Abstract
GPIHBP1, a glycosylphosphatidylinositol-anchored endothelial cell protein of the lymphocyte antigen 6 (Ly6) family, plays a key role in the lipolysis of triglyceride-rich lipoproteins (e.g. chylomicrons). GPIHBP1 is expressed along the luminal surface of endothelial cells of heart, skeletal muscle, and adipose tissue, and GPIHBP1-expressing cells bind lipoprotein lipase (LPL) and chylomicrons avidly. GPIHBP1 contains an amino-terminal acidic domain (amino acids 24-48) that is enriched in aspartate and glutamate residues, and we previously speculated that this domain might be important in binding ligands. To explore the functional importance of the acidic domain, we tested the ability of polyaspartate or polyglutamate peptides to block the binding of ligands to pgsA-745 Chinese hamster ovary cells that overexpress GPIHBP1. Both polyaspartate and polyglutamate blocked LPL and chylomicron binding to GPIHBP1. Also, a rabbit antiserum against the acidic domain of GPIHBP1 blocked LPL and chylomicron binding to GPIHBP1-expressing cells. Replacing the acidic amino acids within GPIHBP1 residues 38-48 with alanine eliminated the ability of GPIHBP1 to bind LPL and chylomicrons. Finally, mutation of the positively charged heparin-binding domains within LPL and apolipoprotein AV abolished the ability of these proteins to bind to GPIHBP1. These studies indicate that the acidic domain of GPIHBP1 is important and that electrostatic interactions play a key role in ligand binding.
Glycosylphosphatidylinositol-anchored high density lipoprotein-binding protein 1 (GPIHBP1),2 a GPI-anchored endothelial cell protein, plays an important role in the lipolytic processing of chylomicrons by lipoprotein lipase (LPL) (1). GPIHBP1 is found exclusively in the lumen of the microvascular endothelial cells of heart, adipose tissue, and muscle, precisely where the lipolytic processing of chylomicrons occurs (1). Cultured cells that express GPIHBP1 bind LPL and chylomicrons avidly (1), suggesting that GPIHBP1 serves as an endothelial cell platform for lipolysis. A deficiency of GPIHBP1 in mice leads to severe chylomicronemia, even on a low-fat diet, with plasma triglyceride levels >2000 mg/dl (1).
GPIHBP1 is a member of a large family of “lymphocyte antigen 6” (Ly6) proteins (2). All members of this family contain a cysteine-rich “Ly6 motif” in which the spacing of 10 cysteine residues is highly conserved (3). Like many other members of the Ly6 family, GPIHBP1 is tethered to the plasma membrane by a GPI anchor (1, 4). GPIHBP1 can be released from the surface of cells by cleaving the GPI anchor with a phosphatidylinositol-specific phospholipase C (PIPLC) (1, 4, 5).
GPIHBP1 is distinguished from other GPI-anchored Ly6 proteins by a strong acidic domain, located immediately after its signal peptide and about 12 amino acids before the Ly6 motif (1, 4, 6). This acidic domain (amino acids 24-48 in the mouse protein) contains a remarkable concentration of aspartate and glutamate residues (17 of 25 consecutive residues in mouse GPIHBP1 are aspartates or glutamates). The corresponding region of human GPIHBP1 contains 21 aspartates or glutamates.
LPL contains two positively charged heparin-binding domains (7-10), and one of these (amino acids 430-441) has been implicated in the binding of LPL to negatively charged heparan sulfate proteoglycans (HSPGs) on the surface of cells (11). The electrostatic interactions between LPL and HSPGs can be disrupted by heparin (strongly anionic) (12-14) or protamine (strongly cationic) (15, 16). Similarly, apolipoprotein (apo) AV, one of the apolipoproteins in triglyceride-rich lipoproteins, contains a positively charged heparin-binding domain that is required for binding to HSPGs (17).
We generated an expression vector for mouse GPIHBP1, and expressed mouse GPIHBP1 in CHO pgsA-745 cells, a mutant line of CHO cells lacking the ability to synthesize HSPGs (18). The GPIHBP1-expressing cells bind avian LPL in a saturable fashion and with high affinity (Kd = 3.6 × 10-8 m) (1). GPIHBP1 also binds a tagged version of human LPL, and this GPIHBP1-bound LPL can be released by heparin. GPIHBP1 binds “chylomicrons” (d < 1.006 g/ml lipoproteins from Gpihbp1-deficient mice) and apoAV-phospholipid disks avidly (1).
We have speculated that the binding of LPL, chylomicrons, and apoAV-phospholipid disks to GPIHBP1 might depend on the acidic domain of GPIHBP1 (1, 6). In the current study, we tested that possibility.
EXPERIMENTAL PROCEDURES
GPIHBP1 Constructs—Expression vectors for mouse GPIHBP1 and an S-protein-tagged version of mouse and human GPIHBP1 have been described previously (1). An S-protein-tagged mouse GPIHBP1 mutant, D,E(24-33)A, in which all of the aspartates and glutamates between residues 24 and 33 were replaced by alanine (EDGDADPEPE to AAGAAAPAPA) was generated by site-directed mutagenesis with the QuikChange kit (Stratagene). Another mouse mutant, D,E(38-48)A, in which all of the aspartates and glutamates between residues 38 and 48 were replaced with alanine (DDDDDEEEEEE to AAAAAAAAAAA) was also generated by site-directed mutagenesis. A mouse GPIHBP1 deletion mutant lacking the acidic domain, Δ(24-58), was generated by flanking the segment of the Gpihbp1 cDNA encoding amino acids 24-58 with ApaI sites. The acidic domain was excised by digestion with ApaI and replaced with a DNA fragment encoding an S-protein tag.
We obtained a human CD59 cDNA from Open Biosystems and cloned it into pTriEx-4 (Novagen). An S-protein tag was inserted between the signal peptide and the Ly6 domain of CD59 by PCR with the In-Fusion 2.0 dry-Down cloning kit (Clontech). We used the same kit to replace the Ly6 domain of GPIHBP1 with the Ly6 domain of CD59 (a GPIHBP1/CD59 chimera). The integrity of all constructs was verified by DNA sequencing.
Cell Transfections—CHO pgsA-745 cells were transfected with GPIHBP1 constructs with Lipofectamine 2000 (Invitrogen). In some cases, stable cell lines were generated by subjecting transfected cells to selection with G418 (0.5 mg/ml). GPIHBP1 expression in the transfected cells was documented by Western blot analysis with a rabbit antibody against GPIHBP1 (1:2500, Novus Biologicals).
To determine whether GPIHBP1 proteins reached the cell surface, we tested whether GPIHBP1 could be released into the medium with PIPLC (1). PIPLC was added to the serum-free culture medium of transiently transfected cells (1 units/ml for 1 h at 37 °C) or stably transfected cells (6 units/ml for 30 min at 37 °C). At the end of the incubation period, the medium was harvested, and cell extracts were collected in RIPA buffer (1% Triton X-100, 0.5% deoxycholate, 0.02% SDS in phosphate buffer saline) containing complete mini EDTA-free protease inhibitors (Roche).
