Abstract
The amyloid β-protein (Aβ), which accumulates abnormally in Alzheimer disease (AD), is degraded by a diverse set of proteolytic enzymes. Aβ-cleaving proteases, largely ignored until only recently, are now known to play a pivotal role in the regulation of cerebral Aβ levels and amyloid plaque formation in animal models, and accumulating evidence suggests that defective Aβ proteolysis may be operative in many AD cases. This review summarizes the growing body of evidence supporting the involvement of specific Aβ-cleaving proteases in the etiology and potential treatment of AD. Recognition of the importance of Aβ degradation to the overall economy of Aβ has revised our thinking about the mechanistic basis of AD pathogenesis and identified a novel class of enzymes that may serve as both therapeutic targets and therapeutic agents.
The hallmark feature of AD2 is the progressive accumulation of aggregated forms of Aβ in brain regions subserving mnemonic and cognitive functions (1). Aβ is a heterogeneous mixture of peptides ranging in size from 37 to 43 amino acids and is excised from APP by proteases known as β- and γ-secretases (1). Aβ production is normally counterbalanced by its elimination via multiple interrelated processes acting in concert, including proteolytic degradation, cell-mediated clearance, active and passive transport out of the brain, and deposition into insoluble aggregates. Although each of these processes contributes to Aβ catabolism, emerging evidence suggests that proteolytic degradation is a particularly important regulator of cerebral Aβ levels and, by extension, AD pathogenesis.
The hypothesis that Aβ plays a causal role in triggering the full spectrum of pathological and behavioral sequelae characterizing AD, once hotly disputed, gained considerable support from analysis of familial forms of AD. Mutations in three separate genes, App and presenilin-1 and -2, were identified that produced a common phenotype consisting of increased production of Aβ, either all forms or specifically the longer, more amyloidogenic forms such as Aβ42 (2). However, there is scant direct evidence that increased Aβ production underlies the vast majority of non-familial forms of AD (3). These facts suggest that reduced degradation of Aβ may represent an alternative cause of many, possibly even most, AD cases.
Despite the obvious appeal of this simple idea, widespread interest in Aβ degradation did not take hold until the turn of the 21st century (4). A key turning point was the publication of a seminal study by Saido and co-workers, the first to examine Aβ degradation in the living animal (5). In addition to identifying NEP as an important Aβ-degrading protease, this study also served to highlight the significance of Aβ degradation to AD pathogenesis generally, thereby igniting interest in a previously underappreciated aspect of Aβ metabolism.
Subsequent growth in this field has been so great that it is now impossible to comprehensively survey even the most seminal papers in a review of this length. A large number of candidate Aβ-degrading proteases have been identified to date (Table 1), and the list will surely grow in coming years. More significant still is the impressive list of conceptual insights that are continuing to emerge from the study of Aβ degradation (4). Accordingly, the primary goal of this review is to convey the principal conceptual advances and to critically evaluate what we have learned from different experimental paradigms. For a more comprehensive discussion of specific Aβ-degrading proteases, the reader is referred to several excellent reviews (6–9).
TABLE 1.
Subcellular localizations of selected Aβ-degrading proteases
ER, endoplasmic reticulum.
|
Protease
|
Class
|
Location
|
|||||
|---|---|---|---|---|---|---|---|
| Extracellular | ER/Golgi | Lysosomes | Cytosol | Mitochondria | Peroxisomes | ||
| NEP | Metallo | + | + | ||||
| ECE-1 | Metallo | + | + | ||||
| ECE-2 | Metallo | + | + | ||||
| MMP-2 | Metallo | + | + | ||||
| MMP-9 | Metallo | + | + | ||||
| IDE | Metallo | + | + | + | + | ||
| PreP | Metallo | + | |||||
| Plasmin | Serine | + | |||||
| CatB | Cysteine | + | + | ||||
Specific Aβ-cleaving Proteases
Zinc Metalloproteases—Most known Aβ-degrading proteases are zinc metalloproteases, which can in turn be sub-divided into vasopeptidases, MMPs, and homologs of IDE (Table 1).
