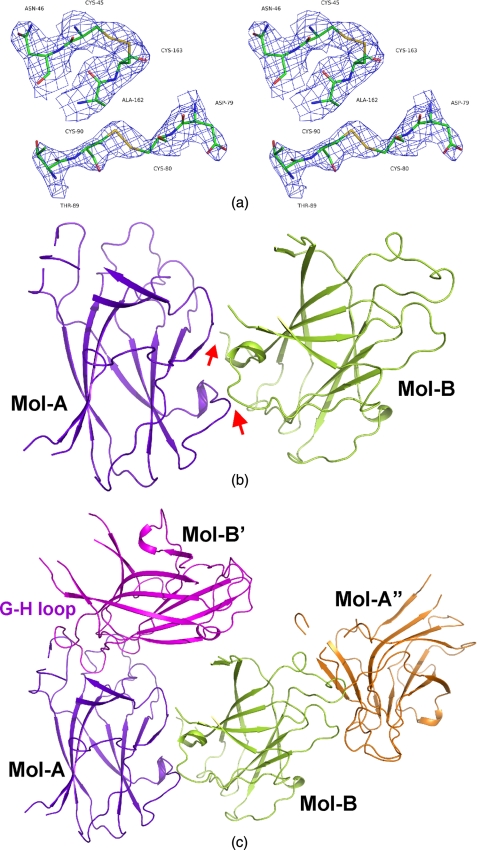
Structure Determination—In this study, we have successfully crystallized the EphA4 ephrin-binding domain without a bound ligand, allowing determination of the crystal structure at 2.8 Å resolution with a final R-factor of 0.2335 (Rfree = 0.2869). Details of the data collection and refinement statistics are summarized in Table 1. In the final model, one asymmetric unit contains two EphA4 molecules designated as A and B (Fig. 1). Because of poor electron density, probably resulting from the inherent flexibility in the absence of bound ligand, some residues were invisible. These residues included the C-terminal seven residues (175-181) for both molecules; Met32, Thr37, Pro38, and Asp133 for molecule A, and Met32-Asn36 and Ile131-Gly132 for molecule B.
FIGURE 1.
Crystal structure of the EphA4 ligand-binding domain. a, stereo view of the two disulfide bridges in the EphA4 ligand-binding domain built into the simulated-annealing 2Fo - Fc electron density map contoured at 1. 5σ. b, ribbon representation of two EphA4 ligand-binding domain molecules A and B (Mol-A and Mol-B) in one asymmetric unit. The red arrows are used to indicate the novel interface between the two molecules. c, ribbon representation of two EphA4 molecules in one asymmetric unit that have differential packing contacts with molecules in other asymmetric units.
As seen in Fig. 1a, there are two conserved disulfide bridges in the EphA4 ligand-binding domain, one within the G-H loop (Cys80-Cys90) and the other between the E-F and L-M loops (Cys45-Cys163). This pattern of disulfide bonds is identical to that observed in the EphB2 and EphB4 structures (9, 14). Interestingly, the two EphA4 molecules appear to pack against each other to form an asymmetric dimer with an interface not observed previously with other Eph receptors, involving residues Ile18-Pro20 and Arg107-Glu111 of molecule A and Val3-Val11 of molecule B (Fig. 1b). Moreover, the two EphA4 molecules in one asymmetric unit pack differently with other EphA4 molecules in neighboring units. The high affinity ligand binding channel of molecule A appears partly occupied by the G-H loop of molecule B′ in a neighboring asymmetric unit, whereas the G-H loop of molecule B inserts into the high affinity ligand binding channel of molecule A″ in another neighboring asymmetric unit (Fig. 1c). Likely because of this differential packing interactions with other EphA4 molecules in neighboring asymmetric units, molecules A and B in the same asymmetric unit show some structural differences over the D-E and J-K loops.
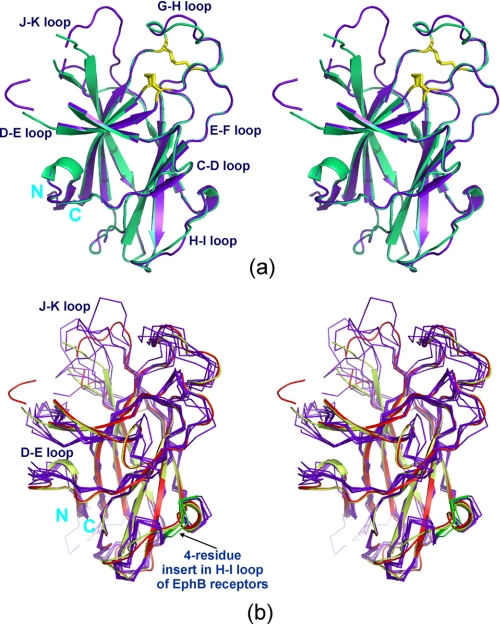
As shown in Fig. 2a, EphA4 molecules A and B adopt the conserved jellyroll folding architecture previously revealed for the EphB2 and EphB4 receptors, composed of 11 antiparallel β-sheets arranged as a compact β-sandwich. The concave sheet is composed of strands C, F, L, H, and I and the convex sheet of strands D, E, A, M, G, K, and J, which are connected by loops of variable length. If only the 11 β-strands are superimposed, the r.m.s.d. between the EphA4 A and B molecules are only 0.074 Å for all atoms and 0.062 Å for backbone atoms. However, molecules A and B have marked differences over the D-E and J-K loops, which are the key components of the high affinity ephrin binding channel. Without considering D-E and J-K loop residues Met32-Ile39 and Asp123-Leu138, the r.m.s.d. between the A and B structures is only 0.4 Å for all atoms. The most distinguishable difference between the A and B molecules involves the J-K loop. The four residues Phe126-Val129, which adopt no regular secondary structure in molecule A, form a short β-strand in molecule B that packs against the extended K-strand residues Met136-Asn139.
FIGURE 2.