Binding of Human LPL to Gpihbp1-transfected CHO Cells—An expression vector for a V5-tagged version of human LPL in pcDNA6 (19) was obtained from Dr. Mark Doolittle (University of California, Los Angeles). A mutant version of this LPL construct, in which all of the lysine and arginine residues from Lys-430 to Lys-441 were replaced with alanines (KIRVKAGETQKK to AIAVAAGETQAA), was generated by site-directed mutagenesis with the QuikChange kit. A mouse LPL cDNA was purchased from Open Biosystem and cloned into pcDNA6 in-frame with the carboxyl-terminal V5 tag.
In some experiments, CHO pgsA-745 cells were cotransfected with the V5-tagged human LPL and the mouse GPIHBP1 constructs or empty vector with Lipofectamine 2000 (Invitrogen). Twenty-four h after the transfection, the cell culture medium was replaced with fresh Ham's F-12 medium, or F-12 medium containing heparin (1 units/ml for 15 min, American Pharmaceutical Partners), polyaspartate peptides (poly(D), 200 μg/ml for 1 h, Sigma), or polyglutamate peptides (poly(E), 200 μg/ml for 1 h, Sigma). At the end of the incubation period, the medium was harvested and cell extracts were collected in RIPA buffer containing complete mini EDTA-free protease inhibitors (Roche).
In other experiments, CHO pgsA-745 cells were transfected with the human GPIHBP1, the human GPIHBP1/CD59 chimera, human CD59, or empty vector with Lipofectamine 2000 (Invitrogen). Twenty-four h after the transfection, the cells were incubated for 2 h at 4°C with conditioned medium (400 μl/well) from nontransfected HeLa cells or HeLa cells that had been transfected with a V5-tagged version of human LPL. In some wells, 100 units/ml of heparin was added to the incubation medium. At the end of the incubation period, cells were washed 6 times in ice-cold 1× PBS containing 1 mm MgCl2 and 1 mm CaCl2, and cell extracts were collected in RIPA buffer containing complete mini EDTA-free protease inhibitors (Roche).
The amount of LPL and GPIHBP1 in cell extracts and the amount of LPL in the cell culture medium were assessed by Western blotting. Proteins were size-fractionated on 4-12% bis-Tris gradient SDS-polyacrylamide gels (cell culture media) or 12% bis-Tris SDS-polyacrylamide gels (cell extracts) and then transferred to nitrocellulose membranes for Western blotting. The antibody dilutions for Western blots were 1:250 for a rabbit polyclonal antibody against the S-protein tag (Abcam); 1:200 for a rabbit polyclonal antibody against the acidic domain of mouse GPIHBP1; 1:100 for a mouse monoclonal antibody against the V5 tag (Invitrogen); 1:500 for a mouse monoclonal antibody against β-actin (Abcam); 1:2000 for an IRdye 680-conjugated goat anti-rabbit IgG (LiCor); and 1:2000 for an IRdye 800-conjugated goat anti-mouse IgG (Li-Cor). Antibody binding was detected with an Odyssey infrared scanner (Li-Cor).
The binding of avian LPL to mouse GPIHBP1 was quantified with CHO pgsA-745 that had been stably transfected with the different mouse GPIHBP1 constructs or empty vector. Stably transfected CHO pgsA-745 cells were grown to confluence in Ham's F-12 medium containing 5% fetal bovine serum. The cells were placed on ice, washed twice with ice-cold PBS, and incubated for 2 h at 4°C with 1.0 ml of Ham's F-12 medium supplemented with 0.5% BSA and varying amounts of purified chicken LPL (20). After a 2-h incubation, the cells were washed once with PBS containing 0.5% BSA and lysed with 400 μl of CHAPS lysis buffer (4 mm CHAPS, 50 mm NH4Cl, and 3 units of heparin/ml). The amount of chicken LPL in the medium and cell lysates was measured by ELISA (21). In parallel, the amount of PIPLC-releasable LPL was measured by ELISA.
Immunofluorescence Microscopy—To detect GPIHBP1 in cultured cells, cells were plated on coverslips at ∼25,000 cells per well in 24-well plates, fixed in 3% paraformaldehyde, blocked with blocking buffer (1× PBS containing 10% bovine serum, 0.2% BSA, 1 mm MgCl2, 1 mm CaCl2), and incubated with either a rabbit antiserum against GPIHBP1 (Novus Biologicals) diluted in blocking buffer (1:5000), or a FITC-conjugated goat antiserum against the S-protein tag (Abcam; 1:400). The cells were not permeabilized with detergents. Bound rabbit immunoglobulin (IgG) was detected with an Alexa 488-conjugated goat anti-rabbit IgG (1:800; Invitrogen). After washing, cells were stained with DAPI to visualize DNA. Images were obtained with an Axiovert 200 MOT microscope (Zeiss) with a ×63/1.25 oil-immersion objective and processed with Axio-Vision 4.2 software (Zeiss).
To evaluate the binding of V5-tagged mouse LPL to mouse GPIHBP1, the cells were incubated for 2 h at 4°C with medium from nontransfected HeLa cells or HeLa cells that had been transfected with a V5-tagged mouse LPL. In some wells, heparin (100 units/ml) was added to the incubation medium. At the end of the incubation period, cells were washed 6 times in ice-cold 1× PBS containing 1 mm MgCl2 and 1 mm CaCl2, and then fixed in ice-cold 3% paraformaldhyde. The binding of LPL to GPIHBP1 was evaluated by immunofluorescence microscopy (as described earlier) with the mouse monoclonal antibody against the V5 tag (Invitrogen; 1:100), and an Alexa 568-conjugated goat anti-mouse IgG (1:800; Invitrogen).
Binding of DiI-labeled Chylomicrons to Transfected Cells—Mouse d < 1.006 g/ml lipoproteins (“chylomicrons”) were labeled with DiI (1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate) and purified from Gpihbp1-deficient mouse plasma as described previously (1).
To evaluate the binding of the DiI-labeled chylomicrons to mouse GPIHBP1, CHO pgsA-745 cells were transiently transfected with the different mouse GPIHBP1 constructs (or vector alone) and incubated with 1 μg/ml of DiI-labeled chylomicrons in PBS containing 1.0 mm CaCl2, 1.0 mm MgCl2, and 0.5% BSA for 2 h at 4 °C. The cells were then washed five times in the same buffer, fixed in 3% paraformaldehyde, and binding of chylomicrons to the cells was assessed by fluorescence microscopy. GPIHBP1 was detected with a rabbit antiserum against GPIHBP1 (Novus Biologicals). In some experiments, the binding of DiI-labeled chylomicrons to GPIHBP1-transfected cells was assessed in the presence or absence of 200 μg/ml poly(D) (Sigma).
