Abstract
For a long period of time, the transcription factor CCAAT/enhancer-binding protein homologous protein (CHOP) has been thought to inhibit transcriptional activity for its ability to interact with CCAAT enhancer-binding protein family factors, thus preventing their binding to DNA. We have previously shown that in human T lymphocytes the CHOP phosphorylation induced by prostaglandin E2 (PGE2)-increased interleukin-8 (IL-8) gene expression. Given the CHOP positive role in the regulation of transcription, here we have investigated the molecular mechanism(s) by which CHOP increases IL-8 gene activity under PGE2 stimulus. Transfection experiments with mutants showed both that the CHOP transactivation domain is essential for IL-8 transcription and that the IL-8/activator protein 1 (AP-1) promoter mutated in NF-κB and NF-IL-6, but not in the AP-1 site, harbors essential CHOP-responsive elements. CHOP silencing confirmed its role in the IL-8 transcriptional regulation and protein production, whereas c-Jun small interfering RNA experiments showed that the PGE2-induced activation of IL-8 promoter is mainly c-Jun-independent. Moreover, PGE2 induced CHOP-DNA complexes only when the entire IL-8/AP-1 promoter or the wild type sequences encompassing the AP-1 upstream region were employed. Mutations introduced in these sequences prevented the DNA-CHOP complex formation. The IL-8/AP-1 mutant promoter lacking the sequence immediately upstream the AP-1 site is PGE2-unresponsive. Finally, chromatin immunoprecipitation data confirmed in vivo that PGE2 induces CHOP binding to the IL-8 promoter. Taken together, our results suggest that the increased expression of CHOP in response to PGE2 exerts a positive transcriptional regulation of the IL-8 promoter mediated by direct binding to a novel consensus site.
We have previously reported that prostaglandin E2 (PGE2)2 stimulates the transcription of IL-8 gene in human T lymphocytes by activating in a protein kinase C-, p38 MAPK-, and phosphatidylinositol 3-kinase-dependent manner the transcription factor C/EBP homologous protein (CHOP) (1). More recently, the positive correlation between CHOP and IL-8 synthesis has been confirmed by experiments showing that the inhibition of CHOP repressed the NF-κB-mediated and proteasome-induced IL-8 activation (2). These results corroborated a positive role for CHOP in the induction of pro-inflammatory genes that was previously indicated on the IL-6 expression in melanoma cells, although with distinct mechanisms (3). The activation of IL-6 transcription was dependent on the CHOP dimerization ability, given that a mutant lacking the leucine zipper domain exhibited a reduced effect on IL-6 promoter activity. CHOP was originally cloned as an inhibitory molecule of the C/EBP family transcriptional factors, to which it binds with its extreme C-terminal leucine zipper region, thus blocking the binding of C/EBPs to DNA (4, 5). Afterward, it has been reported that CHOP can act as transcription factor inducing the gene expression by forming a heterodimer (6). It has also been shown that CHOP can interact with a variety of proteins in a different manner. The gain-of-function of CHOP induces its binding to several nuclear protein from erythroid cells, some of which are not C/EBP family members (7). More recently, it has been reported that truncation of the N-terminal amino acids 1–42 (CHOPΔN) abolishes its co-localization with γ-aminobutyric acid type B1a receptor, indicating that CHOP may also interact with its N-terminal domain (8). On the other hand, a direct activity of CHOP as transcription factor was demonstrated by Ubeda et al. (9), who showed CHOP binding to a specific nucleotide sequence, which is similar to but distinct from the typical C/EBP-binding motif. Moreover, dose-dependent bidirectional effects of CHOP on the monocyte chemo-attractant protein-1 promoter were shown in vascular smooth muscle cells (10). Low expression of CHOP blocked the C/EBP binding to C/EBP-responsive elements, whereas increased expression directly stimulated the activity of monocyte chemo-attractant protein-1. Thus, CHOP seems to exhibit different, and in some ways opposite, mechanisms in regulating gene expression and molecular interactions. In the present study, we addressed the molecular mechanism(s) by which CHOP increases the IL-8 gene expression following its activation by PGE2. Initially we investigated whether PGE2 induced CHOP expression in T cells; successively we demonstrated that CHOP is essential for PGE2-mediated IL-8 transcription, identifying the CHOP domains and the IL-8 promoter sites involved. Finally, a CHOP-responsive element (CHOP-RE) in the IL-8 promoter of human T lymphocytes stimulated by PGE2 was characterized.
EXPERIMENTAL PROCEDURES
Cells and Culture Conditions—Jurkat T cells (clone E6–1) were obtained from European Collection of Cell Cultures (Sigma-Aldrich) and maintained in a humidified atmosphere of 5% CO2 in 95% air (37 °C) in RPMI 1640 (Euroclone, Milan, Italy) supplemented with 50 IU/ml penicillin, 50 μg/ml streptomycin (Euroclone, Milan, Italy), and 20% heat-inactivated fetal calf serum (Euroclone, Milan, Italy). The cells were routinely tested for mycoplasma infection, and the cultures were renewed, from frozen stocks, every 2 months.
RNA Extraction and Reverse Transcriptase—T cells were cultured in 35-mm well plates (5 × 106 cells/well) in the absence or presence of anti-CD3 (1 μg/ml) plus anti-CD28 (250 ng/ml) antibodies (Immnunotech, Marseilles, France), and the cells were then immediately stimulated for the indicated times with 10–5 m PGE2. Total RNA was extracted with TRIzol (Invitrogen) according to the manufacturer's instructions. 1 μg of total RNA was reverse-transcribed in a total volume of 20 μl with an IMProm-IITM reverse transcriptase kit (Promega, Milan, Italy) according to the manufacturer's instructions.
Reverse Transcription PCR—20 μl of reverse transcription products were brought to a volume of 100 μl containing 2 mm MgCl2, 0.2 mm PCR nucleotide mixture, a 1 μm concentration of both the upstream and downstream PCR primers (Sigma), 5 units of Taq DNA polymerase (Transgenomic, Inc., Bergamo, Italy), and 10× PCR buffer (Transgenomic, Inc.). Two pairs of primers were used in this study. The primer sequences were as follows: CHOP, 5′-CAGAACCAGCAGAGGTCACA-3′ (sense) and 5′-AGCTGTGCCACTTTCCTTTC-3′ (antisense); and β-actin, 5′-TGACGGGGTCTACCCACACTGTGCCCCATCTA-3′ (sense) and 5′-CTAGAAGCATTGCGCTGGACGATGGAGGG-3′ (antisense). Amplification was carried in a DNA thermal cycler (Applied Biosystems, Milan) after an initial denaturation at 95 °C for 4 min. This was followed by 35 cycles of PCR using the following temperature and time profile: denaturation at 94 °C for 1 min, primer annealing for 1 min at 60 °C for CHOP and at 62 °C for β-actin, primer extension at 72 °C for 2 min, and a final extension at 72 °C for 7 min. The PCR products were visualized by electrophoresis on 1% agarose gel in 1× buffer containing 89 mm Tris borate and 2 mm EDTA (pH 8.3) after staining with 0.5 μg/ml ethidium bromide. The UV light-illuminated gels were photographed, and the relative sum intensity was calculated by normalizing the sum intensity of the CHOP product to the β-actin mRNA control.
Quantitative Real-time PCR—Quantitative real-time PCR was performed using the ABI Prism 7500 real-time PCR system (Applied Biosystems). We used 5′-GCTTCTCTGGCTTGGCTGACT-3′ (Forward primer), 5′-CTGTTTCCGTTTCCTGGTTCTC-3′ (Reverse primer), and FAM-5′-CACTCTCCAGATTCCAGTCA-3′-MGBNFQ as primers and a TaqMan probe, respectively. Thermal cycling conditions included activation at 95 °C (10 min) followed by 40 cycles each of denaturation at 95 °C (15 s) and annealing/elongation at 55 °C (1 min). Each sample was analyzed in triplicate with β-actin (Applied Biosystems) as an the inner control, and the mean value of CHOP mRNA was calculated.