Structural comparison. a, stereo view of the superimposition of the two EphA4 ligand-binding domain structures observed in the same asymmetric unit (structure A is in purple and structure B is in green). b, stereo view of the superimposition of two EphA4 structures (structure A in red and structure B in lime green) with previously determined EphB2 and EphB4 structures (all in purple).
As shown in Fig. 2b, despite belonging to the EphA subclass, the structure of the EphA4 ligand-binding domain bears a high similarity over the 11 β-stranded regions to the previously determined ligand-binding domains of the EphB2 and EphB4 receptors. The backbone r.m.s.d. of the EphA4 ligand-binding domain over 11 β-strands are 1.05 Å compared with EphB2 in the free state (PDB code 1NUK), 1.07 Å compared with EphB2 in complex with ephrin-B2 (PDB code 1KGY), 0.74 Å compared with EphB2 in complex with ephrin-A5 (PDB code 1SHW), 0.70 Å compared with EphB2 in complex with an antagonistic peptide (PDB code 2QBX), 0.79 Å compared with EphB4 in complex with an antagonistic peptide (PDB code 2BBA), and 0.80 Å compared with EphB4 in complex with ephrin-B2 (PDB code 2HLE). On the other hand, very large variations are observed over the loop regions not only between EphA4 and the EphB receptors but also between EphB receptors, in particular over the D-E and J-K loops, which are critical for ligand binding. Indeed, the structural flexibility of these loops has been well demonstrated in previously determined EphB structures. Interestingly, the additional short β-sheet observed in the J-K loop of molecule B of the EphA4 ligand-binding domain was also observed in the structure of EphB2 in complex with an antagonistic peptide (PDB code 2QBX) (15). In addition, the EphB receptors have a 4-residue insert in the H-I loop, which is absent in the EphA receptors. Although the H-I loop has no regular secondary structure in all the EphB receptor structures examined, the H-I loop of the EphA4 receptor is shorter and residues Glu111-Asn112-Gln113 form a 310-helix (Fig. 2, a and b).
During the preparation of this manuscript, the crystal structure of the EphA2 ligand-binding domain was released by a structural genomics consortium (PDB code 3C8X). The EphA2 crystals have only one molecule in each asymmetric unit, and structural comparison shows that the two EphA4 molecules and EphA2 are highly similar over the 11 β-stranded regions (only ∼0.45 Å for the backbone r.m.s.d.) and have identical patterns of disulfide bridges (supplemental Fig. 1). Additionally, the short 310-helix observed in the H-I loop of EphA4 is also presented in EphA2. Nevertheless, some structural variations exist over the H-I, G-H, and particularly D-E and J-K loops. Although most J-K loop residues (149-159) are completely missing in the EphA2 structure, structural superimposition indicates that the J-K loop of EphA2 is more similar to that of the EphA4 molecule B (supplemental Fig. 1). This observation suggests that EphA4 molecule B may have more properties of the free state, whereas EphA4 molecule A may be more close to the ligand-bound conformation because its ligand binding channel is partly occupied by the G-H loop of the neighboring EphA4 molecules in the other asymmetric unit.
We have assessed the oligomerization state of the EphA4 ligand-binding domain in solution by use of FPLC gel filtration, dynamic light scattering, and analytical ultracentrifugation. The EphA4 ligand-binding domain was constantly eluted as a monomer on an FPLC gel filtration column, even at concentrations of up to 12 mg/ml (HiLoad 16/60 Superdex 200). Dynamic light scattering and analytical ultracentrifugation data also indicate that the EphA4 ligand-binding domain exists in a monomeric state in solution at concentrations of ∼100 μm. Therefore, the EphA4 dimerization observed in the same asymmetric unit and the interactions among EphA4 molecules in the different units likely result from the packing force in the crystals.
Binding Interactions Characterized by Isothermal Titration Calorimetry and Circular Dichroism—Recently, a 2,5-dimethylpyrrolyl benzoic acid derivative has been identified in a high throughput screening for inhibitors of EphA4 ligand binding (54). This small molecule and an isomeric compound were found to antagonize ephrin-induced effects in EphA4-expressing cells. To assess whether the two isomeric small molecules directly interact with the EphA4 ligand-binding domain, we utilized isothermal titration calorimetry to measure their thermodynamic binding parameters. By using a high concentration of the EphA4 ligand-binding domain (70 μm), we succeeded in obtaining these parameters (supplemental Fig. 2 and Table 2), thus clearly confirming that the two small molecules do interact with the ligand-binding domain of EphA4. Interestingly, the two compounds have similar binding affinities for the EphA4 ligand-binding domain (Kd values of 20.4 μm for compound 1 and 26.4 μm for compound 2), but their binding causes different enthalpy changes (ΔH values of -1,001 for compound 1 and -237 cal for compound 2).
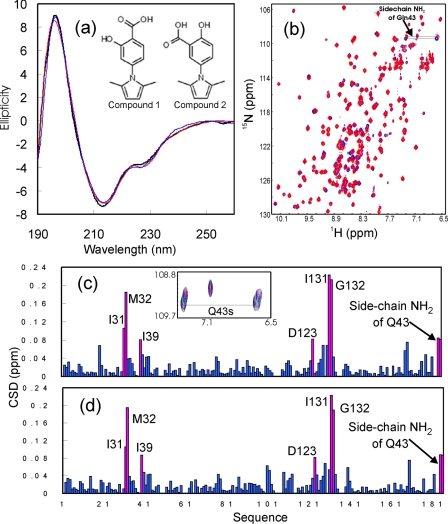
Far-UV circular dichroism spectroscopy was also used to monitor the overall structural changes in the EphA4 ligand-binding domain upon binding of the two molecules. As seen in Fig. 3a, no significant difference was detected between the far-UV CD spectra of EphA4 in the absence and in the presence of the two small molecules at a molar ratio of 1:6 (EphA4/compound). This result implies that no significant changes in secondary structure occurred in the EphA4 ligand-binding domain upon binding, which is consistent with the relatively weak binding affinity of the two molecules.
FIGURE 3.
