Abstract
Phosphoenolpyruvate carboxylase (PEPC) is a tightly regulated enzyme situated at the core of plant C-metabolism. Although its anaplerotic role and control by allosteric effectors, reversible phosphorylation, and oligomerization have been well documented in the endosperm of developing castor oil seeds (COS), relatively little is known about PEPC in germinating COS. The initial phase of COS germination was accompanied by elevated PEPC activity and accumulation of comparable amounts of pre-existing 107-kDa and inducible 110-kDa immunoreactive PEPC polypeptides (p107 and p110, respectively). A 440-kDa PEPC heterotetramer composed of an equivalent ratio of non-phosphorylated p110 and p107 subunits was purified from germinated COS. N-terminal microsequencing, mass spectrometry, and immunoblotting revealed that both subunits arose from the same gene (RcPpc3) that encodes the p107 subunit of a phosphorylated 410-kDa PEPC homotetramer in developing COS but that p110 is a monoubiquitinated form of p107. Tandem mass spectrometry sequencing of a diglycinated tryptic peptide identified Lys-628 as p110's monoubiquitination site. This residue is conserved in vascular plant PEPCs and is proximal to a PEP-binding/catalytic domain. Incubation with a human deubiquitinating enzyme (USP-2 core) converted the p110:p107 PEPC heterotetramer into a p107 homotetramer while significantly reducing the enzyme's Km(PEP) and sensitivity to allosteric activators (hexose-Ps, glycerol-3-P) and inhibitors (malate, aspartate). Monoubiquitination is a non-destructive and reversible post-translational modification involved in the control of diverse processes such as transcription, endocytosis, and signal transduction. The current study demonstrates that tissue-specific monoubiquitination of a metabolic enzyme can also occur and that this modification influences its kinetic and regulatory properties.
Phosphoenolpyruvate (PEP)3 carboxylase (PEPC; EC 4.1.1.31) is an important enzyme situated at a major branch point in plant C-metabolism that catalyzes the irreversible β-carboxylation of PEP to yield oxaloacetate and Pi. PEPC has been intensively studied with regards to its key function in catalyzing the initial fixation of atmospheric CO2 in C4 and crassulacean acid metabolism photosynthesis (1, 2). PEPC also plays essential roles in bacteria and non-green plant cells, particularly the anaplerotic replenishment of tricarboxylic acid cycle intermediates withdrawn for biosynthesis and N-assimilation (3). Most vascular plant PEPCs exist as a homotetrameric “dimer-of-dimer” structure comprised of four identical 100–110-kDa subunits and are controlled by a combination of allosteric effectors and reversible phosphorylation (1, 2). Allosteric activation by glucose-6-P and inhibition by l-malate have routinely been observed, whereas reversible phosphorylation at a conserved N-terminal seryl residue is catalyzed by a Ca2+-independent PEPC protein kinase and protein phosphatase type 2A. PEPC phosphorylation activates the enzyme by reducing sensitivity to malate inhibition while simultaneously enhancing activation by hexose-Ps (1–3).
The molecular and functional properties of two classes of PEPC sharing the same 107-kDa subunit have been investigated in detail from the triglyceride-rich endosperm of developing castor oil seeds (COS) (4–9). Class-1 PEPC is a typical 410-kDa homotetramer composed of 107-kDa plant-type PEPC subunits (p107, encoded by RcPpc3; GenBank™ accession: EF634317), whereas the native Class-2 PEPC 910-kDa heterooligomeric complex arises from a tight interaction between Class-1 PEPC and unrelated 118-kDa bacterial-type PEPC polypeptides (p118, encoded by RcPpc4; GenBank accession: EF634318) that are also phosphorylated in vivo (4, 5, 8, 9). The reversible in vivo seryl phosphorylation of the p107 subunit of Class-1 PEPC in response to sucrose supply indicated that PEPC (p107) protein kinase contributes to the control of carbohydrate partitioning in developing COS at the level of the cytosolic PEP branch point (6, 7). Distinctive Class-1 versus Class-2 PEPC developmental profiles during COS filling, together with Class-2 PEPC's marked insensitivity to allosteric effectors relative to Class-1 PEPC, led to the hypotheses that: (i) Class-1 and Class-2 PEPC respectively support glycolytic carbon flux required for storage protein and storage lipid synthesis in developing COS and (ii) the p118 bacterial-type PEPC functions as a regulatory subunit within the Class-2 PEPC complex (4, 8). Fully mature and germinating COS endosperm contain immunoreactive plant-type PEPC polypeptides that co-migrate with the p107 of developing COS PEPC. However, an additional PEPC polypeptide of ∼110-kDa (p110) appears immediately following COS imbibition and persists throughout germination at an equivalent ratio with the p107 (8, 10). Here we demonstrate that RcPPC3 (p107) subunits become monoubiquitinated at a conserved Lys residue during the initial stages of COS germination, resulting in formation of an unusual p110:p107 Class-1 PEPC heterotetramer. Incubation with a mammalian deubiquitinating enzyme (DUB) converted the p110:p107 heterotetramer into a p107 homotetramer while exerting significant effects on the enzyme's PEP saturation kinetics and response to allosteric effectors. Monoubiquitination has recently emerged as reversible post-translational modification (PTM) involved in the control of diverse processes such as gene expression and endocytosis (11, 12). To our knowledge, however, there have been no reports describing regulatory monoubiquitination of a metabolic enzyme in any biological system studied to date.
EXPERIMENTAL PROCEDURES
Plant Material—Developing or germinating seeds from castor (Ricinus communis L., cv. Baker 296) plants were obtained as described previously (8). Dissected endosperm (free of cotyledon) was frozen in liquid N2 and stored at –80 °C until used.
Enzyme and Protein Assays and Kinetic Studies—PEPC activity was assayed at 340 nm using a Molecular Devices Spectramax Plus microplate reader as described previously (9). One unit of activity is defined as the amount of PEPC catalyzing the formation of 1 μmol of oxaloacetate/min at 25 °C. Protein concentrations were determined using Coomassie Blue G-250 dye binding method using bovine γ-globulin as the protein standard (6). Km, Ka, and I50 values were calculated using the Brooks (13) computer kinetics program. All kinetic parameters are the means of at least four separate experiments and are reproducible to within ±10% (S.E.) of the mean value. Stock solutions of metabolites were made equimolar with MgCl2 and adjusted to pH 7.5.