The cycle threshold (Ct) was used to calculate relative amounts of target DNA. The Ct was determined as the number of PCR cycles required for a given reaction to reach an arbitrary fluorescence value within the linear amplification range.
Plasmid Construction and Transient Transfections—Liposome-mediated transient gene transfer was carried out with DMRIE-C (Invitrogen) as recommended by the manufacturer. Briefly, the cells were seeded at 2 × 106 cells/well and transiently transfected with 1 μg of wt IL-8, IL-8 lacking the AP-1 site (IL-8–97), IL-8–97 mutant C/EBP (IL-8–97/mC/EBP), IL-8–97 mutant NF-κB (IL-8–97/mNF-κB), or IL-8 double mutant for C/EBP and NF-κB (IL-8/AP-1) promoter-driven luciferase reporter vectors developed in collaboration with Dr. Hector R. Wong (Cincinnati Children's Hospital Medical Center, Cincinnati OH); pSV-nlsLacZ DNA, a β-galactosidase expression vector (0.5 μg) and “empty” plasmid DNA (pBSM), at a final concentration of 2 μg/plate. pcDNA 3.1/V5 His expression plasmids for CHOP at a final concentration of 0.2 μg/plate were co-transfected where indicated. A mutant IL-8/AP-1 promoter construct (IL-8/AP-1ΔCHOP-RE) lacking the nucleotides GTGTGATG located upstream from the AP-1 was generated by PCR using forward primer 5′-TCCcccgggACTCAGGTTTGCCCTGAGGGGA and reverse primer 5′-CCGctcgagTGCCTTATGGAGTGCTCCGGTG. PCR conditions were 35 cycles at 94 °C for 45 s, 52 °C for 45 s, 72 °C for 45 s, and a final extension at 72 °C for 10 min. The PCR product was cloned as a SmaI/XhoI fragment into PGL2 basic vector (Promega). The constructs were sequenced before utilization.
wt CHOP and CHOP mutants lacking the basic DNA-binding domain (CHOPΔBR) or the leucine zipper domain (CHOPΔLZ) or mutated into the p38 MAPK-dependent phosphorylation site (S79A,S82A) were kindly provided by Dr. Hidetoshi Hayashi (Nagoya City University, Nagoya, Japan). The CHOP mutant lacking either the DNA-binding or the leucine zipper domains and containing the sole transactivation domain (CHOPTA) was obtained by Stratagene (Stratagene, La Jolla, CA). To create the dominant negative transactivation domain mutant CHOP (TAM-CHOP), the nucleotide sequences were amplified by PCR from the human genome using the forward primer 5′-ATGGGTACCTATGTTTCACCTCCTGG-3′ and the reverse primer 5′-TCATGCTTGGTGCAGATTCACCATTC-3′. The PCR products were cloned into a pcDNA 3.1/V5 His expression vector (TOPO TA; Invitrogen). The sequence was confirmed by DNA sequencing (CEQ 2000; Beckman, Fullerton, CA). wt c-Jun plasmid was provided by Lucia Altucci (Department of General Pathology, Second University of Naples, Naples, Italy).
In all of the experiments, the transfections were stopped after 6 h by adding an equal volume of RPMI 1640 containing 20% fetal calf serum. Twenty-four hours after transfection, the cells were treated with 10 μm PGE2 (Sigma, Milan, Italy). After an additional 6 h, the cells were harvested, and protein extracts were prepared for the luciferase activity using luciferine (Promega, Milan, Italy) as the substrate. Luciferase activity was normalized for β-galactosidase activity produced by co-transfected plasmid pnlsLAC.
siRNA and Transfection—The siRNA sequence targeting CHOP mRNA corresponding to 5′-CAAUUGUUCAUGCUUGGUGUU-3′, the siRNA sequence targeting c-Jun mRNA corresponding to 5′-GGAUCAAGGCGGAGAGGAA-3′ and the nonspecific duplex corresponding to 5′-CAGUGGAGAUCAACGUGCAAGUU-3′ (ctr-siRNA) were obtained from Dharmacon (Lafayette, CO). Concentrations of siRNA and times of incubation were tested. Both CHOP and c-Jun siRNA knockdown reached the maximum at a concentration of 50 nm and at 48 h post-transfection. To assess gene silencing, the protein levels of CHOP and c-Jun were determined by immunoblotting. Jurkat T cells were plated in 35-mm well plates in RPMI 1640 medium without fetal bovine serum and co-transfected with double-stranded siRNA and wt IL-8 promoter (1 μg) using the DMRIE-C reagent according to the manufacturer's instructions. 48 h after the transfection, the cells were incubated in the absence or presence of anti-CD3 (1 μg/ml) plus anti-CD28 (250 ng/ml) antibodies with PGE2 10 μm. After an additional 6 h, the cells were harvested, and protein extracts were prepared for the luciferase activity using luciferine (Promega, Milan, Italy) as the substrate. Luciferase activity was normalized for β-galactosidase activity produced by co-transfected plasmid pnlsLAC.
Western Blot Analysis—The cells were cultured in 35-mm well plates at a concentration of 5 × 106 cells/well. After stimulation, the cells were lysed at 4 °C in 150 μl of lysis buffer M-PER (Pierce) and 1 mm phenylmethylsulfonyl fluoride, 1 mm sodium orthovanadate, 1 mg/ml leupeptin. Where indicated, total extracts were prepared from Jurkat T cells 36 h after transfection with wt or mutant pcDNA 3.1/V5 His CHOP expression plasmids.
Western blotting was carried out as previously described (1). CHOP (Affinity BioReagents™) and c-Jun (Santa Cruz Biotechnology, Santa Cruz, CA) mouse monoclonal antibodies, diluted 1:500, and anti-V5 mouse monoclonal antibody, diluted 1:5000 (Invitrogen), have been employed, as described by the manufacturer. A mouse monoclonal 1:1000 diluted primary anti-β-actin antibody was used to normalize protein loading to that of specific proteins in each lane (Sigma-Aldrich Milan, Italy). After washings in phosphate-buffered saline with 0.3% Tween 20, the membranes were incubated with secondary horseradish peroxidase-conjugated anti-mouse IgG for 1 h. The proteins were visualized with reagents from Pierce (Super-signal West Pico chemiluminescent substrate system).
IL-8 Production Assay—Jurkat T cells were cultured in round-bottom 24-well plates (1 × 106 cells/well) and transiently transfected with IL-8/AP-1 or IL-8/AP-1ΔCHOP-RE promoters, as above reported. Where indicated, the cells were co-transfected with wt CHOP, wt c-Jun plasmids, or both, in the presence or absence of CHOP or c-Jun siRNAs. After transfection, the cells were stimulated with anti-CD3 (1 μg/ml) plus anti-CD28 (250 ng/ml) antibodies and immediately treated with 10–5 m PGE2. 24 h after stimulation, the supernatants were collected and analyzed for IL-8 production using an enzyme-linked immunosorbent assay (ELISA) kit (BioSource International, Nivelles, Belgium) according to the manufacturer's instructions.
Electrophoretic Mobility Shift Assay (EMSA)—Isolation of nuclear proteins from cultures of 1 × 108 Jurkat T cells grown in RPMI 1640 (Euroclone) with 10% fetal calf serum (Euroclone) supplemented with 50 IU/ml penicillin, 50 μg/ml streptomycin (Euroclone) in 5% CO2 at 37 °C was performed according to a recently detailed method (11). Where indicated, the nuclear extracts were prepared from Jurkat T cells 36 h after transfection with c-Jun or TAM-CHOP plasmids.