Characterization of the interactions with two small molecule antagonists. a, far-UV circular dichroism spectra of the EphA4 ligand-binding domain in the absence (black) and in the presence of compound 1 (blue) or compound 2 (red). The chemical structures of the two compounds are presented. b, 1H-15N NMR HSQC spectra of the EphA4 ligand-binding domain in the absence (blue) and in the presence of compound 1 (red). c, residue-specific CSD of the EphA4 ligand-binding domain in the presence of compound 1. d, residue-specific CSD of the EphA4 ligand-binding domain in the presence of compound 2. Violet bars indicate residues with CSD larger than 2.5 times of the standard deviation as described under “Experimental Procedures.” In all experiments the molar ratio of EphA4 to compound was 1:6.
Binding Interactions Characterized by NMR—Because the two small molecules have medium binding affinity for EphA4, it would be difficult to obtain stable receptor-compound complexes for co-crystallization. We therefore decided to probe their binding interactions with EphA4 using NMR spectroscopy, which is highly sensitive to weak binding. We prepared 15N/13C double-labeled EphA4, collected a series of three-dimensional heteronuclear NMR spectra at a protein concentration of 500 μm, and completed the sequential assignments. As evident from the very large dispersions in both dimensions (3.7 ppm for 1H and 25 ppm for 15N) of the HSQC spectrum (Fig. 3b), the EphA4 ligand-binding domain is well folded in solution. Only one set of HSQC peaks was observed for all the EphA4 residues, suggesting that the asymmetric dimer observed in the crystals does not exist in solution on the NMR time scale.
We subsequently used NMR HSQC titrations to detect the EphA4 residues that were perturbed by the binding of two compounds. Because the chemical shift value of an NMR active atom is sensitive to its chemical environment, chemical shift perturbation analysis upon titration of ligands represents a powerful method for identifying residues that directly contact the ligands or that are indirectly affected by the binding event. Two-dimensional 1H-15N HSQC spectra of 15N-labeled EphA4 were recorded to monitor the changes of the HSQC cross-peaks of the amide groups induced by successive additions of the two compounds. We observed a gradual shift of the EphA4 HSQC peaks, correlating with the increased concentrations of the two compounds, which suggests that the free and bound EphA4 molecules undergo a fast exchange on the chemical shift time scale. This allowed assignment of the resonances in the complex by following the shifts in the EphA4 cross-peaks upon gradual addition of increasing amounts of two compounds.
As shown in the isothermal titration calorimetry profiles (supplemental Fig. 2), the binding interaction of EphA4 with the two compounds was largely saturated at molar ratios beyond 1:4 (EphA4/compound). Consistent with this, many HSQC peaks did not exhibit significant further shifts at molar ratios beyond 1:6. Therefore, to identify the interaction surfaces, the chemical shift differences (CSD) between the free state and the bound state in the presence of a 6-fold excess of the two compounds were calculated as described under “Experimental Procedures” and plotted versus the EphA4 sequence (Fig. 3, c and d). The two compounds induced similar shift patterns for the EphA4 residues, and most EphA4 residues did not experience large chemical shift perturbations, indicating that the two compounds did not alter the overall structure of EphA4, consistent with the circular dichroism results shown in Fig. 3a. We have also completed the NMR sequential assignments for the EphA4 ligand-binding domain in the absence and in the presence of compound 1,4 confirming that binding of this compound does not induce significant changes in the secondary structure of EphA4. Interestingly, we observed only eight resonance peaks with significant CSD (deviating more than 2.5 standard deviations from the mean CSD), including residues Ile31-Met32 and Ile39 located in the D-E loop, Gln43 in the E β-strand, and Asp123 and Ile131-Gly132 in the J-K loop. Because the E β-strand and the D-E and J-K loops have been previously shown to be key components of the high affinity ephrin binding channel of the Eph receptors, the NMR titration results thus suggest that the two molecules bind to the high affinity ephrin binding channel of EphA4. We also attempted to estimate the dissociation constants for the binding of the two compounds by fitting the HSQC peak tracings at different compound concentrations (44). However, accurate data fitting was impossible because at high compound concentrations the HSQC peaks for the residues with large shifts disappeared.
Further attempts to identify intermolecular NOE connectivities between EphA4 and the compounds were not successful because the presence of the compounds appeared to induce significant NMR line broadening, which even caused the disappearance of the EphA4 intra- and inter-residue NOEs. On the other hand, with progressive addition of the EphA4 protein, all 1H resonance peaks of the two molecules underwent line broadening and gradual shifting in one-dimensional NMR spectra (data not shown). This indicates that the free and bound forms of the two molecules were in fast exchange on the chemical shift time scale and also suggests that the entire molecules were either directly or indirectly affected by binding to EphA4, consistent with their small size.
Molecular Docking—The absence of intermolecular NOEs between the EphA4 ligand-binding domain and the two molecules made it impossible to calculate the structures of their complexes with NMR distance constraints. As an alternative, we used the HADDOCK docking strategy to construct models of the EphA4 ligand-binding domain in complex with the two molecules. HADDOCK is a recent but well established docking procedure that makes use of NMR chemical shift perturbation data in conjunction with the CNS program to drive the molecular docking of protein-protein and protein-small molecule complexes. Interestingly, as shown in Fig. 1, each crystal asymmetric unit contains two EphA4 molecules A and B, which show large structural differences in the J-K loop. Interestingly, in solution the EphA4 ligand-binding domain is a monomer even at very high concentrations, as demonstrated by FPLC gel filtration, dynamic light scattering, and analytic ultracentrifugation. Analysis of the NMR Cα, Cβ, and Hα chemical shifts for the EphA4 ligand-binding domain in solution shows that the four residues Phe126-Val129 in the J-K loop preferentially form a short β-strand, as observed in molecule B. Furthermore, the NMR structure of the unliganded EphA4 ephrin-binding domain, which we have recently determined, is highly similar to those in the crystal and contains the short β-sheet observed in molecule B (to be published elsewhere). Therefore, it is likely that molecule B in the crystal more closely represents the conformation of EphA4 in solution.