PEPC Purification—All procedures were performed at 4 °C except DEAE-Fractogel and phenyl-Superose FPLC, which were conducted at room temperature. All buffers contained 50 mm Hepes-KOH (pH 8.0), 5 mm MgCl2, 1 mm EDTA, 1 mm dithiothreitol, 5 mm l-malate, 20 mm NaF, and 2 mm 2,2′-dipyridyl disulfide in addition to the following. Buffer A contained 20% (v/v) glycerol, 1 mm EGTA, 20 mm NaF, 0.1% (v/v) Triton X-100, 4% (w/v) PEG 8000, 5 mm thiourea, 1% (w/v) insoluble poly(vinylpolypyrrolidone), 5 mm NaPPi, 2 mm phenylmethylsulfonyl fluoride, and 0.5 mm Na(VO3)4. Buffer B contained 1 mm EGTA and 20% (saturation) (NH4)2SO4. Buffer C was buffer B lacking (NH4)2SO4 but containing 10% (v/v) ethylene glycol. Buffer D contained 15% (v/v) glycerol, 1 mm EGTA, and 100 mm KCl.
Endosperm (300 g) from 4-day germinated COS was homogenized (1:2; w/v) in buffer A using a Polytron. After centrifugation, the supernatant was filtered through two layers of Miracloth. PEG 8000 (50% (w/v) in 50 mm Hepes-KOH, pH 8.0) was added to the solution to a final concentration of 20% (w/v) and stirred at 4 °C for 15 min. After centrifugation, PEG pellets were resuspended in buffer B (lacking (NH4)2SO4) to a final protein concentration of about 30 mg/ml. After centrifugation, a solution of (NH4)2SO4 (100% (saturation), pH 8.0) was added to the supernatant to 20% (saturation). The solution was stirred for 10 min, centrifuged, and absorbed batchwise onto 125 ml of butyl-Sepharose 4 Fast Flow equilibrated with buffer B and packed at 4.5 ml/min into a 3.2-cm diameter column. The column was washed at 5 ml/min with buffer B until the A280 approached base line. PEPC was eluted by 50% buffer C (50% buffer B) (10 ml/fraction). Pooled peak fractions were mixed with an equal volume of 50% (w/v) PEG 8000 and incubated overnight on ice. Following centrifugation, PEG pellets were dissolved in buffer D to a protein concentration of 15 mg/ml, recentrifuged, and loaded at 1.5 ml/min onto a column (1.1 × 13 cm) of Fractogel EMD DEAE-650 (S) connected to anÄKTA FPLC system and equilibrated with buffer D. The column was washed with buffer D, and PEPC activity was eluted with a 100–500 mm KCl gradient (117 ml) in buffer D (3 ml/fraction). Pooled peak fractions were concentrated to 2.5 ml using an Amicon Ultra-15 centrifugal filter unit (30-kDa cutoff), adjusted to 25% (saturation) (NH4)2SO4, and applied at 0.5 ml/min onto a Protein Pak phenyl-5PW column (0.8 × 7.5 cm) pre-equilibrated with buffer B. PEPC was eluted following application of a linear gradient (60 ml) of 0–100% buffer C (100-0% buffer B) (0.5 ml/fraction). Pooled peak fractions were concentrated to 0.4 ml as above, divided into 25-μl aliquots, and stored at –80 °C. PEPC activity was stable for at least 4 months when stored frozen. In some instances, the final preparation was subjected to gel filtration FPLC at 0.25 ml/min on a calibrated Superose-6 HR 10/30 column (4) equilibrated with buffer D.
Electrophoresis and Immunoblotting—SDS and non-denaturing PAGE, subunit and native Mr estimates via SDS- and non-denaturing-PAGE, and immunoblotting were performed as described previously (4, 8). Gels were stained for total protein using Coomassie Blue R-250 or Sypro Red, for phosphoproteins using Pro-Q Diamond (Molecular Probes) or for PEPC activity using the Fast Violet method (4). Sypro-Red- and Pro-Q Diamond-stained gels were scanned using a Typhoon 8600 fluorescence imager (GE Healthcare). For second dimension PAGE, PEPC activity staining bands were excised from a non-denaturing gel and subjected to SDS-PAGE and immunoblotting as described (14). Antigenic polypeptides were visualized using an alkaline phosphatase-conjugated secondary antibody and chromogenic detection (5). All gel and immunoblot results were replicated a minimum of three times, with representative results shown in the various figures.
N-terminal Sequencing—This was performed by automated Edman degradation at the Sheldon Biotechnology Centre (McGill University, Montréal QC, Canada). Similarity searches were performed using the BLAST program available on the NCBI web site (www.ncbi.nlm.nih.gov).