The vasopeptidases, which include NEP, ECE-1, ECE-2, and ACE, are so named because they are implicated in the processing of vasoactive peptides, but they also hydrolyze other substrates involved in diverse physiological functions (10, 11). The vasopeptidases are type 2 integral membrane proteins, with their active site facing the extracellular and/or lumenal space (11), making them well positioned to degrade secreted forms of Aβ (7, 12).
MMPs are related to vasopeptidases, sharing a conserved zinc-binding motif (HEXXH), but they differ in several important respects (13). First, they exist as latent proenzymes that must be proteolytically processed to become fully active (13). Second, their basal expression is low but can be stimulated by pathological insults, including Aβ itself (14). Third, they are optimized for the processing of proteins as opposed to peptides (13) and, as discussed below, show a greater ability to degrade fibrillar forms of Aβ than the vasopeptidases (14).
IDE and a recently identified homolog, PreP (15), belong to a separate superfamily of zinc metalloproteases with distinct evolutionary origins referred to as “inverzincins” because they feature a zinc-binding motif (HXXEH) that is inverted with respect to the canonical one (6). Although functionally similar to vasopeptidases in showing a preference for peptide substrates, IDE and its homologs differ substantially in terms of their subcellular localization (Table 1). IDE is unique among all known Aβ-degrading proteases in being localized to the cytosol and peroxisomes, and both PreP and IDE are also targeted to mitochondria (15, 16), with PreP exclusively so. Although Aβ is not generated within the latter compartments, there is growing evidence that Aβ can nevertheless accumulate in mitochondria and possibly other organelles (17). Like most other Aβ-degrading proteases, IDE is also present in the extracellular space in both secreted and cell-associated forms, although the underlying trafficking pathway remains obscure (6, 16).
Serine Proteases—Three functionally related serine proteases are implicated in Aβ degradation: plasmin, uPA, and tPA. Of these, only plasmin has been shown to directly degrade Aβ and, like MMPs, can degrade both monomeric and fibrillar forms (18, 19). tPA and uPA are, however, responsible for converting the inactive zymogen of plasmin (plasminogen) into its active form (20). tPA is notable because it is stimulated by fibrillar proteins, including Aβ (18). uPA is of interest because of genetic evidence linking uPA to late-onset AD (21).
Cysteine Proteases—Cysteine proteases were initially implicated in Aβ degradation by in vivo pharmacological studies (22). However, only one cysteine protease, CatB, has so far been specifically implicated in the degradation of Aβ in vivo (23). Interestingly, CatB is predominantly present within the endolysosomal protein degradation pathway (24), which is known to degrade Aβ and to be compromised in AD (25). However, enzymatically active CatB is also secreted in certain pathological conditions (24) and is associated with amyloid plaques (23). CatB is notable for having dipeptidyl carboxypeptidase activity, rendering it capable of cleaving Aβ42 to shorter, less amyloidogenic peptides (23).
Categories of Investigation
In Vitro Evidence—At first glance, in vitro studies would seem to be the least informative route to identifying physiologically and pathophysiologically relevant Aβ-degrading proteases. Ironically though, most proteases now validated in vivo were initially discovered through in vitro approaches many years earlier, including (in order of discovery) IDE, MMP-2, MMP-9, NEP, plasmin, and ACE (4, 26). In vitro paradigms have been especially useful for distinguishing Aβ proteases in terms of substrate specificity. Whereas all known Aβ-degrading proteases can cleave monomeric Aβ, aggregated forms can be degraded only by a more limited set. Fibrillar Aβ is degraded by MMP-2 and MMP-9 (27), plasmin (28), and CatB (23). Aβ oligomers naturally secreted from cells (29) or derived from treatment of synthetic Aβ with transglutaminase, both of which potently disrupt long-term potentiation, are readily cleaved by plasmin but not by NEP or IDE (29, 30). On the other hand, oligomers formed non-enzymatically from synthetic Aβ are avidly degraded by NEP (31). Future studies on this topic would appear to offer a novel window into the important question of whether different oligomer preparations are in fact equivalent.