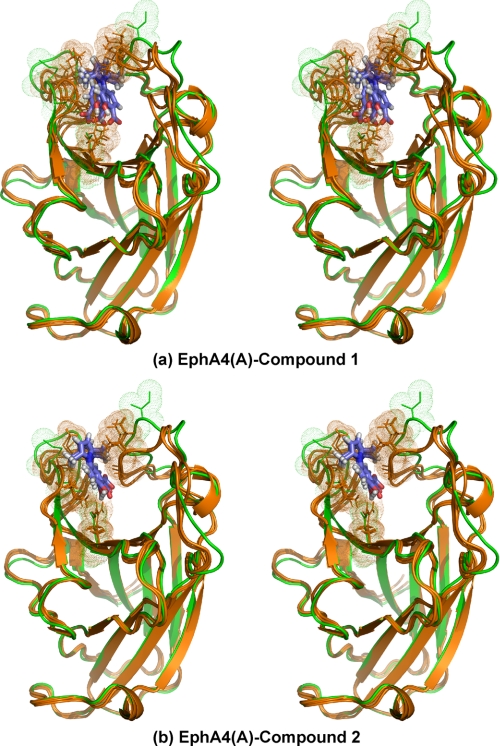
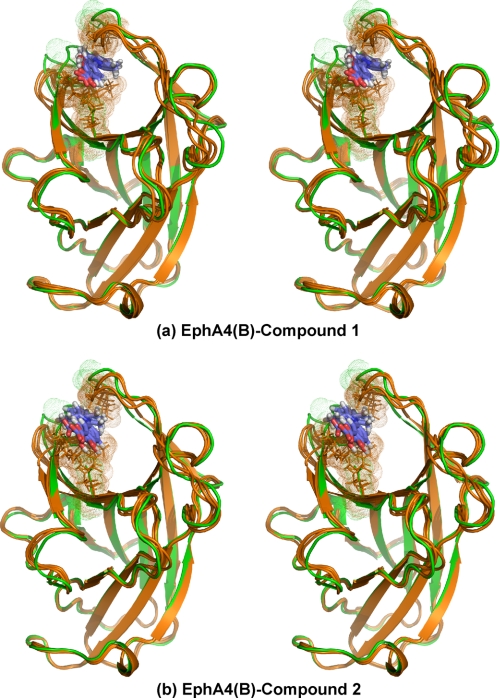
However, here, to better capture the binding properties of the compounds with EphA4, we separately used EphA4 molecules A and B to construct the models of the complexes by using the HADDOCK docking procedure. As a consequence, we obtained four models as follows: EphA4(A)-compound 1, EphA4(A)-compound 2, EphA4(B)-compound 1, and EphA4(B)-compound 2. From the structures generated from each docking running, we selected three with the lowest energies for further display and analysis (Figs. 4 and 5). As revealed from these models of the complexes, the two initial EphA4 A and B structures only need some local conformational rearrangements to accommodate the two small molecules. The average r.m.s.d. between the three selected structures and the initial structure are relatively small as follows: only ∼2.0 (all protein atoms) and 1.1 Å (protein backbone atoms) for EphA4(A)-compound 1; ∼2.1 (all protein atoms) and 1.2 Å (protein backbone atoms) for EphA4(A)-compound 2; ∼1.9 (all protein atoms) and 1.0 Å (protein backbone atoms) for EphA4(B)-compound 1; and ∼1.8 (all protein atoms) and 1.0 Å (protein backbone atoms) for EphA4(B)-compound 2. If not considering the D-E and J-K loops, the r.m.s.d. values reduce to ∼0.8 (all protein atoms) and 0.3 Å (protein backbone atoms) for EphA4(A)-compound 1; ∼0.8 (all protein atoms) and 0.3 Å (protein backbone atoms) for EphA4(A)-compound 2; ∼0.9 (all protein atoms) and 0.4 Å (protein backbone atoms) for EphA4(B)-compound 1; and ∼0.8 (all protein atoms) and 0.3 Å (protein backbone atoms) for EphA4(B)-compound 2.
FIGURE 4.
Models of structure A in complex with small molecule antagonists. a, stereo view of the superimposition of the unbound EphA4 structure A (green) with its three selected docking models in complex with compound 1 (brown). b, stereo view of the superimposition of the unbound EphA4 structure A (green) with its three selected docking models in complex with compound 2 (brown). Both sticks and dots are used to highlight residues Ile31-Met32 in the D-E loop, Gln43 in the E β-strand, and Ile131-Gly132 in the J-K loop.
FIGURE 5.
Models of structure B in complex with small molecule antagonists. a, stereo view of the superimposition of the unbound EphA4 structure B (green) with its three selected docking models in complex with compound 1 (brown). b, stereo view of the superimposition of the unbound EphA4 structure B (green) with its three selected docking models in complex with compound 2 (brown). Both sticks and dots are used to highlight residues Ile31-Met32 in the D-E loop, Gln43 in the E β-strand, and Ile131-Gly132 in the J-K loop.
Strikingly, as seen in Figs. 4 and 5, despite starting from two different EphA4 structures, in all four models the two small molecules occupy a similar cavity of the high affinity ligand binding channel of both EphA4 structures A and B. The two small molecules interact mainly with residues Ile31-Met32 in the D-E loop, Gln43 in the D-E β-strand, and Ile131-Gly132 in the J-K loops, all of which have significant CSDs in the NMR HSQC titration (Fig. 3, c and d). In contrast, despite being set as “active residues” in the docking calculations, residues Ile39 in the D-E loop and Asp123 in the J-K loop do not show direct contact with the two small molecules in any of the models. The HADDOCK docking procedure has been previously reported to correctly identify the residues most likely to form the binding pocket (48, 49, 52). Thus, the chemical shift perturbations observed for Asp123 and Ile39 probably represent a secondary effect of binding-induced rearrangements of the D-E and J-K loops.