Nuclear extracts (20 μg) were incubated for 30 min at room temperature with 50 fmol of biotin-3′-end-labeled, double-strands of different consensus or mutated probes (Table 1) (both complementary oligonucleotides were end-labeled separately and then annealed prior to use). Binding reaction mixture was prepared in a final volume of 20 μl of HEPES buffer containing 1 mg of double-stranded poly(dI/dC), 10% glycerol, 100 mm MgCl2, and 1% Nonidet P-40, performed with the Light-Shift™ Chemiluminescent EMSA kit (Pierce), according to the manufacturer's instructions. For competition assay, 50-fold excess unlabeled double-stranded oligonucleotides used as competitors were incubated with the extracts at room temperature 10 min prior to probe addition. For antibody supershift assays, the nuclear extracts were incubated with the respective antibodies in the same binding reaction volume for an additional hour at 4 °C. The antibodies used were anti-CHOP (Affinity BioReagents™) and anti-c-Jun (Santa Cruz Biotechnology, Santa Cruz, CA). The bound complexes were separated on 7.5% nondenaturating polyacrylamide gels, blotted onto nylon membrane, and visualized on Kodak x-ray film (Kodak) by autoradiography.
TABLE 1.
Sequences of wild type (A, B, and C) and mutated (Mut 1–5) probes employed
The AP-1 site is indicated by the underlined bases. The mutated sequences are shown with bold, italic type.
| Oligonucleotide name | Sequence (5′ → 3′) |
|---|---|
| Aa | GAAGTGTGATGACTCAGGTTGCCCTGAGGGGATGGGCCATC |
| Mut A | GAAGTGTGACTTCTCAGGTTGCCCTGAGGGGATGGGCCATC |
| Bb | ACAAAATAGGAAGTGTGATG |
| Mut B1 | TGGACCGATGAAGTGTGATG |
| Mut B2 | ACAAAATAGGAAGTGTGGCA |
| Mut B3 | ACAAAATAGTCCGTGTGATG |
| Mut B4 | ACAAAATAGGACACGTGATG |
| Mut B5 | ACAAAATAGGAAGTTACATG |
| Cc | TGACTCAGGTTTGCCCTGAGGGATGGGCCATC |
| Mut C | TGACGACGGTTTGCCCTGAGGGATGGGCCATC |
Region of IL-8 promoter (GenBank™ accession number M28130) between bases -141 and -99.
Region of IL-8 promoter between bases -150 and -130.
Region of IL-8 promoter between bases -129 and -99.
Chromatin Immunoprecipitation (ChIP)—ChIP was performed essentially as described by Weinmann and Farnhan (12) with the following modifications. Cultures of 1 × 108 Jurkat T cells were grown in RPMI 1640 (Euroclone) with 10% fetal calf serum (Euroclone) supplemented with 50 IU/ml penicillin, 50 μg/ml streptomycin (Euroclone) in 5% CO2 at 37 °C. Cross-linking was induced by adding 1% (v/v) formaldehyde and incubation for 10 min at room temperature on a shaker. After stopping the cross-linking reaction by adding 0.125 m glycine and incubation for 5 min (with shaking at room temperature), the cells were pelletted and washed twice in ice-cold phosphate-buffered saline including protease inhibitors. The nuclei were isolated by resuspending the cellular pellet in 1 ml of ice-cold swelling buffer (5 mm PIPES, pH 8.0, 85 mm KCl, 0.5% Nonidet P-40, and protease inhibitors), split into two aliquots, and incubated on ice for 10 min. Chromatin was fragmented by subjecting the nuclei to sonication procedure, using four pulses of 15 s each at setting 7 on a Fisher model 60 sonic dismembranator.
The lysate was combined and transferred to a 15-ml conical tube and diluted with 9 ml of dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mm EDTA, 16.7 mm Tris-HCl, pH 8.1, 167 mm NaCl, and protease inhibitors). An aliquot of 500 μl of protein A-Sepharose beads (Pharmacia) was added to the diluted nuclear lysate and incubated for 2 h at 4 °C while rotating. The beads were pelletted for 10 min at 2000 rpm, and the supernatant was divided into 10 aliquots. 25 μl containing 1 μg of the appropriate monoclonal antibody (c-Jun or CHOP; Chemicon International and Affinity BioReagents, respectively) or no antibody was added to the aliquoted supernatant and incubated at 4 °C overnight while rotating. The pellets were washed twice with 1× dialysis buffer (2 mm EDTA, 50 mm Tris-Cl, pH 8.0, phenylmethylsulfonyl fluoride and then four times with IP wash buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mm EDTA, 16.7 mm Tris-Cl, pH 8.1, 167 mm NaCl, protease inhibitors). Elution of antibody-protein-DNA complexes was obtained by the addition of 200 μl of elution buffer (50 mm NaHCO3, 1% SDS). Residual traces of Staph A cells were removed adding 1 μl of high concentration of RNase A (Roche Applied Sciences catalog number 1579681; 10 mg/ml) and 12 μl of 5 m NaCl to a final concentration of 0.3 m. After incubation of samples at 67 °C for 4–5 h to reverse formaldehyde cross-links, 2.5 volumes of ethanol was added, and precipitation was achieved at –20 °C overnight. DNA extraction was performed with phenol/chloroform/isoamyl alcohol protocol. DNA was precipitated adding 30 μl of 5 m NaCl, 5 μg of glycogen (Invitrogen), and 750 μl ethanol for overnight.
DNA was dissolved in 10 μl of TE buffer and quantified using a genomic PCR, as reported by Vij et al. (13). Briefly, the 2-μl DNA sample was mixed with 1 μm IL-8 forward primer (CATACTCCGTATTTGATAAGGAAC), 1 μm IL-8 reverse primer (GGCTCTTGTCCTAGAAGCTT), and 16 μl of PCR supermix (Invitrogen). The PCR was carried out as follows: 95 °C for 10 min, 28 three-step cycles: 95 °C for 15 s, 60 °C for 30 s, and 72 °C for 30 s. Where indicated, DNA was mixed with 1 μm IL-6 forward primer (CCTCAGACATCTCCAGTCCTATA), 1 μm IL-6 reverse primer (TGCTTCTTAGCGCTAGCCTCAAT), and 16 μl of PCR supermix (Invitrogen). The PCR conditions were as follows: 95 °C for 10 min, 38 three-step cycles: 94 °C for 40 s, 68 °C for 50 s, and 72 °C for 60 s. All of the samples were analyzed by 2% agarose gels containing ethidium bromide.
Densitometry and Statistical Analysis—The relative intensities of protein and nucleic acid bands were analyzed using the Digital Sciences one-dimensional program from Kodak Scientific Imaging Systems (New Haven, CT). Standard curves were run, and the data that were obtained were in the linear range of the curve. In addition, all of the values were normalized to their respective lane loading controls. The data are expressed as the means ± S.E. of n determinations. The results were analyzed by two-tailed Student's t test. p values <0.05 were considered significant.
RESULTS
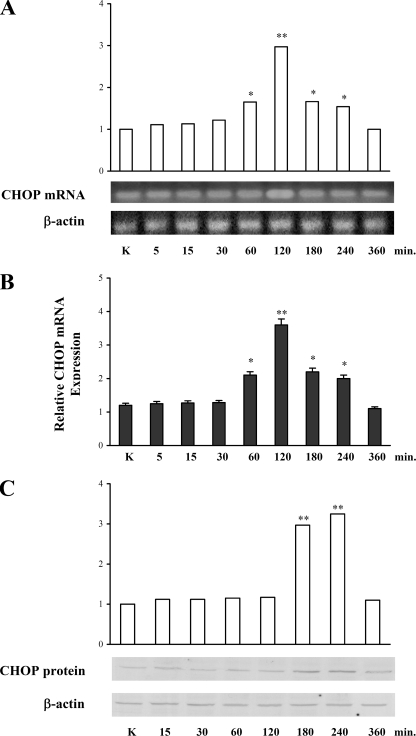
CHOP Expression after Stimulation with PGE2—Because we have previously shown that PGE2 is able to induce IL-8 promoter expression through phosphorylation and activation of CHOP in T lymphocytes (1), we investigated whether PGE2 also affects the expression of this transcription factor. To this end, reverse transcription-PCR, real-time PCR, and Western blot analyses were performed in Jurkat T cells treated with 10–5 m PGE2 for different periods of time. Fig. 1A shows that the mRNA expression of CHOP was induced by PGE2 at 60 min after treatment and decreased at 180 min, with a peak at 120 min (p < 0.01). These results were confirmed and strengthened by data obtained from quantitative PCR experiments, which showed a similar pattern of mRNA synthesis after PGE2 treatment (Fig. 1B). Fig. 1C shows the increase in CHOP protein levels after PGE2 treatment, which reaches the maximum between 180 and 240 min (p < 0.01). Therefore, PGE2 stimulation leads to a time-dependent increase of CHOP at both mRNA and protein levels.