Binding of Apolipoprotein AV-Phospholipid Disks to Gpihbp1-transfected CHO Cells—Human apolipoprotein AV (apoAV) and a mutant apoAV (in which four of the positively charged residues in the principal heparin-binding domain were changed to neutral or negatively charged residues; RKLTLKAK to EQLTLQAE) were expressed in Escherichia coli, purified, and used to generate wild-type or mutant apoAV·dimyristoylphosphatidylcholine (DMPC) disks (22). Previously, it was demonstrated that mutations within the RKLTLKAK sequence interfere with the binding of apoAV to heparin (23). To evaluate the binding of the apoAV·DMPC disks to mouse GPIHBP1, CHO pgsA-745 cells were transiently transfected with a Gpihbp1 cDNA and incubated with apoAV·DMPC disks (5 μg/ml) in PBS containing 1.0 mm CaCl2, 1.0 mm MgCl2, and 0.5% BSA for 2 h at 4°C. The binding of apoAV·DMPC disks was detected with a mouse antibody against apoAV (1:1000; Invitrogen) and an Alexa 568-conjugated goat anti-mouse IgG (1:800; Invitrogen). GPIHBP1 was detected with a rabbit antiserum against GPIHBP1 (Novus Biologicals) diluted in blocking buffer (1:5000) and a FITC-conjugated goat anti-rabbit IgG (1:200; Jackson Immuno-Research). In some experiments, the binding of apoAV·DMPC disks to Gpihbp1-transfected cells was assessed in the presence or absence of 200 μg/ml poly(D) (Sigma).
A Rabbit Antiserum against the Acidic Domain of Mouse GPIHBP1—A peptide corresponding to amino acids 24-48 of mouse GPIHBP1 (Cys-EDGDADPEPENYNYDDDDDEEEEEE) was synthesized (Biomer Technology), and an anti-peptide antibody was generated in rabbits. The ability of the antibody to bind to GPIHBP1 was assessed by Western blot and immunocytochemistry. To generate immunopurified antibodies against the acidic domain, 2.0 ml of rabbit serum was diluted 1:3 with equilibration buffer (0.5 m NaCl, 0.2 m Tris-HCl buffer, pH 8.0) and applied to a Sepharose 4B-peptide affinity column. The column was washed with 120 ml of equilibration buffer and eluted with a glycine buffer (0.5 m NaCl, 0.2 m glycine-HCl, pH 2.8). The eluate (30 ml) was collected into tubes containing sufficient 2 m Tris-HCl, pH 9.0, to adjust the pH of each fraction to 7.4. Purified immunoglobulins were stored at -20 °C in 50% glycerol.
The ability of a rabbit antiserum against the acidic domain of GPIHBP1 to block the binding of LPL to GPIHBP1 was examined with CHO pgsA-745 cells that had been stably transfected with a mouse GPIHBP1 expression vector. The ability of the immunopurified antibodies (0.1 μg/ml) to block the binding of DiI-labeled chylomicrons was examined with transiently transfected CHO pgsA-745 cells. The transfected cells were incubated with either preimmune serum or antibodies against the acidic domain peptide for 1 h at 4°C before adding DiI-labeled chylomicrons.
RESULTS
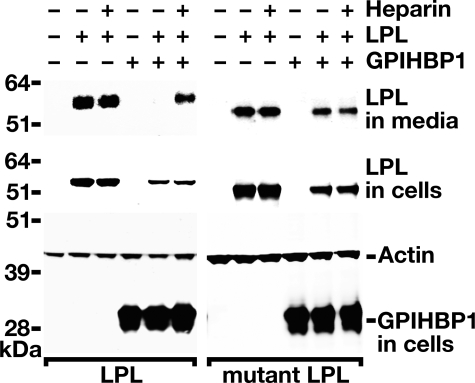
Binding of Wild-type Human LPL and a Mutant LPL Lacking the Main Heparin-binding Domain to GPIHBP1—Previous studies (1) have shown that wild-type LPL binds to cells expressing GPIHBP1, and the bound LPL can be readily released with heparin or PIPLC. To determine whether the binding is dependent on the principal heparin-binding domain of LPL, we examined the binding of wild-type human LPL and a mutant LPL in which the positively charged amino acids of the main heparin-binding domain (Lys-430, Arg-432, Lys-434, Lys-440, and Lys-441) were changed to alanine. The wild-type and mutant LPL constructs were transfected into CHO pgsA-745 cells, along with an expression vector for mouse GPIHBP1 (or empty vector), and Western blots were performed on the cell culture medium and cell extracts. Consistent with our earlier studies (1), substantial amounts of wild-type LPL appeared in the cell culture medium in cells not expressing GPIHBP1 (Fig. 1). In the presence of GPIHBP1, little LPL was detected in the medium of cells (because it is bound by GPIHBP1). The GPIHBP1-bound LPL was readily released by heparin; hence, increased amounts of LPL appeared in the cell culture medium after adding heparin (Fig. 1). When the binding of the mutant LPL was tested, large amounts of the mutant LPL appeared in the culture medium, even in the absence of heparin (Fig. 1). No additional LPL was released into the medium after adding heparin, indicating that the mutant LPL has little or no ability to bind to GPIHBP1 on the surface of cells (Fig. 1). These data indicate that the heparin-binding domain of human LPL is important for binding GPIHBP1.
FIGURE 1.
The principal heparin-binding domain in LPL is required for the binding of LPL to GPIHBP1. Western blot analysis of CHO pgsA-745 cells co-transfected with V5-tagged human LPL expression vectors, along with an S-protein-tagged mouse GPIHBP1 expression vector (or empty vector). Two different human LPLs were examined, a wild-type LPL and a mutant LPL with a modified main heparin-binding domain (Lys-430, Arg-432, Lys-434, Lys-440, and Lys-441 were changed to alanine). Wild-type human LPL bound GPIHBP1 on the cell surface, thus little LPL was found in the medium. Heparin released GPIHBP1-bound LPL into the cell culture medium. The mutant LPL was found in the medium even in the absence of heparin, indicating little or no binding to GPIHBP1, and no additional LPL appeared in the medium after adding heparin.
