Abstract
The forkhead transcription factor Foxo3a is able to inhibit cardiomyocyte hypertrophy. However, its underlying molecular mechanism remains to be fully understood. Our present study demonstrates that Foxo3a can regulate cardiomyocyte hypertrophy through transactivating catalase. Insulin was able to induce cardiomyocyte hypertrophy with an elevated level of reactive oxygen species (ROS). The antioxidant agents, including catalase and N-acetyl-l-cysteine, could inhibit cardiomyocyte hypertrophy induced by insulin, suggesting that ROS is necessary for insulin to induce hypertrophy. Strikingly, we observed that the levels of catalase were decreased in response to insulin treatment. The transcriptional activity of Foxo3a depends on its phosphorylation status with the nonphosphorylated but not phosphorylated form to be functional. Insulin treatment led to an increase in the phosphorylated levels of Foxo3a. To understand the relationship between Foxo3a and catalase in the hypertrophic pathway, we characterized that catalase was a transcriptional target of Foxo3a. Foxo3a bound to the promoter region of catalase and stimulated its activity. The inhibitory effect of Foxo3a on cardiomyocyte hypertrophy depended on its transcriptional regulation of catalase. Finally, we identified that myocardin was a downstream mediator of ROS in conveying the hypertrophic signal of insulin or insulin-like growth factor-1. Foxo3a could negatively regulate myocardin expression levels through up-regulating catalase and the consequent reduction of ROS levels. Taken together, our results reveal that Foxo3a can inhibit hypertrophy by transcriptionally targeting catalase.
Myocardial hypertrophy is a compensatory response to increased hemodynamic load. It is often associated with poor clinical outcomes, including the development of cardiac systolic and diastolic dysfunction and ultimately heart failure (1–7). Hyperinsulinemia and cardiac hypertrophy are closely related. For example, insulin treatment can induce hypertrophy in cultured cardiomyocytes (8). In the animal model, chronic hyperinsulinemia leads to cardiac hypertrophy (9). In particular, hypertensive patients with left ventricular hypertrophy have a higher degree of hyperinsulinemia than hypertensive patients without left ventricular hypertrophy (10). Insulin-like growth factor-1 (IGF-1)3 also is a potent hypertrophic stimulus both in vitro and in vivo (11). To prevent and/or reverse myocardial hypertrophy, it is necessary to identify and characterize the molecules that are involved in the hypertrophic cascades of insulin and IGF-1.
The forkhead family of transcription factors are characterized by the presence of a conserved 100-amino acid DNA binding domain and participate in regulating diverse cellular functions such as apoptosis, differentiation, metabolism, proliferation, and survival (12). The Foxo (Forkhead bOX-containing protein, O sub-family) subgroup contains four members (Foxo1, Foxo3a, Foxo4, and Foxo6). It has been shown that Foxo1 and Foxo3a are expressed in the heart and skeletal muscle (13–15). Foxo3a activation can induce skeletal muscle atrophy by causing transcription of the ubiquitin ligase atrogin-1 promoter (16). Recently, it has been shown that Foxo family members are able to negatively regulate cardiac hypertrophy. For example, the constitutively active form of Foxo3a (caFoxo3a) in which all three Akt sites are replaced by alanine residues can activate the atrogin-1 promoter in both cultured cardiomyocytes and mouse heart, and negatively regulate cardiac hypertrophy (8, 14). Foxo1 or Foxo3 in cardiomyocytes attenuates calcineurin phosphatase activity and inhibits agonist-induced hypertrophic growth (17). In addition, Foxo3a knockout mice show cardiac hypertrophy (17). Given the important role of Foxo3a in controlling cardiac hypertrophy, it necessitates the full elucidation of its underlying molecular mechanisms in regulating cardiac hypertrophy.
Reactive oxygen species (ROS) can convey the hypertrophic signals of a variety of stimuli (18–21). For example, tumor necrosis factor-α causes hypertrophy via the generation of ROS in cardiomyocytes (18). Endothelin-1 can directly stimulate ROS production (19) or mediate leptin-induced ROS production (22). Mechanical stretch-induced hypertrophy is mediated by ROS (20). As second messengers, ROS regulate various intracellular signal transduction cascades and the activity of various transcription factors that regulate cardiac hypertrophy. The activation of nuclear factor-κB and activator protein-1 can be controlled by ROS (19, 23). Another well characterized ROS-sensitive signaling pathway is that of mitogen-activated protein kinases (MAPKs). An increase in ROS production results in the activation of MAPK pathways (24, 25). Administration of the antioxidants can inhibit cardiac hypertrophy (26, 27). Despite these observations, the downstream targets of ROS in the hypertrophic pathways remain to be further identified.
Myocardin is a transcriptional factor expressed in cardiomyocytes. Recently, it has been shown that overexpression of myocardin can induce cardiac hypertrophy (28). Myocardin also can mediate the hypertrophic signal of a variety of stimuli such as phenylephrine, endothelin-1, and serum (28, 29). The expression levels of myocardin can be elevated in response to hypertrophic stimulation (28). Nevertheless, the upstream signals that control the expression of myocardin remain largely unknown. In particular, although both ROS and myocardin are able to convey the hypertrophic signals, it is not yet clear whether they are related in the hypertrophic pathway.
The elevation of ROS levels can result from an increase in their production and/or a decrease in their decomposition. Our recent work shows that catalase, a scavenger of hydrogen peroxide, is down-regulated in response to the hypertrophic stimulation (30). Insulin and insulin-like growth factor-1 are able to induce cardiac hypertrophy (11). It remains enigmatic as to whether catalase expression is altered in response to insulin and IGF-1 stimulation. Furthermore, the molecular mechanism by which catalase is down-regulated in response to hypertrophic stimulation remains unknown.
Our present work aimed to elucidate whether there is an impact between Foxo3a and catalase in the hypertrophic pathway. Our results showed that catalase is a transcriptional target of Foxo3a. Insulin can induce hypertrophy through inactivating Foxo3a and the consequent catalase down-regulation as well as ROS elevation. Furthermore, our results demonstrated that myocardin expression can be stimulated by ROS, and myocardin is necessary for insulin and IGF-1 to induce hypertrophy. Foxo3a negatively regulates myocardin expression through controlling ROS levels. Thus, it appears that Foxo3a and catalase constitute an anti-hypertrophic axis in the heart.
EXPERIMENTAL PROCEDURES
Materials—Insulin, N-acetyl-l-cysteine (NAC), and IGF-1 were purchased from Sigma. 2′,7′-Dichlorofluorescein diacetate (DCFH-DA) was purchased from Molecular Probes Inc. Anti-Foxo3a antibody, anti-catalase antibody, anti-myocardin antibody, anti-phospho Foxo3a (Thr32) antibody, anti-actin antibody, and horseradish peroxidase-conjugated goat anti-rabbit or rabbit anti-goat IgG were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). The adenovirus-harboring catalase was a kind gift from Dr. Joseph J. Cullen. The constitutively active form of human Foxo3a were kindly provided by Dr. Boudewijn M. T. Burgering. The adenovirus harboring dominant negative Akt1 (DN-Akt) was from Vector Biolabs.
Constructions of Adenoviruses Harboring the Constitutively Active Form of Human Foxo3a—The adenovirus harboring the constitutively active form of human Foxo3a (caFoxo3a) was constructed using the Adeno-X™ expression system (Clontech).
