Abstract
The high affinity of IgE for its receptor, FcεRI (Ka ∼ 1010 m–1), is responsible for the persistence of mast cell sensitization. Cross-linking of FcεRI-bound IgE by multivalent allergen leads to cellular activation and release of pro-inflammatory mediators responsible for the symptoms of allergic disease. We previously demonstrated that limiting the IgE-FcεRI interaction to just one of the two Cε3 domains in IgE-Fc, which together constitute the high affinity binding site, results in 1000-fold reduced affinity. Such attenuation, effected by a small molecule binding to part of the IgE:FcεRI interface or a distant allosteric site, rather than complete blocking of the interaction, may represent a viable approach to the treatment of allergic disease. However, the degree to which the interaction would need to be disrupted is unclear, because the importance of high affinity for immediate hypersensitivity has never been investigated. We have incorporated into human IgE a mutation, R334S, previously characterized in IgE-Fc, which reduces its affinity for FcεRI ∼50-fold. We have compared the ability of wild type and R334S IgE to stimulate allergen-induced mast cell activation in vitro and in vivo. We confirmed the expected difference in affinity between wild type and mutant IgE for FcεRI (∼50-fold) and found that, in vitro, mast cell degranulation was reduced proportionately. The effect in vivo was also marked, with a 75% reduction in the passive cutaneous anaphylaxis response. We have therefore demonstrated that the high affinity of IgE for FcεRI is critical to the allergic response, and that even moderate attenuation of this affinity has a substantial effect in vivo.
The binding of IgE to its high affinity receptor, FcεRI, is a critical common step in the allergic reaction (1). IgE, bound to FcεRI expressed on the surface of mast cells and basophils, can be cross-linked by specific multivalent antigen to effect the release of inflammatory mediators. These mediators are responsible for the symptoms associated with an immediate hypersensitivity reaction and the subsequent allergic cascade. The affinity of the complex formed between FcεRI and IgE is extremely high (Ka ∼ 1010 m–1) (2), with a half-life of ∼16 h on cells in suspension (3) and 2 weeks in tissue (4). The crystal structure of this complex has revealed two separate and extensive contact sites for FcεRI on the IgE molecule, one on each chain of the antibody (5). In previous work we showed that interaction with both sites is critical for high affinity binding and that engagement of only one lowers the affinity by 3 orders of magnitude (6). We further suggested that partially blocking the binding of IgE to FcεRI, for example by a small molecule that inhibits binding to one of the two sites, might have a significant effect on allergic sensitization. However, if this is to have therapeutic potential, it is important to determine the degree to which the affinity of IgE for FcεRI must be reduced to impact on the allergic reaction.
Here we describe an investigation into the properties of an IgE molecule with attenuated affinity for FcεRI. Specifically, a recombinant hapten-specific IgE molecule was engineered with a single point mutation (R334S) in the FcεRI-binding site. This mutation has previously been shown to disrupt, but not abrogate, the binding of IgE-Fc fragments to FcεRI (7). In this study, the mutation acts as a model for the effect of partial inhibition on IgE function by a small molecule. There is growing evidence that small molecules can disrupt protein-protein interfaces (8), the binding energies of which are often dominated by a small number of individual amino acid residues or “hot spots” (9, 10). The R334S hapten-specific IgE was compared with wild type IgE for its ability to bind human recombinant sFcεRIα5 by SPR and to initiate degranulation using the RBL-SX38 cell line. It was also tested in vivo in a PCA reaction using a human FcεRI transgenic mouse model.