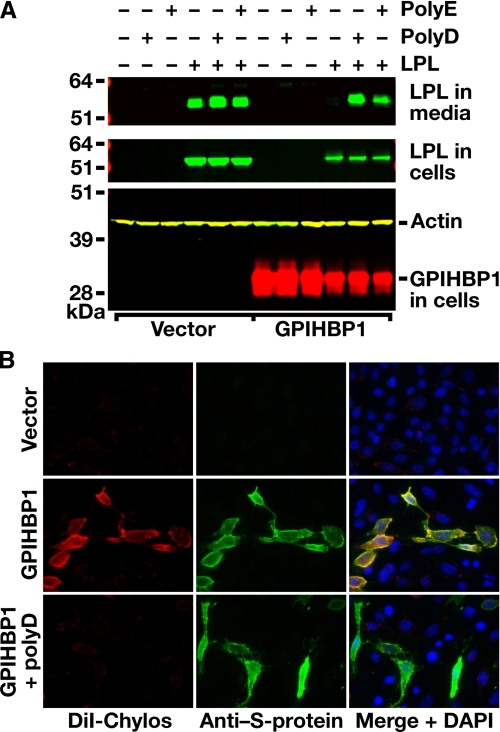
Assessing the Effects of Polyaspartate and Polyglutamate on the Binding of LPL to GPIHBP1—We suspected that anionic peptides such as polyaspartate (poly(D)) or polyglutamate (poly(E)) might interfere with the binding of LPL to GPIHBP1. Indeed, as judged by Western blot analysis, both peptides disrupted LPL-GPIHBP1 interactions, releasing LPL into the cell culture medium (Fig. 2A). Poly(D) also blocked the ability of GPIHBP1-expressing cells to bind DiI-labeled chylomicrons, as judged by immunohistochemistry (Fig. 2B). These experiments suggested that electrostatic interactions are important for ligand binding.
FIGURE 2.
Polyaspartate and polyglutamate peptides block the binding of LPL and chylomicrons to GPIHBP1. A, Western blot studies of LPL binding to GPIHBP1-transfected CHO pgsA-745 cells in the presence and absence of polyaspartate (poly(D)) or polyglutamate (poly(E)). CHO pgsA-745 cells were transfected with a V5-tagged human LPL expression vector and an S-protein-tagged mouse GPIHBP1 expression vector (or empty vector). 24 h after the transfection, cells were incubated in the absence or presence of 200 μg/ml poly(D) or poly(E) for 1 h at 37°C. As before, GPIHBP1 bound wild-type human LPL, allowing little LPL to escape into the medium of GPIHBP1-transfected cells. However, GPIHBP1-bound LPL was released into the medium with poly(D) or poly(E). B, fluorescence microscopy of GPIHBP1-transfected CHO pgsA-745 cells, demonstrating that DiI-labeled chylomicrons cannot bind to GPIHBP1 in the presence of poly(D). CHO pgsA-745 cells were transiently transfected with an S-protein-tagged mouse GPIHBP1 expression vector (or empty vector), and GPIHBP1 expression was assessed by immunocytochemistry with an antibody against the S-protein tag (green); the binding of DiI-labeled chylomicrons to cells was assessed by DiI fluorescence (red). The binding of chylomicrons to cells was assessed in the absence or presence of 200 μg/ml poly(D). DAPI staining (blue) was used to visualize cell nuclei.
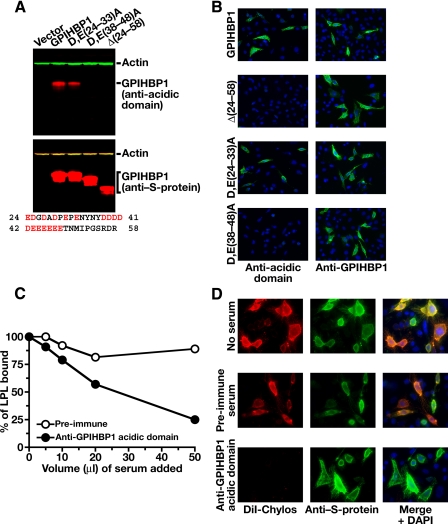
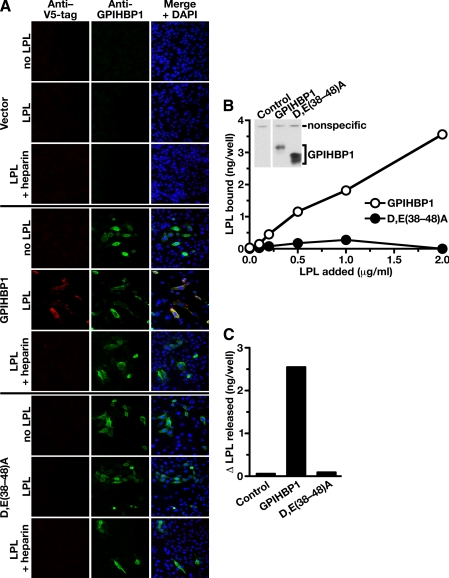
Assessing the Functional Importance of the Acidic Domain of GPIHBP1—To assess the importance of the acidic domain of GPIHBP1 in the binding of LPL and chylomicrons, we generated an antiserum against a synthetic peptide corresponding to the acidic domain of mouse GPIHBP1 (amino acids 24-48). This antiserum bound to wild-type GPIHBP1, but not to a GPIHBP1 mutant, Δ(24-58), lacking the acidic domain, as judged by Western blotting (Fig. 3A). The antiserum bound a GPIHBP1 mutant in which the aspartate and glutamate residues between amino acids 24 and 33 were changed to alanine (D,E(24-33)A) but not to a mutant in which the aspartate and glutamate residues between amino acids 38 and 48 were changed to alanine (D,E(38-48)A); thus, most of the antibodies within the antiserum bound to GPIHBP1 amino acids 38-48 (Fig. 3A). The antiserum bound avidly to wild-type GPIHBP1 on the surface of cells, as judged by immunocytochemistry, but no binding was detected with cells expressing D,E(38-48)A or Δ(24-58) GPIHBP1 (Fig. 3B). The anti-acidic domain antibodies, but not preimmune serum, markedly reduced the binding of avian LPL (Fig. 3C) and chylomicrons (Fig. 3D) to GPIHBP1-expressing cells, suggesting that the acidic domain of GPIHBP1 is important for ligand binding.
FIGURE 3.