Constructions of Adenoviruses Harboring Rat Foxo3a RNAi or Rat Myocardin RNAi—The rat Foxo3a RNAi target sequence was 5′-CAAGTACACCAAGAGCCGA-3′. A nonrelated, scrambled RNAi without any other match in the rat genomic sequence was used as a control (5′-TCAGACAGACAGACAGACC-3′). The rat myocardin RNAi target sequence was 5′-GGTCAGAAACAGATCGGAC-3′. The scrambled myocardin RNAi target sequence was 5′-AGCCTAAGTCAAGGCAGAG-3′. The adenoviruses harboring Foxo3a RNAi or myocardin RNAi were constructed using the pSilencer™ adeno 1.0-CMV System (Ambion) according to the kit's instructions. The effects of these constructs on Foxo3a and myocardin expression were tested. Our results showed that the expression of Foxo3a and myocardin could be significantly inhibited in rat cardiomyocytes by their RNAi constructs, respectively.
Adenovirus Infection—Viruses were amplified in HEK293 cells. The adenovirus (Ad) containing β-galactosidase was as we described elsewhere (31). The adenovirus harboring catalase RNAi was as we described (30). Adenovirus infection of cardiomyocytes was performed as we described (32).
Cardiomyocytes Culture and Treatment—Cardiomyocytes were isolated from 1- to 2-day-old Wistar rats as we described (33). Briefly, after dissection hearts were washed, minced in HEPES-buffered saline solution contained: 130 mm NaCl, 3 mm KCl, 1 mm NaH2PO4, 4 mm glucose, and 20 mm HEPES (pH adjusted to 7.35 with NaOH). Tissues were then dispersed in a series of incubations at 37 °C in HEPES-buffered saline solution containing 1.2 mg/ml pancreatin and 0.14 mg/ml collagenase (Worthington). After centrifugation cells were resuspended in Dulbecco's modified Eagle's medium/F-12 (Amersham Biosciences) containing 5% heat-inactivated horse serum, 0.1 mm ascorbate, insulin-transferring-sodium selenite media supplement, 100 units/ml penicillin, 100 μg/ml streptomycin, and 0.1 mm bromodeoxyuridine. The dissociated cells were preplated at 37 °C for 1 h. The cells were then diluted to 1 × 106 cells/ml and plated in 10 μg/ml laminin-coated different culture dishes according to the specific experimental requirements. For the administration of NAC, cells were pretreated for 1 h with NAC prior to insulin or IGF treatment.
Cell Surface Area Measurement—The cell surface area of F-actin-stained cells or unstained cells was measured after employing hypertrophic stimuli by the computer-assisted planimetry. 100–200 cardiomyocytes in 20–30 fields were examined in each experiment.
Protein/DNA Ratio Detection—For the quantitative analysis of total cellular protein and DNA content, cells were washed twice with phosphate-buffered saline. After wash, 0.2 n perchloric acid (1 ml) was added to the cells. The samples were centrifuged for 10 min at 10,000 × g. The precipitates were incubated for 20 min at 60 °C with 250 μl of 0.3 n KOH. Protein content was analyzed by the Lowry method using human serum albumin as a standard. DNA content was detected using Hoechst dye 33258 with salmon sperm DNA as a standard.
Intracellular ROS Analysis—Intracellular ROS levels were analyzed by employing ROS-sensitive dye, DCFH-DA as we described (30). After DCFH-DA (5 μg/ml) and hypertrophic stimuli incubation, fluorescent images were acquired from a laser confocal microscope (Zeiss LSM 510 META) by employing its ROZ mean function, and the intensity on regions of interest was measured.
Immunoblotting—Immunoblotting was performed as we described (34). In brief, cells were lysed for 1 h at 4 °C in a lysis buffer (20 mm Tris (pH 7.5), 2 mm EDTA, 3 mm EGTA, 2 mm dithiothreitol, 250 mm sucrose, 0.1 mm phenylmethylsulfonyl fluoride, 1% Triton X-100, and a protease inhibitor mixture). Samples were subjected to 12% SDS-PAGE and transferred to nitrocellulose membranes. Equal-protein loading was controlled by Ponceau Red staining of membranes. Blots were probed using antibodies.
Constructions of Rat Catalase Promoter and Its Mutant—Catalase promoter was amplified from rat genome using PCR. The long fragment containing two Foxo3a potential binding sites (wild-type promoter-1, wt-1) was amplified using the forward primer 5′-GTGCTTTCTGCTGCCTCTCCACTTC-3′. The short fragment containing one Foxo3a potential binding (wild-type promoter-2, wt-2) was amplified using the forward primer 5′-CAGATTTCCCCTCTCACAGAGGAGG-3′. Both fragments were amplified using the reverse primer 5′-CATGGTGTAGGATTGCGGAGCTGCA-3′. The promoters were cloned into the reporter plasmid, pGL4.17 (Promega). The introduction of mutations in the putative Foxo3a binding site in wt-1 fragment (–363 to –357 wild type: 5′-ATAAATA-3′, the mutant: 5′-AGCCCTA-3′) was generated using QuikChange II XL site-directed mutagenesis kit (Stratagene). The construct was sequenced to check that only the desired mutations had been introduced.
ChIP Analysis—ChIP was performed following the procedure described with modifications (35). Cells were treated with insulin, and then washed twice with phosphate-buffered saline and cross-linked with 1% formaldehyde at 37 °C for 10 min. After centrifugation, cells were resuspended in 0.3 ml of lysis buffer (1% SDS, 10 mm EDTA, 50 mm Tris-HCl, pH 8.1) containing a protease inhibitor mixture, and sonicated three times for 15 s. Supernatants were collected, and a 20-μl supernatant of sonicated cell lysates was saved as the input sample. The rest supernatants were diluted in buffer (1% Triton X-100, 2 mm EDTA, 150 mm NaCl, 20 mm Tris-HCl, pH 8.1) followed by immunoclearing with 2 μg of sheared salmon sperm DNA and protein A-agarose for 2 h at 4 °C. Immunoprecipitation was performed overnight at 4 °C with 5 μg of anti-Foxo3a antibody, and followed by addition of 45 μl of protein A-agarose and incubation for another hour at 4 °C. Precipitates were washed with buffer-I (0.1% SDS, 1% Triton X-100, 2 mm EDTA, 20 mm Tris-HCl, pH 8.1, 150 mm NaCl), buffer-II (0.1% SDS, 1% Triton X-100, 2 mm EDTA, 20 mm Tris-HCl, pH 8.1, 500 mm NaCl), buffer-III (0.25 m LiCl, 1% Nonidet P-40, 1% deoxycholate, 1 mm EDTA, 10 mm Tris-HCl, pH 8.1), and TE buffer (10 mm Tris-HCl, 1 mm EDTA, pH 8.1). DNA was eluted from the beads with 100 μl of elution buffer (1% SDS, 0.1 m NaHCO3). DNA fragments were purified with a QIAquick Spin Kit (Qiagen). In addition, anti-manganese superoxide dismutase antibody was used for a negative control. The purified DNA was used as a template for PCR amplification. PCR was run under the following conditions: 94 °C for 3 min for 1 cycle; 94 °C for 1 min, 55 °C for 1 min, 72 °C for 1 min for 25 cycles; a final extension at 72 °C for 8 min. The primers were as follows. BS1 (corresponding to a 218-bp fragment): BS1-Forward, 5′-GGTGGACTATTGACAGTGTTGGG-3′; BS1-Reverse, 5′-CGCCATCCAGTTATTTACTCAGG-3′; BS2 (corresponding to a 191-bp fragment): BS2-Forward, 5′-ACCAAATAAATAAGCAAAGTGAG-3′; BS2-Reverse, 5′-GAAACTCTAGAAGGGACAGGATT-3′.
Luciferase Assay—Luciferase activity assay was performed using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer's instructions. Cells were seeded in 24-well plates. They were co-transfected with the expression vectors, the luciferase reporter constructs and the Renilla luciferase plasmids using Lipofectamine 2000 (Invitrogen). Each well contained 0.2 μg of luciferase reporter plasmids, 0.2 μg of expression vectors, 2.5 ng of Renilla luciferase plasmids, respectively. Cells were lysed and assayed for luciferase activity 24 h after transfection. 20 μl of protein extracts was analyzed in a luminometer. Firefly luciferase activities were normalized to Renilla luciferase activity.
