Abstract
CD44 is an important adhesion molecule that functions as the major hyaluronan receptor which mediates cell adhesion and migration in a variety of physiological and pathological processes. Although full activity of CD44 requires binding to ERM (ezrin/radixin/moesin) proteins, the CD44 cytoplasmic region, consisting of 72 amino acid residues, lacks the Motif-1 consensus sequence for ERM binding found in intercellular adhesion molecule (ICAM)-2 and other adhesion molecules of the immunoglobulin superfamily. Ultracentrifugation sedimentation studies and circular dichroism measurements revealed an extended monomeric form of the cytoplasmic peptide in solution. The crystal structure of the radixin FERM domain complexed with a CD44 cytoplasmic peptide reveals that the KKKLVIN sequence of the peptide forms a β strand followed by a short loop structure that binds subdomain C of the FERM domain. Like Motif-1 binding, the CD44 β strand binds the shallow groove between strand β5C and helix α1C and augments the β sheet β5C-β7C from subdomain C. Two hydrophobic CD44 residues, Leu and Ile, are docked into a hydrophobic pocket with the formation of hydrogen bonds between Asn of the CD44 short loop and loop β4C-β5C from subdomain C. This binding mode resembles that of NEP (neutral endopeptidase 24.11) rather than ICAM-2. Our results reveal a characteristic versatility of peptide recognition by the FERM domains from ERM proteins, suggest a possible mechanism by which the CD44 tail is released from the cytoskeleton for nuclear translocation by regulated intramembrane proteolysis, and provide a structural basis for Smad1 interactions with activated CD44 bound to ERM protein.
Hyaluronan is a major ubiquitous glycosaminoglycan component of the extracellular matrix in vertebrates (for reviews, see Refs. 1 and 2). CD44 was the first transmembrane hyaluronan receptor identified, and interest in this receptor stems from the fact that CD44-hyaluronan interactions mediate cell migration in a variety of pathophysiological processes, including tumor metastasis, wound healing, and leukocyte extravasation at inflammation sites (for reviews, see Refs. 3-6). CD44 and its different isoforms retain the link-homology domain in the extracellular domain for hyaluronan binding and common transmembrane and cytoplasmic regions with high sequence conservation among the isoforms and between species (3-4, 7-9).
The CD44 cytoplasmic region, comprising 72 amino acid residues, has been shown to associate with actin filaments in various cells, a process mediated by ERM (ezrin/radixin/moesin) proteins and the closely related protein merlin (also referred to as neurofibromin 2/schwannomin) (10-14), which is the neurofibromatosis type 2 tumor suppressor gene product (15). ERM proteins and merlin play a key role as cross-linkers between adhesion molecules on the plasma membrane and actin filaments (13, 16-19). Increasing evidence has shown that interactions between CD44 and ERM proteins are associated with normal physiological cell adhesion and migration functions in addition to bacterial infection and cancer progression (5-6, 20).
Importantly, a minimal cytoplasmic region projecting from a transmembrane helix is required for efficient hyaluronan binding (21-24), probably stabilizing CD44 at the plasma membrane and facilitating receptor clustering. Moreover, these highly conserved transmembrane and cytoplasmic regions are required for events downstream of hyaluronan binding through regulated intramembrane proteolysis (RIP)5 (25), which produces a CD44 intracellular domain (ICD) fragment that translocates into the nucleus and stimulates transcription via direct interactions with the transcriptional machinery (26). However, the physicochemical and structural features of the ICD fragment, which encompasses the whole cytoplasmic region, are unknown.
ERM proteins comprise three domains, an N-terminal FERM (4.1 and ERM) domain, a central α-helical domain, and a C-terminal tail domain. The FERM domain interacts with the plasma membrane and specifically binds a variety of adhesion molecules, including CD44, whereas the C-terminal tail domain binds to F-actin. The FERM domain is well conserved in all members of ERM proteins and merlin and is believed to bind the same target proteins. The C-terminal tail domain, however, is not well conserved in merlin and shows little association with F-actin (15, 27). Major binding targets of the FERM domain are adhesion molecules classified as type I membrane proteins. Biochemical studies have shown that the FERM domain binds the cytoplasmic regions of intercellular adhesion molecule-1 (ICAM-1), ICAM-2, and ICAM-3 of the immunoglobulin superfamily as well as CD44 and CD43/leukosialin/sialophorin (13). The first crystal of the FERM domain bound to the target adhesion molecule was obtained using the full-length ICAM-2 cytoplasmic peptide comprising 28 residues (28). An x-ray structural study has shown how the radixin FERM domain binds the juxtamembrane region of the ICAM-2 cytoplasmic peptide (28). On the basis of crystal structure and mutation studies, the Motif-1 sequence motif RXXTYXVXXA is proposed as binding to the FERM domain. The ICAM-2 Motif-1 sequence forms a β strand (XXTY) that mediates anti-parallel β-β interactions with the FERM domain and a 310-helix (VXXA) that docks into a hydrophobic pocket. Motif-1 is found in other adhesion molecules of the immunoglobulin superfamily containing VCAM-1 and L1-CAM and proteoglycans such as syndecan and neurexin. All of these adhesion molecules are shown to bind the radixin FERM domain. Recently, PSGL-1 (P-selectin glycoprotein ligand-1) has been shown to maintain a Motif-1-related sequence that binds the FERM domain (29). Interestingly, CD44 retains neither Motif-1 nor Motif-2, MDWXXXXX(L/I)FXX(L/F), which has recently been identified in the FERM-binding region of Na+/H+ exchanger regulatory factor-1 and -2 (NHERF-1 and -2) (30). Thus, the precise binding mode of CD44 to the FERM domain remains unclear.
We report here on physicochemical and hydrodynamic analyses of the CD44 cytoplasmic peptide and the crystal structure of the radixin FERM domain complexed with a CD44 juxtamembrane peptide. We show that the CD44 cytoplasmic peptide is present as a monomeric random coil in solution. In the complex crystal, the CD44 peptide binds subdomain C of the radixin FERM domain. The CD44 binding site overlaps with that of the Motif-1 binding site found in previous complexed structures (28, 29), whereas the binding mode of CD44 to the FERM domain is distinct from that of ICAM-2 and resembles that of the recently reported NEP (neutral endopeptidase 24.11) (31). These results, taken together with analyses of our previous structures, define a characteristic versatility of peptide recognition by the radixin FERM domain, which is distinct from the talin FERM domain (32) and PTB domains. Furthermore, in addressing how phosphorylation may interfere with binding to ERM proteins, we suggest a mechanism by which RIP-mediated cleavage of the CD44 cytoplasmic peptide facilitates nuclear translocation for transcriptional activation. Finally, we suggest a structural basis for Smad1 interactions with activated CD44 bound to ERM protein and linked to actin cytoskeletons.
EXPERIMENTAL PROCEDURES
Protein Preparation—The region of cDNA coding for the cytoplasmic region of mouse CD44 (residues 292-363) was subcloned into pGEX4T-1 or pGEX6P-3 plasmid (GE Healthcare) using the BamHI and SmaI restriction enzyme sites. The CD44 cytoplasmic peptide was expressed in BL21(DE3)RIL cells (Stratagene) as a fusion protein with glutathione S-transferase. Cells were grown at 37 °C in Luria-Bertani medium containing 50 μgml-1 ampicillin and 50 μgml-1 chloramphenicol. When the OD660 of the cell culture reached 0.8, isopropyl β-d-thiogalactopyranoside was added to a concentration of 1 mm to induce expression of the CD44 gene. Cells were grown at 30 °C for an additional 5 h following isopropyl β-d-thiogalactopyranoside induction and then collected by centrifugation at 4000 rpm (Beckman J2-M1 JA10 rotor) for 15 min at 4 °C. Wet cells expressing CD44 peptide were suspended in 50 mm Tris buffer (pH 8.0) containing 500 mm NaCl, 1 mm dithiothreitol (DTT), 1 mm EDTA, and 1.5% Sarkosyl and then disrupted by sonication at 4 °C. The soluble portion of the cell extract was then loaded onto a glutathione S-transferase affinity column comprising glutathione-Sepharose 4B resin (GSH resin) (GE Healthcare) and then washed copiously with 20 mm HEPES buffer (pH 7.3) containing 200 mm NaCl, 1 mm DTT, and 1 mm EDTA. Bound glutathione S-transferase fusion protein was cleaved from the GSH resin using 5 units ml-1 thrombin (Sigma) for 8 h or 2 units ml-1 HRV3C protease (Novagen) for 18 h at 4°C.