A rabbit antiserum against the acidic domain of GPIHBP1 blocks the binding of LPL and chylomicrons to GPIHBP1. A, Western blot studies assessing the ability of a rabbit antiserum against a mouse GPIHBP1 peptide (amino acids 24-48) to bind to wild-type mouse GPIHBP1, a mutant GPIHBP1 lacking amino acids 24-58 (Δ(24-58)), a mutant GPIHBP1 (D,E(24-33)A) in which the aspartate and glutamate residues between amino acids 24 and 33 were changed to alanine, and a mutant GPIHBP1 (D,E(38-48)A) in which the aspartate and glutamate residues between amino acids 38 and 48 were changed to alanine. All GPIHBP1 proteins contained an amino-terminal S-protein tag. Western blots were performed with an antibody against the acidic domain of GPIHBP1 (top panel) or an antibody against the S-protein tag (lower panel). An antibody against actin was used as a loading control. The amino acid sequence of GPIHBP1 residues 24-58 is shown below the Western blots. B, immunocytochemistry demonstrating that the rabbit anti-GPIHBP1 acidic domain peptide antiserum binds to wild-type GPIHBP1 expressed on the cell surface. Duplicate cultures of CHO pgsA-745 cells expressing wild-type GPIHBP1 or mutant GPIHBP1 (D,E(24-33)A; D,E(38-48)A; Δ(24-58)) were incubated with either the anti-acidic domain rabbit antiserum (left column) or the rabbit anti-GPIHBP1 antiserum from Novus Biologicals (right column). Bound antibodies were detected with a FITC-conjugated anti-rabbit antibody (green). DAPI staining (blue) was used to visualize nuclei. Staining with the anti-GPIHBP1 antibody demonstrated that all four GPIHBP1 proteins were expressed on the cell surface. However, the anti-acidic domain antisera bound only to wild-type GPIHBP1 and the D,E(24-33)A GPIHBP1 mutant. C, reduced LPL binding to GPIHBP1 in the presence of a rabbit antiserum against the acidic domain of GPIHBP1. Avian LPL (2.5 μg/ml) was incubated with CHO pgsA-745 cells that had been stably transfected with mouse GPIHBP1 in the presence of rabbit preimmune serum or the rabbit antiserum against the acidic domain of GPIHBP1. After washing the cells, LPL binding to cells was quantified with an ELISA. D, fluorescence microscopy of CHO pgsA-745 cells transfected with a mouse GPIHBP1 expression vector, revealing that the binding of DiI-labeled chylomicrons to GPIHBP1 can be blocked with immunopurified antibodies against the acidic domain of GPIHBP1. GPIHBP1 expression in cells was assessed by immunocytochemistry with a FITC-conjugated goat antibody against the S-protein tag (green); the binding of DiI-labeled chylomicrons to cells was assessed by DiI fluorescence (red). The binding of DiI-labeled chylomicrons was measured in the absence of serum, in the presence of preimmune rabbit serum, or in the presence of immunopurified antibodies against the acidic domain of GPIHBP1. Non-immune rabbit IgG (Sigma) did not block chylomicron binding to GPIHBP1 (not shown). DAPI staining (blue) was used to visualize nuclei.
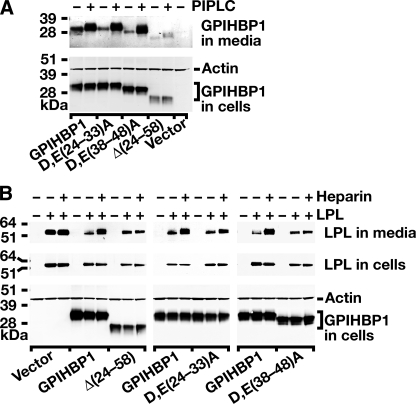
We next tested the ability of the different mutant GPIHBP1 proteins to bind LPL and chylomicrons. Wild-type GPIHBP1, the D,E(24-33)A mutant, the D,E(38-48)A mutant, and the Δ(24-58) mutant were expressed on the cell surface, as judged by the fact that these proteins could be released into the cell culture medium with PIPLC (Fig. 4A). Although lower levels of expression were observed with the Δ(24-58) mutant (Fig. 4, A and B), this protein did reach the cell surface, as judged by immunohistochemistry (Fig. 3B), and could be released with PIPLC (Fig. 4A).
FIGURE 4.
Mutations in the acidic domain of GPIHBP1 prevent LPL binding. A, an S-protein-tagged wild-type GPIHBP1 construct or mutant GPIHBP1 constructs ((D,E(24-33)A, D,E(38-48)A, Δ(24-58)) were transiently transfected into CHO pgsA-745 cells, and Western blots were performed on cell extracts and the cell culture medium with an antibody against the S-protein tag. To determine whether GPIHBP1 reached the surface of the cell, cells were incubated with PIPLC (1 units/ml for 1 h at 37°C), and the appearance of GPIHBP1 in the medium was assessed by Western blot. PIPLC released similar amounts of wild-type GPIHBP1, D,E(24-33)A GPIHBP1, and D,E(38-48)A GPIHBP1 into the medium. Δ(24-58) GPIHBP1 was expressed at lower levels in cells, and PIPLC released smaller amounts of that protein into the medium. B, Western blot analysis of CHO pgsA-745 cells that had been transfected with a V5-tagged human LPL expression vector, along with wild-type or mutant GPIHBP1 constructs. 24 h after the transfection, cell extracts and culture media were harvested and analyzed by Western blotting. After quantifying the bands on an Odyssey infrared scanner (Li-Cor), we determined that heparin treatment led to an 11.75-fold increase in the amount of LPL in the culture medium of cells expressing wild-type GPIHBP1. In the case of Δ(24-58) GPIHBP1 or D,E(38-48)A GPIHBP1, heparin treatment led to negligible changes in the amount of LPL released into the medium (0.97 and 1.1 of the amount at baseline, respectively). In cells expressing D,E(24-33)A GPIHBP1, LPL levels in the medium after heparin increased roughly 2-fold.
We then used Western blotting to test the ability of the different GPIHBP1 mutants to bind LPL, quantifying the intensity of bands with an Odyssey infrared scanner (Li-Cor). As in previous experiments, when CHO pgsA-745 cells were transfected with human LPL alone, a substantial amount of LPL was secreted in the medium, and no additional LPL appeared in the medium after adding heparin, indicating that little LPL is bound to the surface of these cells (Fig. 4B). When the same cells were transfected with both LPL and GPIHBP1, much of the LPL was bound to GPIHBP1 on the cell surface, and little appeared in the medium. However, heparin treatment resulted in an 11.75-fold increase of LPL in the cell culture medium. When the same experiment was performed with the Δ(24-58) GPIHBP1 mutant, LPL was found in the medium, and no additional LPL was released into the medium after heparin (0.97 of that at baseline), suggesting that the mutant GPIHBP1 lacks the capacity to bind LPL (Fig. 4B). However, one could argue that the lower levels of expression with the Δ(24-58) mutant make it difficult to draw definitive conclusions regarding the binding properties of that mutant. We therefore used Western blotting to test the binding properties of the D,E(24-33)A and D,E(38-48)A mutants. When cells were transfected with both LPL and D,E(24-33)A, some LPL appeared in the medium, and this amount was increased 2-fold after heparin, indicating that the D,E(24-33)A GPIHBP1 mutant retains some capacity to bind LPL (Fig. 4B). When cells were transfected with LPL and D,E(38-48)A GPIHBP1, substantial amounts of LPL appeared in the medium, and no additional LPL (1.1-fold baseline) appeared in the medium after heparin, indicating that the D,E(38-48)A GPIHBP1 mutant lacks the ability to bind LPL (Fig. 4B).
To further assess the importance of the acidic domain, we tested the ability of exogenously added mouse LPL to bind to the D,E(38-48)A mutant GPIHBP1. CHO pgsA-745 cells were transiently transfected with wild-type GPIHBP1, the D,E(38-48)A mutant, or empty vector alone. We then incubated the cells for 2 h at 4 °C with medium from nontransfected HeLa cells or HeLa cells that had been transfected with a V5-tagged mouse LPL. Cells that expressed wild-type GPIHBP1 bound the exogenously added LPL avidly, as judged by immunohistochemistry (Fig. 5A). When heparin was added to the incubation medium, LPL binding to GPIHBP1-expressing cells was dramatically reduced (Fig. 5A). In contrast, no LPL binding could be detected in cells transfected with the D,E(38-48)A mutant or the empty vector, either in the presence or absence of heparin (Fig. 5A).