Quantitative Real-time PCR—Quantitative real-time PCR was performed as we described (32). In brief, total RNA was isolated using TRIzol (Invitrogen). RNA was reverse transcribed using Oligo(dT) and amplified using a TaqMan Assays kit (TOYOBO). The samples were run in triplicate using the Applied Biosystems (ABI) 7000 sequence detector according to the manufacturer's instructions. The results were standardized to control values of glyceraldehyde-3-phosphate dehydrogenase. The sequences of catalase primers were: Forward, 5′-CCTCCTCGTTCAAGATGTGGTTTTC-3′; Reverse, 5′-CGTGGGTGACCTCAAAGTATCCAAA-3′; glyceraldehyde-3-phosphate dehydrogenase forward primer, 5′-GCTAACATCAAATGGGGTGATGCTG-3′; Reverse, 5′-GAGATGATGACCCTTTTGGCCCCAC-3′. PCR was run under the following conditions: 95 °C for 60 s for 1 cycle; 95 °C for 15 s, 55 °C for 15 s, and 72 °C for 45 s for 40 cycles. The specificity of the PCR amplification was confirmed by agarose gel electrophoresis.
Statistical Analysis—The results are expressed as means ± S.E. The statistical comparison among different groups was performed by one-way analysis of variance. Paired data were evaluated by Student's t test. p < 0.05 was considered statistically significant.
RESULTS
ROS Participates in Mediating the Hypertrophic Signal of Insulin—Although insulin is able to induce cardiomyocyte hypertrophy (8, 9), and ROS can mediate the hypertrophic signals (19, 20, 36), it remains unknown as to whether ROS participates in conveying the hypertrophic signal of insulin. We explored whether ROS is necessary for insulin to induce hypertrophy. To this end, ROS levels in response to insulin treatment were analyzed. As shown in Fig. 1 (A and B), treatment of cells with insulin led to an increase in ROS levels. Either the antioxidant NAC or exogenous catalase was able to attenuate ROS levels upon insulin treatment. These data suggest that insulin is able to induce an elevated level of ROS.
FIGURE 1.
ROS mediates the hypertrophic signal of insulin. A, NAC reduces ROS levels upon treatment with insulin. The neonatal rat cardiomyocytes were incubated with 5 μm DCFH-DA for 30 min at 37 °C, and then treated with 20 μg/ml insulin. ROS was analyzed 1 h after insulin treatment. *, p < 0.05 versus control; #, p < 0.05 versus insulin alone. B, insulin-induced ROS elevation is attenuated by catalase. Cardiomyocytes were infected with Ad-β-galactosidase or Ad-catalase at an m.o.i. of 50. 24 h after infection cells were incubated with 5 μm DCFH-DA for 30 min at 37 °C, and then treated with 20 μg/ml insulin. ROS was analyzed 1 h after insulin treatment. *, p < 0.05 versus control; #, p < 0.05 versus insulin alone. C, NAC attenuates insulin-induced hypertrophic responses analyzed by cell surface area measurement. Cardiomyocytes were treated as described for A. 48 h after treatment cell surface area was measured. *, p < 0.05 versus control; #, p < 0.05 versus insulin alone. D, NAC attenuates insulin-induced hypertrophic responses revealed by protein/DNA ratio. Cardiomyocytes were treated as described for C. 48 h after treatment cells were harvested for the detection of protein/DNA ratio. *, p < 0.05 versus control; #, p < 0.05 versus insulin alone. E and F, catalase inhibits hypertrophy induced by insulin. Cardiomyocytes were treated as described for B. 48 h after treatment cells were collected for the analysis of cell surface area (E) and protein/DNA ratio (F). *, p < 0.05 versus control; #, p < 0.05 versus insulin alone. G, insulin treatment led to sarcomere organization blocked by NAC and catalase. Representative photos show sarcomere organization. The neonatal rat cardiomyocytes were treated as described for A and B. 48 h after treatment cells were collected for the staining with TRITC-conjugated phalloidin. The nuclei were stained with 4′,6-diamidino-2-phenylindole. Bar = 10 μm. Data in Fig. 1 are expressed as the mean ± S.E. of three independent experiments.
To understand whether ROS plays a functional role in insulin-induced hypertrophy, we detected whether the inhibition of ROS is able to influence hypertrophy. Administration of insulin resulted in an increase in cardiomyocyte surface area and protein/DNA ratio. NAC could attenuate the increase in cell surface area (Fig. 1C) and protein/DNA ratio (Fig. 1D) induced by insulin. A similar result was obtained by the employment of catalase (Fig. 1, E and F). Insulin treatment led to sarcomere organization. NAC and catalase could inhibit sarcomere organization (Fig. 1G). Thus, it appears that ROS participates in conveying the hypertrophic signals of insulin.
Insulin Induces a Decrease in Catalase and an Increase in Foxo3a Phosphorylation Levels—Catalase decomposes hydrogen peroxide, and our recent work has shown that catalase can be down-regulated in the hypertrophic pathway (30). We detected catalase levels in response to insulin treatment. As shown in Fig. 2A, catalase mRNA levels were decreased after treatment with insulin. The protein levels of catalase also were decreased upon insulin treatment (Fig. 2B). These results indicate that catalase is down-regulated in response to insulin treatment. Insulin is an upstream stimulus of Foxo3a (11). We tested whether Foxo3a could be influenced by insulin. Insulin treatment led to no significant alterations in Foxo3a protein levels, but an increase in the levels of phosphorylated form of Foxo3a (p-Foxo3a, Fig. 2C). Thus, it appears that insulin could lead to an elevated level of non-functional form of Foxo3a and a decrease of catalase expression.
FIGURE 2.
Insulin induces a decrease in catalase expression levels and an increase in the levels of phosphorylated Foxo3a. A and B, insulin treatment leads to a reduction in catalase expression levels. The neonatal rat cardiomyocytes were treated with 20 μg/ml insulin. Cells were harvested at the indicated time for the analysis of catalase mRNA levels by real-time PCR (A) or catalase protein levels by immunoblot (B). *, p < 0.05 versus control. Data are expressed as the mean ± S.E. of three independent experiments. C, the phosphorylation levels of Foxo3a were increased in response to insulin treatment. Cardiomyocytes were treated as described for A. Cells were harvested at the indicated times for the analysis of phosphorylated and total Foxo3a levels by immunoblot. A representative blot of three independent experiments is shown.
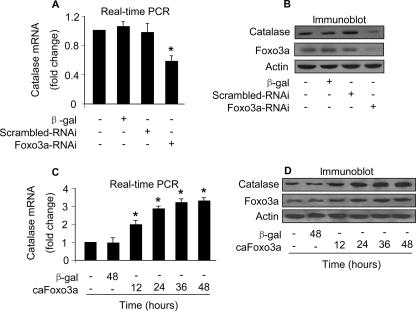
Foxo3a Is Able to Alter Catalase mRNA and Protein Levels—Foxo3a is a transcription factor, and its transcriptional activity is dependent on its phosphorylation status. The reduction in the levels of functional Foxo3a as well as catalase encouraged us to consider whether Foxo3a is related to catalase expression. We tested whether catalase expression is regulated by endogenous Foxo3a. Knockdown of endogenous Foxo3a led to a reduction in the levels of catalase mRNA (Fig. 3A) and protein (Fig. 3B). To understand whether exogenous Foxo3a can influence catalase expression, we detected catalase levels in cells enforced expression of exogenous caFoxo3a. caFoxo3a could induce an elevated level of catalase mRNA (Fig. 3C) and protein (Fig. 3D). These data suggest that Foxo3a is able to influence catalase expression levels.