FIGURE 1.
CHOP synthesis in Jurkat T cells treated with 10–5m PGE2. A, CHOP and β-actin (as internal control) mRNA were measured by reverse transcription-PCR. The expected product sizes of CHOP and β-actin are 198 and 391 bp, respectively. The relative density was calculated by dividing the density of CHOP band by the density of the β-actin band at the same time point. The data are the means ± S.D. of five independent experiments. *, p < 0.05; **, p < 0.01 versus control untreated cells, based on Student's two-tailed t test. B, quantification of CHOP mRNA was performed using real-time PCR, and the β-actin gene was used as an internal control. The data are the means ± S.D. of three independent experiments. *, p < 0.05; **, p < 0.01 versus control untreated cells, based on Student's two-tailed t test. C, total cell lysates of T cells were loaded on gels (60 μg of protein/lane) and subjected to SDS-PAGE and immunoblotted using monoclonal antibody anti-CHOP. The mean densitometry values are depicted as the means ± S.D. of five independent experiments. All of the densitometry values were normalized to the endogenous β-actin protein. **, p < 0.01 versus control untreated cells based on Student's two-tailed t test.
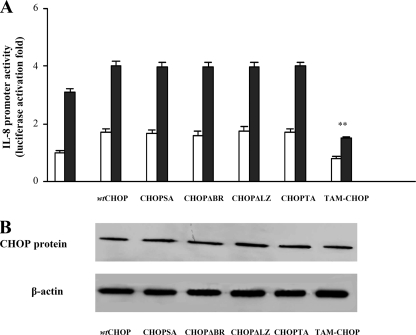
CHOP Domains Involved in the IL-8 Promoter Induction—To identify the CHOP domain(s) responsible for the IL-8 promoter activation in untreated (Fig. 2A, white bars) as well as in PGE2-treated (Fig. 2A, black bars) cells, co-transfection with the wt IL-8 promoter along with constructs overexpressing different CHOP mutants was performed, and luciferase activity in the cell lysates was measured. CHOP mutants that either (i) lacked the basic DNA-binding domain (CHOPΔBR), (ii) lacked the leucine zipper domain (CHOPΔLZ), (iii) lacked both (CHOP-TA), or (iv) were mutated into the p38 MAPK-dependent phosphorylation sites (S79A,S82A) were still able to activate IL-8 promoter activity similarly to wt CHOP (Fig. 2A). The CHOP mutant lacking the transactivation domain (TAM-CHOP) exhibited a slight inhibition of the IL-8 promoter in untreated cells, thus suggesting that the basal activity of IL-8 gene is largely CHOP-independent. In PGE2-treated cells overexpressing the CHOPΔBR, CHOPΔLZ, CHOP-TA, or CHOP (S79A,S82A), the IL-8 promoter activity is up-regulated to the same extent of cells expressing wt CHOP. In contrast, expression of TAM-CHOP highly reduced IL-8 promoter activation when compared with wt CHOP (Fig. 2A, black bars). Thus, PGE2 induces IL-8 gene expression mainly through the CHOP transactivating domain. Of note, all mutants resulted in similar expression levels after transfection (Fig. 2B), thus confirming that the described regulations are specific.
FIGURE 2.
CHOP domains responsible for the activation of IL-8 promoter. A, Jurkat T cells were transfected with wt IL-8 promoter and, where indicated, co-transfected with wt or mutants CHOP expression plasmids. After transfection, Jurkat T cells were treated with anti-CD3 (1 μg/ml) plus anti-CD28 (250 ng/ml) antibodies in the absence (white bars) or presence (black bars) of 10–5m PGE2. The data are the means ± S.D. of five independent experiments and are expressed as fold induction. β-Galactosidase levels were determined for transfection efficiency. **, p < 0.01 versus wt CHOP co-transfected cells, based on two-tailed Student's t test. B, Western blot of total extracts prepared from Jurkat T cells 36 h after transfection with pcDNA 3.1/V5 His wt or mutant CHOP expression vectors. The total proteins were resolved by 10% SDS-PAGE and immunoblotted using anti-V5 mouse monoclonal antibody.
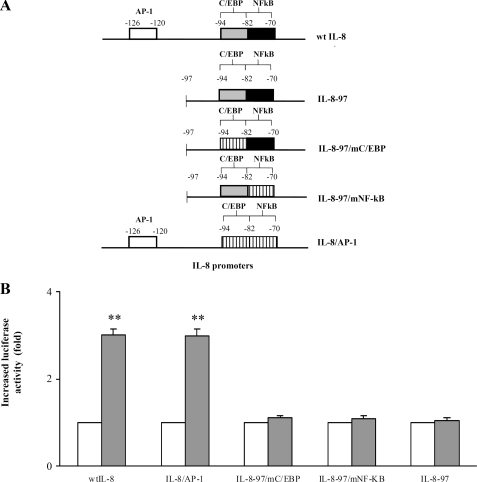
IL-8 Promoter Sites Responsive to PGE2—To identify the IL-8 promoter region involved in PGE2 activation, promoter regions named wt IL-8, IL-8/AP-1, IL-8–97/mC/EBP, IL-8–97/mNF-κB, and IL-8–97 (Fig. 3A) were transfected in PGE2-treated cells, and their luciferase reporter activities were measured. Up-regulation of luciferase expression upon PGE2 stimuli was observed only when wt IL-8 or IL-8/AP-1 promoters were respectively employed (Fig. 3B). Thus, deletion of AP-1 impaired the activation by PGE2, given that either IL-8–97, IL-8–97/mC/EBP, or IL-8–97/mNF-κB promoters failed to increase the luciferase activity. Therefore, the element involved in PGE2 IL-8 promoter regulation is distinct from C/EBP and NF-κB sites and lies in the IL-8/AP-1 promoter region.
FIGURE 3.
IL-8 promoter sites involved in the PGE2-induced activation. A, the IL-8 promoter mutants used are shown: white, gray, and black bars, wild type sites; striped bars, mutated sites. B, Jurkat T cells were transfected with wt IL-8, IL-8/AP-1, IL-8–97/mC/EBP, IL-8–97/mNF-κB, IL-8–97 promoters, and, subsequently, stimulated with PGE2 10–5m for 6 h. The graph shows fold increase of luciferase activity in cells treated with PGE2 (black boxes) by comparing with those without PGE2 treatment (white boxes). The data are the means ± S.D. of five independent experiments and are expressed as fold induction. β-Galactosidase levels were determined for transfection efficiency. **, p < 0.01 wt IL-8 and IL-8/AP-1 transfected plus PGE2-treated cells versus the relative controls based on Student's two-tailed t test.
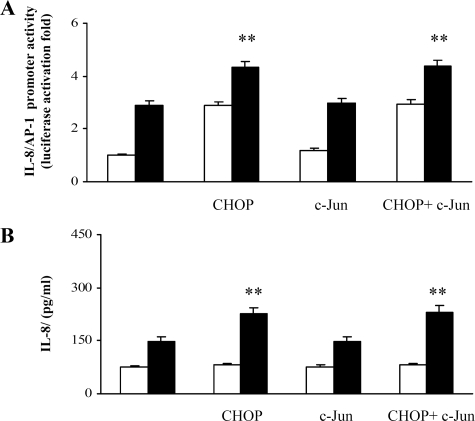
Activation of IL-8/AP-1 promoter by PGE2 is dependent on CHOP and independent of c-Jun—Given that the IL-8/AP-1 promoter region contains the consensus sequence for c-Jun, we tested the effect of PGE2 in the IL-8/AP-1 region of IL-8 promoter in cells overexpressing CHOP, c-Jun, or both. Fig. 4A shows that although c-Jun overexpression did not significantly modify either the basal or the PGE2-induced levels of IL-8 promoter, CHOP overexpression was able to up-regulate IL-8 promoter activity. CHOP and c-Jun overexpression data were similar to those obtained with the single CHOP, thereby reinforcing the concept that CHOP drives IL-8 promoter activity upon PGE2 stimuli. In agreement with these observations, IL-8 protein synthesis shown in Fig. 4B is only enhanced by CHOP overexpression.