FIGURE 5.
Mutations in the acidic domain of GPIHBP1 prevent the binding of exogenously added LPL. A, binding of a V5-tagged mouse LPL to GPIHBP1-transfected cells. CHO pgsA-745 cells were transiently transfected with wild-type mouse GPIHBP1, the D,E(38-48)A GPIHBP1 mutant, or empty vector. The transfected cells were incubated for 2 h at 4°C with V5-tagged mouse LPL in the presence or absence of heparin (100 units/ml). After washing the cells with ice-cold 1× PBS containing 1 mm MgCl2 and 1 mm CaCl2, the expression of GPIHBP1 and the binding of LPL was assessed by immunofluorescence microscopy with an antibody against GPIHBP1 (green) and an antibody against the V5 tag (red). DAPI (blue) was used to visualize cell nuclei. LPL binding was observed in cells expressing wild-type GPIHBP1 but not in cells that had been transfected with the D,E(38-48)A mutant or empty vector. B, binding of avian LPL to CHO pgsA-745 cells stably transfected with a wild-type GPIHBP1 expression vector or a D,E(38-48)A GPIHBP1 expression vector. After incubating the cells for 2 h with different concentrations of avian LPL, the cells were washed extensively, and the amount of LPL bound to cells was quantified with a sandwich-based ELISA (21). Inset, Western blot showing that both cell lines expressed GPIHBP1. In this experiment, cells from 12-well plates were harvested in 250 μl of lysis buffer, and equal amounts of cell extracts (50 μl) were loaded onto the gel (note the similar intensity of a nonspecific band in all lanes). C, change in the amount of avian LPL in the cell culture medium after PIPLC. Cells that had been stably transfected with wild-type GPIHBP1 or the D,E(38-48)A mutant GPIHBP1 were incubated with avian LPL (0.5 μg/ml). The amount of avian LPL present in the culture medium before and after PIPLC treatment (6 units/ml) for 30 min at 37 °C was quantified by ELISA, and the difference in these values was plotted.
We also generated stably transfected cells expressing wild-type GPIHBP1 or D,E(38-48)A GPIHBP1. We incubated the cells with purified avian LPL and assessed the amount of LPL binding with an ELISA. Substantially more LPL bound to the cells expressing wild-type GPIHBP1 than to cells expressing the D,E(38-48)A mutant (Fig. 5B). In parallel, we incubated the cells with LPL, washed the cells, then treated the cells with PIPLC, and quantified the amount of LPL released into the medium. The amount of PIPLC-releasable LPL was far lower in cells expressing the D,E(38-48)A mutant (Fig. 5C). These data support the notion that the acidic domain, and in particular the acidic amino acids within residues 38-48, are important for binding LPL.
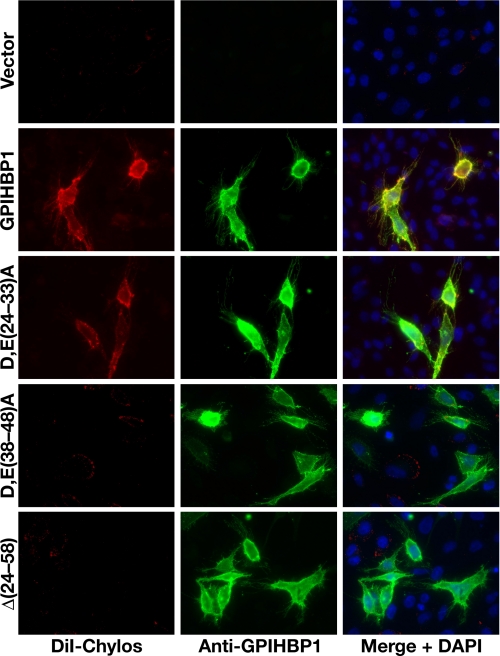
We also assessed the ability of the mutant GPIHBP1 proteins to bind chylomicrons by immunohistochemistry. DiI-labeled chylomicrons bound avidly to cells expressing wild-type GPIHBP1 but not to cells expressing D,E(38-48)A GPIHBP1 or the Δ(24-58) GPIHBP1 mutant (Fig. 6). Chylomicrons did bind to cells expressing the D,E(24-33)A mutant, although less avidly than wild-type GPIHBP1, as judged by the intensity of fluorescence (Fig. 6). Again, these data indicate that the acidic domain of GPIHBP1, and in particular the acidic amino acids within residues 38-48, are important for binding DiI-labeled chylomicrons.
FIGURE 6.
Mutations in the acidic domain of GPIHBP1 interfere with the binding of DiI-labeled chylomicrons. CHO pgsA-745 cells were transiently transfected with different GPIHBP1 constructs. The expression of GPIHBP1 on the surface of cells was assessed by immunofluorescence microscopy with an antibody against GPIHBP1 (green); binding of DiI-labeled chylomicrons was detected by DiI fluorescence (red). DAPI (blue) was used to visualize cell nuclei. The DiI-labeled chylomicrons bound to cells expressing wild-type GPIHBP1 but not to cells expressing D,E(38-48)A GPIHBP1 or Δ(24-58) GPIHBP1. Chylomicron binding to cells transfected with the D,E(24-33)A GPIHBP1 was detectable, but was less intense than with wild-type GPIHBP1 (in each of six separate experiments).
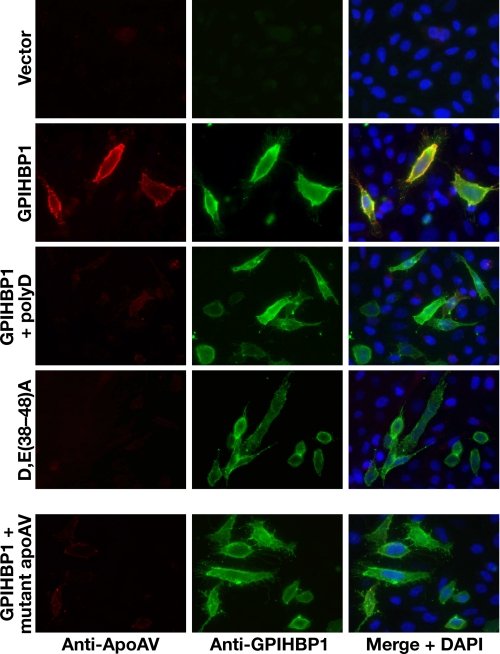
The Heparin-binding Domain of ApoAV and the Acidic Domain of GPIHBP1 Are Required for the Interaction of These Molecules—ApoAV-phospholipid disks bind GPIHBP1 (1). ApoAV contains a heparin-binding domain, and we suspected that electrostatic interactions involving this domain were important for binding to GPIHBP1. We found that the binding of apoAV-phospholipid disks to GPIHBP1 was disrupted by poly(D) (Fig. 7), as judged by immunohistochemistry. Also, the D,E(38-48)A mutant GPIHBP1 failed to bind apoAV-phospholipid disks, indicating that the interaction requires acidic amino acids between residues 38 and 48 (Fig. 7). The interaction between apoAV and GPIHBP1 also depends on the heparin-binding domain of apoAV; an apoAV mutant in which the heparin-binding domain was mutated did not bind to GPIHBP1 (Fig. 7).