FIGURE 3.
Foxo3a is able to regulate mRNA and protein levels of catalase. A and B, knockdown of endogenous Foxo3a leads to a reduction in catalase expression levels. The neonatal rat cardiomyocytes were infected with Ad-Foxo3a-RNAi or its scrambled form (Ad-scrambled-RNAi) at an m.o.i. of 100. Cells were harvested 48 h after infection for the analysis of catalase mRNA levels by Real-time PCR (A) or catalase protein levels by immunoblot (B). *, p < 0.05 versus control. Data are expressed as the mean ± S.E. of 3 independent experiments. C and D, caFoxo3a stimulates catalase expression at mRNA and protein levels. Cardiomyocytes were infected with Ad-β-galactosidase or Ad-caFoxo3a at an m.o.i. of 50. Cells were harvested at the indicated time for the analysis of catalase mRNA levels by Real-time PCR (C) or catalase protein levels by immunoblot (D). *, p < 0.05 versus control. Data are expressed as the mean ± S.E. of three independent experiments. A representative blot of three independent experiments is shown.
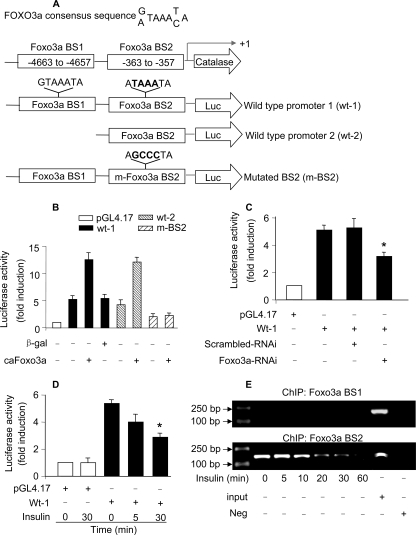
Catalase Is a Transcriptional Target of Foxo3a—The ability of Foxo3a to influence catalase mRNA and protein levels led us to consider whether the regulation of Foxo3a on catalase occurs in a transcriptional manner. To this end, we analyzed the promoter region of catalase and observed that it has two potential Foxo3a binding sites (Fig. 4A). We first tested whether Foxo3a can regulate catalase promoter activity. A fragment containing the two potential binding sites (wt-1) and a fragment containing only BS2 (wt-2) were constructed and cloned into the luciferase vector, respectively. As shown in Fig. 4B, caFoxo3a led to a marked elevation of wt-1 promoter activity. wt-2 promoter had a comparable activity as wt-1 in response to caFoxo3a stimulation, suggesting that BS2 is the binding site. To confirm this conclusion, we introduced mutations in BS2 in wt-1, and the data showed that the mutated promoter (m-BS2) could not be stimulated by caFoxo3a. Thus, it appears that BS2 in the promoter region of catalase is the binding site of Foxo3a.
FIGURE 4.
Foxo3a binds to catalase promoter and stimulates its activity. A, catalase promoter contains two potential Foxo3a binding sites. The two potential Foxo3a binding sites are indicated as BS1 and BS2. Two fragments of catalase promoter were synthesized and linked to luciferase (Luc) reporter gene. The mutations were introduced to BS2 in wt-1. B, Foxo3a activates catalase promoter activity. Cardiomyocytes were infected with Ad-β-galactosidase or Ad-caFoxo3a at an m.o.i. of 50. 24 h after infection cells were transfected with the constructs of the empty vector (pGL-4) or catalase promoter constructs, respectively. Firefly luciferase activities were normalized to Renilla luciferase activities. C, knockdown of endogenous Foxo3a leads to a reduction of catalase promoter activity. Cardiomyocytes were infected with Ad-Foxo3a-RNAi or Ad-scrambled-RNAi at an m.o.i. of 100. 24 h after infection cells were transfected with the constructs of the empty vector (pGL-4) or wt-1 catalase promoter constructs, respectively. Firefly luciferase activities were normalized to Renilla luciferase activities. *, p < 0.05 versus wt-1 alone. D, insulin treatment results in a decrease in catalase promoter activity. Cardiomyocytes were transfected with the constructs of the empty vector (pGL-4) or wt-1 catalase promoter constructs, respectively. 24 h after transfection cells were treated with 20 μg/ml insulin. Cells were harvested at the indicated time for the analysis of luciferase activity. *, p < 0.05 versus wt-1 alone. E, ChIP analysis of in vivo Foxo3a binding to catalase promoter. Cardiomyocytes were treated with 20 μg/ml insulin. Cells were harvested at the indicated times for ChIP analysis. Chromatin-bound DNA was immunoprecipitated with the anti-Foxo3a antibody. The anti manganese-superoxide dismutase antibody was used as a negative control (neg). Immunoprecipitated DNA was analyzed by PCR using a primer combination that encompassed Foxo3a BS1 (upper panel) or BS2 (lower panel). Data in Fig. 4 are expressed as the mean ± S.E. of three independent experiments.
Subsequently, we tested whether endogenous Foxo3a is able to influence the promoter activity of catalase. Knockdown of Foxo3a led to a reduced level of catalase promoter activity (Fig. 4C). We further detected whether insulin is able to influence catalase promoter activity. Insulin treatment resulted in a reduction of catalase promoter activity (Fig. 4D).
Finally, we detected whether endogenous Foxo3a binds to catalase promoter in vivo. As shown in Fig. 4E, ChIP assay revealed that Foxo3a bound to the BS2 but not the BS1 of catalase promoter. Insulin treatment led to a reduced associating level of endogenous Foxo3a with catalase promoter. Taken together, these data suggest that Foxo3a can transcriptionally regulate catalase.
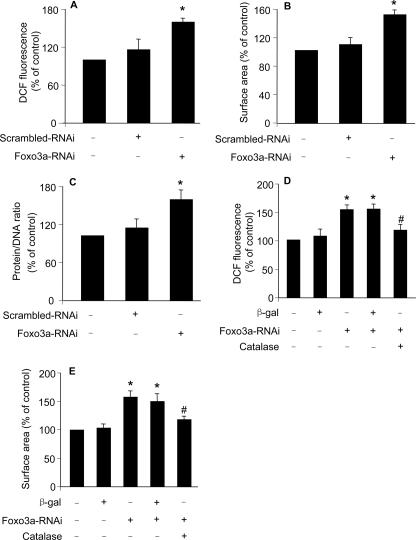
Foxo3a and Catalase Are Functionally Related in Inhibiting Hypertrophy—Because catalase and the functional form of Foxo3a are down-regulated in hypertrophy induced by insulin, and Foxo3a can transcriptionally regulate catalase, we asked whether catalase and Foxo3a are functionally related in hypertrophy. To answer this question, we first tested whether Foxo3a deficiency can influence ROS levels. Knockdown of Foxo3a resulted in an elevated level of ROS (Fig. 5A). Concomitantly, cells underwent hypertrophic responses as assessed by cell surface area measurement (Fig. 5B) and protein/DNA ratio assay (Fig. 5C). These data suggest that Foxo3a knockdown is able to induce ROS elevation and hypertrophy.
FIGURE 5.