Mass spectrometry has been widely employed to identify the specific peptide bonds within Aβ cleaved by different proteases (32). Based on these analyses, it is often assumed that specific cleavage sites can be used to predict the involvement of individual proteases. However, it is becoming increasingly likely that this assumption is invalid as the list of known Aβ-degrading proteases (and the cleavages effected by each) continues to expand.
Cell Culture Studies—Experiments using cultured cells have been instrumental in identifying several physiologically relevant Aβ-degrading proteases. As shown initially by Selkoe and co-workers (33), IDE appears to be the major Aβ-cleaving protease secreted into the medium of a wide range of cultured cells. Confirming these initial findings, primary neurons cultured from IDE knock-out mice showed >90% reductions in the initial rate of degradation of physiological levels of exogenously applied Aβ (34). However, quite a different picture emerges when one considers the effects of Aβ-cleaving proteases when expressed in cells actively producing Aβ. Using this paradigm, several proteases, including NEP, ECE-1, ACE, have been shown to markedly decrease net levels of secreted Aβ (8). Pharmacological studies performed in cultured cells have also shown that multiple proteases, principally zinc metalloproteases, are present within the secretory pathway and normally catabolize substantial amounts of Aβ prior to its secretion (35, 36).
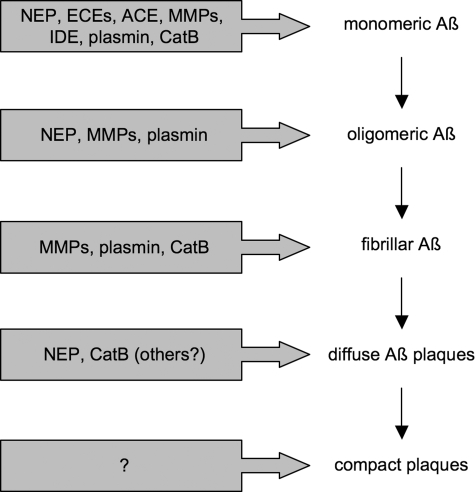
IDE is involved in degrading Aβ from intracellular sites as well. Recalling that IDE cannot degrade cell-derived Aβ oligomers once they are formed, it is notable that overexpression of IDE nonetheless lowers the net production of oligomers by cultured cells (37). This finding is of special importance both pathologically and therapeutically because it positions Aβ degradation upstream (as well as downstream) of Aβ aggregation (Fig. 1).
FIGURE 1.
Action of different Aβ-cleaving proteases on monomeric and aggregated forms of Aβ.
Animal Modeling Studies—Animal models have been indispensable for establishing the in vivo relevance of Aβ-cleaving proteases. Some of the earliest and most significant studies used pharmacological inhibitors to interrogate the importance of Aβ degradation in vivo, yielding mixed results. For example, an early study tested the effect of different inhibitors on the deposition of Aβ administered intracerebroventricularly to rats (22). Although some enhancement of amyloid deposition and associated cytopathology was observed, the impact of this study was limited as the inhibitors used were relatively nonselective. The seminal study of Saido and co-workers (5) also used pharmacological inhibitors in vivo, in this case testing their ability to slow the degradation of radiolabeled Aβ42 superfused into rat hippocampus. The significance of the latter study over the former lay in the fact that it implicated a specific Aβ-degrading protease, NEP. This conclusion was based on the observation that thiorphan, an inhibitor thought to be selective for NEP, was the most effective from among several inhibitors tested at slowing Aβ degradation (5). This same study showed that chronic intracerebroventricular administration of thiorphan led to the development of amyloid plaques in normal rats.