FIGURE 4.
IL-8/AP-1 promoter activity in PGE2-treated cells overexpressing CHOP and c-Jun. A, Jurkat T cells were transfected with IL-8/AP-1 promoter and, where indicated, co-transfected with wt CHOP, wt c-Jun, or both the expression vectors. After transfection, Jurkat T cells were treated with anti-CD3 (1 μg/ml) plus anti-CD28 (250 ng/ml) antibodies in the absence (white bars) or presence (black bars) of 10–5 PGE2. The data are the means ± S.D. of five independent experiments and are expressed as fold induction. β-Galactosidase levels were determined for transfection efficiency. **, p < 0.01 versus control untreated cells, based on two-tailed Student's t test. B, Jurkat T cells, transfected with IL-8/AP-1 promoter or, where indicated, co-transfected with wt CHOP, wt c-Jun plasmids, or both, were stimulated with anti-CD3 (1μg/ml) plus anti-CD28 (250 ng/ml) antibodies and treated for 24 h with (black bars) or without (white bars)10–5 m PGE2. IL-8 protein was measured in cell supernatants by ELISA assay. The data are depicted as the means ± S.D. of five independent experiments. **, p < 0.01 versus control untreated cells, based on two-tailed Student's t test.
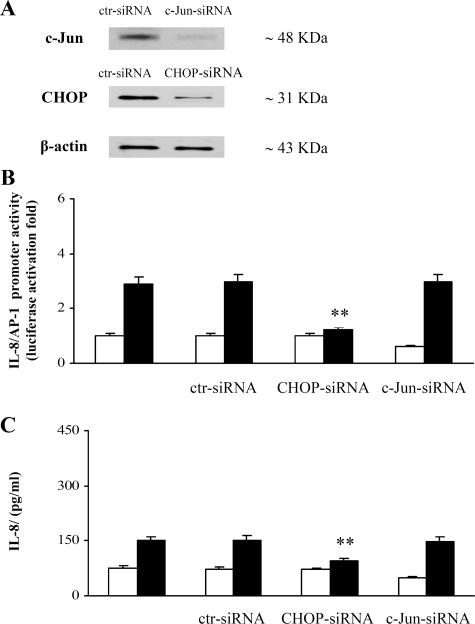
To further elucidate the relative roles of CHOP and c-Jun in the PGE2-induced IL-8/AP-1 promoter activation, we performed gene silencing experiments using synthetic CHOP and c-Jun siRNAs (Fig. 5 and “Experimental Procedures”). Under optimal conditions (50 nm, 48 h post-transfection), we achieved 80 and 88% knockdown of CHOP and c-Jun proteins, respectively (Fig. 5A). We found that the inhibition of the endogenous CHOP, but not c-Jun, suppressed the PGE2-induced IL-8 promoter activity (Fig. 5B) as well as IL-8 protein production (Fig. 5C). Therefore, these data provide the definitive evidence of the functional role of CHOP in PGE2-induced IL-8 gene expression and exclude any involvement of c-Jun. Thereby to explain this evidence, we postulate that CHOP may act in the region of the AP-1 site on the IL-8 promoter.
FIGURE 5.
CHOP, but not c-Jun, inhibition, suppresses the PGE2-induced IL-8 transcriptional activation. A, representative immunoblot of CHOP and c-Jun proteins in Jurkat T cells transfected with control (ctr), CHOP, or c-Jun siRNAs (50 nm), activated with anti-CD3 (1 μg/ml) plus anti-CD28 (250 ng/ml) antibodies, and treated with 10–5 m PGE2. B, Jurkat T cells were transfected with IL-8/AP-1 promoter and, where indicated, co-transfected with control, CHOP, or c-Jun siRNAs (50 nm). After transfection, Jurkat T cells were treated with anti-CD3 (1 μg/ml) plus anti-CD28 (250 ng/ml) antibodies in the absence (white bars) or presence (black bars) of 10–5 m PGE2. the data are the means ± S.D. of five independent experiments and are expressed as fold induction. β-Galactosidase levels were determined for transfection efficiency. **, p < 0.01 versus IL-8/AP-1 transfected cells, based on two-tailed Student's t test. C, Jurkat T cells, transfected with IL-8/AP-1 promoter or, where indicated, co-transfected with control, CHOP, or c-Jun siRNAs were stimulated with anti-CD3 (1μg/ml) plus anti-CD28 (250 ng/ml) antibodies and treated for 24 h with (black bars) or without (white bars)10–5 m PGE2. IL-8 protein was measured in cell supernatants by ELISA assay. The data are depicted as the means ± S.D. of five independent experiments. **, p < 0.01 versus control untreated cells, based on two-tailed Student's t test.
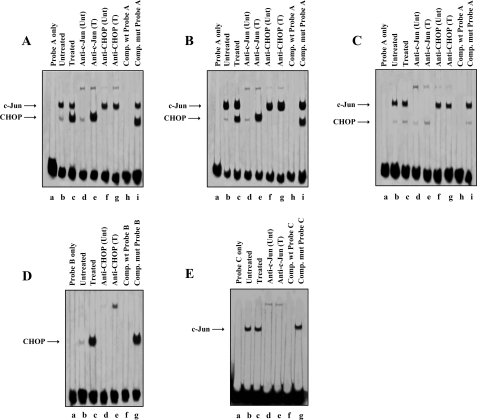
In Vitro DNA-Protein Complex Formation in Untreated and PGE2-treated T Cells—Looking for the CHOP-binding element(s) within the IL-8/AP-1 promoter region, nuclear extracts prepared from untreated and PGE2-treated cells were analyzed by electrophoretic mobility shift and supershift assays. Three double-stranded oligonucleotide probes reproducing (i) the sequence from –141 to –99, encompassing the sequences upstream and downstream of the AP-1 site (probe A), (ii) the sequence from –150 to –130, which encompasses the first base of the AP-1 site and the upstream region (probe B), and (iii) the sequences from –129 to –99, which encompass the AP-1 site and the downstream region (probe C), respectively, were employed (Table 1). We found that two nucleoprotein complexes were generated with cell lysates from PGE2-treated cells after incubation with the probe A (Fig. 6A, lane c), with the lower complex being very weak in the nuclear lysates from untreated cells (Fig. 6A, lane b). The antibody anti-c-Jun supershifted the high molecular weight nucleoprotein complex that does not show changes in treated versus untreated nuclear extracts (Fig. 6A, lanes d and e). The antibody against CHOP supershifted the PGE2 inducible lower molecular weight nucleoprotein complex (Fig. 6A, lanes f and g). The formation of the protein-DNA complexes could be competed out by 50-fold excess of unlabeled wild type probe (Fig. 6A, lane h) but not by a competing mutant probe (Fig. 6A, lane i), thus confirming DNA binding activity specificity. We extended these observations by testing the electrophoretic mobility of protein-DNA complexes obtained by nuclear extracts from c-Jun (Fig. 6B) or TAM-CHOP (Fig. 6C) overexpressing cells, in the absence or presence of PGE2. A specific DNA-protein complex was clearly formed independently from PGE2 treatment (Fig. 6B, lanes b and c) in the c-Jun overexpressing cells. The mobility could be retarded by incubation with the anti-c-Jun antibody to generate a supershifted complex (Fig. 6B, lanes d and e). An evident DNA-protein complex (Fig. 6B, lane c), supershifted by the anti-CHOP antibody (Fig. 6B, lane g), was clearly formed only after PGE2 treatment. Nuclear extracts prepared from PGE2-treated or untreated Jurkat T cells transfected with the TAM-CHOP expression vector were still able to form the high molecular weight specific complex (Fig. 6C, lanes b and c) supershifted by the anti-c-Jun antibody (Fig. 6C, lanes d and e), whereas only a very weak complex was formed as lower molecular weight that was supershifted by the anti-CHOP antibody (Fig. 6C, lanes f and g).