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification of NP-IgE and R334S NP-IgE—A new vector, termed pSV-VNP Hε R334S, encoding an NP-specific human IgE heavy chain incorporating an R334S mutation, was created using the Quickchange XL site-directed mutagenesis kit (Stratagene) and the vector pSV-VNP Hε (11) as a template (R334S forward primer, gattccaacccgagcggggtgagcgcc; R334S reverse primer, ggcgctcaccccgctcgggttggaatc). pSV-VNP Hε R334S was transfected by electroporation into the J558L cell line (ECACC 88032902) and positive transfectants selected (11). Cells expressing R334S NP-IgE and NP-IgE (ECACC 87080706) were grown in Dulbecco's modified Eagle's medium (Sigma) supplemented with 10% fetal bovine serum (Invitrogen), and IgE was purified by affinity chromatography (11, 12). After affinity purification, the eluted IgE was concentrated by microfiltration, and antibody monomer was isolated from the aggregate by gel filtration chromatography using a S200 10/300 GL Superdex column (GE Biosciences) run at 0.75 ml/min in a Tris-saline buffer (0.5 m Tris, 0.25 m NaCl, pH 7.2). The integrity of the protein was further confirmed by nonreducing SDS-PAGE.
SPR Analysis of NP-IgE and R334S NP-IgE Using Biacore—SPR assays were performed using a Biacore 3000 instrument (GE Healthcare), as described previously (6, 7). Data were analyzed using the BIAevaluation package (version 4.1, GE Healthcare) with double referencing applied (13).
In Vitro Degranulation Assays—The ability of NP-IgE and R334S NP-IgE to cause degranulation was measured in vitro using the RBL-SX38 cell line; this expresses the human tetramer form of FcεRI (14). Cells were maintained under standard culture conditions. For degranulation assays, cells were plated overnight (2 × 104 per well in flat-bottomed 96-well tissue culture plates) and the following day sensitized for 2 h with IgE at indicated concentrations. Cells were then washed twice (Hanks' balanced salt solution plus 1% BSA) and triggered for 30 min with 100 μl of NIP5-BSA (Biosearch Technologies) at 100 ng/ml in wash buffer. Degranulation was terminated by placing the cells on ice. Supernatants were collected, including controls (cells treated with no antigen for background release or wash buffer plus 1% Triton X for total release). Degranulation was measured by release of β-hexosaminidase, assayed using a fluorogenic substrate (4-methylumbelliferyl-N-acetyl-β-d-glucosaminide) (15). Background release, subtracted from all values, was always <10% of total release. Data were fitted to a nonlinear regression model using Prism (GraphPad software) to obtain EC50 values.
Stimulation with Immobilized Antigen-IgE Complexes—Cells were stimulated with immobilized antigen-IgE complexes according to a method adapted from Yamasaki et al. (16). Bone marrow-derived mast cells prepared from hFcεRI Tg mice were loaded with [3H]serotonin (used as a measure of degranulation) (17). Resuspended cells (1 × 106/ml in Hanks' balanced salt solution, 1% BSA) were centrifuged at 400 × g onto 24-well tissue culture plates coated with antigen-IgE complexes, or antigen alone as a control (200 μl per well), prepared as described (16). After centrifugation, degranulation was left to proceed for 30 min at 37 °C; controls and reaction termination were carried out as for RBL-SX38 assays. Radioactivity in the supernatant was assessed by scintillation counting.
Detection of IgE Binding to RBL-SX38 Cells by Flow Cytometry—Cells sensitized according to the degranulation protocol were washed and treated, on ice, with the protein-protein cross-linker bis(sulfosuccinimidyl) suberate (Pierce) for 15 min, harvested by trypsinization, followed by washing with FACS buffer (PBS with 2% normal goat serum). Cells were then stained with fluorescein isothiocyanate-conjugated goat polyclonal anti-IgE (Vector Laboratories), washed, and analyzed for IgE binding relative to untreated controls using a FACSCalibur and Cellquest Pro software (BD Biosciences).
Titration of NP-IgE in a hFcεRI Tg Mouse Model of PCA—The minimal amount of NP-IgE required to give maximal PCA in hFcεRI Tg mice (17, 18) was measured, using the extravasation of Evans blue dye into IgE sensitized ears following antigen challenge as an indicator. Eighteen mice were divided into six groups of 3; for each mouse one ear was sensitized with a specific concentration of NP-IgE in PBS by intradermal injection (total volume 20 μl) and the other ear with PBS alone. Forty eight hours after sensitization, animals were injected in the tail vein with 200 μl of PBS containing 100 μg of NIP5-BSA and 2% Evans blue dye. Animals were sacrificed 1.5 h after intravenous antigen challenge, and ears amputated, cut to identical sizes, and Evans blue dye extracted into 1 ml of formamide by mincing followed by incubation at 80 °C; dye was measured by absorbance at 620 nm after filtration to remove ear debris.