Ironically, the main conclusion of this study, that NEP is the major Aβ42-degrading protease in vivo, was in fact predicated on an erroneous assumption, that thiorphan is selective for NEP. That this is not the case is evident from the subsequent finding that, in the absence of overexpressed APP, convincing evidence of amyloid deposition is not observed in rodents genetically engineered to lack NEP (38) or even those lacking NEP together with other known Aβ-degrading proteases (39). Contrariwise, this same study also concluded that IDE did not play role in vivo based on the finding that insulin, a competitive inhibitor of IDE, failed to slow the degradation of Aβ in this superfusion paradigm (5). In this case, the failure of insulin to slow Aβ catabolism likely had less to do with the lack of involvement of IDE than it did with deficiencies of the inhibitor, insulin (34). As this analysis shows, it is generally inadvisable to infer the involvement of specific proteases solely on the basis of effects produced by pharmacological inhibitors.
Pharmacological studies in animals should not be universally proscribed, however, as the following counterexample illustrates. Concerns have been raised recently that ACE inhibitors, widely prescribed for the treatment of hypertension, could increase the risk for AD by inhibiting ACE-mediated Aβ degradation (39). Here, it was entirely appropriate to test ACE inhibitors in animal models of AD. In this particular case, several independent studies failed to show any effect of ACE inhibitors on Aβ accumulation (39, 40). Together with other findings from knock-out animals (39), these pharmacological studies were instrumental in establishing that ACE is not important for Aβ degradation in vivo (at least in rodents). In general, in vivo pharmacological testing should be considered appropriate in cases in which the independent variable is the drug itself rather than the drug's inferred target.
Analysis of protease knock-out mice is widely regarded as the preferred method of determining whether or not a given protease is relevant to Aβ degradation in vivo. Mice lacking one or both alleles of NEP, ECE-2, MMP-2, MMP-9, or IDE or one allele of ECE-1 have all been shown to have significant elevations in endogenous cerebral Aβ levels (7, 27). These findings highlight two key interrelated points: (i) that multiple Aβ-degrading proteases act in parallel to regulate steady-state Aβ levels and (ii) that Aβ degradation pathways are normally fully engaged, with no reserve capacity. However, the merits of quantifying steady-state Aβ levels in protease knock-out mice need to be tempered by several important qualifications. First, this approach will not necessarily identify all relevant proteases, particularly those that are operative only in a pathological context. Significant increases in brain Aβ levels have not been observed in mice lacking plasminogen, tPA, uPA, or CatB (23, 41), yet evidence from other paradigms supports the involvement of each (7, 23, 42). Second, measurements of brain-wide Aβ levels fail to account for differential effects on distinct pools of Aβ, some of which might be more pathogenic than others (4). This point is especially relevant given substantial differences in the regional and, perhaps more importantly, subcellular distributions of individual proteases (Table 1). Third, knock-out mice are prone to compensatory changes that might influence Aβ levels indirectly. For example, IDE knock-out mice develop an age-dependent diabetic phenotype (34), raising the question of whether elevated Aβ levels result from the absence of IDE per se instead of or in addition to the resulting compensatory changes.
Crosses of protease knock-out mice with APP transgenic mice have only recently begun to emerge. As predicted from the prior analysis, proteases that do not affect endogenous Aβ levels have been shown to exert significant changes in amyloid plaque formation. For example, deletion of CatB in APP transgenic mice led to increases in thioflavin-positive plaque formation while showing no significant changes in steady-state Aβ levels (23). In other cases, qualitative rather than merely quantitative changes have emerged. For example, NEP knock-out mice crossed with APP transgenic mice were unexpectedly found to develop cerebral amyloid angiopathy (43). The higher levels of Aβ in APP transgenic mice have also permitted more sophisticated analyses not feasible in the absence of APP overexpression. An especially interesting development is the use of microdialysis to measure interstitial Aβ levels in real time in vivo. This approach was recently used to monitor Aβ levels in APP transgenic mice with or without NEP in real time (43); critically, by administering a γ-secretase inhibitor to halt ongoing Aβ production, it was possible to monitor the clearance of Aβ in real time. This analysis is significant because it represents the first direct measurement of the influence of a protease on Aβ catabolism itself rather than on a surrogate marker such as steady-state Aβ levels. Interestingly, although altered significantly, the half-life of Aβ was changed only incrementally in absolute terms in the absence of NEP (43). Using the same microdialysis paradigm, a similar result was obtained with a broad-spectrum MMP inhibitor (27). These results underscore the point that specific proteases or even whole classes of proteases contribute only fractionally to the overall catabolism of Aβ.