Mass Spectrometry—Excised gel bands were destained with 100 mm NH4HCO3/acetonitrile (ACN) (1:1, v/v) until colorless, dehydrated in 100% ACN, and dried using a SpeedVac. Following reduction with 10 mm dithiothreitol (in 100 mm NH4HCO3) at 56 °C for 1 h, proteins were alkylated with an equal volume of 55 mm iodoacetamide at 23 °C for another 45 min. Subsequent digestion was performed using 10 ng of sequencing grade trypsin (Calbiochem) in 25 mm NH4HCO3 (pH 7.6) at 37 °C overnight. The resulting peptides were extracted by successive sonication with 0.1% trifluoroacetic acid, 0.1% trifluoroacetic acid in 50% ACN, and 100% ACN followed by a C18 ZipTip (Millipore) cleanup step. The peptides were deposited on a matrix-assisted laser desorption ionization (MALDI) target by mixing with an equal volume (0.5 μl) of 2,5-dihydroxybenzoic acid matrix (100 mg/ml in 50% ACN). MALDI data were acquired using an Applied Biosystems/MDS Sciex QStar XL Q-TOF mass spectrometer equipped with an oMALDI II source and a nitrogen laser operating at 337 nm. Peptide sequencing of the selected ions was carried out by MALDI quadrupole time-of-flight (QqTOF) MS/MS measurements using argon as the collision gas. All peptide fingerprinting masses and MS/MS ion searches were performed with the Mascot search engine (MatrixScience) using the NCBI non-redundant data base (NCBInr released 10/10/2007; containing 5,507,867 protein sequences). These searches take into account one missed cleavage and the modifications carbamidomethylation, asparagine and glutamine deamidation to aspartate acid and glutamic acid, as well as N-terminal pyroglutamation and methionine oxidation. The mass tolerance between calculated and observed masses used for the data base search was considered at the range of ±100 ppm for mass spectrometry (MS) peaks and ±0.1 Da for MS/MS fragment ions. Ubiquitin (UB) conjugation site identification was performed as described by Ref. 37 using MS/MS analyses of both tagged (sulfonated) and non-tagged tryptic peptides. N-terminal sulfonation was performed with 10 mg/ml sulfophenyl isothiocyanate in 20 mm NaHCO3 (pH 9.0) (15). Following C18 Ziptip cleanup, MS/MS analysis of sulfonated peptides by low energy collision induced dissociation yields a set of C-terminal y fragment ions in the positive ionization mode, which enabled a reliable and rapid sequence identification of the UB-conjugation site.
Deubiquitination—All DUBs were obtained from Progenra Inc. (Malvern, PA). Purified PEPC (6 μg) was incubated at 37 °C for up to 60 min with 1 μm of each DUB in 20 mm Tris-HCl (pH 7.5), 50 mm KCl, 5 mm MgCl2, 20% (v/v) glycerol, and 2 mm dithiothreitol (final volume = 50-μl).
Statistics—Data were analyzed using the Student's t test and deemed significant if p < 0.05.
RESULTS AND DISCUSSION
Clarified Extract Studies—Preliminary experiments corroborated earlier reports (8, 10) that: (i) PEPC increases following imbibition from about 0.25 units/g of fresh weight in ungerminated COS endosperm to a maximum of about 0.5 units/g of fresh weight 3–4 days later, (ii) this is paralleled by the accumulation of comparable amounts of immunoreactive 110- and 107-kDa plant-type PEPC polypeptides (p110 and p107, respectively) (supplemental Fig. S1A), and (iii) a single Class-1 PEPC activity staining or immunoreactive band is obtained following non-denaturing PAGE of clarified COS extracts (supplemental Fig. S2, A and B). The p110 and p107 were detected in an equal ratio when the 440-kDa PEPC activity staining band obtained following non-denaturing PAGE of a germinated COS extract was denatured and subjected to second dimension SDS-PAGE and immunoblotting (supplemental Fig. S2C). However, no anti-(p107 phosphorylation-site specific)-IgG (6) immunoreactive polypeptides were detected on immunoblots of clarified germinated COS extracts (supplemental Fig. S1B), suggesting that p110 and p107 were not phosphorylated at PEPC's conserved N-terminal seryl residue. This contrasts with the phosphorylated 410-kDa Class-1 PEPC p107 homotetramer from developing COS (supplemental Fig. S1B) (6, 7), as well as PEPCs from germinated cereal (wheat, sorghum, and barley) seeds (16–18).
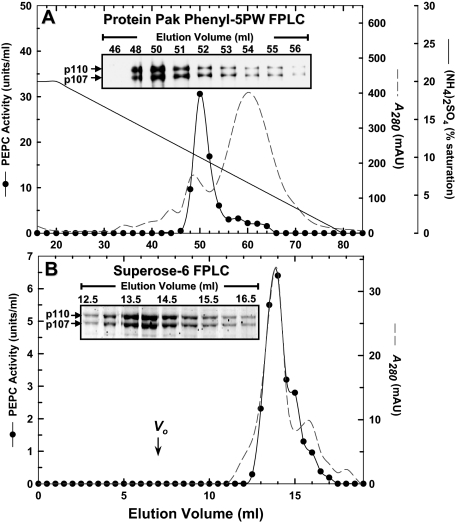
PEPC Purification from Germinating Castor Oil Seeds—About 6 mg of PEPC from 4-day-old germinating COS endosperm was purified 1,920-fold and a final specific activity of 9.6 units/mg with an overall yield of 37% (Table 1). The single peak of PEPC activity that was resolved during all chromatography steps co-eluted with a 1:1 ratio of immunoreactive or protein-staining p110 and p107 (Figs. 1 and 2). Although purified to near homogeneity (Fig. 2), the final preparation demonstrated no detectable phosphorylation when subjected to SDS-PAGE followed by Pro-Q Diamond phosphoprotein staining or anti-(p107 phosphorylation site specific)-IgG immunoblotting (not shown). The enzyme's native Mr as estimated by FPLC on a calibrated Superose-6 column (Fig. 1) was 440 ± 10 kDa (mean ± S.E., n = 3). Thus, the native PEPC exists as a non-phosphorylated heterotetramer comprised of an equivalent ratio of p110 and p107 subunits. Similar to the Class-1 PEPC homotetramer from developing COS (4), the purified enzyme was relatively heat-labile, losing 100% of its activity when preincubated at 55 °C for 3 min.
TABLE 1.
Purification of PEPC from 300 g of 4-day-old germinating COS endosperm
| Step | Activity | Protein | Specific activity | Purification | Yield |
|---|---|---|---|---|---|
| units | mg | units/mg | -fold | % | |
| Clarified extract | 143 | 29,700 | 0.005 | 1 | 100 |
| PEG fractionation | 186 | 16,355 | 0.011 | 2 | 130 |
| Butyl-Sepharose | 117 | 887 | 0.13 | 26 | 82 |
| DEAE-Fractogel | 109 | 61 | 1.8 | 360 | 76 |
| Protein Pak Phenyl-5PW | 53 | 5.5 | 9.6 | 1,920 | 37 |
FIGURE 1.