Effect of the R334S Mutation on PCA—Once the appropriate dose of NP-IgE was determined by titration in the PCA model, a further experiment was carried out to assess the effect of the R334S mutation. Intensity of PCA was compared between the ears of mice sensitized with 20 μg of NP-IgE and with 20 μg of R334S NP-IgE in opposite ears. Antigen challenge was then carried out at 24, 48, or 96 h (n = 2–7), and samples were processed as described for the NP-IgE titration. Significant differences between R334S IgE and the wild type were determined for each time point on the basis of a two-tailed t test.
Rate of Clearance of NP-IgE from Mouse Ear Tissue—For experiments measuring the rate of IgE diffusion out of ears, 14 μg of NP-IgE or R334S NP-IgE in a 20-μl volume was injected into the dorsal surface of the mouse ear. Mice (n = 9) were sacrificed either 24, 48, or 96 h later without antigen challenge or Evans blue dye injection, and ears were removed and cut to identical sizes. IgE was extracted from ear tissue by homogenization of each individual ear in Lysis Buffer (10 mm Hepes, pH 7.2, 100 mm NaCl, 1.5 mm MgCl2, 10 mm KCl, 25% glycerol, 0.5%Nonidet P-40). Supernatants containing IgE extracts were stored at –80 °C until ELISA analysis.
IgE ELISA—ELISA was used for detection of human IgE according to a standard method (19). For detection of NP-IgE and R334S NP-IgE in mouse ear homogenates, known concentrations of both NP-IgE and R334S NP-IgE were used to confirm equal detection of both and to enable adjustment for differences in signal between plates.
RESULTS
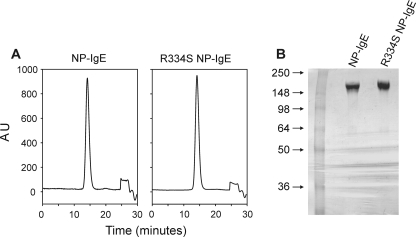
Expression and Characterization of R334S NP-IgE—R334S NP-IgE was expressed and purified and found to be identical to NP-IgE as judged by SDS-PAGE and gel filtration chromatography (Fig. 1, A and B).
FIGURE 1.
Biochemical analysis of wild type and R334S NP-IgE. A, analytical S200 10/300 GL Superdex gel filtration traces of NP-IgE and R334S NP-IgE after purification; run at 0.75 ml/min in 0.5 m Tris, 0.25 m NaCl, pH 7.2. B, 4–20% Tris glycine SDS-PAGE of NP-IgE and R334S NP-IgE after purification. AU, absorbance at 280 nm (normalized).
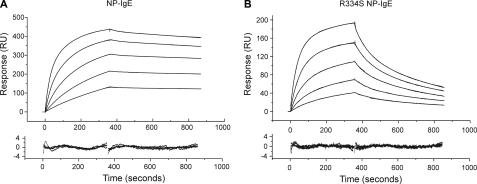
SPR Analysis of NP-IgE and R334S NP-IgE Using Biacore—Biacore analysis of NP-IgE and R334S NP-IgE showed distinct differences in their kinetics of binding to sFcεRIα (Fig. 2, A and B), with the values for NP-IgE in good agreement with those previously published for full-length IgE, and the R334S IgE values in agreement with those for an R334S IgE-Fc fragment (7). NP-IgE affinity constants were Ka1 = 0.40 × 107 m–1, Ka2 = 1.53 × 109 m–1, R1/R0 = 0.03 ± 0.01, and for R334S NP-IgE were Ka1 = 0.96 × 107 m–1, Ka2 = 4.68 × 107 m–1, R1/R0 = 0.31 ± 0.04; see Table 1 for complete kinetic data. Two association constant values are presented because IgE binding to sFcεRIα is fitted to a biphasic kinetic model (7). The relative contributions of these higher and lower affinity components are described by the R1/R0 ratio (being the fractional contribution of the first, Ka1 component). The R334S mutation has a significant effect on both the principal component (Ka2), mainly because of a faster off-rate (see Table 1), and the R1/R0 ratio. Overall, this means that the R334S mutation decreases the affinity of IgE for FcεRI by ∼50-fold if an average weighted affinity is calculated (based on the different contributions of the two affinity constants according to the R1/R0 values) or 33-fold if only the major component is compared (Ka2).