The cleaved sample was collected and purified by chromatography using HiTrap SP (GE Healthcare) and HiPrep 26/10 desalting (GE Healthcare) with 10 mm HEPES buffer (pH 7.5) containing 50 mm NaCl and 0.5 mm DTT. Purified CD44 peptide was concentrated using Microsep™ centrifugal devices 1K (Pall Corp.). The peptide sample was divided into 50-μl aliquots in 0.5-ml tubes (Eppendorf) and immediately frozen in liquid nitrogen. Frozen samples were stored at -80 °C until use. The radixin FERM domain was expressed in E. coli cells and purified as described previously (33).
Gel Filtration—The molecular size of the recombinant CD44 cytoplasmic peptide was examined using gravity flow gel filtration techniques. CD44 was chromatographed at 4 °C through a Superdex 75 HR 10/30 column (GE Healthcare) with buffer C containing 20 mm HEPES buffer (pH 7.5), 150 mm KCl, 1 mm DTT, and 1 mm EDTA. The molecular mass was determined on the basis of the elution volume from a plot of log (molecular mass) of standard proteins, comprising bovine γ-globulin (158 kDa), chicken ovalbumin (44 kDa), equine myoglobin (17 kDa), and vitamin B-12 (1.4 kDa) (Bio-Rad), versus the elution volume.
Circular Dichroism Spectroscopy—CD spectra of the purified CD44 cytoplasmic peptide were recorded at 4 °C using a Jasco J720W spectropolarimeter. The CD44 cytoplasmic peptide was dissolved in 5 mm Tris buffer (pH 8.0) containing 50 mm NaCl, 0.7 mm EDTA, and 0.5 mm 2-mercaptoethanol or containing 150 mm NaCl, 0.7 mm EDTA, and 0.5 mm 2-mercaptoethanol. The CD44 cytoplasmic peptide and the FERM domain were mixed at a 1:1 molar ratio (13 μm:13 μm) and dissolved in 5 mm Tris buffer (pH 8.0) containing 150 mm NaCl, 0.7 mm EDTA, and 0.5 mm 2-mercaptoethanol. Secondary structure estimations were calculated using the Jasco secondary structure estimation software.
Analytical Ultracentrifugation—Sedimentation velocity ultracentrifugation experiments were performed at 10 °C using a Beckman Coulter Optima XLA analytical ultracentrifuge equipped with an An-60 Ti rotor and double sector centerpieces. Purified samples of the CD44 cytoplasmic peptide were dissolved in 5 mm Tris buffer (pH 7.8) containing 50 mm NaCl and 2 mm DTT (TSD buffer) at a sample concentration of 0.5 mg ml-1 and then centrifuged at 42,000 rpm. Radial absorbance scans were measured every 15 min at a wavelength of 230 nm. The resultant data were analyzed using the programs Sedfit and Sednterp. To glean further insight into the CD44 conformation in the FERM-bound state, similar experiments were performed for the FERM domain in the free and the CD44-bound forms in 5 mm Tris buffer (pH 7.4) containing 150 mm NaCl at 20 °C.
Sedimentation equilibrium ultracentrifugation experiments were performed at 10 °C using the same ultracentrifuge and rotor as described above. Six-sector centerpieces were used. The CD44 cytoplasmic peptide was dissolved in TSD buffer at sample concentrations of 0.0625, 0.125, and 0.25 mg ml-1 and then centrifuged at 32,000, 38,000, 40,000, and 48,000 rpm. Radial absorbance scans were measured at 230 nm after 22 h, at which time equilibrium had been achieved. The resultant data were analyzed using XLA/XL-I data analysis software.
Structural Determination of the FERM-CD44 Complex—Preparation and crystallization of the radixin FERM domain complexed with the CD44 cytoplasmic peptide were as previously described (34). Confirmation that the resulting crystals contained the FERM domain and CD44 peptide was achieved using matrix-assisted laser desorption/ionization time-of-flight mass spectroscopy (PerSeptive Inc.). Diffraction tests of crystals were performed at 100 K using a Rigaku R-Axis IV detector equipped with a Rigaku FR-E x-ray generator. Rodlike crystals grown in 0.1 m Tris (pH 8.6) containing 15% polyethylene glycol 3350 and 0.2 m potassium thiocyanate were shown to diffract to 2.1 Å resolution using a Rigaku MSC Jupiter 210 detector installed on beamline BL38B1 at SPring-8 (Harima, Japan). The crystal data and the intensity data statistics are summarized in Table 1.
TABLE 1.
Crystallographic data for the radixin FERM domain-CD44 complex
| Parameter | Value |
|---|---|
| X-ray data | |
| Space group | P212121 |
| Cell parameters, (Å) | a = 62.70, b = 66.18, c = 86.22 |
| Resolution (Å)a | 20-2.10 (2.17-2.10) |
| Mosaicity | 0.5-0.8 |
| Reflections, total/unique | 156,148/21,492 |
| Completeness (%) | 100.0 (100.0) |
| <I/σ1> | 12.2 (4.9) |
| Rmerge (%) | 5.0 (36.1) |
| Refinement | |
| No. of residues included | |
| FERM (residues 1-310) | 295 (residues 3-297) |
| CD44 (residues 2-21) | 9 (residues 8-16) |
| Water | 171 |
| No. of atoms | 2,714 |
| Rwork/Rfree (%)b | 23.1/25.6 |
| Average B-factor (Å2) | |
| FERM | 37.0 |
| CD44 | 60.8 |
| Water | 42.6 |
| Root mean square bond length (Å), angles (degrees) | 0.006, 1.2 |
Numbers in parentheses refer to statistics for the outer resolution shell.
Rwork = ∑||Fo| - |Fc||/∑|Fo|. Rfree is the same as Rwork except for a 5% subset of all reflections that were never used in the crystallographic refinement.
Diffracted x-ray intensities were processed using the HKL-2000 program suite (35). Phases were determined by molecular replacement using the program PHASER (36) with the free form structure of the FERM domain as a search model (37). The procedure gave a clear solution corresponding to one FERM molecule in the asymmetric unit of the crystal. The calculated molecular replacement maps showed definite residual electron densities for the CD44 peptide at the groove between strand β5C and helix α1C of subdomain C of the FERM domain in both 2Fo-Fc and Fo-Fc maps. CD44 peptide residues were modeled manually using O (38). The complex structure was refined by simulated annealing, followed by restrained individual B-factor refinement performed using the program CNS (39). The refinement statistics are summarized in Table 1. The stereochemical quality of the model was assessed using the program PROCHECK. In the Ramachandran plot, 89.8 and 9.9% of residues were located within the most favored and additional allowed regions, respectively. One exceptional outlier was flagged in the plot, that of Asp252 located within a type II reverse turn between strands β5C and β6C. This outlier repeatedly appeared in the FERM domain structures of radixin (28, 30, 37), moesin (40), and merlin (41). We also checked our structure with MolProbity (42). In the Ramachandran plot, 96.3, 3.3, and 0.3% (Lys296 but not Asp252) of the residues fell in favored, allowed, and outlier regions. Thus, judgment of outliers in our structure is subtle. Molecular illustrations were prepared using the program PyMOL (DeLano Scientific). Superposition of the FERM domains and peptides were performed using the program Lsqkab (43).
Pull-down Assay of the Radixin FERM Domain with Wild-type, S2D, and S2p CD44 Peptides—N-terminal biotinylated CD44 cytoplasmic peptides were purchased from Toray Research Center (Tokyo, Japan) for the in vitro binding assay. The wild-type peptide is the same as that used in the x-ray structural work. The S2p peptide contains phosphoserine at position 2, and the S2D mutant peptide contains a negatively charged aspartic acid that mimics the phosphorylated state of Ser2. Pull-down assays were performed using Streptavidin-Sepharose high performance resin (GE Healthcare). For each reaction, 25 μl of the resin was mixed with 25 pmol of each N-terminal biotinylated peptide and suspended in 1 ml of 10 mm HEPES buffer (pH 7.4) containing 70 mm KCl and 1 mm DTT (pull-down buffer) in a 1.5-ml tube (Eppendorf). Resin free from bound peptide was used as the control. The resin was harvested as a pellet by centrifugation (2000 × g for 1 min). After removing the supernatant, the resin was suspended in 1 ml of pull-down buffer again, and this wash was repeated two times. 25 μl of 100 μm FERM domain dissolved in pull-down buffer was added to the resin. The resin was incubated for 2 h at room temperature with occasional mixing and then washed two times with pull-down buffer by centrifugation. To elute the streptavidin-bound peptide and its associated FERM domain, 25 μl of SDS-sample buffer was added to recovered resin, and then each sample was incubated for 5 min at 96 °C. The amount of streptavidin in each eluate was determined by SDS-PAGE, and eluted proteins were visualized using SimplyBlue™SafeStain (Invitrogen). An appropriate amount of each eluate containing the same amount of streptavidin was then subjected to SDS-PAGE. The amount of bound FERM domain was determined by densitometric scanning using the software Image J 1.36b. The relative amount of the FERM domain bound per streptavidin was calculated, and the amount of FERM domain bound to the control resin was subtracted from each eluate. This pull-down assay was performed three times, and the average amount of FERM domain binding to each peptide was estimated.