Knockdown of Foxo3a leads to hypertrophy inhibited by catalase. A, knockdown of Foxo3a leads to an elevated level of ROS. The neonatal rat cardiomyocytes were infected with Ad-Foxo3a-RNAi or its scrambled form (Ad-scramble RNAi) at an m.o.i. of 100. 24 h after infection, cells were incubated with 5 μm DCFH-DA for 30 min at 37 °C. *, p < 0.05 versus control. B and C, knockdown of Foxo3a leads to hypertrophy. Cardiomyocytes were treated as described for A. 48 h after treatment cells were harvested for the detection of cell surface area (B) or protein/DNA ration (C). *, p < 0.05 versus control. D, catalase attenuates ROS elevation induced by Foxo3a knockdown. Cardiomyocytes were infected with Ad-catalase at an m.o.i. of 50. 24 h after infection, cells were infected with Ad-Foxo3a-RNAi or Ad-scrambled-RNAi at an m.o.i. of 100. Cells were incubated with 5 μm DCFH-DA for 30 min at 37 °C. *, p < 0.05 versus control; #, p < 0.05 versus Foxo3a-RNAi alone. E, catalase attenuates hypertrophy induced by Foxo3a knockdown. Cardiomyocytes were treated as described for D. Cell surface area was measured 48 h after Foxo3a-RNAi treatment. Data in Fig. 5 are expressed as the mean ± S.E. of three independent experiments.
Next, we tested whether catalase can influence hypertrophy upon Foxo3a knockdown. Catalase could attenuate ROS levels (Fig. 5D) and hypertrophy (Fig. 5E) upon Foxo3a knockdown. Thus, it appears that Foxo3a and catalase are functionally related in inhibiting hypertrophy.
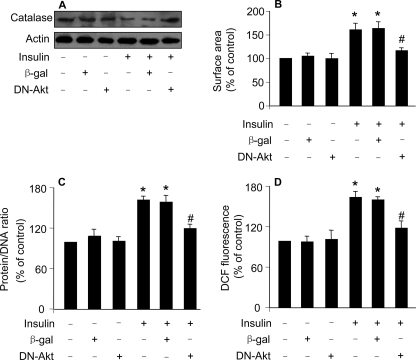
Catalase Is Regulated by Akt in the Hypertrophic Pathway of Insulin—Akt is an upstream regulator of Foxo3a (11). To further confirm the role of Foxo3a-catalase in controlling hypertrophy, we attempted to test whether Akt participates in regulating catalase in the hypertrophic pathway of insulin. As shown in Fig. 6A, the insulin-induced decrease in catalase levels could be attenuated by dominant negative Akt (DN-Akt). Concomitantly, insulin-induced hypertrophic responses as assessed by cell surface area measurement (Fig. 6B) and protein/DAN ratio (Fig. 6C) could be inhibited by DN-AKt. DN-Akt also led to a reduction in ROS levels upon insulin treatment (Fig. 6D). These data suggest that Akt is involved in regulating catalase expression and hypertrophy upon treatment with insulin.
FIGURE 6.
Dominant negative Akt inhibits the decrease of catalase expression and hypertrophy induced by insulin. A, dominant negative Akt (DN-Akt) attenuates the reduction of catalase levels upon treatment with insulin. The neonatal rat cardiomyocytes were infected with Ad-β-galactosidase or Ad-DN-Akt at an m.o.i. of 100. 24 h after infection they were treated with 20 μg/ml insulin. 1 h after treatment cells were collected for the detection of catalase levels by immunoblot. B and C, DN-Akt attenuates hypertrophy upon treatment with insulin. Cardiomyocytes were treated as described for A. 48 h after treatment cells were harvested for the detection of cell surface area (B) or protein/DNA ration (C). *, p < 0.05 versus control; #, p < 0.05 versus insulin alone. D, DN-Akt attenuates ROS elevation induced by insulin. Cardiomyocytes were treated as described for A. 24 h after infection cells were incubated with 5 μm DCF-DA for 30 min at 37 °C, and then treated with 20 μg/ml insulin. ROS was analyzed 1 h after insulin treatment. *, p < 0.05 versus control; #, p < 0.05 versus insulin alone. Data in Fig. 6 are expressed as mean ± S.E. of three independent experiments.
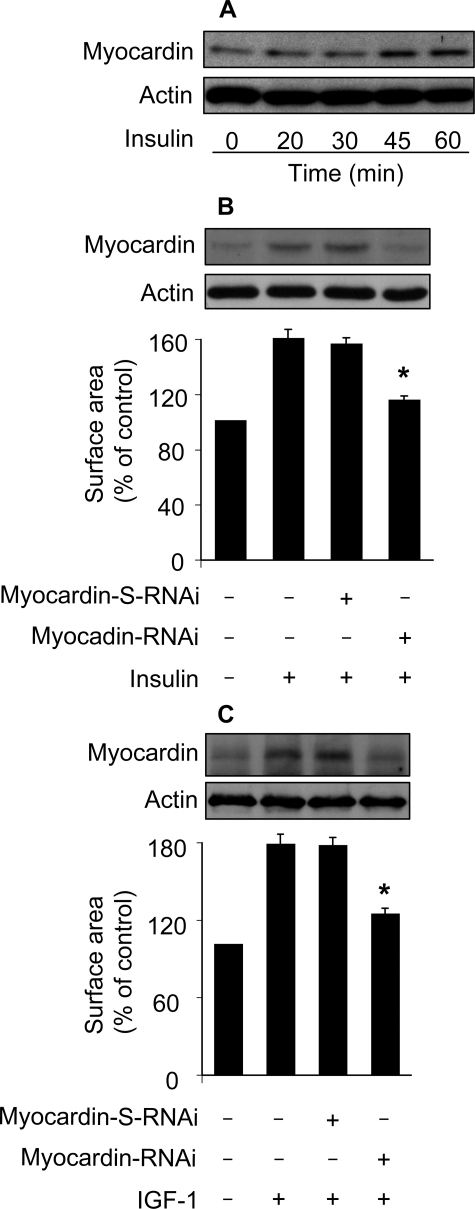
Myocardin Participates in Conveying the Hypertrophic Signal of Insulin and IGF-1—Our above results demonstrate that Foxo3a and catalase constitute an anti-hypertrophic axis. Although this anti-hypertrophic axis is inactivated in insulin-induced hypertrophy, which pro-hypertrophic factor is activated and initiates the hypertrophic program? Myocardin initiates cardiac hypertrophy through a transcription-dependent manner (28), and ROS is able to stimulate the transcriptional machinery (24, 38–40). This encouraged us to test whether myocardin is involved in the hypertrophic pathway of insulin. Insulin treatment led to an elevated level of myocardin (Fig. 7A). We tested whether myocardin is necessary for insulin to regulate hypertrophy. To this end, myocardin RNAi construct was produced, and it could decrease myocardin protein levels (upper panel in Fig. 7B). Knockdown of myocardin could attenuate the hypertrophic responses induced by insulin (lower panel, Fig. 7B). Insulin and IGF-1 have been shown to employ a similar pathway to initiate hypertrophy (11). We thus tested whether myocardin is necessary for IGF-1 to induce hypertrophy. IGF-1 treatment led to an increase in myocardin protein levels. Knockdown of myocardin reduced hypertrophy in response to IGF-1 treatment (Fig. 7C). These results suggest that myocardin is a mediator of insulin and IGF-1 in initiating hypertrophy.
FIGURE 7.
Myocardin is required for insulin and IGF-1 to induce hypertrophy. A, myocardin is up-regulated in response to insulin treatment. The neonatal rat cardiomyocytes were treated with 20 μg/ml insulin, and then harvested at the indicated times for the detection of myocardin using immunoblot. A representative result of three independent experiments is shown. B, myocardin is required for insulin to induce hypertrophy. Cardiomyocytes were infected with Ad-myocardin-RNAi or Ad-myocardin-S-RNAi at an m.o.i. of 80. 30 h after infection cells were treated with insulin. Analysis of myocardin levels by immunoblot was performed 1 h after insulin treatment (upper panel). Cell surface area measurement was performed 48 h after insulin treatment (lower panel). *, p < 0.05 versus insulin alone. C, myocardin is required for IGF-1 to induce hypertrophy. Cardiomyocytes were infected with Ad-myocardin-RNAi or Ad-myocardin-S-RNAi as described for B, except that 30 ng/ml IGF-1 was used. Analysis of myocardin levels by immunoblot was performed 1 h after IGF-1 treatment (upper panel). Cell surface area measurement was performed 48 h after IGF-1 treatment (lower panel). *, p < 0.05 versus IGF-1 alone. Data in B and C are expressed as the mean ± S.E. of three independent experiments.