FIGURE 2.
SPR analysis of NP-IgE (A) and R334S NP-IgE (B) binding to sFcεRIα in the concentration range 12–200 nm. The top panel shows binding curves overlaid with biphasic fits that best describe the interaction; the bottom panel shows a plot of the residuals for the fit. See Table 1 for full kinetic parameters derived from the fits.
TABLE 1.
Kinetic parameters and affinity constants derived from the SPR analysis of NP-IgE and R334S NP IgE binding to immobilized FcεRIα
Both IgEs were analyzed using a biphasic interaction model from which association and dissociation constants were derived for each component (shown ± S.D. for at least five determinations in the concentration range 12.5–100 nm). R1/R0 describes the fractional contribution of the first component to the overall fit.
| Constant | NP-IgE | R334S NP-IgE | Deviation of R334S NP-IgE from NP-IgE |
|---|---|---|---|
| ka1 (m-1 s-1) | (0.85 ± 2.7) × 105 | (1.27 ± 0.24) × 105 | Within error |
| kd1 (s-1) | (2.14 ± 0.48) × 10-2 | (1.32 ± 0.16) × 10-2 | Within error |
| ka2 (m-1 s-1) | (1.88 ± 0.51) × 105 | (8.25 ± 2.3) × 104 | ∼2× slower |
| kd2 (s-1) | (1.23 ± 0.31) × 10-4 | (1.71 ± 0.12) × 10-3 | ∼10× faster |
| Ka1 (m-1) | 0.40 × 107 | 0.96 × 107 | Within error |
| Ka2 (m-1) | 1.53 × 109 | 4.68 × 107 | ∼33× lower |
| R1/R0 | 0.03 ± 0.01 | 0.31 ± 0.04 |
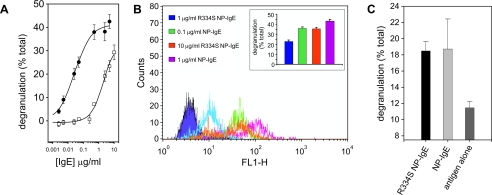
Comparison of NP-IgE and R334S NP-IgE in Vitro Degranulation—The ability of the R334S NP-IgE to initiate the allergic cascade was first measured in vitro using the RBL-SX38 cell line, which expresses the human form of FcεRI (14). In these experiments a clear difference in the EC50 was observed for the R334S NP-IgE (2.5 μg/ml) when compared with NP-IgE (0.025 μg/ml); see Fig. 3A for a representative plot. The range observed for the difference between R334S NP-IgE and NP-IgE was 40–100-fold, with the mean EC50 (concentration at which 50% of the maximum effect is observed) for NP-IgE 0.11 ± 0.04 nm and for R334S NP-IgE 6.7 ± 5.4 nm (both n = 5).
FIGURE 3.
In vitro analysis of R334S and wild type NP-IgE-mediated effector cell activation. A, titration of (•) wild type and (□) R334S IgE-induced degranulation from sensitized RBL-SX38 cells. Values are an average of three measurements corrected for background release ± S.D. and are expressed as a percentage of total release. B, flow cytometry of anti-IgE-stained RBL-SX3 cells sensitized in an identical manner to those used in the degranulation experiments. Post-sensitization cells were treated, on ice, with protein cross-linker to immediately fix the cells. Cells were then harvested and stained with fluorescein isothiocyanate anti-IgE. The points at which traces overlap indicate equivalent IgE binding, and these points correlate with equivalent levels of degranulation in identically treated cells (inset; values calculated as A). C, degranulation triggered by surface immobilized R334S and wild type IgE. Tg hFcεRI bone marrow-derived mast cells were spun onto tissue culture plates pre-treated with antigen and then IgE, and degranulation was measured by the release of [3H]serotonin into the culture medium. Values are an average of three measurements ± S.D. and are expressed as a percentage of total release.