Co-elution of PEPC activity with 110- and 107-kDa PEPC polypeptides (p110 and p107, respectively) during Protein Pak Phenyl-5PW and Superose-6 FPLC of PEPC from germinating COS endosperm. A, Protein Pak Phenyl-5PW elution profile. Inset, aliquots (1-μl each) from various fractions were subjected to SDS-PAGE and immunoblotting using anti-(COS p107)-IgG (5). B, Superose-6 HR 10/30 elution profile. Vo denotes the column's void volume. Inset, aliquots (5-μl each) from various fractions were subjected to SDS-PAGE followed by protein staining with Sypro-Red.
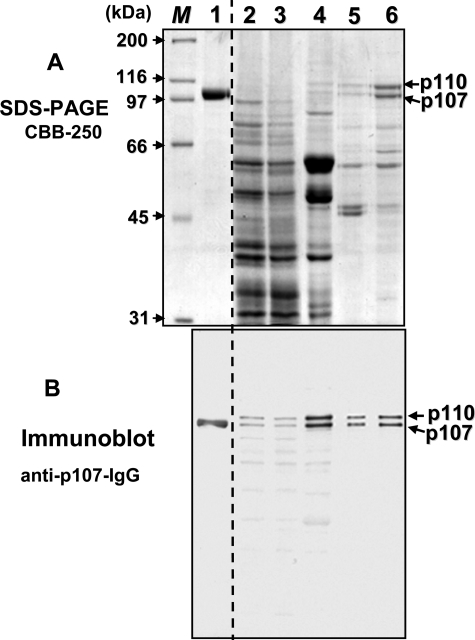
FIGURE 2.
SDS-PAGE and immunoblot analysis of various fractions obtained during the purification of germinating COS PEPC. A, SDS-PAGE was followed by protein staining with Coomassie Blue R-250 (CBB-250). B, immunoblot analysis was performed using anti-(COS p107)-IgG (5). Lane 1 contains 2.5 μg (A) or 50 ng (B) of homogeneous, non-proteolyzed, Class-1 PEPC (p107) from stage VII (full cotyledon) developing COS endosperm (4, 5). Lanes 2 and 3 contain 45 (A) or 15 μg (B) of protein from the clarified extract and 4–20% PEG fractions, respectively. Lane 4 contains 4 (A) or 2 μg (B) of the butyl-Sepharose fraction. Lanes 5 and 6 contain 2 (A) or 0.1 μg (B) of the DEAE-Fractogel and Protein Pak Phenyl-5PW fractions, respectively. M denotes various protein Mr standards.
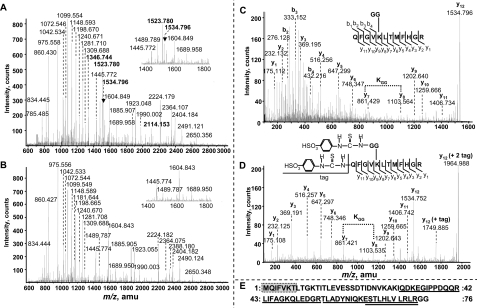
Mass Spectrometry and N-terminal Sequencing—Coomassie Blue R-250-stained p110 and p107 were excised from SDS gels of the final preparation and subjected to in-gel tryptic digestion, MALDI QqTOF MS peptide mapping (Fig. 3, A and B, and supplemental Table S1) and N-terminal microsequencing (Figs. 3E and 4). The results demonstrate that both subunits originated from the same gene (RcPpc3) that encodes the p107 subunit of the Class-1 PEPC homotetramer from developing COS and whose transcripts are abundant in both developing and germinated COS endosperm (8). Apart from displaying virtually identical peptide mass fingerprints (Fig. 3, A and B, and supplemental Table S1), p110 and p107 also shared an identical N-terminal sequence (VPAKVSE) (Fig. 4). Comparison with RcPPC3's cDNA-deduced sequence indicated that both subunits were N-terminally truncated by 19 amino acid residues (Fig. 4). This was likely due to a specific in vivo post-translational processing since: (i) 2 mm 2,2′-dipyridyl disulfide (included in all purification buffers) negates in vitro truncation of RcPPC3's N terminus by endogenous thiol proteases (5) and (ii) p110 and p107 were both observed on immunoblots of clarified extracts prepared under denaturing conditions in the presence of 10% trichloroacetic acid (supplemental Fig. S1) (10). As RcPPC3's N-terminal phosphorylation site is removed a result of this processing (Fig. 4), this provides a rationale for the absence of detectable p110 or p107 phosphorylation in germinated COS PEPC. By contrast, the N terminus of the p107 subunit of the phosphorylated Class-1 PEPC of developing COS is truncated by only 5 amino acids and thus retains its conserved N-terminal seryl phospho-site (Fig. 4) (5, 6, 8). Differential in vivo processing of PEPC's N terminus may represent an additional, but little studied, PTM that influences the enzyme's functional and regulatory properties. Interestingly, a second N-terminal sequence (MQIFVKT) was obtained when the p110 subunit of the purified germinated COS PEPC was analyzed by Edman degradation. Analysis of this sequence demonstrated that it corresponded to the N terminus of UB (Fig. 3E). The peptide mass fingerprints also revealed several p110 peptides (Fig. 3A, highlighted in bold) that were not detected in p107 tryptic digests (Fig. 3B). QqTOF MS/MS sequencing of four of these peptides identified them as originating from UB (Fig. 3E and supplemental Table S2), which corroborated the N-terminal sequencing results while providing a rationale for the size discrepancy between p110 and p107 on SDS-PAGE. Probing immunoblots of the final PEPC preparation with anti-UB-IgG resulted in monospecific detection of p110 (Fig. 5A). No additional higher Mr immunoreactive PEPC or UB bands indicative of polyubiquitination were observed.
FIGURE 3.