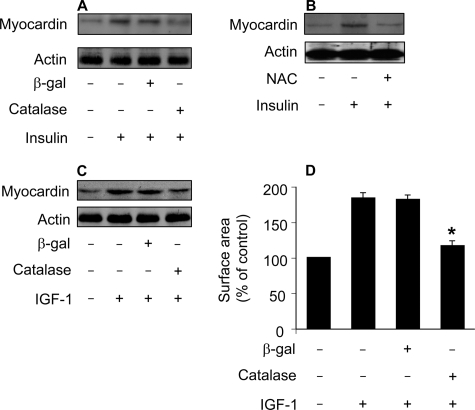
ROS Is Responsible for Myocardin Up-regulation in the Hypertrophic Pathways of Insulin and IGF-1—Because both ROS and myocardin are necessary for insulin to initiate hypertrophy, this raises a question as to what the relationship between ROS and myocardin is. To answer this question, we tested whether inhibition of ROS can influence myocardin expression levels. Enforced expression of catalase could attenuate myocardin levels upon insulin treatment (Fig. 8A). Administration of NAC led to a reduction in myocardin levels (Fig. 8B). IGF-1-induced myocardin up-regulation could be attenuated by catalase (Fig. 8C). Concomitantly, catalase inhibited hypertrophy upon treatment with IGF-1 (Fig. 8D). These data indicate that myocardin up-regulation is a downstream event of ROS in the hypertrophic pathways of insulin and IGF-1.
FIGURE 8.
Myocardin up-regulation induced by insulin and IGF-1 can be attenuated by the antioxidant agents. A, catalase is able to attenuate myocardin levels upon treatment with insulin. The neonatal rat cardiomyocytes were infected with Ad-β-galactosidase or Ad-catalase at an m.o.i. of 50. 24 h after infection cells were treated with 20 μg/ml insulin. Cells were harvested for immunoblot analysis of myocardin 1 h after insulin treatment. B, the antioxidant NAC is able to attenuate myocardin levels upon treatment with insulin. Cardiomyocytes were pretreated with 10 mm NAC for 1 h and then treated with 20 μg/ml insulin. Cells were harvested for immunoblot analysis of myocardin 1 h after insulin treatment. C, catalase attenuates myocardin levels upon treatment with IGF-1. Cardiomyocytes were treated as described for A, except that 30 ng/ml IGF-1 was used. D, catalase inhibits hypertrophy upon treatment with IGF-1. Cardiomyocytes were treated as described for C. Cell surface area measurement was performed 48 h after IGF-1 treatment. *, p < 0.05 versus IGF-1 alone. Data are expressed as the mean ± S.E. of three independent experiments. A representative blot of three independent experiments is shown in Fig. 8.
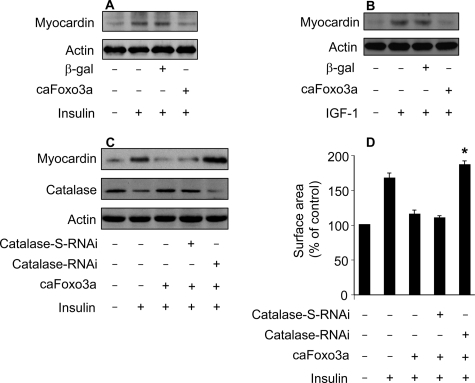
Foxo3a Negatively Regulates Myocardin Levels through Catalase—Finally, we carried out experiments to understand the relationship between Foxo3a and myocardin in the hypertrophic cascades of insulin and IGF-1. We tested whether Foxo3a is able to influence myocardin levels upon treatment with insulin or IGF-1. caFoxo3a could attenuate the elevation of myocardin levels upon insulin treatment (Fig. 9A). IGF-1-induced up-regulation of myocardin also could be inhibited by caFoxo3a (Fig. 9B). To understand whether the effect of caFoxo3a on myocardin expression is related to catalase, we tested whether knockdown of catalase can influence the ability of caFoxo3a to regulate myocardin levels. caFoxo3a could attenuate insulin-induced myocardin up-regulation depending on the presence of catalase (Fig. 9C). Concomitantly, insulin-induced hypertrophy could be attenuated by caFoxo3a in the absence but not presence of catalase RNAi (Fig. 9D). A similar result was obtained in the hypertrophic model of IGF-1 in which caFoxo3a could attenuate myocardin levels and hypertrophy upon IGF-1 treatment in the absence but not presence of catalase RNAi (data not shown). Thus, it appears that Foxo3a negatively regulates myocardin levels depending on catalase.
FIGURE 9.
Foxo3a negatively regulates myocardin levels through catalase. A, caFoxo3a is able to attenuate myocardin levels upon treatment with insulin. The neonatal rat cardiomyocytes were infected with Ad-β-galactosidase or Ad-caFoxo3a at an m.o.i. of 50. 24 h after infection cells were treated with 20 μg/ml insulin. Cells were harvested for immunoblot analysis of myocardin 1 h after insulin treatment. B, caFoxo3a is able to attenuate myocardin levels in response to IGF-1 treatment. Cardiomyocytes were treated as described for A, except that 30 ng/ml IGF-1 was used. C and D, knockdown of catalase abolishes the effect of caFoxo3a on myocardin expression and hypertrophy. Cardiomyocytes were co-infected with Ad-catalase-RNAi (30 m.o.i.), Ad-catalase-S-RNAi (30 m.o.i.), or Ad-caFoxo3a (50 m.o.i.). 24 h after infection cells were treated with 20 μg/ml insulin. Cells were harvested for immunoblot 1 h after insulin treatment (C), or for cell surface area measurement 48 h after insulin treatment (D). *, p < 0.05 versus insulin plus caFoxo3a. Data are expressed as the mean ± S.E. of three independent experiments.
DISCUSSION
Our present work demonstrates that Foxo3a is a modulator of hypertrophy induced by insulin and IGF-1. Foxo3a exerts its anti-hypertrophic effect through transcriptionally activating catalase, thereby reducing ROS levels. Our results further reveal that ROS can stimulate the expression of myocardin, and the latter mediates the hypertrophic signal of insulin and IGF-1. In addition, Foxo3a can negatively regulate myocardin levels through catalase. Our data reveal a novel anti-hypertrophic axis composed of Foxo3a catalase, and the downstream target of this axis is myocardin.
Insulin has been shown to be able to elevate ROS levels in hepatic cell line HepG2 and cardiac cell line H9c2 (41). However, it is not yet clear whether insulin can elevate ROS levels in primary cardiomyocytes. Our present work demonstrates that insulin is able to stimulate ROS elevation during hypertrophy. In particular, ROS is a prerequisite for insulin to induce cardiomyocyte hypertrophy. Previous studies have shown that insulin may initiate hypertrophy through Akt, the latter then controls the function of hypertrophic regulators such as nuclear factor of activated T cells, GATA-4, and atrogin-1 (11). Our data for the first time demonstrate that ROS can be a downstream mediator of insulin in the induction of cardiomyocyte hypertrophy.