According to SPR measurements the R334S IgE dissociates from FcεRI on the time scale of the degranulation experiments (compare Fig. 2, A with B). Therefore the differences observed in maximum degranulation, measured for R334S NP-IgE compared with wild type NP-IgE, may be due to R334S IgE-FcεRI complexes dissociating prior to the completion of downstream signaling cascades necessary for full stimulation. To address this, the amount of FcεRI-bound IgE present at the time of antigen challenge was estimated by flow cytometry, to correlate it with the level of degranulation produced. Cells used for flow cytometric analyses were treated in an identical manner to those used for degranulation experiments, except that instead of antigen challenge, they were treated with a chemical cross-linker to covalently couple receptor-bound IgE to the cell, but not IgE free in solution (Fig. 3B and inset). In the flow cytometry experiment 0.1 μg/ml NP-IgE resulted in a level of receptor occupancy equivalent to sensitization with 10 μg/ml R334S NP-IgE, and these concentrations yielded similar degranulation values (compare orange and green columns and flow cytometry plots in Fig. 3B).
Finally, it was investigated whether indeed R334S NP-IgE was capable of producing equivalent levels of degranulation as NP-IgE when receptor occupancy was equal. This was achieved by using avidity to overcome the effect of the R334S mutation. To ensure that all antigen was engaged simultaneously, cells were spun onto either wild type or R334S IgE that had been captured on a tissue culture plate coated with NIP-BSA. In these experiments the R334S NP-IgE and NP-IgE were found to be equivalent; see Fig. 3C.
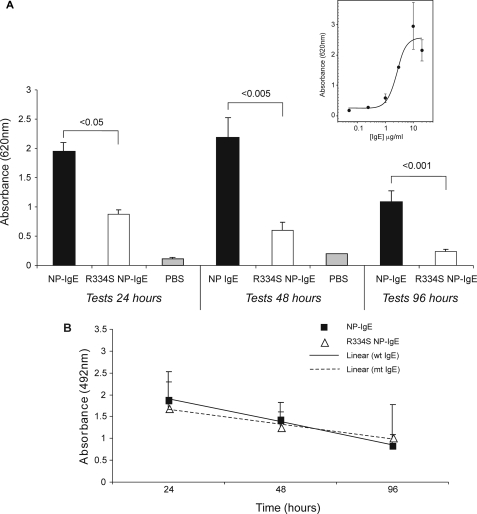
Comparison of Wild Type NP-IgE and R334S NP-IgE in Vivo—The in vitro assays indicate that the lower affinity of R334S NP-IgE hinders its ability to induce an allergic response compared with wild type NP-IgE. To discover whether these results are translated in vivo, the mutant and wild type IgE were compared in a transgenic human FcεRI mouse model of PCA. First, the wild type molecule was titrated to establish the level at which maximal degranulation was achieved after 48 h of sensitization (10 μg of IgE per ear; see Fig. 4A, inset panel). Mice were then injected with either NP-IgE or R334S NP-IgE in excess of this concentration (20 μg), and the PCA reaction was monitored after 24, 48, and 96 h (see Fig. 4A, main panel). At doses in excess of those identified as necessary to yield maximal degranulation 48 h after sensitization, the R334S mutant was only ∼25% as effective as the wild type IgE in eliciting a PCA reaction (differences were significant to p = 0.002). Significant differences were also noted at 24 h (p = 0.02) and 96 h (p = 0.001).
FIGURE 4.