Aβ-cleaving proteases have also been overexpressed in APP transgenic mice using a range of approaches, yielding a bounty of fresh insights. Transgenic overexpression of IDE by 1-fold produced a >50% reduction in steady-state Aβ levels, amyloid plaque burden, and associated cytopathology and also reduced premature lethality present in APP transgenic mice (30). In addition, 7-fold overexpression of NEP reduced Aβ levels by >90% and eliminated plaque formation entirely (30). These results are impressive given that Aβ levels are many orders of magnitude higher in this animal model relative to non-transgenic mice and illustrate the important point that Aβ-degrading proteases act catalytically to remove Aβ.
Viral overexpression paradigms have permitted the investigation of the effects of Aβ-degrading proteases not only on on-going amyloid deposition but also on pre-existing amyloid deposits (44). Such studies have shown that pre-existing amyloid deposits can be reduced to a certain degree by protease treatment, even by peptidases such as NEP, implying that plaques may be more labile than previously thought (44).
Whereas therapies based on blocking secretase activity must necessarily act locally to affect Aβ production, therapies based on increasing Aβ catabolism can, in principle, act at sites widely separated from the sites of Aβ production. Illustrating this point, Hemming et al. (45) recently showed that Aβ accumulation in APP transgenic mice could be attenuated by transplantation of murine astrocytes engineered to overexpress NEP. Significantly, reductions in Aβ were observed not merely adjacent to the transplanted astrocytes but at distal sites as well (45).
Human Studies—Analyses of post-mortem human brain tissue have lent additional credibility to the hypothesis that defects in specific Aβ-cleaving proteases may underlie some cases of AD. Multiple studies have documented reductions in NEP or IDE protein levels in an age- and brain region-dependent manner (7, 8). Significantly, oxidative damage to these proteases has also been demonstrated in some cases (9, 46). Collectively, these studies implicate impaired Aβ degradation as a plausible mechanism linking the risk of AD to aging and other known environmental risk factors.
Human molecular genetic studies represent another large category of analysis linking Aβ-degrading proteases to AD pathogenesis. In general, positive studies have emerged implicating specific Aβ-degrading proteases, including IDE, NEP, uPA, and ECE-1 (8), only to be followed by studies that alternately confirm or confute the original studies. To date, AD-causing missense mutations affecting specific proteases have not been definitively demonstrated. This result may be a reflection of the larger number of processes involved in Aβ catabolism vis-à-vis Aβ production. Reductions in Aβ catabolism might be caused by large defects in individual catabolic processes, but the stochastically more probable scenario is the accrual of multiple, more subtle changes to multiple catabolic processes, which is more difficult to detect by genetic analysis.
If some cases of AD are in fact attributable to defects in Aβ degradation, this may provide new ways of diagnosing the disease and/or detecting it early. For example, mass spectrometric profiling of Aβ catabolites in cerebrospinal fluid has been reported to distinguish AD patients from age-matched controls with high selectivity and specificity (47). Another promising line of research is the development of methods to monitor Aβ catabolism in real time in humans (48). Using these and other approaches, it is possible to envision an era in which individual AD cases can be ascribed to different underlying etiologies and then treated with therapeutics tailored to address the biochemical defect(s) specific to each.
Concluding Remarks
Testifying to the rapid development of this field, two recently published studies deserve special mention. Jiang et al. (49) have provided evidence that apoE promotes the proteolytic degradation of Aβ in an isoform-specific manner, with the AD risk-associated apoEε4 isoform showing a relative deficiency in this function. Given the strong influence of apoE status in determining risk for AD, this intriguing finding suggests that altered Aβ degradation may be operative in a very large number of AD cases. A second study (50) describes the development of a novel drug that promotes plasmin-mediated Aβ degradation and is effective in lowering Aβ levels and reversing memory defects in animal models. The drug, developed by Wyeth, works by inhibiting plasminogen activator inhibitor 1, an endogenous inhibitor that normally prevents the conversion of inactive plasminogen to plasmin. This work represents an important step for the field because it demonstrates that Aβ-cleaving proteases can in fact be modulated pharmacologically, a finding that will hopefully encourage the development of other drugs targeting Aβ degradation. Future work will surely identify new Aβ-cleaving proteases and provide new insights into the many ways they impact the pathogenesis, detection, and treatment of AD.