Binding Assay—The binding affinity for the 23-residue CD44 peptide was examined by using equilibrium surface plasmon resonance measurements, which were carried out on a Biacore Biosensor instrument (Biacore 3000; GE Healthcare), as previously described (30). The human merlin FERM domain was purified as described previously (41). The biotinylated peptide of the juxtamembrane region (23 residues of mouse CD44; see Fig. 5A) was purchased from Sawady Technology (Tokyo, Japan). The peptide was coupled via the N-terminal biotin moiety to a streptavidin-coated sensor chip (sensor chip SA Biacore AB). The purified FERM domain (5-1280 nm) was injected into both peptide-linked and nonlinked sensor chips for correction of background signals. All binding experiments were performed at 25 °C with a flow rate of 20 μl/min in buffer consisting of 10 mm HEPES (pH 7.4), 150 mm NaCl, 1 mm EDTA, 1 mm DTT, and 0.005% surfactant P20. The kinetic parameters were evaluated by using the BIA evaluation software (GE Healthcare). The KD values were obtained by averaging of at least three independent measurements. The obtained KD values for FERM binding to the 23-residue CD44 peptide are 110 ± 9 nm (melin) and 120 ± 9 nm (radixin).
FIGURE 5.
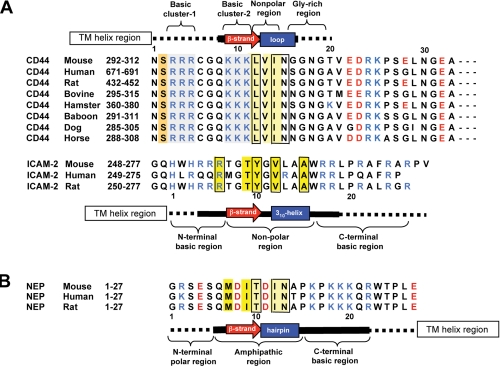
Comparison of FERM-binding peptide sequences. A, alignment of the juxtamembrane regions of CD44 cytoplasmic tails is compared with that of ICAM-2. The secondary structures found in our FERM-CD44 complexed structure are shown at the top with those found in the FERM-ICAM-2 complex (28). The solid boldface lines indicate peptide residues defined in the maps, and the dotted lines indicate residues that were not defined in the maps. Basic and acidic residues are shown in blue and red, respectively, and key residues for binding are marked with boxes and highlighted in yellow. Two conserved basic clusters of CD44 are highlighted in blue. B, alignment of the juxtamembrane regions of NEP cytoplasmic tails is shown together with the secondary structures found in the FERM-NEP complex (31).
RESULTS
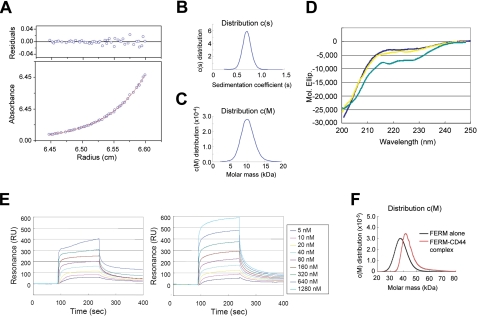
Conformational Properties of the CD44 Cytoplasmic Peptide in Solution—The structural features of the CD44 cytoplasmic region, comprising 72 amino acid residues, are largely unknown. One intriguing question relates to whether this longer cytoplasmic peptide forms a stable compact domain that possesses the ability of self-association to form dimeric or oligomeric structures. In an effort to address these uncertainties, analytical ultracentrifugation methods were employed to investigate the assembly state of the whole cytoplasmic peptide and to determine the approximate shape of the molecule/assembly in solution. Sedimentation equilibrium analyses resulted in an excellent fit of the observed absorbance data with the calculated curve based on an ideal single-species model (Fig. 1A). The obtained molecular mass of 8.8 kDa is close to the theoretical value (8.382 kDa) and suggests that the CD44 peptide adopts a monomeric form in solution. Sedimentation velocity measurements showed the presence of a single boundary, which suggests monodispersity of the sample containing the molecular species with a sedimentation coefficient of 0.73 S and an estimated molecular mass of 10.4 kDa, which is similar to that of the sedimentation equilibrium experiments (Fig. 1, B and C). Thus, it was demonstrated that the CD44 peptide adopts a monomeric form. Interestingly, the CD44 peptide adopts an elongated shape, as estimated from the obtained translational frictional ratio (f/f0) of 1.89, which suggests a major/minor axial ratio (a/b) of 16.0. Supposing a diameter of 1.5 nm for a peptide chain in a random coiled state, the obtained a/b value suggests that the cytoplasmic peptide extends by ∼24 nm out from the inner cell membrane toward the cytoplasmic region or otherwise parallel to the membrane. The juxtamembrane region that is rich in basic residues might run parallel to the membrane and interact with the negatively charged inner membrane surfaces. The rest of the peptide residues, however, may not be parallel to the membrane because of many negatively charged residues; it contains 11 acidic but only 6 basic residues.
FIGURE 1.
Results of analytical ultracentrifugation of the CD44 cytoplasmic peptide. A, sedimentation equilibrium data with a fitted curve for the 0.25 mg ml-1 CD44 cytoplasmic peptide solution at 38,000 rpm. The upper panel shows residuals representing the difference between the calculated fit and the experimental data using XLA/XL-I data analysis software. The molecular mass of the CD44 cytoplasmic tail was estimated to be 8.8 kDa. This result shows that the CD44 cytoplasmic tail adopts a monomeric form in the solution state. B, the distribution of sedimentation coefficients obtained from sedimentation velocity analysis of the CD44 cytoplasmic tail. The single peak is found at 0.73 S. C, the distribution of apparent molecular mass obtained from sedimentation velocity analysis of the CD44 cytoplasmic tail. The single peak is found at 10.4 kDa. D, CD spectra of the 72-residue CD44 cytoplasmic tail. Spectra of the CD44 peptide at 50 mm NaCl (blue) and 150 mm (yellow) are shown with a difference spectrum between the FERM domain and the 1:1 complex of the radixin FERM domain and CD44 (cyan). E, sensorgrams for each concentration of the merlin (left) or radixin (right) FERM domain. The signals from the control surface were subtracted. F, distribution of apparent molecular mass obtained from sedimentation velocity analysis of the FERM domain in the free and the CD44-bound forms. Estimated molecular masses are 38.7 kDa (free form) and 44.1 kDa (CD44-bound form), suggesting monomers.
The results of the sedimentation analyses are consistent with the results obtained from the use of conventional gel filtration, where the hydrodynamic properties of proteins are affected by the molecular shape and surface properties of the sample molecule. Our gel filtration analysis using a Superdex column yielded a single peak that corresponded to an apparent molecular mass of 18 kDa, which is ∼2-fold larger than the theoretical molecular mass (date not shown), suggesting an elongated form in solution.
Further insight into the secondary structure was gleaned by examination of the CD spectra of the CD44 cytoplasmic peptide. The spectra obtained clearly suggested the absence of typical secondary structures, such as the α-helix and β-sheet, at a sample concentration of 0.1 mg/ml (13 μm). Titration of the FERM domain induced no significant spectral changes, suggesting that the CD44 peptide is present largely as a random coil without global secondary structural changes when bound to the FERM domain (Fig. 1D). Thus, the interaction between radixin and CD44 could be more appropriately described as a protein-peptide interaction rather than a protein-protein interaction. To verify that notion, we again performed analytical ultracentrifugation with the free and CD44-bound FERM domain. Sedimentation velocity measurements showed estimated molecular masses indicating monomers (Fig. 1F). Quantitative analysis revealed an increase of the sedimentation coefficient (2.82 S) by complex formation and suggested an a/b ratio of 8.2 (the f/f0 ratio of 1.65). This large ellipticity implies the lack of structure of most of the CD44 peptide in the complex, which is consistent with the lack of changes in CD spectra in the titration experiment.