The p110 subunit of purified PEPC from germinated COS is monoubiquitinated at Lys-628. A and B, MALDI-TOF MS spectra of p110 and p107 tryptic peptides. Peptide mass fingerprinting of the p110 subunit (A) revealed several tryptic peptides (labeled with bold font) that were not observed in the p107 spectra (B). A, the singly charged ion at m/z 1534.796 matched a diglycinated (i.e. ubiquitin-conjugated) p110 peptide, whereas the remaining peptides unique to p110 originated from ubiquitin (supplemental Table S2). C and D, MS/MS analysis of the ubiquitin-conjugated peptide (C) and corresponding sulfonated derivative p110 tryptic peptide (D). C- and N-terminal fragment ions are denoted by y and b, respectively. E, alignment of amino acid sequences obtained during MALDI QqTOF MS/MS sequencing of p110 tryptic peptides or N-terminal microsequencing of p110, with the deduced sequence of Arabidopsis ubiquitin (gi 136666). MALDI QqTOF MS/MS sequenced peptides isolated from the p110 tryptic digest (supplemental Table S2) are underlined, whereas a 7-amino-acid sequence obtained during p110's N-terminal microsequencing is enclosed in a shaded box.
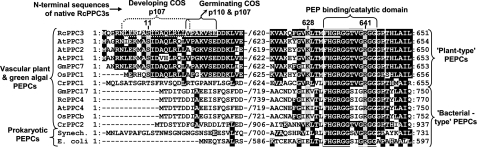
FIGURE 4.
Amino acid sequence alignment of COS RcPPC3 domains subject to in vivo post-translational modification with representative plant and prokaryotic PEPCs. Identical amino acids are indicated by black or gray shading. N-terminal sequences obtained by Edman degradation of native Class-1 PEPC from developing COS (p107) (4) and germinated COS (p110 and p107) are outlined by dotted and solid boxes, respectively. The Ser-11 phosphorylation and Lys-628 monoubiquitination sites of RcPPC3 (as occurs in Class-1 PEPC from developing and germinated COS, respectively) are indicated. The 12-amino-acid sequence of the diglycine-branched tryptic peptide of the p110 subunit (Fig. 3) is underlined and overlaps with a conserved flexible loop region (enclosed in a box) essential for PEP binding and PEPC catalysis; Arg-641 is indispensable for catalytic activity (2). The abbreviated species name for each sequence follows: Rc, R. communis (castor); At, Arabidopsis thaliana; Os, Oryza sativa (rice); Gm, Glycine max (soybean); Cr, Chlamydomonas reinhardtii (green alga); Synech., Synechocystis sp. PCC6803 (cyanobacteria).
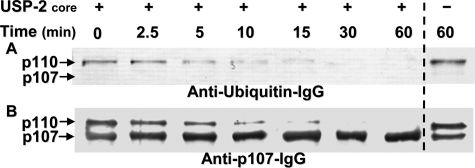
FIGURE 5.
Incubation with the deubiquitinating enzyme USP-2 core converts the germinated COS p110:p107 PEPC heterotetramer into a p107 homotetramer. Purified germinated COS PEPC was incubated in the presence and absence of 1 μm USP-2 core. Aliquots were removed at various times and subjected to immunoblot analysis using anti-(bovine ubiquitin)-IgG (Millipore, catalog number 05-944) (1 μg of PEPC/lane) (A) or anti-(COS p107)-IgG (5) (0.1 μg of PEPC/lane) (B).
UB is a highly conserved globular protein of eukaryotic cells that modifies target proteins via its covalent attachment through an isopeptide bond between the C-terminal Gly residue of UB and the ε-amino group of a Lys residue on a target protein. A multienzyme system consisting of activating (E1), conjugating (E2), and ligating (E3) enzymes attach UB to cellular proteins. Polyubiquitination tags many proteins for their proteolytic elimination by the 26 S proteasome (11, 19, 20). Indeed, degradation of plant PEPC and PEPC protein kinase by the polyubiquitin-proteasome pathway has been reported (21, 22). However, non-destructive functions for protein monoubiquitination in yeast and mammals have recently emerged. Monoubiquitination often mediates protein-protein interactions and localization to help control processes such as endocytosis, DNA repair, transcription and translation, and signal transduction (11, 12). UB-related pathways are also believed to be of widespread importance in the plant kingdom. Genomic analyses indicated that the UB-related pathway alone comprises over 6% of the Arabidopsis or rice proteome with thousands of different proteins being probable targets (19). A crucial goal for current research is the characterization of the monoubiquitinated proteome of eukaryotic cells, the UB-binding domain proteins that they interact with, and the influence of this PTM on the target protein's subcellular localization and functional properties (11, 20). There are very few reports describing monoubiquitination of metabolic enzymes. Phosphoglycerate mutase is monoubiquitinated in colorectal cancer tissues (23), but there was no indication what function this plays or whether this PTM altered the enzyme's kinetic properties. High throughput proteomic screens have identified numerous ubiquitinated metabolic (including glycolytic) enzymes in Arabidopsis cell cultures and seedlings (24, 25). However, neither group reported PEPC ubiquitination nor determined whether the various targets were poly- or monoubiquitinated (24, 25). The current study provides a precedent for tissue-specific monoubiquitination of a key enzyme of central metabolism and that this PTM influences its kinetic/regulatory properties (see below). However, additional research is required to assess: (i) what UB-binding domain proteins may interact with germinated COS PEPC in vivo, (ii) whether this PTM influences its microcompartmentation within the COS cytosol, and (iii) the specific E3 ligase and related signaling pathways that result in RcPPC3's monoubiquitination early in the germination process.
Lys-628 Is Monoubiquitinated in the p110 Subunit of Germinated COS PEPC—MS was employed to identify the site of UB conjugation to p110 using both untagged and 4-sulfophenyl isothiocyanate-tagged tryptic digests (Fig. 3, C and D) (15). Trypsin digestion of a UB-conjugated protein produced a signature peptide at the ubiquitination site containing a diglycine remnant derived from the C terminus of UB that remains attached to the target Lys residue via an isopeptide bond (15). The singly charged ion at m/z 1534.796 matched a diglycinated p110 peptide (Fig. 3B). MALDI QqTOF MS/MS sequencing of this peptide demonstrated that Lys-628 is ubiquitinated in the peptide sequence QFGVKLTMFHGR of p110 since an abnormal mass of 242.1 Da resulting from the diglycinated Lys was observed between fragments y7 and y8 (Fig. 3C). A characteristic mass increment of 114.1 Da at Lys-628 indicated a Gly-Gly attachment, and the doubly tagged modification after N-terminal sulfonation revealed an additional mass of 430.2 Da in the modified peptide of m/z 1964.988 (Fig. 3D). Its low energy collision dissociation by MS/MS measurements yielded a complete set of y fragment ions and further demonstrated the branched site-specific UB-conjugated peptide containing Lys(628)-Gly(UB-75)-Gly(UB-76).