Supplementary Material
This is the sixth article of eleven in the Thematic Minireview Series on the Molecular Basis of Alzheimer Disease. This minireview will be reprinted in the 2008 Minireview Compendium, which will be available in January, 2009.
Footnotes
The abbreviations used are: AD, Alzheimer disease; Aβ, amyloid β-protein; APP, amyloid precursor protein; NEP, neprilysin; MMP, matrix metalloproteinase; IDE, insulin-degrading enzyme; ECE, endothelin-converting enzyme; ACE, angiotensin-converting enzyme; PreP, presequence protease; uPA, urokinase-type plasminogen activator; tPA, tissue-type plasminogen activator; CatB, cathepsin B.
References
- 1.Selkoe, D. J., and Schenk, D. (2003) Annu. Rev. Pharmacol. Toxicol. 43 545–584 [DOI] [PubMed] [Google Scholar]
- 2.Selkoe, D. J. (1997) Science 275 630–631 [DOI] [PubMed] [Google Scholar]
- 3.Selkoe, D. J. (2001) Neuron 32 177–180 [DOI] [PubMed] [Google Scholar]
- 4.Leissring, M. A. (2006) Curr. Alzheimer Res. 3 431–435 [DOI] [PubMed] [Google Scholar]
- 5.Iwata, N., Tsubuki, S., Takaki, Y., Watanabe, K., Sekiguchi, M., Hosoki, E., Kawashima-Morishima, M., Lee, H. J., Hama, E., Sekine-Aizawa, Y., and Saido, T. C. (2000) Nat. Med 6 143–150 [DOI] [PubMed] [Google Scholar]
- 6.Hersh, L. B. (2006) CMLS Cell. Mol. Life Sci. 63 2432–2434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leissring, M. A., and Saido, T. C. (2007) in Alzheimer's Disease: Advances in Genetics, Molecular and Cellular Biology (Sisodia, S., and Tanzi, R., eds) pp. 157–178, Springer-Verlag New York Inc., New York
- 8.Turner, A. J., and Nalivaeva, N. N. (2007) Int. Rev. Neurobiol. 82 113–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang, D. S., Dickson, D. W., and Malter, J. S. (2006) J. Biomed. Biotechnol. 2006 58406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roques, B. P. (1998) Pathol. Biol. 46 191–200 [PubMed] [Google Scholar]
- 11.Turner, A. J., Isaac, R. E., and Coates, D. (2001) BioEssays 23 261–269 [DOI] [PubMed] [Google Scholar]
- 12.Turner, A. J., and Nalivaeva, N. N. (2006) Cell. Mol. Biol. (Noisy-Le-Grand) 52 40–48 [PubMed] [Google Scholar]
- 13.Vartak, D. G., and Gemeinhart, R. A. (2007) J. Drug Target. 15 1–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miners, J. S., Baig, S., Palmer, J., Palmer, L. E., Kehoe, P. G., and Love, S. (2008) Brain Pathol. 18 240–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Falkevall, A., Alikhani, N., Bhushan, S., Pavlov, P. F., Busch, K., Johnson, K. A., Eneqvist, T., Tjernberg, L., Ankarcrona, M., and Glaser, E. (2006) J. Biol. Chem. 281 29096–29104 [DOI] [PubMed] [Google Scholar]
- 16.Leissring, M. A., Farris, W., Wu, X., Christodoulou, D. C., Haigis, M. C., Guarente, L., and Selkoe, D. J. (2004) Biochem. J. 383 439–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen, X., and Yan, S. D. (2006) IUBMB Life 58 686–694 [DOI] [PubMed] [Google Scholar]
- 18.Van Nostrand, W. E., and Porter, M. (1999) Biochemistry 38 11570–11576 [DOI] [PubMed] [Google Scholar]