Crystal Structure Determination—We set out to determine the crystal structure of the complex between the mouse radixin FERM domain (residues 1-310) and the CD44 peptide. As expected from the flexible nature of the CD44 cytoplasmic tail in solution, crystallization trials carried out using the full-length CD44 cytoplasmic peptide were unsuccessful. A previous biochemical study has shown that moesin binds 19- and 32-residue juxtamembrane regions of CD44 cytoplasmic tails, whereas deletion of the 19 juxtamembrane residues from the cytoplasmic tail almost completely abolished binding (13). Our quantitative binding assay using surface plasmon resonance measurements showed that both the marlin and radixin FERM domain bind a CD44 peptide comprising 23 juxtamembrane residues with dissociation constant (KD) values of 110 and 120 nm, respectively (Fig. 1E). These values are consistent with a reported quantitative binding assay involving surface plasmon resonance measurements using a longer (37-residue) CD44 peptide (28). Accordingly, the crystallization trials that followed made use of shorter CD44 juxtamembrane peptides of different lengths. We found that a FERM-CD44 complex crystal suitable for structure determination was obtained using the 20-residue juxtamembrane peptide of mouse CD44 (residues 293-312; sequence SRRRCGQKKKLVINGGNGTV). The peptide residues encompass the previously reported 19-residue region that was shown to directly interact with moesin (13). For convenience, the peptide residues are numbered from 2 to 21, corresponding to the 72 cytoplasmic residues. The crystals contained one FERM-CD44 complex in the asymmetric unit. The structure of the FERM-CD44 complex was determined by molecular replacement and subsequently refined to 2.1 Å resolution with an R-value of 23.1% (and a free R-value of 25.6%). The crystallographic statistics are summarized in Table 1. On the current electron density map, the CD44 peptide model contains 9 residues (positions 8-16) of 20. No models were built for two N-terminal and 13 C-terminal residues of the FERM domain, which were not observed in the electron density map.
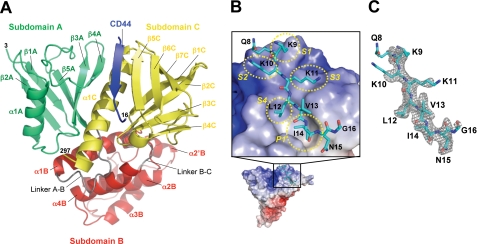
Structure of the Radixin FERM Domain in the Complex—The radixin FERM domain bound to CD44 comprises three subdomains: subdomain A (N-terminal residues 3-82; green) having a typical ubiquitin fold, subdomain B (residues 95-195; red) folded into an α-helix bundle, and subdomain C (residues 204-297; yellow) folded into a standard seven-stranded β-sandwich with a long capping α-helix (Fig. 2A). These structural characteristics are identical to previously reported structures of the radixin FERM domain (28, 29, 31, 37). The overall root mean square deviation between the FERM domain bound to the CD44 peptide and the free radixin FERM domain is 0.76 Å for 292 Cα carbon atoms (residues 5-296), suggesting no significant structural changes in the global structure.
FIGURE 2.
Crystal structure of the FERM-CD44 complex. A, ribbon representations of the radixin FERM domain complexed with the CD44 cytoplasmic peptide (blue). The radixin FERM domain comprises three subdomains: A (residues 3-82 in green), B (residues 96-195 in red), and C (residues 204-297 in yellow). Linkers A-B (residues 83-95) and B-C (residues 196-203) are colored gray. B, surface electrostatic potentials of the FERM domain and a close-up view of the CD44 cytoplasmic peptide docked into the groove formed between helix α1C and strand β5C of subdomain C. The peptide is shown as a stick model (labeled with one-letter codes), and the four side chain-binding sites (S1--S4) for the bound β strand of CD44 and the deep hydrophobic pocket (P1) are labeled and indicated with yellow dashed circles. Site S4 adjoins pocket P1. C, a stick model of the CD44 cytoplasmic peptide is shown in the omit electron density map for the CD44 cytoplasmic peptide at the contour level of 1σ.
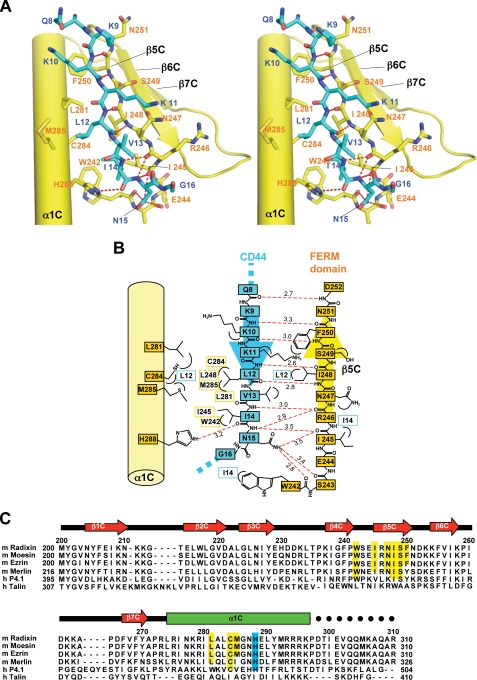
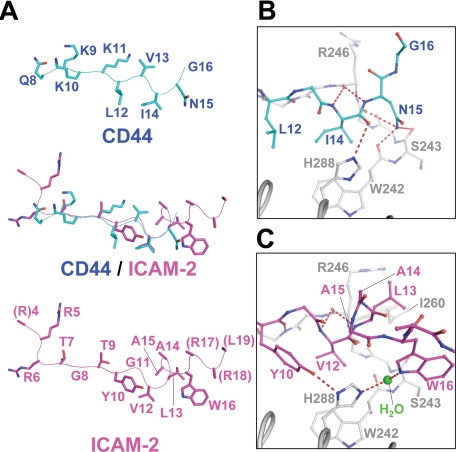
Structure of the CD44 Peptide and Its Recognition by the Radixin FERM Domain—The CD44 peptide binds subdomain C, whose fold could be classified as a phosphotyrosine-binding (PTB) domain (37) (Fig. 2A). The binding region of CD44 encompasses residues 8-16, which contains one of the basic clusters, KKK, followed by a nonpolar region (Fig. 2B). Along the hydrophobic shallow groove formed by helix α1C and strand β5C from subdomain C, the peptide forms a short β strand structure (residues 9-12; Lys-Lys-Lys-Leu), which augments the β sheet formed by strands β5C-β7C from subdomain C. The groove creates side chain binding sites, S1-S4, that interact with side chains of the CD44 peptide (Fig. 2B). The CD44 β strand forms five regular main chain-main chain hydrogen bonds with strand β5C (Fig. 3, A and B). A short loop structure (residues 13-16; Val-Ile-Asn-Gly) follows the β strand and docks into a pocket P1 connected to the hydrophobic groove. Two hydrophobic residues of the CD44 peptide, Leu12 and Ile14, position the aliphatic side chains into the deep hydrophobic S4 site and P1 pocket, respectively (Fig. 2B). The main chain of Ile14 is hydrogen-bonded to His288 from helix α1C. Importantly, the CD44 loop residues form three main chain-main chain hydrogen bonds with the end of strand β5C and the following loop β4C-β5C residues (Fig. 3, A and B). In addition to the main chain-main chain interactions, the side chain of CD44 Asn15 forms a hydrogen bond with the main-chain carbonyl group of Trp242 from loop β4C-β5C and probably with those of other loop residues (Ile245 and Ser243).
FIGURE 3.
CD44 binding to subdomain C of the FERM domain and sequence alignment of subdomain C of ERM and related proteins. A, the CD44 cytoplasmic peptide (blue stick model) binds the groove between helix α1C (yellow cylinder) and strand β5C (yellow stick) of subdomain C of the FERM domain. Hydrogen bonds are shown as red dashed lines. B, schematic representation of the interactions between the CD44 cytoplasmic peptide (blue) and subdomain C (yellow). The nonpolar contacts involving side chains are shown with contacted residues. Two hydrogen bonds formed by the main chain carbonyl group of FERM Ile245 with the main and side chains of CD44 Asn15 are relatively long (3.45 Å). C, sequence alignment of subdomain C of mouse (m) radixin, ezrin, moesin, human band 4.1 protein (h P 4.1), and talin (h Talin). The secondary structure of the radixin FERM subdomain C is shown at the top. Green rectangle, α-helix; red arrows, β-strands. Residues that participate in nonpolar and polar interactions with the CD44 peptide are highlighted in yellow and cyan, respectively.