FIGURE 7.
The acidic domain of GPIHBP1 and the heparin-binding domain of apoAV are required for the binding of apoAV to GPIHBP1. CHO pgsA-745 cells were transiently transfected with different GPIHBP1 constructs, and the expression of GPIHBP1 was assessed by immunofluorescence microscopy with an antibody against GPIHBP1 (green); binding of apoAV·DMPC disks to cells was detected with an antibody against apoAV (red). Rows 1-3, immunofluorescence microscopy showing that the binding of apoAV·DMPC disks to cells expressing wild-type GPIHBP1 was eliminated by poly(D) (200 μg/ml). Row 4, immunofluorescence microscopy showing that apoAV·DMPC disks do not bind to cells expressing D,E(38-48)A GPIHBP1. Row 5, immunofluorescence microscopy examining the binding of mutant apoAV·DMPC disks to cells expressing wild-type GPIHBP1. In the mutant apoAV, four of the positively charged residues in the principal heparin-binding domain were replaced with other amino acids (R210E, K211Q, K215Q, K217E).
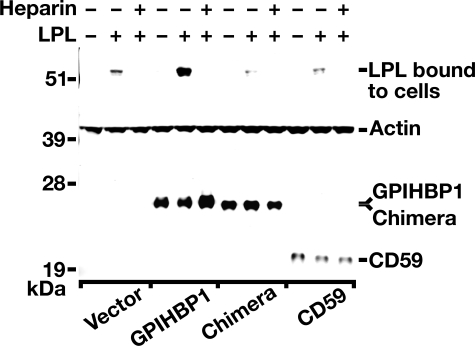
The Acidic Domain of GPIHBP1 Is Required but Not Sufficient for LPL Binding—To determine whether the acidic domain of GPIHBP1 is solely responsible for the interaction of GPIHBP1 with LPL, we generated a GPIHBP1/CD59 chimera in which the Ly6 domain of GPIHBP1 was replaced with the Ly6 domain of human CD59. We then transiently transfected CHO pgsA-745 cells with that mutant and used Western blotting to test the ability of those cells to bind exogenously added V5-tagged human LPL. The intensity of the LPL, GPIHBP1, and actin bands were quantified on an Odyssey infrared scanner (Li-Cor). As expected, GPIHBP1-transfected cells bound substantially more LPL (7.1-fold more) than cells that had been transfected with empty vector (Fig. 8). This binding was blocked with heparin. In contrast, the GPIHBP1/CD59 chimera lacked the ability to bind LPL, despite the fact that this protein contained the entire acidic domain of human GPIHBP1 (Fig. 8). These data suggest that the acidic domain of GPIHBP1 is required but not sufficient to support the interaction of GPIHBP1 and LPL.
FIGURE 8.
The Ly6 domain of GPIHBP1 is also required for binding LPL. CHO pgsA-745 cells were transiently transfected with human GPIHBP1, human CD59, empty vector, or a GPIHBP1/CD59 chimera in which the Ly6 domain of human GPIHBP1 was replaced with the Ly6 domain of human CD59. The transfected cells were incubated for 2 h at 4°C with V5-tagged human LPL in the presence or absence of 100 units/ml of heparin. After washing the cells with ice-cold 1× PBS containing 1 mm MgCl2 and 1 mm CaCl2, cell extracts were prepared and the amount of LPL binding was assessed by Western blotting. Quantification of the bands on an Odyssey infrared scanner (Li-Cor) revealed a 7.1-fold increase in LPL binding to cells that had been transfected with wild-type human GPIHBP1 (compared with cells that had been transfected with empty vector). This binding was blocked by heparin. In contrast, cells that had been transfected with the GPIHBP1/CD59 chimera bound very little LPL, similar to the amount bound by cells that had been transfected with CD59 or empty vector.
DISCUSSION
Arguably the most striking structural feature of GPIHBP1, and the feature that distinguishes it from other Ly6 family members, is an amino-terminal acidic domain. This domain, 25 amino acids in length, is highly enriched in aspartate and glutamate residues. Previously, we speculated that this domain might be important for binding LPL and chylomicrons (1, 6). In the current study, we show the acidic domain is indeed functionally important. First, polyaspartate peptides interfere with the ability of GPIHBP1 to bind LPL, chylomicrons, and apoAV-phospholipids disks. Second, a rabbit antiserum against the acidic domain blocks the binding of LPL and chylomicrons to GPIHBP1. Third, mutating the acidic domain eliminates the ability of the GPIHBP1 to bind LPL and chylomicrons. A GPIHBP1 mutant lacking the acidic amino acids between residues 28 and 38 could not bind LPL, either avian LPL in a quantitative ELISA-based assay or a V5-tagged human LPL in a Western blot assay, nor did this mutant have any ability to bind DiI-labeled chylomicrons.
Positively charged heparin-binding domains within LPL and apoAV are important for the binding of these molecules to GPIHBP1. Mutating the principal heparin-binding domain of human LPL eliminated its ability to bind to GPIHBP1. Similarly, mutating the heparin-binding domain within apoAV prevented apoAV-phospholipid disks from binding to GPIHBP1. Previous studies established that the heparin-binding domains of LPL and apoAV are crucial for the binding of those proteins to heparin and HSPGs (7-11, 17). The current study shows that these same regions are important for binding GPIHBP1, supporting the notion that electrostatic interactions are important for GPIHBP1-ligand interactions.
Using Western blot assays, we consistently and repeatedly observed binding of LPL to wild-type GPIHBP1 and the release of GPIHBP1-bound LPL into the medium with heparin. Deleting the acidic domain, or mutating a portion of the acidic domain, eliminated the ability of GPIHBP1 to bind LPL. The acidic domain was also important for binding chylomicrons. Although the current studies show that the acidic domain of GPIHBP1 is important for binding ligands, our studies also revealed that the interaction of GPIHBP1 and LPL is complex and requires both the acidic and Ly6 domains of GPIHBP1. The fact that the Ly6 domain is also important is not particularly surprising given that, in the case of other Ly6 family members, the Ly6 motif is important for binding ligands. For example, the Ly6 motifs in UPAR (urokinase-type tissue plasminogen activator receptor) are required for binding tissue plasminogen activator (24-26). In the case of GPIHBP1, it is tempting to speculate that the acidic domain could function as the “magnet” for heparin-binding proteins such as LPL, whereas the Ly6 motif might constitute a crucial ligand-binding domain. Interestingly, specific regions of the Ly6 motif of GPIHBP1 have been highly conserved in mammalian evolution. We would not be surprised if future studies were to uncover important functions for the Ly6 motif of GPIHBP1.