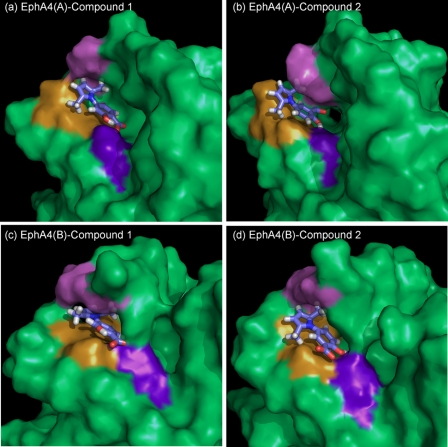
As shown in Fig. 6, a close examination of all the model structures reveals that the pyrrole and benzene rings of the two small molecules stack onto the hydrophobic surface formed by residues Ile31 and Met32 in the E-E loop. Moreover, the pyrrole ring is sandwiched by the hydrophobic side chains of Ile31-Met32 in the D-E loop and those of Ile131 in the J-K loop. On the other hand, one of the methyl groups on the pyrrole ring inserts into the hydrophobic patch between the Ile31 and Met32 side chains, and the other methyl group is in close contact with the Ile131 side chain. These interactions emphasize the importance of the two methyl groups on the pyrrole ring, which is completely consistent with the structure-activity relationship analysis of a series of small molecules with a pyrrolyl benzene scaffold (see Ref. 54).
FIGURE 6.
EphA4 binding cavity for the small molecule antagonists. Surface representation of the EphA4 binding cavity of the docking model with the lowest energy. a, EphA4 structure A with compound 1; b, EphA4 structure A with compound 2; c, EphA4 structure B with compound 1; d, EphA4 structure B with compound 2. The small molecule antagonists are represented by sticks and oxygen atoms are colored in red. EphA4 residues Ile31-Met32 in the D-E loop are in brown; residue Gln43 in the E β-strand is in blue/purple, and residues Ile131-Gly132 in the J-K loop are in violet.
In all 12 selected models, the carboxylic and hydroxyl groups on the benzene ring always orient toward the side chain of the EphA4 residue Gln43. Detailed analysis indicates that in all these models at least one hydrogen bond forms between the oxygen atoms of the carboxylic or hydroxyl groups and the side chain amide protons of Gln43. In some structures, even two hydrogen bonds can be identified between them. This observation may explain why removal of either the carboxylic or the hydroxyl group causes a dramatic loss in the activity of the modified compounds (54). Taken together, the docking results imply that the pyrrole and benzene rings, the two methyl groups on the pyrrole ring, and the carboxylic and hydroxyl groups on the benzene ring are all critical for the binding of small molecules with a 2,5-dimethylpyrrolyl benzene scaffold to the EphA4 ligand-binding domain.
DISCUSSION
The extensive involvement of the Eph receptor-ephrin interaction in various pathologies suggests that the main interface between the two proteins may serve as a promising new target for drug development. Previous studies reveal that the Eph receptor-ephrin interaction is mediated by two binding sites in the ligand-binding domain of the Eph receptor. One is a high affinity binding site, which includes a hydrophobic channel that is mainly constituted by the convex sheet of four β-strands and the D-E and J-K loops and that accommodates the protruding G-H loop of the ephrin. The other is a separate low affinity binding site (10-12, 14). In particular, the high affinity hydrophobic channel of the receptor appears to be highly amenable for targeting by small molecule antagonists. However, previously identified small molecules, including a natural product from green tea (30-33, 53), all seem to target the intracellular kinase domain of the Eph receptors. Only now two small molecules with a 2,5-dimethylpyrrolyl benzene scaffold have been successfully identified in a high throughput screen (see Ref. 54). The fact that the two compounds competitively inhibit ephrin binding to EphA4 strongly suggests that the two compounds occupy the ephrin binding channel, thus directly competing with ephrins in binding with the EphA4 receptor. Therefore, it was of significant interest to define the structural mechanism by which the two compounds interact with the EphA4 receptor.
To achieve this, in this study we have crystallized the EphA4 ligand-binding domain in the free state and determined its structure. This represents the first structure determined for the ligand-binding domain of an Eph receptor of the A subclass. In the crystal, each asymmetric unit contains two EphA4 molecules that show some large structural differences in the J-K loop because of their differential packing interactions with other EphA4 molecules in the neighboring asymmetric units. In solution, however, the EphA4 ligand-binding domain was found to be monomeric. The EphA4 ligand-binding domain adopts the same jellyroll β-sandwich architecture that was previously reported for the EphB2 and EphB4 ligand-binding domains. Interestingly, despite belonging to the Eph receptor A subclass, the core β-stranded regions of EphA4 bear a high similarity to those of the EphB2 and EphB4 receptors. Nevertheless, large variations do exist in the loop regions. For example, a short 310-helix is formed in the H-I loop of EphA4. This helix has not been observed in the EphB receptors, which have a 4-residue insert in this loop. There are also dramatic differences in the D-E and J-K loops. Because large variations in the positioning of the D-E and J-K loops have also been observed in the different EphB structures previously determined in the free state or in complex with an ephrin or peptide ligands, this may reflect the intrinsic flexibility of the D-E and J-K loops, which may be required to accommodate the binding of different ligands.
We have used isothermal titration calorimetry, circular dichroism, NMR, and computational docking to characterize the possible binding interactions of the EphA4 ligand-binding domain with the two small molecules that inhibit the binding of peptide and ephrin ligands. The isothermal titration calorimetry results show that both small molecules bind to the EphA4 ligand-binding domain with similar Kd values in the micromolar range. On the other hand, consistent with the modest binding affinity of the compounds, the circular dichroism results indicate that binding of the two small molecules does not induce significant structural changes in the EphA4 ligand-binding domain. To identify the EphA4 residues involved in the binding of the two small molecules, we have collected a large set of NMR spectra and succeeded in obtaining sequential assignments. This allowed us to identify the EphA4 residues that are significantly perturbed upon binding of the two small molecules by performing NMR HSQC titrations. Interestingly, only a few EphA4 residues showed significant perturbations upon binding, which include residues Ile31-Met32 in the D-E loop, Gln43 in the E β-strand, and Ile131-Gly132 in the J-K loop, in agreement with the small sizes of the two small molecules.