The side chains of Gln8 and three lysines (Lys9, Lys10, and Lys11) of the β strand appear as partially disordered in the electron density map (Fig. 2C) and are obviously flexible, exposing the side chain end groups toward the solvent region in the absence of direct contacts with the FERM domain. Three lysines, however, have the aliphatic bases of their side chains positioned on the S1-S3 sites (Fig. 2B). Notably, the positively charged terminal groups are oriented toward the proposed plasma membrane (Fig. 4). The radixin residues that participate in interactions with the CD44 peptide are well conserved in other ERM members, such as ezrin and moesin, and merlin, suggesting that the observed binding mode in our structure could also be expected in the case of interactions with other ERM proteins as well as merlin (Fig. 3C).
FIGURE 4.
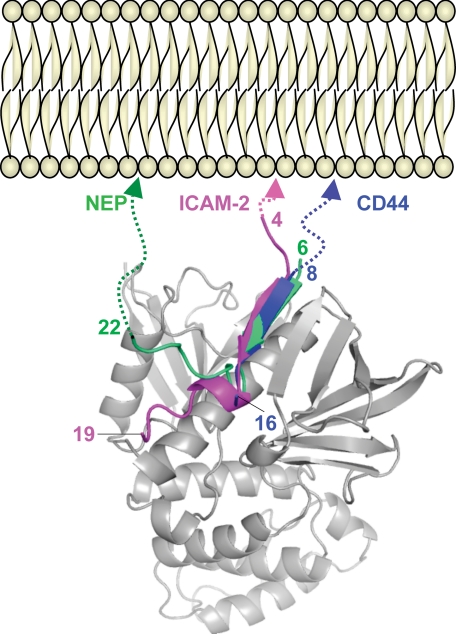
Comparison of ICAM-2, NEP, and CD44 peptides bound to the FERM domain. Shown is superposition of ICAM-2 (magenta) from the FERM-ICAM-2 complex (28) and NEP (green) from the FERM-NEP complex (31) on the FERM-CD44 complex. The N-terminal extensions of ICAM-2 and CD44 or the C terminus of NEP that would be linked to the transmembrane helix in the plasmamembrane are indicated with dotted lines.
Comparison with the FERM-ICAM-2 Complex—The CD44-binding site on subdomain C in our complex overlaps with that of the ICAM-2 peptide found in this FERM-ICAM-2 complex (28); both the CD44 and ICAM-2 peptides bind the groove between helix α1C and strand β5C of subdomain C and involve antiparallel β-β interactions with strand β5C (Fig. 4). This similarity was unexpected given the lack of sequence homology between the two peptides; the juxtamembrane region of CD44 contains two clusters of basic residues followed by a nonpolar region and a glycine-rich stretch, whereas that of ICAM-2 contains a Motif-1 nonpolar region that is sandwiched between two basic regions (Fig. 5A). Structure-based sequence alignment suggests that the QKKKLVINGG sequence of CD44 corresponds to the Motif-1 sequence of ICAM-2. With this alignment, CD44 replaces the Motif-1 RXXTY and XVXXA sequence stretches with QKKKL and XINGG sequences, respectively. Structural alignment revealed that conserved CD44 Ile14 corresponds to ICAM-2 Val12, and these 2 residues dock into the same P1 pocket and are completely overlapped (Fig. 6). Contrary to this excellent overlap, the side chain of CD44 Leu12 is shifted from that of ICAM-2 Tyr10, although both dock into the S4 site. This is a consequence of the site not being large enough to accommodate the large tyrosine side chain of ICAM-2. Therefore, ICAM-2 orients the side chain of Tyr10 toward His288 of the FERM domains and forms a hydrogen bond (Fig. 6C). The VXXA stretch of the ICAM-2 peptide forms a 310 helix, whereas the CD44 peptide fails to form a 310 helix, probably due to the glycine-rich sequence that displays structural flexibility. CD44 lacks an Ala residue that is essential for docking the 310 helix into the P1 pocket, which contributes toward stabilization of the helix. ICAM-2 possesses Leu13 instead of CD44 Asn15, which forms multiple hydrogen bonds to the main chains of loop β4C-β5C as described. In the FERM-ICAM-2 complex, ICAM-2 Leu13 is a component of the 310 helix and projects the side chain from the pocket toward the side chain of Ile260 from loop β6C-β7C of the FERM domain (Fig. 6C). The ICAM-2 310 helix also enables Trp16 to form a water-mediated hydrogen bond to His288. The large aromatic ring of Trp16 is located at the side of the P1 pocket and replaces Asn15 of CD44.
FIGURE 6.
Comparison of CD44 and ICAM-2 peptides bound to the radixin FERM domain. A, comparison of the FERM-bound peptide conformations of the CD44 (cyan) and ICAM-2 (magenta) peptides. Two peptide structures are overlaid in the middle. The side chains and main chains are shown as stick and line-tracing models, respectively. B, close-up view of the loop region of the CD44 cytoplasmic peptide (cyan) bound to the FERM domain (gray). The CD44 loop comprising Val13-Ile14-Asn15-Gly16 is docked into pocket P1 on subdomain C. Hydrogen bonds are shown as red dashed lines. The side chain of Val13 is not shown for clarity. CD44 Asn15 forms three hydrogen bonds to the main chain carbonyl groups of Trp242, Ser243, and Ile245 of the FERM domain. C, close-up view of the 310 helix of the ICAM-2 peptide (magenta) bound to the FERM domain (gray) in the FERM-ICAM-2 complex (28). The 310 helix comprising Val12-Leu13-Ala14-Ala15 is docked into pocket P1 on subdomain C. The side chain of Ala15 is not shown for clarity. The water molecule mediating the hydrogen bond between ICAM-2 Trp16 and FERM His288 is shown as a green sphere.
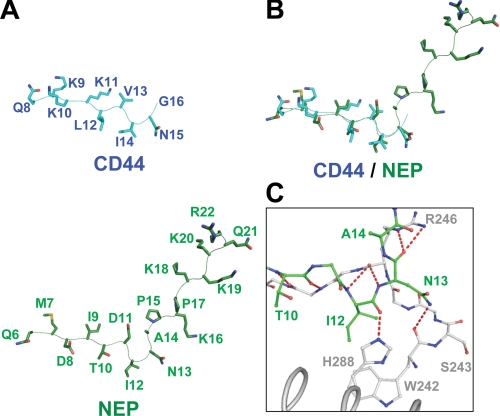
Comparison with the FERM-NEP Complex—Recently, our group reported on the crystal structures of the radixin FERM domain bound to the cytoplasmic peptide of type II membrane protein NEP (31). Despite the opposite chain polarity of the cytoplasmic tails, the NEP peptide binds the groove between helix α1C and strand β5C of subdomain C by forming antiparallel β-β associations with strand β5C that overlaps the observed binding site for the CD44 peptide of the current structure. Notably, structure alignment between the CD44 and NEP peptides reveals a better overlap of the corresponding side chain pairs than structural alignment between CD44 and ICAM-2 (Figs. 7, A and B). This unexpected close overlap is reflected by the CD44 Leu12 and NEP Thr10 pair and the CD44 Ile14 and NEP Ile12 pair, which bind the S4 site and P1 pocket, respectively, and the CD44 Asn15 and NEP Asn13 pair. In the case of NEP, the β strand formed by the MDIT sequence is followed by a hairpin-like structure of the DINA sequence (Fig. 5). The sharp hairpin structure of NEP slightly shifts the Asn13 side chain from a position occupied by CD44 Asn15 away from the FERM domain (Fig. 7B). This shift results in fewer hydrogen bonds being formed between the NEP Asn13 side chain and loop β6C-β7C of the FERM domain (Fig. 7C). Instead, the NEP hairpin is stabilized by formation of an additional hydrogen bond between the main chain of NEP Asn13 and the side chain of Arg246 from strand β5C (Fig. 7C). Notwithstanding the aforementioned deviations in the intermolecular interactions between the FERM-CD44 and FERM-NEP complexes, the overall binding mode of the CD44 peptide resembles more closely that of the NEP peptide rather than the ICAM-2 peptide. Our previous mutation studies based on the complexed structure identified the NEP signature sequence MXITXIN (Motif-1β), which is distinct from Motif-1 of ICAM-2 (31). The sequence of this motif is less conserved in CD44, whereas the Ile-Asn sequence is conserved in CD44 and plays a role in the binding as mentioned above (Fig. 5B).
FIGURE 7.
Comparison of CD44 and NEP peptides bound to the FERM domain. A, comparison of the FERM-bound peptide conformations of the CD44 (cyan) and NEP (green) peptides. The side chains and main chains are shown as stick and line-tracing models, respectively. B, two peptide structures are overlaid. C, close-up view of the hairpin of the NEP peptide (green) bound to the FERM domain (gray). The hairpin comprising Asp11-Ile12-Asn13-Ala14 is docked into pocket P1 on subdomain C. NEP Asn13 forms a hydrogen bond to the main chain carbonyl group of FERM Trp242.