- 19.Tucker, H. M., Kihiko-Ehmann, M., Wright, S., Rydel, R. E., and Estus, S. (2000) J. Neurochem. 75 2172–2177 [DOI] [PubMed] [Google Scholar]
- 20.Myohanen, H., and Vaheri, A. (2004) CMLS Cell. Mol. Life Sci. 61 2840–2858 [DOI] [PubMed] [Google Scholar]
- 21.Serretti, A., Olgiati, P., and De Ronchi, D. (2007) J. Alzheimer's Dis. 12 73–92 [DOI] [PubMed] [Google Scholar]
- 22.Frautschy, S. A., Horn, D. L., Sigel, J. J., Harris-White, M. E., Mendoza, J. J., Yang, F., Saido, T. C., and Cole, G. M. (1998) J. Neurosci. 18 8311–8321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mueller-Steiner, S., Zhou, Y., Arai, H., Roberson, E. D., Sun, B., Chen, J., Wang, X., Yu, G., Esposito, L., Mucke, L., and Gan, L. (2006) Neuron 51 703–714 [DOI] [PubMed] [Google Scholar]
- 24.Mort, J. S., and Buttle, D. J. (1997) Int. J. Biochem. Cell Biol. 29 715–720 [DOI] [PubMed] [Google Scholar]
- 25.Glabe, C. (2001) J. Mol. Neurosci. 17 137–145 [DOI] [PubMed] [Google Scholar]
- 26.Hu, J., Igarashi, A., Kamata, M., and Nakagawa, H. (2001) J. Biol. Chem. 276 47863–47868 [DOI] [PubMed] [Google Scholar]
- 27.Yin, K. J., Cirrito, J. R., Yan, P., Hu, X., Xiao, Q., Pan, X., Bateman, R., Song, H., Hsu, F. F., Turk, J., Xu, J., Hsu, C. Y., Mills, J. C., Holtzman, D. M., and Lee, J. M. (2006) J. Neurosci. 26 10939–10948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tucker, H. M., Kihiko, M., Caldwell, J. N., Wright, S., Kawarabayashi, T., Price, D., Walker, D., Scheff, S., McGillis, J. P., Rydel, R. E., and Estus, S. (2000) J. Neurosci. 20 3937–3946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walsh, D. M., Klyubin, I., Fadeeva, J. V., Cullen, W. K., Anwyl, R., Wolfe, M. S., Rowan, M. J., and Selkoe, D. J. (2002) Nature 416 535–539 [DOI] [PubMed] [Google Scholar]
- 30.Leissring, M. A., Farris, W., Chang, A. Y., Walsh, D. M., Wu, X., Sun, X., Frosch, M. P., and Selkoe, D. J. (2003) Neuron 40 1087–1093 [DOI] [PubMed] [Google Scholar]
- 31.Kanemitsu, H., Tomiyama, T., and Mori, H. (2003) Neurosci. Lett. 350 113–116 [DOI] [PubMed] [Google Scholar]
- 32.Eckman, E. A., and Eckman, C. B. (2005) Biochem. Soc. Trans. 33 1101–1105 [DOI] [PubMed] [Google Scholar]
- 33.Qiu, W. Q., Ye, Z., Kholodenko, D., Seubert, P., and Selkoe, D. J. (1997) J. Biol. Chem. 272 6641–6646 [DOI] [PubMed] [Google Scholar]
- 34.Farris, W., Mansourian, S., Chang, Y., Lindsley, L., Eckman, E. A., Frosch, M. P., Eckman, C. B., Tanzi, R. E., Selkoe, D. J., and Guenette, S. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 4162–4167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eckman, E. A., Reed, D. K., and Eckman, C. B. (2001) J. Biol. Chem. 276 24540–24548 [DOI] [PubMed] [Google Scholar]
- 36.White, A. R., Du, T., Laughton, K. M., Volitakis, I., Sharples, R. A., Xilinas, M. E., Hoke, D. E., Holsinger, R. M., Evin, G., Cherny, R. A., Hill, A. F., Barnham, K. J., Li, Q. X., Bush, A. I., and Masters, C. L. (2006) J. Biol. Chem. 281 17670–17680 [DOI] [PubMed] [Google Scholar]