We further used the well established HADDOCK docking procedure to construct models of the EphA4 ligand-binding domain in complex with the two small molecules. The docking results indicate that both molecules occupy a cavity of the high affinity ephrin binding channel of EphA4 in a similar manner, by interacting mainly with EphA4 residues in the E strand and the D-E and J-K loops. The results also reveal that all three building blocks of the 2,5-dimethylpyrrolyl benzene scaffold, namely the dimethylpyrrole ring, the benzene ring, and the carboxylic/hydroxyl groups on the benzene ring, are crucial for binding to the EphA4 ligand-binding domain. The pyrrole and benzene rings appear to play a key role in establishing stacked aromatic-hydrophobic interactions with Ile31-Met32 on the D-E loop and Ile131 on the J-K loop. The two methyl groups on the pyrrole ring further anchor the small molecules in between the D-E and J-K loops by using one methyl group to interact with the hydrophobic side chains of Ile31-Met32 and the other to interact with the hydrophobic side chain of Ile131. Furthermore, the carboxylic and hydroxyl groups on the benzene ring are involved in hydrogen bonding to the side-chain amide protons of Gln43 in EphA4, thus providing additional contacts with EphA4 as well as dictating the orientation of the small molecules in the complexes. Consequently, the docking models provide the structural rationale for the results of an extensive study on the structure-activity relationship of small molecules with a pyrrolyl benzene scaffold as EphA4 ligand-binding antagonists (see Ref. 54).
Our results shed light on how such small molecules are capable of selectively targeting only EphA4 and the closely related EphA2 receptor (see accompanying article, Ref. 54). Sequence alignment reveals that some of the EphA4 residues that are perturbed by the binding are not conserved in other Eph receptors (supplemental Fig. 3). In particular, residues Ile31-Met32 are only presented in EphA4 and EphA2 but not other Eph receptors, which may be at least partly responsible for the high binding selectivity of the two molecules for the EphA4 and EphA2 receptors.
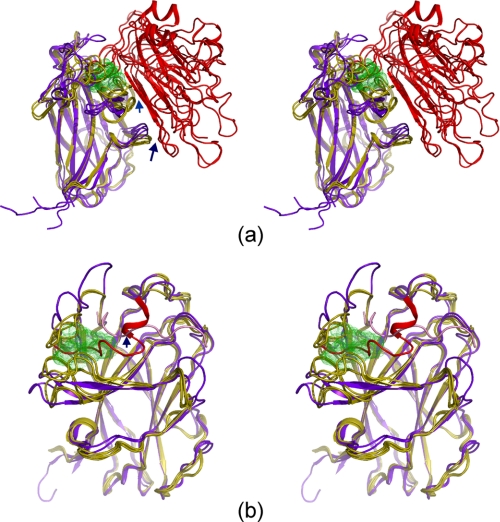
Our results may also explain why the two small molecules bind to EphA4 with a relatively weak affinity. First, EphA4 residues Ile31-Met32 and Ile131, which are critical for binding, are from the D-E and J-K loops. These loops are relatively flexible, as indicated by previous crystal structures and our NMR 15N heteronuclear NOE data (to be published). Second, as shown in Fig. 7a, the two small molecules only occupy a portion of the EphA4 ligand binding channel, which in EphB2 and EphB4 is occupied by the tip of the G-H loop of the ephrin ligands, corresponding to residues 122PNLWGL127 for ephrin-B2 and Pro127PFSLGF132 for ephrin-A5 (10-12, 14). In contrast, interactions occurring outside of the high affinity binding pocket of the Eph receptor are totally absent in the case of the small molecules. These interactions include those between the ephrin G β-strand and the Eph receptor D and E β-strands and A-C loop (10-12, 14). Even within the high affinity binding channel, a large portion of the key Eph receptor-ephrin interactions is absent in the EphA4-small molecule complexes because of the small size of the dimethylpyrrole derivatives. For example, NMR titrations did not detect strong interactions between the two small molecules and the EphA4 G and M β-strands. Furthermore, as shown in Fig. 7b, the interaction interface between EphA4 and the two compounds is also smaller than the interaction interfaces between the EphB2 and EphB4 receptors and their respective peptide ligands (13, 15). For example, the two small molecules do not interact with the EphA4 disulfide bridge linking Cys45 and Cys53, whereas this interaction was found to be conserved in all the EphB structures in complex with either ephrins or antagonistic peptides (15).
FIGURE 7.
Comparison of the EphA4 small molecule models with EphB receptors in complex with ephrins or peptides. a, stereo view of the superimposition of four selected EphA4 small molecule models with previously determined structures of EphB-ephrin complexes (PDB codes 1KGY, 1SHW, and 2HLE). EphA4 is represented by a yellow ribbon and the small molecules by green dots. The EphB receptors are in purple, and ephrin-B2/ephrin-A5 are in red. The blue arrows indicate the contact regions outside of the ligand binding channel that contribute to the high affinity Eph receptor-ephrin binding interface. b, stereo view of the superimposition of four selected EphA4 small molecule models with previously determined structures of EphB-peptide complexes (PDB codes 2QBX and 2BBA). EphA4 is in yellow; EphB receptors are in purple; one peptide is in red and another in pink. The blue arrow indicates a conserved binding motif identified in all the EphB structures in complex with either ephrins or antagonistic peptides (see Ref. 15).
In conclusion, our studies confirm the binding interaction between the EphA4 ligand-binding domain and two novel small molecule antagonists with a 2,5-dimethylpyrrolyl benzene scaffold. Furthermore, we utilized NMR titrations to map out the residues involved in the interaction and used this information to construct models of the EphA4 ligand-binding domain in complex with the two small molecules. These models provide a structural rationale for the results of an extensive structure-activity study on a large set of small molecules with a pyrrolyl benzene scaffold and for the high binding selectivity but relatively weak affinity of the compounds. Based on our model, we propose that modifications to enhance interactions with the EphA4 G and M β-strands may represent a promising direction to improve the binding activity and specificity of the EphA4 antagonists with a 2,5-dimethylpyrrolyl benzene scaffold.