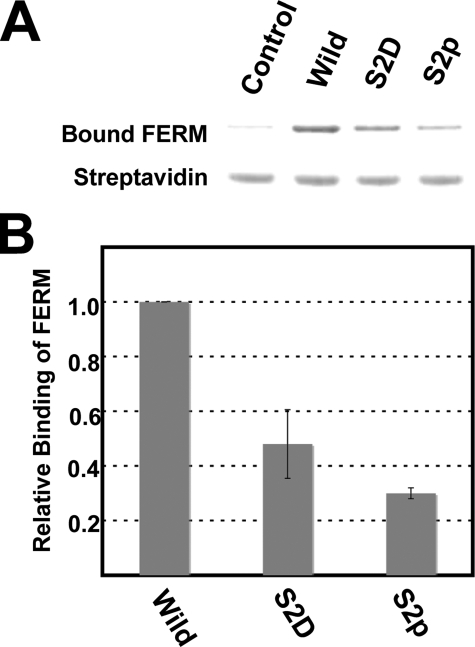
Influence of Ser2 Phosphorylation on the Binding of CD44 Peptide to the Radixin FERM Domain—Previously, Ser291 (Ser2 in our numbering) of the human CD44 cytoplasmic tail was found to be phosphorylated by protein kinase C (44). Interestingly, a point mutation to aspartic acid, which mimics the phosphorylated side chain, was also shown to reduce the interaction with ezrin in vitro using cell lysates and in vivo, as determined by fluorescence lifetime imaging microscopy. We attempted pull-down assays using purified radixin FERM domain and CD44 peptides to test whether phosphorylation of Ser2 interferes with the FERM-CD44 interaction. In our pull-down assay, the binding affinity of the radixin FERM domain to the CD44 peptide having an S2D mutation was reduced by 52%, and that of the radixin FERM domain to the Ser2-phosphorylated CD44 peptide was reduced by 70% in comparison with the affinity to wild-type CD44 peptide (Fig. 8), demonstrating that Ser2 phosphorylation indeed interferes with the interaction between the CD44 peptide and the radixin FERM domain, although Ser2 exhibits no direct interaction with the FERM domain in our structure.
FIGURE 8.
Pull-down assays of the FERM domain with wild-type and phosphorylated CD44. A, eluted samples analyzed by SDS-PAGE. Ser2 is replaced with Asp (S2D) and phosphoserine (S2p). B, summary of pull-down assays of wild-type and phosphorylated CD44 to the radixin FERM domain. The bar graph documents the relative binding affinity.
DISCUSSION
Our structural and biophysical characterization of the CD44 cytoplasmic peptide provides several clues concerning the physiological role of the CD44 cytoplasmic tail and ERM proteins. Our hydrodynamic studies clearly show that the CD44 cytoplasmic tail is present as an extended monomeric form in solution. Projection of the extended cytoplasmic tail from the inner plasma membrane allows for effective binding to multiple proteins containing ERM proteins, ankyrin, and guanine nucleotide exchange factors of the Rho family, such as Tiam1/2 (T-lymphoma invasion and metastasis 1 and 2) (45-47).
Our structure reveals that CD44 binds the same binding site on subdomain C of the FERM domain as that of Motif-1, whereas the CD44 sequence of the binding site, KKKLVIN, is distinct from Motif-1. This versatility of peptide recognition by subdomain C is in sharp contrast with that observed for the PTB domains. Most of the PTB domains recognize the NPXY (Y is usually a phosphotyrosine) motif (48, 49), and some recognize the GPY or QVTVS motifs (50, 51), whereas none of the PTB domains recognize both the NPXY and GPY or QVTVS motifs. In some other PTB domains, the peptide wraps around the domain by sitting on the β-sheet comprising β5-β6-β7-β1 strands (32, 51). However, no such interaction has been found to date in the FERM domains of ERM proteins and merlin.
Several factors influence the ability of CD44 to bind hyaluronan of extracellular matrix. These include the expression level of CD44 and posttranslational modifications, such as glycosylation of the extracellular domain. However, a frequently asked question with respect to CD44 activation concerns whether intracellular events can modulate ligand binding, referred to as “inside-out signaling.” Colocalization of CD44 with activated ERM proteins correlates with hyaluronan binding (24). This binding activity requires the CD44 cytoplasmic tail and its ERM-binding site (21, 24, 52). Interestingly, artificial dimerization abolishes this requirement, suggesting that the role of the cytoplasmic tail may be to promote CD44 clustering (53). Thus, it has been a fascinating question to consider whether CD44 possesses an intrinsic ability to form dimers and/or oligomers that contribute to localization and clustering, a process believed to have physiological importance in regulating hyaluronan binding avidity (54). Our hydrodynamic studies, however, are substantial enough for us to speculate that the cytoplasmic tail might possess no such ability to initiate dimerization or clustering by self-association. Clustering and oligomerization of CD44 are probably induced by interactions with ERM and other proteins that mediate a mechanical link of the tail to actin cytoskeletons. Notably, the extracellular hyaluronan-binding domain (HABD) of CD44 adopts a monomeric form, which is able to bind hyaluronan (55). We speculate that clustering of CD44 by ERM proteins could accelerate HABD dimerization, which facilitates increased hyaruronan binding. HABD dimerization is also induced by superagonist antibodies whose epitopes were mapped on the HABD surface (56).
As in the case of amyloid β precursor and Notch, it has been shown that CD44 is subject to regulated proteolytic cleavage via an RIP pathway to initiate the CD44-mediated intracellular signaling pathway (57). CD44 cleavage by presenilin-1/γ-secretase can generate the ICD fragment encompassing the whole cytoplasmic tail in addition to the secreted extracellular domain fragment (26, 58, 59). Translocation of the ICD fragment into the nucleus is an essential step for transcriptional activation, which provides a feedback mechanism for regulating CD44 expression (26). Since the CD44 cytoplasmic tail is anchored to actin cytoskeletons by binding to ERM proteins, the cleaved cytoplasmic tail should be released from ERM proteins prior to nuclear transport, implying that the RIP pathway of CD44 may be coupled with the regulation of ERM proteins. One possible mechanism of release from ERM proteins involves phosphorylation of Ser2 of the cytoplasmic tail by protein kinase C, which reduces CD44 binding to ERM proteins. Protein kinase C is activated by phorbol esters, and, in turn, the transcription of genes controlled by phorbol ester-responsive elements is mediated by the ICD fragment. Thus, positive feedback involving CD44 phosphorylation may regulate CD44 outside-in signaling. In our complex structure, Ser2 is located at the disordered N-terminal 6 residues and is not expected to be in direct contact with the FERM domain. However, we point out that Ser2 is located near the two basic clusters RRR and KKK and speculate that the negative charges of phosphorylated Ser2 would strongly interact with the positive charges of the basic residues (Fig. 5A). Since the second basic cluster KKK is part of the FERM-binding site, the postulated electrostatic interactions may destroy the KKK strand structure, thereby resulting in diminished CD44-FERM binding.
Another possible mechanism for ICD fragment release from the ERM protein-mediated link to cytoskeletons may involve inactivation of ERM proteins. ERM proteins adopt two states, masked and unmasked, and are inactive as a linker in the masked state (60-62). Unmasking is triggered at the plasma membrane by binding of phosphatidylinositol 4,5-bisphosphate to the FERM domain (37, 63, 64), which allows for subsequent phosphorylation at the C-terminal tail domain by Rho-kinase (65) or protein kinase C (66). All of these cues relating to the activation of ERM proteins, phosphatidylinositol 4,5-bisphosphate production induced by Ras, and phosphorylation by Rho-kinase and protein kinase C are in parallel with the stimulation of CD44 cleavage, since the reported experimental data show that CD44 cleavage is induced by the activation of Ras and Rho signaling as well as protein kinase C activation (67, 68). Thus, these signaling pathways would not contribute toward triggering ERM inactivation, and it is unlikely that selective inactivation of ERM proteins occurs during the RIP process.
CD44 is a major component of cartilage and modulates Smad1 activation in chondrocytes in embryonic and adult tissues (69). In this process, a functional link exists between CD44 and the signaling cascade of BMP-7 (bone morphogenetic protein-7), a member of the transforming growth factor-β superfamily (70-73). In the BMP-7 pathway, CD44 recruits Smad1 to transforming growth factor receptor, ALK2, and ActR-II at the plasmamembrane by direct binding. Since the Smad1-binding region is mapped to the C-terminal 54 residues of the CD44 cytoplasmic tail, there is no overlap between the ERM and Smad1 binding sites, suggesting that the CD44 cytoplasmic tail bound to ERM proteins is able to bind Smad1.