In vivo analysis of NP-IgE- and R334S NP-IgE-mediated effector cell activation. A, inset, titration of NP IgE in ears of hFcεRI Tg mice to determine IgE dose required for maximal PCA. Data are expressed as a mean of all measurements (±S.E.). A, main panel, comparison of R334S NP-IgE and NP-IgE in hFcεRI Tg PCA model. All data are expressed as a mean of all measurements (±S.E.); controls are mock sensitizations carried out with an equivalent volume of PBS alone (24 h; 48 h). B, ELISA to determine rate of clearance of injected NP-IgE and R334S NP-IgE from mouse ears. All values are shown ± S.D.
To determine whether this reduction was the result of an increased rate of clearance of the R334S NP-IgE from the ear, resulting from its lower affinity for FcεRI, or the result of a difference in the ability of the IgE to sensitize the effector cells in the ear, dosing was repeated, and the relative amount of IgE present in the ear was assayed at each time point (Fig. 4B). ELISA analysis from unchallenged sensitized ears indicated that, within a 96-h time frame, the rate of loss of IgE from the ear did not differ between the wild type and R334S molecules; values for each time point were compared on the basis of a t test (not shown).
DISCUSSION
The experiments described in this study were carried out to ascertain the degree to which the allergic response is susceptible to changes in the IgE:FcεRI affinity constant. We engineered an anti-NP-specific IgE molecule containing the R334S mutation and demonstrated a reduction in affinity for FcεRI of between 33- and 50-fold (SPR data, Fig. 2). This is consistent with earlier studies in which the R334S mutation in IgE-Fc led to a decrease in affinity of between 15-fold (cell binding data) and 120-fold (SPR data) (7). The creation of a full-length R334S IgE enabled us to study the functional consequences of a reduction in IgE-FcεRI interaction affinity following allergen challenge both in vitro and in vivo. In vitro degranulation assays demonstrated that the presence of the R334S mutation led to an ∼60-fold increase in the EC50 compared with wild type IgE (Fig. 3A). The difference in potency between R334S and wild type IgE simply reflects a difference in FcεRI occupancy at the time of allergen challenge, rather than the degree of cross-linking, as the change in affinity constant parallels that for the reduction in effector cell response. This was confirmed by in vitro experiments that showed a correlation between levels of R334S and wild type IgE bound to the cell, and subsequent degranulation (Fig. 3B). Indeed, when equal FcεRI occupancy by R334S to wild type IgE was forced (Fig. 3C), degranulation responses were equal. However, this situation does not reflect how allergen would be encountered in vivo.
Using an in vivo model of PCA, a 75% reduction in cellular activation was observed with R334S IgE relative to its wild type counterpart (Fig. 4A). This is despite the fact that equivalent amounts of IgE were present in the tissue at the time of antigen challenge (Fig. 4B). The similarity in the slow clearance rates of wild type and mutant IgE is unsurprising, given that the half-life of free immunoglobulin injected into skin is ∼2.5 days (20), a function of the size of the molecule and restricted diffusion in tissue. Therefore, at all times within the time frame of our experiments, the reduced PCA mediated by R334S IgE is not the result of preferential clearance from the tissue but is reduced receptor occupancy.
Our data thus demonstrate that the effect of the R334S mutation is to shift the response curve for effector cell stimulation to the right. Critically, a mere 50-fold reduction in affinity leads to a marked reduction in an in vivo allergic response. The importance of these observations is that they demonstrate that the allergic response is titratable. It is well established that IgE: FcεRI-stimulated effector mechanisms are sensitive to blocking by antibodies (21), as exemplified by the successful introduction of an anti-IgE monoclonal antibody therapy, omalizumab. Anti-IgE functions by lowering the amount of IgE that is free in serum to bind FcεRI. As a consequence, the number of IgE-FcεRI complexes present on effector cells is reduced, such that they are below the critical level for effective antigenic stimulation (22). (Indeed, reduced IgE occupancy of FcεRI leads to down-regulation of surface-expressed receptor, further amplifying this effect (23).) We now show that an IgE molecule with reduced affinity for receptor has a similar outcome. A small molecule inhibitor of the FcεRI-IgE interaction would create such a population of attenuated molecules, thereby presenting a route to low cost allergy therapy.