Catalase has been shown to participate in the regulation of cardiac hypertrophy. Leptin-induced cardiomyocyte hypertrophy is mediated by ROS and can be blocked by catalase (22). Our recent work demonstrates that catalase is a target of a variety of hypertrophic stimuli such as tumor necrosis factor-α, phenylephrine, and angiotensin II. These stimuli induce a decrease in catalase levels and an increase in ROS levels (30). Our present work reveals that catalase also is a target of insulin and IGF-1 in exerting their hypertrophic effect. Although a recent study has shown that catalase expression can be up-regulated by Foxo1 (42), it is not yet clear whether such regulation is through a direct or indirect manner. Our present study shows that Foxo3a can regulate catalase through a direct and transcription-dependent way. Furthermore, insulin and IGF-1 initiate hypertrophy through inactivating Foxo3a and down-regulating catalase. Thus, it appears that Foxo3a and catalase may form an axis that participates in antagonizing cardiac hypertrophy. Foxo family contains four members (Foxo1, Foxo3a, Foxo4, and Foxo6). It would be interesting to detect whether other members can regulate hypertrophy under physiological and pathological conditions and, if so, whether they control hypertrophy by transcriptionally targeting catalase.
A growing body of evidence has shown that Akt plays an important role in triggering cardiac hypertrophy (11). For example, insulin and IGF-1 can stimulate cardiac hypertrophy via the Akt pathway (14, 43). Overexpression of activated forms of Akt1 or Akt3 causes cardiac hypertrophy (44–46). Nevertheless, the downstream targets of Akt in cardiac hypertrophy remain to be fully understood. Foxo3a is a substrate of Akt (47). Our present study shows that Foxo3a transactivates catalase expression. It would be interesting to elucidate the role of catalase in the hypertrophic pathway of Akt. Hitherto, it remains unknown as to whether Akt requires ROS to mediate its hypertrophic signal. Our present study in the cellular model reveals that the dominant negative Akt is able to suppress insulin-induced ROS elevation, suggesting that there is a link between Akt and ROS in hypertrophic cascades. Future studies are required to reveal whether ROS can be a molecular messenger of Akt in induction of hypertrophy in animal model.
ROS is a signaling molecule, and it itself cannot execute hypertrophy. What are the downstream targets of ROS in triggering hypertrophy? Our results demonstrate that myocardin is up-regulated in the hypertrophic pathway of insulin and IGF-1. Strikingly, the up-regulation of myocardin is a consequent event of ROS elevation in response to insulin and IGF-1 treatment. Myocardin is a transcriptional factor that is able to stimulate cardiac hypertrophy (28). Also, it can mediate the hypertrophic signal of a variety of stimuli such as phenylephrine and endothelin-1 (28, 29). Phenylephrine, mechanical stress, and endothelin-1 require ROS to convey their hypertrophic signals (19, 21, 48, 49). The stimulation of myocardin expression by ROS as revealed by our results may provide a clue in understanding the role of myocardin in ROS-related hypertrophy.
Our present work reveals that Foxo3a catalase constitutes an anti-hypertrophic axis, and this axis can be a target of insulin and IGF-1 in induction of hypertrophy. Thus, our data may shed new light in understanding the molecular mechanism by which insulin and IGF-1 induce hypertrophy. However, there is a large amount of stimuli that is able to regulate cardiac hypertrophy (5, 26, 37, 50, 51). In particular, a variety of hypertrophic stimuli require ROS to initiate hypertrophy. The role of the Foxo3a catalase axis in the hypertrophic pathway of these molecules has not been studied in our present work. Nevertheless, our results may lead to future studies to explore not only the implications of this axis in hypertrophy, but also the beneficial effect of this axis as a biological target for the potential novel therapy for maladaptive cardiac hypertrophy as well as heart failure.
This work was supported by the National Natural Science Foundation of China (Grant 30730045), the National Basic Research Program of China (973 Program, Grant 2007CB512000), and the American Heart Association. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: IGF-1, insulin-like growth factor-1; DN-Akt, dominant negative Akt; NAC, N-acetyl-l-cysteine; RNAi, RNA interference; ROS, reactive oxygen species; MAPK, mitogen-activated protein kinase; DCFH-DA, 2′,7′-dichlorofluorescein diacetate; ChIP, chromatin immunoprecipitation; caFoxo3a, constitutively active form of Foxo3a; CMV, cytomegalovirus; m.o.i., multiplicity of infection; TRITC, tetramethylrhodamine isothiocyanate.
References
- 1.Sugden, P. H. (2003) Circ. Res. 93 1179–1192 [DOI] [PubMed] [Google Scholar]
- 2.Kass, D. A., Bronzwaer, J. G., and Paulus, W. J. (2004) Circ. Res. 94 1533–1542 [DOI] [PubMed] [Google Scholar]
- 3.Hardt, S. E., and Sadoshima, J. (2002) Circ. Res. 90 1055–1063 [DOI] [PubMed] [Google Scholar]
- 4.Nadal-Ginard, B., Kajstura, J., Leri, A., and Anversa, P. (2003) Circ. Res. 92 139–150 [DOI] [PubMed] [Google Scholar]
- 5.Wilkins, B. J., and Molkentin, J. D. (2004) Biochem. Biophys. Res. Commun. 322 1178–1191 [DOI] [PubMed] [Google Scholar]
- 6.De Windt, L. J., Lim, H. W., Taigen, T., Wencker, D., Condorelli, G., Dorn, G. W., 2nd, Kitsis, R. N., and Molkentin, J. D. (2000) Circ. Res. 86 255–263 [DOI] [PubMed] [Google Scholar]
- 7.Satoh, M., Ogita, H., Takeshita, K., Mukai, Y., Kwiatkowski, D. J., and Liao, J. K. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 7432–7437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li, H. H., Willis, M. S., Lockyer, P., Miller, N., McDonough, H., Glass, D. J., and Patterson, C. (2007) J. Clin. Invest. 117 3211–3223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Samuelsson, A. M., Bollano, E., Mobini, R., Larsson, B. M., Omerovic, E., Fu, M., Waagstein, F., and Holmang, A. (2006) Am. J. Physiol. 291 H787–H796 [DOI] [PubMed] [Google Scholar]
- 10.Paolisso, G., Galzerano, D., Gambardella, A., Varricchio, G., Saccomanno, F., D'Amore, A., Varricchio, M., and D'Onofrio, F. (1995) Eur. J. Clin. Invest. 25 529–533 [DOI] [PubMed] [Google Scholar]
- 11.Shiojima, I., and Walsh, K. (2006) Genes Dev. 20 3347–3365 [DOI] [PubMed] [Google Scholar]
- 12.Accili, D., and Arden, K. C. (2004) Cell 117 421–426 [DOI] [PubMed] [Google Scholar]