Another interesting viewpoint concerns the notion that transforming growth factor-β signaling is somehow functionally coupled with the RIP pathway of CD44, since both the IDC fragment and Smad1 act with p300/CBP to regulate transcriptional activation by bone morphogenetic proteins (26, 74). In this case, Smad1 bound to the CD44 ICD fragment may translocate into the nucleus and function as accessory modulators of transcriptional regulation. It has been shown that phosphorylated Smad1 is released from transforming growth factor receptor and binds Smad4, which then translocates into the nucleus. We speculate that the Smad1-CD44 ICD complex might bind Smad4 and cooperatively function as a transcriptional activator. Further investigations including the structural analysis of complexes will be required to address this issue.
In conclusion, our biophysical studies indicate that the 72-residue cytoplasmic region of CD44 is present as a flexible tail that possesses no intrinsic ability to self-associate to form dimers/oligomers. Crystal structure investigation of the FERM-CD44 complex reveals a distinct peptide binding mode of the radixin FERM domain compared with that of ICAM-2 and other Ig family adhesion molecules. Based on our structure and reported phosphorylation of the N-terminal Ser of the cytoplasmic tail, we suggest a possible mechanism by which CD44 is released from ERM-mediated links to the cytoskeleton for nuclear translocation in the RIP pathway. The identified FERM binding site is located away from the binding region for Smad1, which allows for Smad1 interactions with activated CD44 bound to ERM protein and linked to actin cytoskeletons.
Acknowledgments
We thank J. Tsukamoto for technical support in performing the matrix-assisted laser desorption/ionization time-of-flight mass spectroscopy analysis and Dr. S. Sakurai for calculation of the rotation function. We gratefully acknowledge Drs. Sachiko Tsukita and Shoichiro Tsukita for providing mouse radixin cDNA.
The atomic coordinates and structure factors (code 2ZPY) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
This work was supported in part by a grant-in-aid for scientific research (A) and scientific research on priority areas from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) of Japan (to T. H.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: RIP, regulated intramembrane proteolysis; ICAM, intercellular adhesion molecule; PTB, phosphotyrosine binding; ICD, intracellular domain; DTT, dithiothreitol; HABD, hyaluronan-binding domain.
References
- 1.Toole, B. P., Wight, T. N., and Tammi, M. I. (2002) J. Biol. Chem. 277 4593-4596 [DOI] [PubMed] [Google Scholar]
- 2.Tammi, M. I., Day, A. J., and Turley, E. A. (2002) J. Biol. Chem. 277 4581-4584 [DOI] [PubMed] [Google Scholar]
- 3.Bajorath, J. (2000) Proteins 39 103-111 [PubMed] [Google Scholar]
- 4.Lesley, J., Hyman, R., and Kincade, P. W. (1993) Adv. Immunol. 54 271-335 [DOI] [PubMed] [Google Scholar]
- 5.Martin, T. A., Harrison, G., Mansel, R. E., and Jiang, W. G. (2003) Crit. Rev. Oncol. Hematol. 46 165-186 [DOI] [PubMed] [Google Scholar]
- 6.Ponta, H., Sherman, L., and Herrlich, P. A. (2003) Nat. Rev. Mol. Cell. Biol. 4 33-45 [DOI] [PubMed] [Google Scholar]
- 7.Naor, D., Sionov, R. V., and Ish-Shalom, D. (1997) Adv. Cancer Res. 71 241-319 [DOI] [PubMed] [Google Scholar]
- 8.Cichy, J., and Pure, E. (2003) J. Cell Biol. 161 839-843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thorne, R. F., Legg, J. W., and Isacke, C. M. (2004) J. Cell Sci. 117 373-380 [DOI] [PubMed] [Google Scholar]
- 10.Tsukita, S., Oishi, K., Sato, N., Sagara, J., Kawai, A., and Tsukita, S. (1994) J. Cell Biol. 126 391-401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hirao, M., Sato, N., Kondo, T., Yonemura, S., Monden, M., Sasaki, T., Takai, Y., Tsukita, S., and Tsukita, S. (1996) J. Cell Biol. 135 37-51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sainio, M., Zhao, F., Heiska, L., Turunen, O., den Bakker, M., Zwarthoff, E., Lutchman, M., Rouleau, G. A., Jääskeläinen, J., Vaheri, A., and Carpén, O. (1997) J. Cell Sci. 110 2249-2260 [DOI] [PubMed] [Google Scholar]
- 13.Yonemura, S., Hirao, M., Doi, Y., Takahashi, N., Kondo, T., Tsukita, S., and Tsukita, S. (1998) J. Cell Biol. 140 885-895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsukita, S., and Yonemura, S. (1999) J. Biol. Chem. 274 34507-34510 [DOI] [PubMed] [Google Scholar]
- 15.McClatchey, A. I. (2003) Nat. Rev. Cancer 3 877-883 [DOI] [PubMed] [Google Scholar]
- 16.Bonilha, V. L., Finnemann, S. C., and Rodriguez-Boulan, E. (1999) J. Cell Biol. 147 1533-1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yonemura, S., Tsukita, S., and Tsukita, S. (1999) J. Cell Biol. 145 1497-1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zohar, R., Suzuki, N., Suzuki, K., Arora, P., Glogauer, M., McCulloch, C. A., and Sodek, J. (2000) J. Cell. Physiol. 184 118-130 [DOI] [PubMed] [Google Scholar]
- 19.Lee, J. H., Katakai, T., Hara, T., Gonda, H., Sugai, M., and Shimizu, A. (2004) J. Cell Biol. 167 327-337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cywes, C., and Wessels, M. R. (2001) Nature 414 648-652 [DOI] [PubMed] [Google Scholar]
- 21.Lesley, J., He, Q., Miyake, K., Hamann, A., Hyman, R., and Kincade, P. W. (1992) J. Exp. Med. 175 257-266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomas, L., Byers, H. R., Vink, J., and Stamenkovic, I. (1992) J. Cell Biol. 118 971-977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mori, H., Tomari, T., Koshikawa, N., Kajita, M., Itoh, Y., Sato, H., Tojo, H., Yana, I., and Seiki, M. (2002) EMBO J. 21 3949-3959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brown, K. L., Birkenhead, D., Lai, J. C., Li, L., Li, R., and Johnson, P. (2005) Exp. Cell Res. 303 400-414 [DOI] [PubMed] [Google Scholar]
- 25.Brown, M. S., Ye, J., Rawson, R. B., and Goldstein, J. L. (2000) Cell 100 391-398 [DOI] [PubMed] [Google Scholar]
- 26.Okamoto, I., Kawano, Y., Murakami, D., Sasayama, T., Araki, N., Miki, T., Wong, A. J., and Saya, H. (2001) J. Cell Biol. 155 755-762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bretscher, A., Edwards, K., and Fehon, R. G. (2002) Nat. Rev. Mol. Cell. Biol. 3 586-599 [DOI] [PubMed] [Google Scholar]
- 28.Hamada, K., Shimizu, T., Yonemura, S., Tsukita, S., Tsukita, S., and Hakoshima, T. (2003) EMBO J. 22 502-514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takai, Y., Kitano, K., Terawaki, S., Maesaki, R., and Hakoshima, T. (2007) Genes Cells 12 1329-1338 [DOI] [PubMed] [Google Scholar]
- 30.Terawaki, S., Maesaki, R., and Hakoshima, T. (2006) Structure 14 777-789 [DOI] [PubMed] [Google Scholar]
- 31.Terawaki, S., Kitano, K., and Hakoshima, T. (2007) J. Biol. Chem. 282 19854-19861 [DOI] [PubMed] [Google Scholar]