The crystal structure of the complex between a sub-fragment of IgE-Fc (consisting of a dimer of the Cε3 and Cε4 domains, Fcε3–4) with FcεRI shows that the interface consists of two distinct binding sites, each comprising residues from the Cε3 domain of each chain (5). The extensive surface area (∼1850 Å2) is very much greater than that occupied by a typical small molecule inhibitor (∼300 Å2) (25), which might therefore be expected to be incapable of blocking IgE binding to FcεRI completely. However, if the small molecule could restrict IgE binding to a single sub-site, we have shown in previous studies that the affinity of the complex would be reduced 1000-fold (6). This is far in excess of the effect of the R334S mutation and reveals the potential for inhibitors that prevent full engagement of IgE with FcεRI, an effect that could also be achieved by allosteric mechanisms, given the known requirement for conformational changes within IgE and the receptor (5, 24, 26–28).
In summary, we have demonstrated that even a moderate attenuation of the interaction between IgE and FcεRI is sufficient to cause a physiological effect and that this encourages a small molecule strategy for inhibition of this interaction. If such a molecule were to create a population of R334S-like IgE molecules, this would raise the threshold for cellular activation sufficiently to dampen the allergic response.
Acknowledgments
We thank Natalie McCloskey for critical reading of the manuscript, Kate Kirwan for expert help in preparing figures, and Professors Jean-Pierre Kinet and Michael S. Neuberger for the kind gifts of the RBL-SX38 cell line and the vector pSV-VNP Hε, respectively.
This work was supported by Asthma UK, Biotechnology and Biological Sciences Research Council (UK), Medical Research Council (UK), and Marie Curie Cancer Care. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: sFcεRIα, soluble fragment of the high affinity IgE receptor FcεRI α-chain; ELISA, enzyme-linked immunosorbent assay; hFcεRI Tg mouse, human FcεRI transgenic mouse; NP, nitrophenylacetyl; NIP, 5-iodo-4-hydroxy-3-nitrophenylacetyl; PCA, passive cutaneous anaphylaxis; SPR, surface plasmon resonance; PBS, phosphate-buffered saline; BSA, bovine serum albumin.
References
- 1.Gould, H. J., and Sutton, B. J. (2008) Nat. Rev. Immunol. 8 205–217 [DOI] [PubMed] [Google Scholar]
- 2.Miller, L., Blank, U., Metzger, H., and Kinet, J. P. (1989) Science 244 334–337 [DOI] [PubMed] [Google Scholar]
- 3.Ishizaka, K., and Ishizaka, T. (1971) in Progress in Immunology I (Amos, B., ed) pp. 859–874, Academic Press, New York
- 4.Geha, R. S., Helm, B., and Gould, H. (1985) Nature 315 577–578 [DOI] [PubMed] [Google Scholar]
- 5.Garman, S. C., Wurzburg, B. A., Tarchevskaya, S. S., Kinet, J. P., and Jardetzky, T. S. (2000) Nature 406 259–266 [DOI] [PubMed] [Google Scholar]
- 6.Hunt, J., Beavil, R. L., Calvert, R. A., Gould, H. J., Sutton, B. J., and Beavil, A. J. (2005) J. Biol. Chem. 280 16808–16814 [DOI] [PubMed] [Google Scholar]
- 7.Henry, A. J., Cook, J. P., McDonnell, J. M., Mackay, G. A., Shi, J., Sutton, B. J., and Gould, H. J. (1997) Biochemistry 36 15568–15578 [DOI] [PubMed] [Google Scholar]