Acknowledgments
We thank Jennifer A. Beckstead for technical assistance.
This work was supported by a Scientist Development Award from the American Heart Association, National Office (to A. P. B.), BayGenomics Grants HL66621 and HL66600, R01HL087228 (to S.G.Y.), and HL073061 (to R.O.R.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: GPIHBP1, glycosylphosphatidylinositol-anchored high density lipoprotein-binding protein 1; GPI, glycosylphosphatidylinositol; LPL, lipoprotein lipase; PIPLC, phosphatidylinositol-specific phospholipase C; DMPC,dimyristoylphosphatidylcholine; CHO,Chinesehamsterovary;DiI,1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate; HSPG, heparan sulfate proteoglycans; PBS, phosphate-buffered saline; BSA, bovine serum albumin; ELISA, enzyme-linked immunosorbent assay; FITC, fluorescein iso-thiocyanate; DAPI, 4′,6-diamidino-2-phenylindole; bis-Tris, 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol; CHAPS, 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid.
References
- 1.Beigneux, A. P., Davies, B., Gin, P., Weinstein, M. M., Farber, E., Qiao, X., Peale, P., Bunting, S., Walzem, R. L., Wong, J. S., Blaner, W. S., Ding, Z. M., Melford, K., Wongsiriroj, N., Shu, X., de Sauvage, F., Ryan, R. O., Fong, L. G., Bensadoun, A., and Young, S. G. (2007) Cell Metab. 5 279-291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.LeClair, K. P., Rabin, M., Nesbitt, M. N., Pravtcheva, D., Ruddle, F. H., Palfree, R. G. E., and Bothwell, A. (1987) Proc. Natl. Acad. Sci. U. S. A. 84 1638-1642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang, Y., Dang, J., Johnson, L. K., Selhamer, J. J., and Doe, W. F. (1995) Eur. J. Biochem. 227 116-122 [DOI] [PubMed] [Google Scholar]
- 4.Ioka, R. X., Kang, M.-J., Kamiyama, S., Kim, D.-H., Magoori, K., Kamataki, A., Ito, Y., Takei, Y. A., Sasaki, M., Suzuki, T., Sasano, H., Takahashi, S., Sakai, J., Fujino, T., and Yamamoto, T. T. (2003) J. Biol. Chem. 278 7344-7349 [DOI] [PubMed] [Google Scholar]
- 5.Low, M. G. (1992) Phospholipases That Degrade the Glycosylphosphatidylinositol Anchor of Membrane Proteins, Oxford University Press, Oxford, UK
- 6.Young, S. G., Davies, B. S., Fong, L. G., Gin, P., Weinstein, M. M., Bensadoun, A., and Beigneux, A. P. (2007) Curr. Opin. Lipidol. 18 389-396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berryman, D. E., and Bensadoun, A. (1993) J. Biol. Chem. 268 3272-3276 [PubMed] [Google Scholar]
- 8.Hata, A., Ridinger, D. N., Sutherland, S., Emi, M., Shuhua, Z., Myers, R. L., Ren, K., Cheng, T., Inoue, I., and Wilson, D. E. (1993) J. Biol. Chem. 268 8447-8457 [PubMed] [Google Scholar]
- 9.Lookene, A., Nielsen, M. S., Gliemann, J., and Olivecrona, G. (2000) Biochem. Biophys. Res. Commun. 271 15-21 [DOI] [PubMed] [Google Scholar]
- 10.Ma, Y., Henderson, H. E., Liu, M. S., Zhang, H., Forsythe, I. J., Clarke-Lewis, I., Hayden, M. R., and Brunzell, J. D. (1994) J. Lipid Res. 35 2049-2059 [PubMed] [Google Scholar]
- 11.Sendak, R. A., and Bensadoun, A. (1998) J. Lipid Res. 39 1310-1315 [PubMed] [Google Scholar]
- 12.Hahn, P. F. (1943) Science 98 19-20 [DOI] [PubMed] [Google Scholar]
- 13.Korn, E. D. (1955) J. Biol. Chem. 215 15-26 [PubMed] [Google Scholar]
- 14.Korn, E. D. (1955) J. Biol. Chem. 215 1-14 [PubMed] [Google Scholar]
- 15.Bragdon, J. H., and Havel, R. J. (1954) Science 120 113-114 [DOI] [PubMed] [Google Scholar]
- 16.Hultin, M., Olivecrona, G., and Olivecrona, T. (1994) Biochem. J. 304 959-966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lookene, A., Beckstead, J. A., Nilsson, S., Olivecrona, G., and Ryan, R. O. (2005) J. Biol. Chem. 280 25383-25387 [DOI] [PubMed] [Google Scholar]
- 18.Esko, J. D., Stewart, T. E., and Taylor, W. H. (1985) Proc. Natl. Acad. Sci. U. S. A. 82 3197-3201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ben-Zeev, O., Mao, H. Z., and Doolittle, M. H. (2002) J. Biol. Chem. 277 10727-10738 [DOI] [PubMed] [Google Scholar]
- 20.Bensadoun, A., Hsu, J., and Hughes, B. (1999) Methods Mol. Biol. 109 145-150 [DOI] [PubMed] [Google Scholar]
- 21.Cisar, L. A., Hoogewerf, A. J., Cupp, M., Rapport, C. A., and Bensadoun, A. (1989) J. Biol. Chem. 264 1767-1774 [PubMed] [Google Scholar]
- 22.Beckstead, J. A., Oda, M. N., Martin, D. D., Forte, T. M., Bielicki, J. K., Berger, T., Luty, R., Kay, C. M., and Ryan, R. O. (2003) Biochemistry 42 9416-9423 [DOI] [PubMed] [Google Scholar]
- 23.Nilsson, S. K., Lookene, A., Beckstead, J. A., Gliemann, J., Ryan, R. O., and Olivecrona, G. (2007) Biochemistry 46 3896-3904 [DOI] [PubMed] [Google Scholar]
- 24.Behrendt, N., Ploug, M., Patthy, L., Houen, G., Blasi, F., and Dano, K. (1991) J. Biol. Chem. 266 7842-7847 [PubMed] [Google Scholar]
- 25.Llinas, P., Le Du, M. H., Gardsvoll, H., Dano, K., Ploug, M., Gilquin, B., Stura, E. A., and Menez, A. (2005) EMBO J. 24 1655-1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ploug, M. (1998) Biochemistry 37 16494-16505 [DOI] [PubMed] [Google Scholar]