FIGURE 6.
CHOP and c-Jun in vitro binding to the IL-8/AP-1 promoter region. A, biotin-labeled oligonucleotide probe A was incubated with nuclear extracts from untreated (lane b) and PGE2-treated Jurkat T cells (lane c). In supershift analyses, the antibodies against c-Jun (lanes d and e) and CHOP (lanes f and g) were incubated with the nuclear extract-probe mixtures for an additional hour at 4 °C. For competition assays 50-fold excess of unlabeled wt or mutant probe A was incubated with nuclear extracts at room temperature 10 min before the addition of labeled probe A (lanes h and i). B, biotin-labeled oligonucleotide probe A was incubated with nuclear extracts from untreated (lane b) and PGE2-treated cells (lane c) transiently transfected with the c-Jun plasmid. In supershift analyses, the antibodies against c-Jun (lanes d and e) and CHOP (lanes f and g) were incubated with the nuclear extract-probe mixtures for an additional hour at 4 °C. For competition assays 50-fold excess of unlabeled wt or mutant probe A was incubated with nuclear extracts at room temperature 10 min before the addition of labeled probe A (lanes h and i). C, biotin-labeled oligonucleotide probe A was incubated with nuclear extracts from untreated (lane b) and PGE2-treated cells (lane c) transiently transfected with the TAM-CHOP plasmid. In supershift analyses, the antibodies against c-Jun (lanes d and e) and CHOP (lanes f and g) were incubated with the nuclear extract-probe mixtures for an additional hour at 4 °C. For competition assays 50-fold excess of unlabeled wt or mutant probe A was incubated with nuclear extracts at room temperature 10 min before the addition of labeled probe A (lanes h and i). D, biotin-labeled probe B was incubated with nuclear extracts from untreated (lane b) and PGE2-treated cells (lane c). In supershift analyses, the antibody against CHOP (lanes d and e) was incubated with the nuclear extract-probe mixtures for an additional hour at 4 °C. For competition assays 50-fold excess of unlabeled wt or mutant probe B was incubated with nuclear extracts at room temperature 10 min before the addition of labeled probe B (lanes f and g). E, biotin-labeled probe C was incubated with nuclear extracts from untreated (lane b) and PGE2-treated cells (lane c). In supershift analyses, the antibody against c-Jun (lanes d and e) was incubated with the nuclear extract-probe mixtures for an additional hour at 4 °C. For competition assays 50-fold excess of unlabeled wt or mutant probe C was incubated with nuclear extracts at room temperature 10 min before the addition of labeled probe C (lanes f and g). Wild type or mutant probes were described in Table 1. Unt, untreated; T, treated.
Only one low marked nucleoprotein complex was generated upon PGE2 stimulation after incubation with probe B (Fig. 6D, lane c), whereas this nucleoprotein complex was much weaker in untreated cells (Fig. 6D, lane b). In supershift assays, the antibody against CHOP supershifted the nucleoprotein complex (Fig. 6D, lanes d and e). Again formation of the protein-DNA complex could be competed out by 50-fold excess of unlabeled wt probe (Fig. 6D, lane f) but not by a competing mutant (Fig. 6D, lane g), thus strengthening the specificity of the results.
One nucleoprotein complex was generated from either nonstimulated or PGE2-stimulated cells after incubation with probe C (Fig. 6E, lanes b and c). In supershift assays, the anti-c-Jun antibody supershifted the nucleoprotein complex (Fig. 6E, lanes d and e). Also in this case, c-Jun DNA binding activity was clearly specific, because the complex formation could be competed out by 50-fold excess unlabeled wt probe (Fig. 6E, lane f) and not by competing with an AP-1 mutant probe (Fig. 6E, lane g).
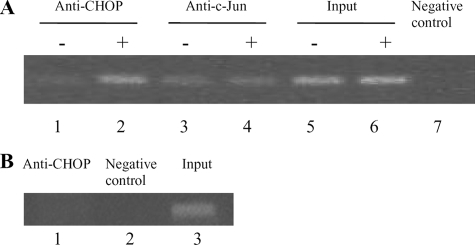
In Vivo Binding of CHOP to the IL-8 Promoter Following PGE2 Treatment—To elucidate the mechanism of CHOP-mediated and PGE2-induced IL-8 activation, we used ChIP assay to detect the in vivo binding of CHOP transcription factor to the IL-8 promoter. The binding of c-Jun in PGE2-treated and untreated cells was also studied. We found that PGE2 induced a marked interaction of CHOP on the IL-8 promoter (Fig. 7A, lane 2), whereas the binding of CHOP detected in untreated cells was very weak (Fig. 7A, lane 1). On the contrary, c-Jun was bound to the IL-8 promoter either in untreated (Fig. 7A, lane 3) or in PGE2-treated cells (Fig. 7A, lane 4). Thus, it can be argued that CHOP directly binds in vivo to the IL-8 promoter in response to the pro-inflammatory stimulus of PGE2. To verify whether in T cells the PGE2-induced activation of CHOP is followed by in vivo binding to the promoter of another target pro-inflammatory gene, such as IL-6, we next performed a further ChIP assay. Cross-linked chromatin fragments were isolated and analyzed for the presence of specific DNA sequence corresponding to the IL-6 promoter. As shown in Fig. 7B, IL-6 promoter sequences were not immunoprecipitated with anti-CHOP antibody (lane 1).
FIGURE 7.
PGE2-induced in vivo binding of CHOP to the IL-8 promoter. A, the interaction of CHOP and c-Jun with IL-8 DNA sequence was evaluated using a ChIP assay. Aliquots of ChIPs immunoprecipitated with CHOP and c-Jun antibodies from untreated (–)(lanes 1 and 3) and PGE2-treated (+) (lanes 2 and 4) cells were amplified through PCR of the IL-8 promoter. Lanes 5 and 6 are “input” lanes where no immunoprecipitation was performed prior to PCR (positive control). Lane 7 contained IgG antibody for immunoprecipitation (negative control). B, cross-linked chromatin isolated from T lymphocytes treated with 10–5 m PGE2 was immunoprecipitated with the antibody against CHOP. Immunoprecipitates were analyzed by PCR to confirm the presence of the promoter sequence of the human IL-6 gene (lane 1). Lanes 2 and 3 are the negative and positive controls, respectively.
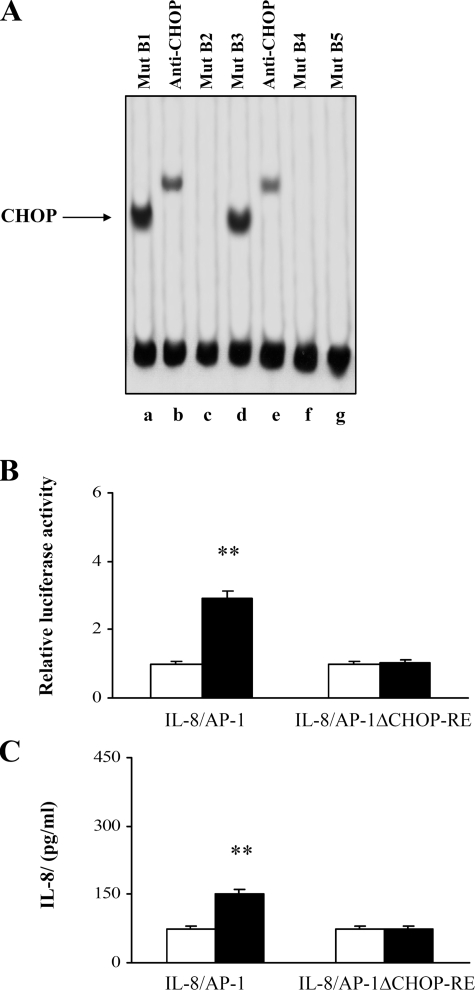
Binding of CHOP to Its Responsive Element—To identify the CHOP-RE at the 5′ upstream region of the IL-8/AP-1 promoter, EMSA experiments were performed with probe B mutated at different nucleotides, as reported in Table 1. As shown in Fig. 8A, following PGE2 treatment, a DNA-protein complex was detected only when Mut B1 and B3 probes were tested (lanes a and d). The anti-CHOP antibody supershifted the complexes (lanes b and e). No complexes were detected when other types of mutated B probes were employed (lanes c, f, and g). Therefore, the CHOP-RE (GTGTGATG) was found to lie between bases –130 and –137 of the IL-8 promoter and to share 1 bp with the AP-1 site.