- 32.Wegener, K. L., Partridge, A. W., Han, J., Pickford, A. R., Liddington, R. C., Ginsberg, M. H., and Campbell, I. D. (2007) Cell 128 171-182 [DOI] [PubMed] [Google Scholar]
- 33.Hamada, K., Matsui, T., Tsukita, S., Tsukita, S., and Hakoshima, T. (2000a) Acta Crystallogr. Sect. D Biol. Crystallogr. 56 922-923 [DOI] [PubMed] [Google Scholar]
- 34.Mori, T., Kitano, K., Terawaki, S., Maesaki, R., and Hakoshima, T. (2007) Acta Crystallogr. F Struct. Biol. Crystalliz. Comm. 63 844-847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Otwinowski, Z., and Minor, W. (1997) Methods Enzymol. 276 307-326 [DOI] [PubMed] [Google Scholar]
- 36.McCoy, A. J., Grosse-Kunstleve, R. W., Storoni, L. C., and Read, R. J. (2005) Acta Crystallogr. Sect. D Biol. Crystallogr. 61 458-464 [DOI] [PubMed] [Google Scholar]
- 37.Hamada, K., Shimizu, T., Matsui, T., Tsukita, S., and Hakoshima, T. (2000) EMBO J. 19 4449-4462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jones, T. A., Zou, J. Y., Cowan, S. W., and Kjeldgaard, M. (1991) Acta Crystallogr. Sect. A 47 110-119 [DOI] [PubMed] [Google Scholar]
- 39.Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T., and Warren, G. L. (1998) Acta Crystallogr. Sect. D Biol. Crystallogr. 54 905-921 [DOI] [PubMed] [Google Scholar]
- 40.Pearson, M. A., Reczek, D., Bretscher, A., and Karplus, P. A. (2000) Cell 101 259-270 [DOI] [PubMed] [Google Scholar]
- 41.Shimizu, T., Seto, A., Maita, N., Hamada, K., Tsukita, S., Tsukita, S., and Hakoshima, T. (2002) J. Biol. Chem. 277 10332-10336 [DOI] [PubMed] [Google Scholar]
- 42.Davis, I. W., Leaver-Fay, A., Chen, V. B., Block, J. N., Kapral, G. J., Wang, X., Murray, L. W., Arendall, W. B., 3rd, Snoeyink, J., Richardson, J. S., and Richardson, D. C. (2007) Nucleic Acids Res. 35 W375-W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Collaborative Computational Project, N. (1994) Acta Crystallogr. Sect. D Biol. Crystallogr. 50 760-76315299374 [Google Scholar]
- 44.Legg, J. W., Lewis, C. A., Parsons, M., Ng, T., and Isacke, C. M. (2002) Nat. Cell Biol. 4 399-407 [DOI] [PubMed] [Google Scholar]
- 45.Lokeshwar, V. B., and Bourguignon, L. Y. (1991) J. Biol. Chem. 266 17983-17989 [PubMed] [Google Scholar]
- 46.Bourguignon, L. Y., Zhu, H., Shao, L., and Chen, Y. W. (2000) J. Biol. Chem. 275 1829-1838 [DOI] [PubMed] [Google Scholar]
- 47.Oliferenko, S., Kaverina, I., Small, J. V., and Huber, L. A. (2000) J. Cell Biol. 148 1159-1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eck, M. J., Dhe-Paganon, S., Trüb, T., Nolte, R. T., and Shoelson, S. E. (1996) Cell 85 695-705 [DOI] [PubMed] [Google Scholar]
- 49.Garcia-Alvarez, B., de Pereda, J. M., Calderwood, D. A., Ulmer, T. S., Critchley, D., Campbell, I. D., Ginsberg, M. H., and Liddington, R. C. (2003) Mol. Cell 11 49-58 [DOI] [PubMed] [Google Scholar]
- 50.Li, S.-C., Zwahlen, C., Vincent, S. J., McGlade, C. J., Kay, L. E., Pawson, T., and Forman-Kay, J. D. (1998) Nat. Struct. Biol. 5 1075-1083 [DOI] [PubMed] [Google Scholar]
- 51.Dhalluin, C., Yan, K. S., Plotnikova, O., Lee, K. W., Zeng, L., Kuti, M., Mujtaba, S., Goldfarb, M. P., and Zhou, M. M. (2000) Mol. Cell 6 921-929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu, D., Zhang, D., Mori, H., and Sy, M.-S. (1996) Cell. Immunol. 174 73-83 [DOI] [PubMed] [Google Scholar]
- 53.Perschl, A., Lesley, J., English, N., Trowbridge, I., and Hyman, R. (1995) Eur. J. Immunol. 25 495-501 [DOI] [PubMed] [Google Scholar]
- 54.Lesley, J., Hascall, V., Tammi, M., and Hyman, R. (2000) J. Biol. Chem. 275 26967-26975 [DOI] [PubMed] [Google Scholar]
- 55.Banerji, S., Wright, A. J., Noble, M., Mahoney, D. J., Campbell, I. D., Day, A. J., and Jackson, D. G. (2007) Nat. Struct. Mol. Biol. 14 234-239 [DOI] [PubMed] [Google Scholar]
- 56.Teriete, P., Banerji, S., Noble, M., Blundell, C. D., Wright, A. J., Pickford, A. R., Lowe, E., Mahoney, D. J., Tammi, M. I., Kahmann, J. D., Campbell, I. D., Day, A. J., and Jackson, D. G. (2004) Mol. Cell 13 483-496 [DOI] [PubMed] [Google Scholar]
- 57.Nagano, O., and Saya, H. (2004) Cancer Sci. 95 930-935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lammich, S., Okochi, M., Takeda, M., Kaether, C., Capell, A., Zimmer, A. K., Edbauer, D., Walter, J., Steiner, H., and Haass, C. (2002) J. Biol. Chem. 277 44754-44759 [DOI] [PubMed] [Google Scholar]
- 59.Murakami, D., Okamoto, I., Nagano, O., Kawano, Y., Tomita, T., Iwat-subo, T., de Strooper, B., Yumoto, E., and Saya, H. (2003) Oncogene 22 1511-1516 [DOI] [PubMed] [Google Scholar]
- 60.Gary, R., and Bretscher, A. (1995) Mol. Biol. Cell 6 1061-1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ishikawa, H., Tamura, A., Matsui, T., Sasaki, H., Hakoshima, T., Tsukita, S., and Tsukita, S. (2001) J. Mol. Biol. 310 973-978 [DOI] [PubMed] [Google Scholar]
- 62.Li, Q., Nance, M. R., Kulikauskas, R., Nyberg, K., Fehon, R., Karplus, P. A., Bretscher, A., and Tesmer, J. J. (2007) J. Mol. Biol. 365 1446-1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fievet, B. T., Gautreau, A., Roy, C., Del Maestro, L., Mangeat, P., Louvard, D., and Arpin, M. (2004) J. Cell Biol. 164 653-659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yonemura, S., Matsui, T., Tsukita, S., and Tsukita, S. (2002) J. Cell Sci. 115 2569-2580 [DOI] [PubMed] [Google Scholar]
- 65.Matsui, T., Maeda, M., Doi, Y., Yonemura, S., Amano, M., Kaibuchi, K., Tsukita, S., and Tsukita, S. (1998) J. Cell Biol. 140 647-657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pietromonaco, S. F., Simons, P. C., Altman, A., and Elias, L. (1998) J. Biol. Chem. 273 7594-7603 [DOI] [PubMed] [Google Scholar]
- 67.Okamoto, I., Kawano, Y., Tsuiki, H., Sasaki, J., Nakao, M., Matsumoto, M., Suga, M., Ando, M., Nakajima, M., and Saya, H. (1999) Oncogene 18 1435-1446 [DOI] [PubMed] [Google Scholar]
- 68.Kawano, Y., Okamoto, I., Murakami, D., Itoh, H., Yoshida, M., Ueda, S., and Saya, H. (2000) J. Biol. Chem. 275 29628-29635 [DOI] [PubMed] [Google Scholar]
- 69.Peterson, R. S., Andhare, R. A., Rousche, K. T., Knudson, W., Wang, W., Grossfield, J. B., Thomas, R. O., Hollingsworth, R. E., and Knudson, C. B. (2004) J. Cell Biol. 166 1081-1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Moustakas, A., Souchelnytskyi, S., and Heldin, C.-H. (2001) J. Cell Sci. 114 4359-4369 [DOI] [PubMed] [Google Scholar]
- 71.Tsukazaki, T., Chiang, T. A., Davison, A. F., Attisano, L., and Wrana, J. L. (1998) Cell 95 779-791 [DOI] [PubMed] [Google Scholar]
- 72.Wu, G., Chen, Y. G., Ozdamar, B., Gyuricza, C. A., Chong, P. A., Wrana, J. L., Massagué, J., and Shi, Y. (2000) Science 287 92-97 [DOI] [PubMed] [Google Scholar]
- 73.Sasaki, A., Masuda, Y., Ohta, Y., Ikeda, K., and Watanabe, K. (2001) J. Biol. Chem. 276 17871-17877 [DOI] [PubMed] [Google Scholar]
- 74.Pouponnot, C., Jayaraman, L., and Massagué, J. (1998) J. Biol. Chem. 273 22865-22868 [DOI] [PubMed] [Google Scholar]