Supplementary Material
Acknowledgments
We thank Dr. Alexandre M. J. J. Bonvin at Utrecht University for the HADDOCK software and suggestions on the docking setup.
The atomic coordinates and structure factors (code 3CKH) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
This work was supported by National Medical Research Council of Singapore Grant R-154-000-382-213 (to J. S.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1-3.
Footnotes
The abbreviations used are: Eph, erythropoietin-producing hepatocellular; HSQC, heteronuclear single quantum coherence; NOE, nuclear Overhauser effect; NOESY, nuclear Overhauser effect spectroscopy; r.m.s.d., root mean square deviation; CSD, chemical shift difference; FPLC, fast protein liquid chromatography; PDB, Protein Data Bank.
H. Qin, J, Shi, and S. Song, manuscript in preparation.
References
- 1.Adams, R. H. (2002) Semin. Cell Dev. Biol. 13 55-60 [DOI] [PubMed] [Google Scholar]
- 2.Pasquale, E. B. (2005) Nat. Rev. Mol. Cell. Biol. 6 462-475 [DOI] [PubMed] [Google Scholar]
- 3.Egea, J., and Klein, R. (2007) Trends Cell Biol. 17 230-238 [DOI] [PubMed] [Google Scholar]
- 4.Luo, L., and Flanagan, J. G. (2007) Neuron 56 284-300 [DOI] [PubMed] [Google Scholar]
- 5.Heroult, M., Schaffner, F., and Augustin, H. G. (2006) Exp. Cell Res. 312 642-650 [DOI] [PubMed] [Google Scholar]
- 6.Pasquale, E. B. (2008) Cell 133 38-52 [DOI] [PubMed] [Google Scholar]
- 7.Cowan, C. A., and Henkemeyer, M. (2001) Nature 413 174-179 [DOI] [PubMed] [Google Scholar]
- 8.Song, J. (2003) J. Biol. Chem. 278 24714-24720 [DOI] [PubMed] [Google Scholar]
- 9.Himanen, J. P., Henkemeyer, M., and Nikolov, D. B. (1998) Nature 396 486-491 [DOI] [PubMed] [Google Scholar]
- 10.Himanen, J. P., Saha, N., and Nikolov, D. B. (2007) Curr. Opin. Cell Biol. 19 534-542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Himanen, J. P., Rajashankar, K. R., Lackmann, M., Cowan, C. A., Henkemeyer, M., and Nikolov, D. B. (2001) Nature 414 933-938 [DOI] [PubMed] [Google Scholar]
- 12.Himanen, J. P., Chumley, M. J., Lackmann, M., Li, C., Barton, W. A., Jeffrey, P. D., Vearing, C., Geleick, D., Feldheim, D. A., Boyd, A. W., Henkemeyer, M., and Nikolov, D. B. (2004) Nat. Neurosci. 7 501-509 [DOI] [PubMed] [Google Scholar]
- 13.Chrencik, J. E., Brooun, A., Recht, M. I., Kraus, M. L., Koolpe, M., Kolatkar, A. R., Bruce, R. H., Martiny-Baron, G., Widmer, H., Pasquale, E. B., and Kuhn, P. (2006) Structure (Lond.) 14 321-330 [DOI] [PubMed] [Google Scholar]
- 14.Chrencik, J. E., Brooun, A., Kraus, M. L., Recht, M. I., Kolatkar, A. R., Han, G. W., Seifert, J. M., Widmer, H., Auer, M., and Kuhn, P. (2006) J. Biol. Chem. 281 28185-28192 [DOI] [PubMed] [Google Scholar]
- 15.Chrencik, J. E., Brooun, A., Recht, M. I., Nicola, G., Davis, L. K., Abagyan, R., Widmer, H., Pasquale, E. B., and Kuhn, P. (2007) J. Biol. Chem. 282 36505-36513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pasquale, E. B. (2004) Nat. Neurosci. 7 417-418 [DOI] [PubMed] [Google Scholar]
- 17.Klein, R. (2004) Curr. Opin. Cell Biol. 16 580-589 [DOI] [PubMed] [Google Scholar]
- 18.Yamaguchi, Y., and Pasquale, E. B. (2004) Curr. Opin. Neurobiol. 14 288-296 [DOI] [PubMed] [Google Scholar]
- 19.Bourgin, C., Murai, K. K., Richter, M., and Pasquale, E. B. (2007) J. Cell Biol. 178 1295-1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Richter, M., Murai, K. K., Bourgin, C., Pak, D. T., and Pasquale, E. B. (2007) J. Neurosci. 27 14205-14215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gallarda, B. W., Bonanomi, D., Müller, D., Brown, A., Alaynick, W. A., Andrews, S. E., Lemke, G., Pfaff, S. L., and Marquardt, T. (2008) Science 320 233-236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prevost, N., Woulfe, D. S., Jiang, H., Stalker, T. J., Marchese, P., Ruggeri, Z. M., and Brass, L. F. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 9820-9825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goldshmit, Y., Galea, M. P., Wise, G., Bartlett, P. F., and Turnley, A. M. (2004) J. Neurosci. 24 10064-10073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benson, M. D., Romero, M. I., Lush, M. E., Lu, Q. R., Henkemeyer, M., and Parada, L. F. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 10694-10699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Du, J., Fu, C., and Sretavan, D. W. (2007) Curr. Pharm. Des. 13 2507-2518 [DOI] [PubMed] [Google Scholar]
- 26.Easty, D. J., Mitchell, P. J., Patel, K., Florenes, V. A., Spritz, R. A., and Bennett, D. C. (1997) Int. J. Cancer 71 1061-1065 [DOI] [PubMed] [Google Scholar]