FIGURE 8.
The CHOP-RE is necessary for PGE2-induced activation of the IL-8 promoter. A, EMSAs of nuclear extracts from PGE2-treated cells incubated with mutated B probes. In supershift analysis, the antibody against CHOP (lanes b and e) was incubated with the nuclear extract-probe mixtures for an additional hour at 4 °C. Mutant B probes were described in Table 1. B, Jurkat T cells were transfected with IL-8/AP-1 and IL-8/AP-1ΔCHOP-RE IL-8 promoter plasmids. After transfection, the cells were treated with anti-CD3 (1 μg/ml) plus anti-CD28 (250 ng/ml) antibodies in the absence (white bars) or presence (black bars) of 10–5 m PGE2. The data are the means ± S.D. of results of five independent experiments and are expressed in fold induction. β-Galactosidase levels were determined to establish transfection efficiency. **, p < 0.01 versus the relative untreated controls, based on two-tailed Student's t test. C, Jurkat T cells, transfected with IL-8/AP-1 or IL-8/AP-1ΔCHOP-RE IL-8 promoter plasmids, were stimulated with anti-CD3 (1 μg/ml) plus anti-CD28 (250 ng/ml) antibodies and treated for 24 h with (black bars) or without (white bars)10–5 m PGE2. IL-8 protein was measured in cell supernatants by ELISA. The data are depicted as the means ± S.D. of results of five independent experiments. **, p < 0.01 versus control untreated cells, based on two-tailed Student's t test.
Finally, to firmly establish the CHOP-DNA binding functional role in the regulation of the IL-8 promoter, we introduced a deletion into the IL-8/AP-1 promoter at the CHOP-RE to determine the effect of this mutation on PGE2-induced IL-8/AP-1 promoter activity. PGE2-treated and untreated cells were transfected with IL-8/AP-1ΔCHOP-RE plasmid, and the luciferase activity as well as the IL-8 protein concentration were detected. The lack of the CHOP-RE prevents any activation of the IL-8 promoter by PGE2 (Fig. 8B) and as well as IL-8 protein synthesis (Fig. 8C), thus clearly indicating that CHOP binding to its consensus sequence is a necessary event for IL-8 promoter up-regulation induced by PGE2.
DISCUSSION
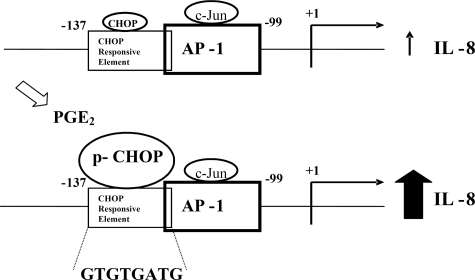
From the results here shown we may hypothesize that in basal conditions CHOP is weakly bound to its RE located at IL-8 promoter, which is diverse, but slightly overlapping the AP-1/c-Jun-binding site. Differently, when PGE2 is added to the cells, the CHOP expression level gets up-regulated inducing a reorganization of the transcription factors binding at the IL-8/AP-1 promoter region. This hypothesis is clearly demonstrated by the experiments in which CHOP silencing completely abolished the PGE2-induced IL-8 promoter activity. In agreement with our previous observations that CHOP-induced IL-8 activation is dependent on CHOP phosphorylation at sites different from the target of p38 MAPK (1), the mutant CHOPSA still activates IL-8 gene. Only staurosporine, a protein kinase C inhibitor, abolished CHOP transactivation activity, whereas SB, a well known p38 MAPK inhibitor, reduced it only ∼40–45% (1). Surprisingly, from the data shown in Fig. 2, we found that the CHOP transactivation domain is necessary and sufficient for the full activation of IL-8 promoter by PGE2. This aspect shades a new light on the ability of CHOP domains to bind DNA, given that only very few reports have shown that CHOP directly binds to DNA (6, 9, 14). A recent work confirms the positive involvement of CHOP in the PGE2-induced activation of IL-8 gene expression shown by us (1) in cystic fibrosis cells (13) and reports the direct binding of this transcription factor to the IL-8 promoter in vivo. The data hereby presented are the first supporting the direct binding of CHOP transactivation domain to DNA and describe the CHOP consensus sequence on IL-8 promoter. Our and previously reported data (9, 15, 16) indicate that depending on the mechanism by which CHOP exerts its function as transcription factor, the DNA-binding domain regions may vary. Note to this respect that both EMSA and functional analysis performed by us with mutated probes or IL-8 promoter constructs fully support the CHOP direct binding induced by PGE2 to a novel and well defined responsive element on the IL-8 promoter. From the data shown in Fig. 4, we deduce that CHOP induces a significant rise of the basal as well as PGE2-induced activation of IL-8/AP-1 promoter and IL-8 synthesis. On the contrary, c-Jun does not modify the response of IL-8/AP-1 promoter to PGE2 treatment. Furthermore, c-Jun is unable to increase the basal levels of IL-8 gene activity and of IL-8 protein synthesis, thus indicating it to be dispensable in modulating the IL-8 gene expression. Accordingly, the quantity of c-Jun bound to IL-8 promoter is consistent either in treated or in untreated cells, whereas the amount of CHOP is very low in basal conditions increasing under PGE2 stimulation (Fig. 6). Thus, it could be hypothesized that the constitutive levels of IL-8 protein in T cells are dependent on c-Jun, and the increase upon exposition to PGE2 is CHOP-mediated.
Although we fully demonstrated CHOP/DNA direct binding by in vitro and in vivo experiments, we cannot exclude that, apart from the shown DNA binding, at the same time protein complexes containing CHOP and c-Jun may also take place. Note in this respect that CHOP transactivation domain is the DNA-binding domain, given that is the only CHOP domain able to activate IL-8 promoter in both basal and PGE2-induced conditions (Fig. 2). For this reason, we consider it a weak possibility that protein-protein interactions between CHOP and c-Jun may also occur. Our hypothesis supports a model by which the PGE2-mediated induction of CHOP influences the transcriptional regulation of IL-8, favoring the CHOP binding to the IL-8 promoter. These results are very interesting considering that we found a novel CHOP-responsive element on IL-8 promoter and a new mechanism of CHOP regulation of gene expression. On the base of the obtained results, we propose the model shown in Fig. 9 in which PGE2 alters the c-Jun/CHOP equilibrium on IL-8 promoter, thereby shifting the c-Jun-dependent basal IL-8 expression into CHOP-dependent increased IL-8 activation. The fact that CHOP is up-regulated in diverse, but specific, cellular stress responses (9, 14, 17–23) and that it plays a role in differentiation, adipogenesis (24–26), and interleukin gene expression (1, 3) suggests that additional factors may determine specificity. Those factors may involve CHOP associates, such as C/EBPβ (4, 6, 27–29), ATF3 (20), and AP-1 (9), or the nature of the signaling pathway that is activated in response to various stimuli. In the case of IL-8 gene expression induced by PGE2 hereby shown, a different mechanism of CHOP gene regulation is highlighted. In fact, by introducing several mutations either in the CHOP protein or in the involved region of IL-8/AP-1 promoter, we demonstrated the CHOP binding, via its transactivation domain, to the novel consensus sequence GTGTGATG. Note that this notion is fully supported by the fact that the IL-8/AP-1 promoter construct lacking this region (IL-8/AP-1ΔCHOP-RE) is unable to be activated by PGE2, thus showing that the PGE2-induced and CHOP-driven activation of IL-8 promoter needs the direct binding of CHOP to a specific responsive element located 5′ upstream the AP-1 site. The newly identified CHOP-RE on IL-8 promoter shows only a modest similarity to the residues TGCAATC recognized by the CHOP/C/EBP dimer previously reported (6, 9). The difference in four of eight bases may depend on either the absence of a CHOP dimerization partner or on the different domain of CHOP responsible for the DNA binding. The higher similarity of the nucleotides ATGTGACG within the IL-6 promoter to the CHOP-RE found by us was not sufficient to induce a physical association of CHOP with this promoter upon PGE2 treatment, thus showing that the recruitment of CHOP to the IL-8 promoter is gene-specific and/or requires the complete consensus sequence for binding. These findings are in agreement with Hattori et al. (30), who reported that CHOP plays an important role in IL-6 production without binding to its promoter. The results reported here are most consistent with two novel aspects of the molecular mechanisms by which CHOP acts as transcription factor. They are the first to show a direct positive role for CHOP in inducing the IL-8 gene expression in T cells exposed to the inflammation mediator PGE2 and, thus, regulating the immune homeostasis. Moreover, evidence has been provided for a new consensus region on IL-8 promoter for CHOP transactivation domain. These issues represent interesting topics for further investigation in other cell systems.