- 8.Wells, J. A., and McClendon, C. L. (2007) Nature 450 1001–1009 [DOI] [PubMed] [Google Scholar]
- 9.Bogan, A. A., and Thorn, K. S. (1998) J. Mol. Biol. 280 1–9 [DOI] [PubMed] [Google Scholar]
- 10.Moreira, I. S., Fernandes, P. A., and Ramos, M. J. (2007) Proteins 68 803–812 [DOI] [PubMed] [Google Scholar]
- 11.Bruggemann, M., Williams, G. T., Bindon, C. I., Clark, M. R., Walker, M. R., Jefferis, R., Waldmann, H., and Neuberger, M. S. (1987) J. Exp. Med. 166 1351–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi, J., Ghirlando, R., Beavil, R. L., Beavil, A. J., Keown, M. B., Young, R. J., Owens, R. J., Sutton, B. J., and Gould, H. J. (1997) Biochemistry 36 2112–2122 [DOI] [PubMed] [Google Scholar]
- 13.Myszka, D. G. (2000) Methods Enzymol. 323 325–340 [DOI] [PubMed] [Google Scholar]
- 14.Wiegand, T. W., Williams, P. B., Dreskin, S. C., Jouvin, M. H., Kinet, J. P., and Tasset, D. (1996) J. Immunol. 157 221–230 [PubMed] [Google Scholar]
- 15.Hammond, G., and Koffer, A. (2006) in Cell Biology (Celis, J. E., Carter, N., Simons, K., Small, J. V., Hunter, T., and Shotton, D., eds) pp. 221–227, Academic Press, Orlando, FL
- 16.Yamasaki, S., Ishikawa, E., Kohno, M., and Saito, T. (2004) Blood 103 3093–3101 [DOI] [PubMed] [Google Scholar]
- 17.Dombrowicz, D., Brini, A. T., Flamand, V., Hicks, E., Snouwaert, J. N., Kinet, J. P., and Koller, B. H. (1996) J. Immunol. 157 1645–1651 [PubMed] [Google Scholar]
- 18.Dombrowicz, D., Flamand, V., Brigman, K. K., Koller, B. H., and Kinet, J. P. (1993) Cell 75 969–976 [DOI] [PubMed] [Google Scholar]
- 19.McCloskey, N., Hunt, J., Beavil, R. L., Jutton, M. R., Grundy, G. J., Girardi, E., Fabiane, S. M., Fear, D. J., Conrad, D. H., Sutton, B. J., and Gould, H. J. (2007) J. Biol. Chem. 282 24083–24091 [DOI] [PubMed] [Google Scholar]
- 20.Tada, T., Okumura, K., Platteau, B., Beckers, A., and Bazin, H. (1975) Int. Arch. Allergy Appl. Immunol. 48 116–131 [DOI] [PubMed] [Google Scholar]
- 21.Chang, T. W. (2000) Nat. Biotechnol. 18 157–162 [DOI] [PubMed] [Google Scholar]
- 22.MacGlashan, D., Jr. (2005) Ann. N. Y. Acad. Sci. 1050 73–88 [DOI] [PubMed] [Google Scholar]
- 23.MacGlashan, D., Jr., Lichtenstein, L. M., Kenzie-White, J., Chichester, K., Henry, A. J., Sutton, B. J., and Gould, H. J. (1999) J. Allergy Clin. Immunol. 104 492–498 [DOI] [PubMed] [Google Scholar]
- 24.Harwood, N. E., and McDonnell, J. M. (2007) Biomed. Pharmacother. 61 61–67 [DOI] [PubMed] [Google Scholar]
- 25.Cheng, A. C., Coleman, R. G., Smyth, K. T., Cao, Q., Soulard, P., Caffrey, D. R., Salzberg, A. C., and Huang, E. S. (2007) Nat. Biotechnol. 25 71–75 [DOI] [PubMed] [Google Scholar]
- 26.Garman, S. C., Sechi, S., Kinet, J. P., and Jardetzky, T. S. (2001) J. Mol. Biol. 311 1049–1062 [DOI] [PubMed] [Google Scholar]
- 27.Wurzburg, B. A., Garman, S. C., and Jardetzky, T. S. (2000) Immunity 13 375–385 [DOI] [PubMed] [Google Scholar]
- 28.Wan, T., Beavil, R. L., Fabiane, S. M., Beavil, A. J., Sohi, M. K., Keown, M., Young, R. J., Henry, A. J., Owens, R. J., Gould, H. J., and Sutton, B. J. (2002) Nat. Immunol. 3 681–686 [DOI] [PubMed] [Google Scholar]