Overall, our results indicate that 50% of RcPPC3 (p107) subunits are monoubiquitinated at Lys-628 at the outset of COS germination, resulting in formation of a novel p110:p107 Class-1 PEPC heterotetramer (supplemental Fig. S3). It is notable that Lys-628 is conserved in vascular plant (but not prokaryotic) PEPCs (Fig. 4) and that a pair of equal intensity-staining immunoreactive plant-type PEPC polypeptides in the range of 100–110 kDa have been observed on immunoblots of clarified extracts from all germinating seeds examined to date, including COS, sorghum, barley, and wheat (10, 16–18, 26). It will be interesting to establish whether a universal feature of germinated seed PEPCs is the formation of a monoubiquitinated Class-1 PEPC heterotetramer.
In Vitro Deubiquitination Converts the Germinated COS p110:p107 PEPC Heterotetramer into a p107 Homotetramer—Nine different members of recombinant human DUB families were screened for their ability to hydrolyze UB from the p110 subunit of the purified COS PEPC (supplemental Fig. S4). The only DUB capable of catalyzing p110's deubiquitination over the 60-min time course was the UB-specific protease-2 (USP-2) core, in which p110's disappearance was correlated with a progressive increase in the amount of p107 (Fig. 5 and supplemental Fig. S4). USP-2 is expressed as two distinct isozymes sharing a common catalytic core but differing in their N-terminal sequences (27). These divergent N-terminal sequences modulate USP-2's substrate specificity (28). As the catalytic core lacks an N-terminal regulatory domain, it can deubiquitinate a large number of substrates in vitro (28). The other DUBs tested were full-length enzymes predicted to display more stringent substrate specificity than USP-2 core. As the Lys-628 monoubiquitination site is proximal to a PEP-binding/catalytic domain of PEPC (Fig. 4 and supplemental Fig. S5), we next compared the kinetic properties of the ubiquitinated versus in vitro deubiquitinated PEPC (UB-PEPC and deUB-PEPC, respectively).
Kinetic Properties—Similar to other Class-1 PEPCs, including the enzyme from developing COS endosperm (4), PEPC from germinated COS displayed a broad pH activity profile with optimal activity occurring in the range of pH 8.0–8.5. Hyperbolic PEP saturation kinetics were observed at pH 8.0 and physiological pH (7.3) for both the UB-PEPC and the deUB-PEPC. Although their Vmax values were identical, deUB-PEPC displayed a significantly lower Km(PEP) value at pH 8.0 (0.039 ± 0.002 mm) relative to UB-PEPC (0.056 ± 0.003 mm). This trend was more pronounced at pH 7.3 where deUB-PEPC's Km(PEP) value (0.093 ± 0.006 mm) was about 50% of that obtained for UB-PEPC (0.211 ± 0.014 mm). Thus, monoubiquitination at Lys-628 interferes with PEP binding to the adjacent catalytic domain (Fig. 4 and supplemental Fig. S5).
Various compounds were tested as potential effectors at pH 7.3 and 8.0 with subsaturating concentrations of PEP (Table 2). The following metabolites exerted little or no influence (±20% of the control rate) on UB- or deUB-PEPC activity: AMP, ADP, citrate, Asn, Gln, and Glu (2 mm each). Similar to other plant PEPCs, UB-PEPC displayed pH-dependent modulation by several metabolites that were generally more effective at pH 7.3 than pH 8.0 (Table 2). Glucose-6-P, glucose-1-P, fructose-6-P, ribose-5-P, and glycerol-3-P functioned as activators, whereas malate, isocitrate, Asp, and ATP were inhibitory. Although UB-PEPC was markedly activated at pH 7.3 by the hexose-Ps, ribose-5-P, and glycerol-3-P, deUB-PEPC was only weakly activated by these compounds (Tables 2 and 3). Similarly, UB-PEPC was significantly more sensitive to inhibition by malate and Asp, relative to deUB-PEPC. The PEPC exhibited a unique response to isocitrate, which was a more effective inhibitor as assay pH was raised, and at pH 7.3 exerted more pronounced inhibition on UB-PEPC relative to deUB-PEPC (Tables 2 and 3). Overall, the kinetic studies indicate that monoubiquitination of COS PEPC interferes with its ability to bind PEP, while simultaneously enhancing sensitivity to the majority of its metabolite effectors.
TABLE 2.
Influence of various metabolites on the activity of PEPC purified from germinated COS endosperm
Assays were conducted at pH 8.0 and/or 7.3 using a subsaturating concentration of PEP (0.1 and 0.25 mm, respectively). DeUB-PEPC was prepared by incubating UB-PEPC for 30 min with 1 μm of USP-2 as described in the Experimental Procedures. PEPC activity in the presence of 2 mm of each effector is expressed relative to the control set at 100%. All values represent means of four independent experiments and are reproducible to within ±10% (S.E.) of the mean value.
|
Addition
|
Relative activity
|
||
|---|---|---|---|
|
pH 7.3
|
pH 8.0
|
||
| UB-PEPC | DeUB-PEPC | UB-PEPC | |
| Glucose-6-P | 154 | 125 | 123 |
| Glucose-1-P | 142 | 133 | 120 |
| Fructose-6-P | 155 | 132 | 130 |
| Ribose-5-P | 134 | 110 | 112 |
| Glycerol-3-P | 160 | 122 | 126 |
| Malate | 20 | 35 | 74 |
| Isocitrate | 56 | 44 | 27 |
| Aspartate | 49 | 65 | 92 |
| ATP | 64 | 75 | 56 |
TABLE 3.