FIGURE 9.
Schematic draw of CHOP and c-Jun occupancy on IL-8 promoter in unstimulated and PGE2-exposed T cells. In basal conditions, the constitutive levels of IL-8 are c-Jun-dependent. Following exposure to PGE2, overexpression as well as activation of CHOP occurs. These events induce the binding of CHOP through its transactivation domain to the sequence GTGTGATG 5′ upstream from the AP-1 site, resulting in an increase of IL-8 synthesis.
This work was supported by grants from the the Dottorato di Ricerca in Patologia della proliferazione cellulare e del differenziamento, the Progetto di Ricerca di Ateneo (to D. T.), and the School of Specialization in Clinical Pathology (University of Messina). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: PGE2, prostaglandin E2; IL, interleukin; C/EBP, CCAAT enhancer-binding protein; CHOP, C/EBP homologous protein; NF, nuclear factor; AP-1, activator protein 1; siRNA, small interfering RNA; wt, wild type; MAPK, mitogen-activated protein kinase; CHOP-RE, CHOP-responsive element; ELISA, enzyme-linked immunosorbent assay; EMSA, electrophoretic mobility shift assay; ChIP, chromatin immunoprecipitation; PIPES, 1,4-piperazinediethanesulfonic acid.
References
- 1.Caristi, S., Piraino, G., Cucinotta, M., Valenti, A., Loddo, S., and Teti, D. (2005) J. Biol. Chem. 280 14433–14442 [DOI] [PubMed] [Google Scholar]
- 2.Vij, N., Fang, S., and Zeitlin, P. L. (2006) J. Biol. Chem. 281 17369–17378 [DOI] [PubMed] [Google Scholar]
- 3.Hattori, T., Itoh, S., Hayashi, H., Chiba, T., Takii, T., Yoshizaki, K., and Onozaki, K. (2001) J. Interferon Cytokine Res. 21 323–332 [DOI] [PubMed] [Google Scholar]
- 4.Ron, D. and Habener, J. F. (1992) Genes Dev. 6 439–453 [DOI] [PubMed] [Google Scholar]
- 5.Shirakawa, K., Maeda, S., Gotoh, T., Hayashi, M., Shinomiya, K., Ehata, S., Nishimura, R., Mori, M., Onozaki, K., Hayashi, H., Uematsu, S., Akira, S., Ogata, E., Miyazono, K., and Imamura, T. (2006) Mol. Cell Biol. 26 6105–6116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao, Q., Wang, J., Levichkin, I. V., Stasinopoulos, S., Ryan, M. T., and Hoogenraad, N. J. (2002) EMBO J. 21 4411–4419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cui, K., Coutts, M., Stahl, J., and Sytkowski., A. J. (2000) J. Biol. Chem. 275 7591–7596 [DOI] [PubMed] [Google Scholar]
- 8.Sauter, K., Grampp, T., Fritschy, J. M., Kaupmann, K., Bettler, B., Mohler, H., and Benke, D. (2005) J. Biol. Chem. 280 33566–33572 [DOI] [PubMed] [Google Scholar]
- 9.Ubeda, M., Wang, X. Z., Zinszner, H., Wu, I., Habener, J. F., and Ron, D. (1996) Mol. Cell. Biol. 16 1479–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kodama, K., Nishio, Y., Sekine, O., Sato, Y., Egawa, K., Maegawa, H., and Kashiwagi, A. (2005) Am. J. Physiol. 289 C582–C590 [DOI] [PubMed] [Google Scholar]
- 11.Monici, M. C., Aguennouz, M., Mazzeo, A., Messina, C., and Vita, G. (2003) Neurology 60 993–997 [DOI] [PubMed] [Google Scholar]
- 12.Weinmann, A. S. and Farnham, P. J. (2002) Methods 26 37–47 [DOI] [PubMed] [Google Scholar]
- 13.Vij, N., Amoako, O. M., Mazur, S., and Zeitlin, P. M. (2008) Am. J. Respir. Cell Mol. Biol. 38 176–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fornace, A. J., Jr., Nebert, D. W., Hollander, M. C., Luethy, J. D., Papathanasiou, M., Fargnoli, J., and Holbrook, N. J. (1989) Mol. Cell. Biol. 9 4196–4203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chipitsyna, G., Sawaya, B. E., Khalili, K., and Amini, S. (2006) J. Cell. Physiol. 207 605–613 [DOI] [PubMed] [Google Scholar]
- 16.Ubeda, M., Vallejo, M., and Habener, J. F. (1999) Mol. Cell Biol. 19 7589–7599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bartlett, J. D., Luethy, J. D., Carlson, S. G., Sollott, S. J., and Holbrook, N. J. (1992) J. Biol. Chem. 267 20465–20470 [PubMed] [Google Scholar]
- 18.Bruhat, A., Jousse, C., Wang, X. Z., Ron, D., Ferrara, M., and Fafournoux, P. (1997) J. Biol. Chem. 272 17588–17593 [DOI] [PubMed] [Google Scholar]
- 19.Carlson, S. G., Fawcett, T. W., Bartlett, J. D., Bernier, M., and Holbrook, N. J. (1993) Mol. Cell. Biol. 13 4736–4744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen, Q., Yu, K., Holbrook, N. J., and Stevens, J. L. (1992) J. Biol. Chem. 267 8207–8212 [PubMed] [Google Scholar]
- 21.Marten, N. W., Burke, E. J., Hayden, J. M., and Straus, D. S. (1994) FASEB J. 8 538–544 [DOI] [PubMed] [Google Scholar]
- 22.Price, B. D. and Calderwood, S. K. (1992) Cancer Res. 52 3814–3817 [PubMed] [Google Scholar]
- 23.Schmitt-Ney, M. and Habener, J. F. (2000) J. Biol. Chem. 275 40839–40845 [DOI] [PubMed] [Google Scholar]
- 24.Maytin, E. V. and Habener, J. F. (1998) J. Investig. Dermatol. 110 238–246 [DOI] [PubMed] [Google Scholar]
- 25.Pereira, R. C., Delany, A. M., and Canalis, E. (2004) Endocrinology 145 1952–1960 [DOI] [PubMed] [Google Scholar]
- 26.Tang, Q. Q. and Lane, M. D. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 12446–12450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fawcett, T. W., Eastman, H. B., Martindale, J. L., and Holbrook, N. J. (1996) J. Biol. Chem. 271 14285–14289 [DOI] [PubMed] [Google Scholar]
- 28.Friedman, A. D. (1996) Cancer Res. 56 3250–3256 [PubMed] [Google Scholar]
- 29.Friedman, A. D. and McKnightm, S. L. (1990) Genes Dev. 4 1416–1426 [DOI] [PubMed] [Google Scholar]
- 30.Hattori, T., Ohoka, N., Hayashi, H., and Onozaki, K. (2003) FEBS Lett. 541 33–39 [DOI] [PubMed] [Google Scholar]