- 13.Furuyama, T., Nakazawa, T., Nakano, I., and Mori, N. (2000) Biochem. J. 349 629–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Skurk, C., Izumiya, Y., Maatz, H., Razeghi, P., Shiojima, I., Sandri, M., Sato, K., Zeng, L., Schiekofer, S., Pimentel, D., Lecker, S., Taegtmeyer, H., Goldberg, A. L., and Walsh, K. (2005) J. Biol. Chem. 280 20814–20823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anderson, M. J., Viars, C. S., Czekay, S., Cavenee, W. K., and Arden, K. C. (1998) Genomics 47 187–199 [DOI] [PubMed] [Google Scholar]
- 16.Sandri, M., Sandri, C., Gilbert, A., Skurk, C., Calabria, E., Picard, A., Walsh, K., Schiaffino, S., Lecker, S. H., and Goldberg, A. L. (2004) Cell 117 399–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ni, Y. G., Berenji, K., Wang, N., Oh, M., Sachan, N., Dey, A., Cheng, J., Lu, G., Morris, D. J., Castrillon, D. H., Gerard, R. D., Rothermel, B. A., and Hill, J. A. (2006) Circulation 114 1159–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakamura, K., Fushimi, K., Kouchi, H., Mihara, K., Miyazaki, M., Ohe, T., and Namba, M. (1998) Circulation 98 794–799 [DOI] [PubMed] [Google Scholar]
- 19.Hirotani, S., Otsu, K., Nishida, K., Higuchi, Y., Morita, T., Nakayama, H., Yamaguchi, O., Mano, T., Matsumura, Y., Ueno, H., Tada, M., and Hori, M. (2002) Circulation 105 509–515 [DOI] [PubMed] [Google Scholar]
- 20.Pimentel, D. R., Amin, J. K., Xiao, L., Miller, T., Viereck, J., Oliver-Krasinski, J., Baliga, R., Wang, J., Siwik, D. A., Singh, K., Pagano, P., Colucci, W. S., and Sawyer, D. B. (2001) Circ. Res. 89 453–460 [DOI] [PubMed] [Google Scholar]
- 21.Aikawa, R., Nagai, T., Tanaka, M., Zou, Y., Ishihara, T., Takano, H., Hasegawa, H., Akazawa, H., Mizukami, M., Nagai, R., and Komuro, I. (2001) Biochem. Biophys. Res. Commun. 289 901–907 [DOI] [PubMed] [Google Scholar]
- 22.Xu, F.-P., Chen, M.-S., Wang, Y.-Z., Yi, Q., Lin, S.-B., Chen, A. F., and Luo, J.-D. (2004) Circulation 110 1269–1275 [DOI] [PubMed] [Google Scholar]
- 23.Brasier, A. R., Jamaluddin, M., Han, Y., Patterson, C., and Runge, M. S. (2000) Mol. Cell. Biochem. 212 155–169 [PubMed] [Google Scholar]
- 24.Kuster, G. M., Pimentel, D. R., Adachi, T., Ido, Y., Brenner, D. A., Cohen, R. A., Liao, R., Siwik, D. A., and Colucci, W. S. (2005) Circulation 111 1192–1198 [DOI] [PubMed] [Google Scholar]
- 25.Nishida, M., Maruyama, Y., Tanaka, R., Kontani, K., Nagao, T., and Kurose, H. (2000) Nature 408 492–495 [DOI] [PubMed] [Google Scholar]
- 26.Ago, T., and Sadoshima, J. (2007) Antioxid. Redox Signal. 9 679–687 [DOI] [PubMed] [Google Scholar]
- 27.Kuster, G. M., Siwik, D. A., Pimentel, D. R., and Colucci, W. S. (2006) Antioxid. Redox Signal. 8 2153–2159 [DOI] [PubMed] [Google Scholar]
- 28.Xing, W., Zhang, T. C., Cao, D., Wang, Z., Antos, C. L., Li, S., Wang, Y., Olson, E. N., and Wang, D. Z. (2006) Circ. Res. 98 1089–1097 [DOI] [PubMed] [Google Scholar]
- 29.Badorff, C., Seeger, F. H., Zeiher, A. M., and Dimmeler, S. (2005) Circ. Res. 97 645–654 [DOI] [PubMed] [Google Scholar]
- 30.Murtaza, I., Wang, H. X., Feng, X., Alenina, N., Bader, M., Prabhakar, B. S., and Li, P. F. (2008) J. Biol. Chem. 283 5996–6004 [DOI] [PubMed] [Google Scholar]
- 31.Li, P. F., Dietz, R., and von Harsdorf, R. (1999) EMBO J. 18 6027–6036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li, Y. Z., Lu, D. Y., Tan, W. Q., Wang, J. X., and Li, P. F. (2008) Mol. Cell. Biol. 28 564–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.von Harsdorf, R., Li, P. F., and Dietz, R. (1999) Circulation 99 2934–2941 [DOI] [PubMed] [Google Scholar]
- 34.Li, P. F., Li, J., Muller, E. C., Otto, A., Dietz, R., and von Harsdorf, R. (2002) Mol. Cell 10 247–258 [DOI] [PubMed] [Google Scholar]
- 35.Shang, Y., Hu, X., DiRenzo, J., Lazar, M. A., and Brown, M. (2000) Cell 103 843–852 [DOI] [PubMed] [Google Scholar]
- 36.Giordano, F. J. (2005) J. Clin. Invest. 115 500–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang, J., Shelton, J. M., Richardson, J. A., Kamm, K. E., and Stull, J. T. (2008) J. Biol. Chem. 283 19748–19756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wollert, K. C., Heineke, J., Westermann, J., Ludde, M., Fiedler, B., Zierhut, W., Laurent, D., Bauer, M. K. A., Schulze-Osthoff, K., and Drexler, H. (2000) Circulation 101 1172–1178 [DOI] [PubMed] [Google Scholar]
- 39.Higuchi, Y., Otsu, K., Nishida, K., Hirotani, S., Nakayama, H., Yamaguchi, O., Matsumura, Y., Ueno, H., Tada, M., and Hori, M. (2002) J. Mol. Cell. Cardiol. 34 233–240 [DOI] [PubMed] [Google Scholar]
- 40.Sen, C. K., and Roy, S. (2005) Am. J. Physiol. 289 H17–H19 [DOI] [PubMed] [Google Scholar]
- 41.Biswas, S., Gupta, M. K., Chattopadhyay, D., and Mukhopadhyay, C. K. (2007) Am. J. Physiol. 292 H758–H766 [DOI] [PubMed] [Google Scholar]
- 42.Alcendor, R. R., Gao, S., Zhai, P., Zablocki, D., Holle, E., Yu, X., Tian, B., Wagner, T., Vatner, S. F., and Sadoshima, J. (2007) Circ. Res. 100 1512–1521 [DOI] [PubMed] [Google Scholar]
- 43.McMullen, J. R., Shioi, T., Huang, W.-Y., Zhang, L., Tarnavski, O., Bisping, E., Schinke, M., Kong, S., Sherwood, M. C., Brown, J., Riggi, L., Kang, P. M., and Izumo, S. (2004) J. Biol. Chem. 279 4782–4793 [DOI] [PubMed] [Google Scholar]
- 44.Condorelli, G., Drusco, A., Stassi, G., Bellacosa, A., Roncarati, R., Iaccarino, G., Russo, M. A., Gu, Y., Dalton, N., Chung, C., Latronico, M. V. G., Napoli, C., Sadoshima, J., Croce, C. M., and Ross, J., Jr. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 12333–12338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matsui, T., Li, L., Wu, J. C., Cook, S. A., Nagoshi, T., Picard, M. H., Liao, R., and Rosenzweig, A. (2002) J. Biol. Chem. 277 22896–22901 [DOI] [PubMed] [Google Scholar]
- 46.Taniyama, Y., Ito, M., Sato, K., Kuester, C., Veit, K., Tremp, G., Liao, R., Colucci, W. S., Ivashchenko, Y., Walsh, K., and Shiojima, I. (2005) J. Mol. Cell. Cardiol. 38 375–385 [DOI] [PubMed] [Google Scholar]
- 47.Greer, E. L., and Brunet, A. (2005) Oncogene 24 7410–7425 [DOI] [PubMed] [Google Scholar]
- 48.Tanaka, K., Honda, M., and Takabatake, T. (2001) J. Am. Coll. Cardiol. 37 676–685 [DOI] [PubMed] [Google Scholar]
- 49.Ruwhof, C., and van der Laarse, A. (2000) Cardiovasc. Res. 47 23–37 [DOI] [PubMed] [Google Scholar]
- 50.Kassiri, Z., Zobel, C., Nguyen, T. T., Molkentin, J. D., and Backx, P. H. (2002) Circ. Res. 90 578–585 [DOI] [PubMed] [Google Scholar]
- 51.Vega, R. B., Rothermel, B. A., Weinheimer, C. J., Kovacs, A., Naseem, R. H., Bassel-Duby, R., Williams, R. S., and Olson, E. N. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 669–674 [DOI] [PMC free article] [PubMed] [Google Scholar]