- 27.Ashida, S., Nakagawa, H., Katagiri, T., Furihata, M., Iiizumi, M., Anazawa, Y., Tsunoda, T., Takata, R., Kasahara, K., Miki, T., Fujioka, T., Shuin, T., and Nakamura, Y. (2004) Cancer Res. 64 5963-5972 [DOI] [PubMed] [Google Scholar]
- 28.Iiizumi, M., Hosokawa, M., Takehara, A., Chung, S., Nakamura, T., Katagiri, T., Eguchi, H., Ohigashi, H., Ishikawa, O., Nakamura, Y., and Nakagawa, H. (2006) Cancer Sci. 97 1211-1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yao, V. J., Ozawa, M. G., Trepel, M., Arap, W., McDonald, D. M., and Pasqualini, R. (2005) Am. J. Pathol. 166 625-636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Caligiuri, M., Molz, L., Liu, Q., Kaplan, F., Xu, J. P., Majeti, J. Z., Ramos-Kelsey, R., Murthi, K., Lievens, S., Tavernier, J., and Kley, N. (2006) Chem. Biol. 13 711-722 [DOI] [PubMed] [Google Scholar]
- 31.Karaman, M. W., Herrgard, S., Treiber, D. K., Gallant, P., Atteridge, C. E., Campbell, B. T., Chan, K. W., Ciceri, P., Davis, M. I., Edeen, P. T., Faraoni, R., Floyd, M., Hunt, J. P., Lockhart, D. J., Milanov, Z. V., Morrison, M. J., Pallares, G., Patel, H. K., Pritchard, S., Wodicka, L. M., and Zarrinkar, P. P. (2008) Nat. Biotechnol. 26 127-132 [DOI] [PubMed] [Google Scholar]
- 32.Miyazaki, Y., Maeda, Y., Sato, H., Nakano, M., and Mellor, G. W. (2008) Bioorg. Med. Chem. Lett. 18 1967-1971 [DOI] [PubMed] [Google Scholar]
- 33.Kolb, P., Kipouros, C. B., Huang, D., and Caflisch, A. (2008) Proteins 73 11-18 [DOI] [PubMed] [Google Scholar]
- 34.Ran, X., Qin, H., Liu, J., Fan, J. S., Shi, J., and Song, J. (2008) Proteins 72 1019-1029 [DOI] [PubMed] [Google Scholar]
- 35.Ran, X., and Song, J. (2005) J. Biol. Chem. 280 19205-19212 [DOI] [PubMed] [Google Scholar]
- 36.Pace, C. N., Vajdos, F., Fee, L., Grimsley, G., and Gray, T. (1995) Protein Sci. 4 2411-2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi, J., Sivaraman, J., and Song, J. (2008)) J. Virol. 82 4620-4629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Otwinowski, Z., and Minor, W. (1997) Methods Enzymol. 276, 307-326 [DOI] [PubMed] [Google Scholar]
- 39.McCoy, A. J., Grosse-Kunstleve, R. W., Storoni, L. C., and Read, R. J. (2005) Acta Crystallogr. Sect. D Biol. Crystallogr. 61 458-464 [DOI] [PubMed] [Google Scholar]
- 40.Emsley, P., and Cowtan, K. (2004) Acta Crystallogr. Sect. D Biol. Crystallogr. 60 2126-2132 [DOI] [PubMed] [Google Scholar]
- 41.Adams, P. D., Grosse-Kunstleve, R. W., Hung, L.-W., Ioerger, T. R., McCoy, A. J., Moriarty, N. W., Read, R. J., Sacchettini, J. C., Sauter, N. K., and Terwilliger, T. C. (2002) Acta Crystallogr. Sect. D Biol. Crystallogr. 58 1948-1954 [DOI] [PubMed] [Google Scholar]
- 42.Laskowski, R. A., MacArthur, M. W., Moss, D. S., and Thornton, J. M. (1993) J. Appl. Crystallogr. 26 283-291 [Google Scholar]
- 43.Shi, J., Lua, S., Du, N., Liu, X., and Song, J. (2008) Biomaterials 29 2820-2828 [DOI] [PubMed] [Google Scholar]
- 44.Liu, J., Li, M., Ran, X., Fan, J. S., and Song, J. (2006) Biochemistry 45 7171-7184 [DOI] [PubMed] [Google Scholar]
- 45.Sattler, M., Schleucher, J., and Griesinger, C. (1999) Prog. NMR Spectrosc. 34 93-158 [Google Scholar]
- 46.Delaglio, F., Grzesiek, S., Vuister, G. W., Zhu, G., Pfeifer, J., and Bax, A. (1995) J. Biomol. NMR 6 277-293 [DOI] [PubMed] [Google Scholar]
- 47.Johnson, B. A., and Blevins, R. A. (1994) J. Biomol. NMR 4 603-614 [DOI] [PubMed] [Google Scholar]
- 48.Dominguez, C., Boelens, R., and Bonvin, A. M. (2003) J. Am. Chem. Soc. 125 1731-1737 [DOI] [PubMed] [Google Scholar]
- 49.de Vries, S. J., van Dijk, A. D., Krzeminski, M., van Dijk, M., Thureau, A., Hsu, V., Wassenaar, T., and Bonvin, A. M. (2007) Proteins 69 726-733 [DOI] [PubMed] [Google Scholar]
- 50.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T., and Warren, G. L. (1998) Acta Crystallogr. Sect. D Biol. Crystallogr. 54 905-921 [DOI] [PubMed] [Google Scholar]
- 51.Schuettelkopf, A. W., and van Aalten, D. M. F. (2004) Acta Crystallogr. Sect. D Biol. Crystallogr. 60 1355-1363 [DOI] [PubMed] [Google Scholar]
- 52.Zhang, N., Liu, L., Liu, F., Wagner, C. R., Hanna, P. E., and Walters, K. J. (2006) J. Mol. Biol. 363 188-200 [DOI] [PubMed] [Google Scholar]
- 53.Tang, F. Y., Chiang, E. P., and Shih, C. J. (2007) J. Nutr. Biochem. 18 391-399 [DOI] [PubMed] [Google Scholar]
- 54.Noberini, R., Koolpe, M., Peddibhotla, S., Dahl, R., Su, Y., Cosford, N. D. P., Rothe, G. P., and Pasquale, E. B. (2008) J. Biol. Chem. 283 29461-29472 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.