Influence of USP-2 mediated deubiquitination on allosteric effector sensitivity of purified PEPC from 4-day old germinated COS endosperm
DeUB-PEPC was prepared by incubating UB-PEPC for 30 min with 1 μm of USP-2 as described in the Experimental Procedures. I50 and Ka values were determined with 0.25 mm PEP at pH 7.3. All parameters represent means four independent experiments and are reproducible to within ±10% (S.E.) of the mean value.
| Kinetic parametera | UB-PEPC | deUB-PEPC |
|---|---|---|
| mm | ||
| I50 (malate) | 0.7 | 1.3 |
| I50 (isocitrate) | 1.6 | 1.1 |
| I50 (Asp) | 2.6 | 4.1 |
| Ka (glucose-6-P) | 0.11 (1.7)b | NDc (1.2) |
| Ka (fructose-6-P) | 0.80 (1.6) | ND (1.3) |
| Ka (glycerol-3-P) | 0.48 (1.6) | ND (1.3) |
I50 and Ka values represent inhibitor and activator concentrations yielding 50% inhibition and activation, respectively.
Values in parentheses indicate maximal fold-activation by saturating levels of these activators.
ND, not determined (weak activation of deUB-PEPC by these effectors precluded accurate Ka determinations).
Concluding Remarks—As summarized in Fig. 4 and supplemental Fig. S3, it is remarkable that expression of the RcPpc3 gene in COS results in a plant-type PEPC that is subject to at least three different types of PTMs (including differential processing of its N terminus) and three different oligomeric states. In developing endosperm, RcPPC3 exists as a typical 410-kDa Class-1 PEPC homotetramer that is activated in vivo by phosphorylation of its p107 subunit at Ser-11 but also tightly interacts with the bacterial-type PEPC RcPPC4 (p118) to form the 910-kDa Class-2 PEPC hetero-octameric complex (4, 6–9). COS maturation is accompanied by disappearance of the Class-2 PEPC complex and RcPPC4/p118 polypeptides and RcPpc4 transcripts, a marked reduction in the activity of the Class-1 PEPC and the amount of its RcPPC3/p107 subunits, as well as conversion of p107 into its dephosphorylated form (4, 6, 8). Subsequent COS imbibition and germination results in elevated RcPpc3 transcripts and PEPC activity/amount and monoubiquitination of 50% of p107 at Lys-628 to form the p110:p107 heterotetrameric Class-1 PEPC (supplemental Figs. S1 and S3–5) (8, 10).
The metabolism of germinating castor seedlings is dominated by the mobilization of storage lipid and protein reserves in the endosperm to support the needs of the growing hypocotyls and roots. This process includes the massive conversion of reserve triacylglycerols into sucrose, which is absorbed by the cotyledons of the growing seedling (29, 30). Part of the sucrose is used by the endosperm to fuel anabolic processes and to generate ATP through respiration. PEPC has been suggested to fulfill a crucial function early in the germination process to build up cellular pools of C4 acids needed to trigger subsequent Krebs and glyoxylate cycle activity (10). An ensuing role for PEPC in germinating oil seeds is to replenish dicarboxylic acids required as substrates for the substantial transamination reactions that follow storage protein hydrolysis (29), as well as for glutamine synthetase's reassimilation of NH +4 released during the oxidation of glutamate to 2-oxoglutarate via glutamate dehydrogenase (31). The labeling of metabolic intermediates to isotopic steady state and the modeling of the tricarboxylic acid cycle in germinating lettuce seeds indicated that 70% of glycolytic flux from carbohydrate oxidation enters the tricarboxylic acid cycle via the PEPC reaction (32). At the same time, PEP carboxykinase catalyzes the initial step in the gluconeogenic conversion of storage lipids into sucrose as it uses ATP to decarboxylate and phosphorylate oxaloacetate derived from the glyoxylate cycle into PEP (29). PEP carboxykinase is abundant in the cytosol of germinating COS endosperm (33) but is incompatible with PEPC since their combined reactions result in a futile cycle, i.e. the wasteful hydrolysis of ATP to ADP and Pi. In their classic study of gluconeogenesis in germinating COS endosperm, Kobr and Beevers (30) determined that the maximum rate of glycolysis is about one-tenth that of gluconeogenesis and that the pools of glycolytic and gluconeogenic intermediates likely occur in separate intracellular regions and appear to be independently regulated. Several studies have provided evidence for glycolytic or gluconeogenic multienzyme complexes (metabolons) in the plant cytosol (3), including the metabolically sequential: (i) PEPC, malate dehydrogenase, and malic enzyme in sugar cane leaves (34) and (ii) cytosolic isozymes of fructose-1,6-P2 aldolase and fructose-1,6-bisphosphatase in germinated COS (35). Furthermore, seven different glycolytic enzymes formed a metabolon on the mitochondrial surface of Arabidopsis suspension cells during periods of increased respiration so as to channel carbon from cytosolic metabolite pools into the mitochondria while restricting substrate use by competing metabolic pathways (36). It will therefore be important to establish whether PEPC monoubiquitination contributes to formation of a glycolytic metabolon while minimizing its futile cycling with PEP carboxykinase in the cytosol of germinated COS.
Supplementary Material
Acknowledgments
We are grateful to Prof. Linda Hicke (Northwestern University) for helpful discussions. The MS and MS/MS analyses were performed at the Queen's University MassSpec Facility, which is funded by the Canada Foundation for Innovation.
This work was supported by grants from the Natural Sciences and Engineering Research Council of Canada and the Queen's Research Chairs program (to W. C. P.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains two supplemental tables, five supplemental figures, and references.
This article was selected as a Paper of the Week.
Footnotes
The abbreviations used are: PEP, phosphoenolpyruvate; PEPC, PEP carboxylase; ACN, acetonitrile; COS, castor oil seed; UB, ubiquitin; deUB, deubiquitinated; DUB, deubiquitinating enzyme; USP, ubiquitin-specific protease; MALDI QqTOF, matrix-assisted laser desorption ionization quadrupole time-of-flight; MS, mass spectrometry; MS/MS, tandem MS; FPLC, fast protein liquid chromatography; p107 and p110; 107- and 110-kDa polypeptides, respectively; PEG, polyethylene glycol; PTM, post-translational modification.