- 37.Vekrellis, K., Ye, Z., Qiu, W. Q., Walsh, D., Hartley, D., Chesneau, V., Rosner, M. R., and Selkoe, D. J. (2000) J. Neurosci. 20 1657–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iwata, N., Tsubuki, S., Takaki, Y., Shirotani, K., Lu, B., Gerard, N. P., Gerard, C., Hama, E., Lee, H. J., and Saido, T. C. (2001) Science 292 1550–1552 [DOI] [PubMed] [Google Scholar]
- 39.Eckman, E. A., Adams, S. K., Troendle, F. J., Stodola, B. A., Kahn, M. A., Fauq, A. H., Xiao, H. D., Bernstein, K. E., and Eckman, C. B. (2006) J. Biol. Chem. 281 30471–30478 [DOI] [PubMed] [Google Scholar]
- 40.Hemming, M. L., Selkoe, D. J., and Farris, W. (2007) Neurobiol. Dis. 26 273–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tucker, H. M., Simpson, J., Kihiko-Ehmann, M., Younkin, L. H., McGillis, J. P., Younkin, S. G., Degen, J. L., and Estus, S. (2004) Neurosci. Lett. 368 285–289 [DOI] [PubMed] [Google Scholar]
- 42.Melchor, J. P., Pawlak, R., and Strickland, S. (2003) J. Neurosci. 23 8867–8871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Farris, W., Schutz, S. G., Cirrito, J. R., Shankar, G. M., Sun, X., George, A., Leissring, M. A., Walsh, D. M., Qiu, W. Q., Holtzman, D. M., and Selkoe, D. J. (2007) Am. J. Pathol. 171 241–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marr, R. A., Guan, H., Rockenstein, E., Kindy, M., Gage, F. H., Verma, I., Masliah, E., and Hersh, L. B. (2004) J. Mol. Neurosci. 22 5–11 [DOI] [PubMed] [Google Scholar]
- 45.Hemming, M. L., Patterson, M., Reske-Nielsen, C., Lin, L., Isacson, O., and Selkoe, D. J. (2007) PLoS Med. 4 e262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Neant-Fery, M., Garcia-Ordonez, R. D., Logan, T. P., Selkoe, D. J., Li, L., Reinstatler, L., and Leissring, M. A. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 9582–9587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Portelius, E., Zetterberg, H., Andreasson, U., Brinkmalm, G., Andreasen, N., Wallin, A., Westman-Brinkmalm, A., and Blennow, K. (2006) Neurosci. Lett. 409 215–219 [DOI] [PubMed] [Google Scholar]
- 48.Bateman, R. J., Wen, G., Morris, J. C., and Holtzman, D. M. (2007) Neurology 68 666–669 [DOI] [PubMed] [Google Scholar]
- 49.Jiang, Q., Lee, C. Y., Mandrekar, S., Wilkinson, B., Cramer, P., Zelcer, N., Mann, K., Lamb, B., Willson, T. M., Collins, J. L., Richardson, J. C., Smith, J. D., Comery, T. A., Riddell, D., Holtzman, D. M., Tontonoz, P., and Landreth, G. E. (2008) Neuron 58 681–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jacobsen, J. S., Comery, T. A., Martone, R. L., Elokdah, H., Crandall, D. L., Oganesian, A., Aschmies, S., Kirksey, Y., Gonzales, C., Xu, J., Zhou, H., Atchison, K., Wagner, E., Zaleska, M. M., Das, I., Arias, R. L., Bard, J., Riddell, D., Gardell, S. J., Abou-Gharbia, M., Robichaud, A., Magolda, R., Vlasuk, G. P., Bjornsson, T., Reinhart, P. H., and Pangalos, M. N. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 8754–8759 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.