References
- 1.Chollet, R., Vidal, J., and O'Leary, M. H. (1996) Annu. Rev. Plant Physiol. Plant Mol. Biol. 47 273–298 [DOI] [PubMed] [Google Scholar]
- 2.Izui, K., Matsumura, H., Furumoto, T., and Kai, Y. (2004) Annu. Rev. Plant Biol. 55 69–84 [DOI] [PubMed] [Google Scholar]
- 3.Plaxton, W. C., and Podestá, F. E. (2006) CRC Crit. Rev. Plant Sci. 25 159–198 [Google Scholar]
- 4.Blonde, J. D., and Plaxton, W. C. (2003) J. Biol. Chem. 278 11867–11873 [DOI] [PubMed] [Google Scholar]
- 5.Crowley, V., Gennidakis, S., and Plaxton, W. C. (2005) Plant Cell Physiol. 46 1855–1862 [DOI] [PubMed] [Google Scholar]
- 6.Tripodi, K. E., Turner, W. L., Gennidakis, S., and Plaxton, W. C. (2005) Plant Physiol. 139 969–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murmu, J., and Plaxton, W. C. (2007) Planta 226 1299–1310 [DOI] [PubMed] [Google Scholar]
- 8.Gennidakis, S., Rao, S., Greenham, K., Uhrig, R. G., O'Leary, B., Snedden, W. A., Lu, C., and Plaxton, W. C. (2007) Plant J. 52 839–849 [DOI] [PubMed] [Google Scholar]
- 9.Uhrig, R. G., O'Leary, B., Spang, H. E., MacDonald, J. A., She, Y. M., and Plaxton, W. C. (2008) Plant Physiol. 146 1346–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sangwan, R. S., Singh, N., and Plaxton, W. C. (1992) Plant Physiol. 99 445–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schnell, J. D., and Hicke, L. (2003) J. Biol. Chem. 278 35857–35860 [DOI] [PubMed] [Google Scholar]
- 12.Mukhopadhyay, D., and Riezman, H. (2007) Science 315 201–205 [DOI] [PubMed] [Google Scholar]
- 13.Brooks, S. (1992) BioTechniques 13 906–911 [PubMed] [Google Scholar]
- 14.Law, R. D., and Plaxton, W. C. (1995) Biochem. J. 307 807–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang, D., Kalume, D., Pickart, C., Pandey, A., and Cotter, R. J. (2006) Anal. Chem. 78 3681–3687 [DOI] [PubMed] [Google Scholar]
- 16.Osuna, L., Pierre, J. N., Gonzalez, M. C., Alvarez, R., Cejudo, F. J., Echevarria, C., and Vidal, J. (1999) Plant Physiol. 119 511–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Osuna, L., Gonzalez, M. C., Cejudo, F. J., Vidal, J., and Echevarria, C. (1996) Plant Physiol. 111 551–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nhiri, M., Bakrim, N., El Hachimi-Messouak, Z., Echevarría, C., and Vidal, J. (2000) Plant Sci. 151 29–37 [Google Scholar]
- 19.Smalle, J., and Viestra, R. D. (2004) Annu. Rev. Plant Biol. 55 555–590 [DOI] [PubMed] [Google Scholar]
- 20.Downes, B., and Vierstra, R. D. (2005) Biochem. Soc. Trans. 33 393–399 [DOI] [PubMed] [Google Scholar]
- 21.Agetsuma, M., Furumoto, T., Yanagisawa, S., and Izui, K. (2005) Plant Cell Physiol. 46 389–398 [DOI] [PubMed] [Google Scholar]
- 22.Schulz, M., Klockenbring, T., Hunte, C., and Schnabl, H. (1993) Bot. Acta 106 143–145 [Google Scholar]
- 23.Usuba, T., Ishibashi, Y., Okawa, Y., Hirakawa, T., Takada, K., and Ohkawa, K. (2001) Int. J. Cancer 94 662–668 [DOI] [PubMed] [Google Scholar]
- 24.Maor, R., Jones, A., Nuhse, T. S., Studholme, D. J., Peck, S. C., and Shirasu, K. (2007) Mol. Cell. Proteomics 6 601–610 [DOI] [PubMed] [Google Scholar]
- 25.Manzano, C., Abraham, Z., Lopez-Torrejon, G., and Del Pozo, J. C. (2008) Plant Mol. Biol. 68 145–158 [DOI] [PubMed] [Google Scholar]
- 26.Gonzalez, M. C., Osuna, L., Echevarria, C., Vidal, J., and Cejudo, F. J. (1998) Plant Physiol. 116 1249–1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gousseva, N., and Baker, R. T. (2003) Gene Expr. 11 163–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin, H., Yin, L., Reid, J., Wilkinson, K. D., and Wing, S. S. (2001) J. Biol. Chem. 276 20357–203563 [DOI] [PubMed] [Google Scholar]
- 29.Botha, F., Potgieter, G., and Botha, A.-M. (1992) Plant Growth Regul. 11 211–244 [Google Scholar]
- 30.Kobr, M. J., and Beevers, H. (1971) Plant Physiol. 47 48–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Glevarec, G., Bouton, S., Jaspard, E., Riou, M.-T., Cliquet, J.-B., Suzuzki, A., and limami, A. M. (2004) Planta 219 286–297 [DOI] [PubMed] [Google Scholar]
- 32.Salon, C., Raymond, P., and Pradet, A. (1988) J. Biol. Chem. 263 12278–12287 [PubMed] [Google Scholar]
- 33.Martín, M., Plaxton, W., and Podestá, F. (2007) Physiol. Plant 130 484–494 [Google Scholar]
- 34.Baret, P., Cesari, M., Queiroz, C., Rouch, C., Meunier, J. C., and Cadet, F. (1999) C R Acad. Sci. III (Paris) 322 29–34 [DOI] [PubMed] [Google Scholar]
- 35.Moorhead, G. B. G., Hodgson, R. J., and Plaxton, W. C. (1994) Arch. Biochem. Biophys. 312 326–335 [DOI] [PubMed] [Google Scholar]
- 36.Graham, J. W., Williams, T. C., Morgan, M., Fernie, A. R., Ratcliffe, R. G., and Sweetlove, L. J. (2007) Plant Cell 19 3723–3738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang, D., and Cotter, R. J. (2005) Anal Chem. 77 1458–1466 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.