Abstract
Neutral sphingomyelinases (SMases) are involved in the induction of ceramide-mediated proapoptotic signaling under heat stress conditions. Although ceramide is an important mediator of apoptosis, the neutral SMase that is activated under heat stress has not been identified. In this study, we cloned an Mg2+-dependent neutral SMase from a zebrafish embryonic cell cDNA library using an Escherichia coli expression-cloning vector. Screening of the clones using an SMase activity assay with C6-7-nitro-2–1,3-benzoxadiazol-4-yl-sphingomyelin as the substrate resulted in the isolation of one neutral SMase cDNA clone. This cDNA encoded a polypeptide of 420 amino acids (putative molecular weight: 46,900) containing two predicted transmembrane domains in its C-terminal region. The cloned neutral SMase 1 acted as a mediator of stress-induced apoptosis. Bacterially expressed recombinant neutral SMase 1 hydrolyzed [choline-methyl-14C]sphingomyelin optimally at pH 7.5 in the presence of an Mg2+ ion. In zebrafish embryonic cells, the endogenous SMase enzyme was localized in the microsomal fraction. In FLAG-tagged SMase-overexpressing cells, neutral SMase 1 colocalized with a Golgi marker in a cytochemical analysis. Inactivation of the enzyme by an antisense phosphorothioate oligonucleotide repressed the induction of ceramide generation, caspase-3 activation, and apoptotic cell death by heat stress. Thus, neutral SMase 1 participates in an inducible ceramide-mediating, proapoptotic signaling pathway that operates in heat-induced apoptosis in zebrafish embryonic cells.
Sphingolipids have been implicated as bioactive molecules in animal cells. Specifically, ceramide is a mediator of apoptosis (1, 2), differentiation (3), and senescence (4, 5). Ceramide is accumulated in response to heat shock in U937 (6), BAE (6), HL-60 (7), and NIH WT-3T3 (8) cells. Ceramide initiates apoptosis through the stress-activated protein kinase (SAPK/JNK) cascade in U937 and BAE cells (6). The exogenous cell-permeable ceramide analog N-acetylsphingosine (C2-ceramide)2 also induces αB-crystallin transcription, mirroring the effect of heat shock-produced ceramide in vivo (8). In heat-shocked HL-60 cells, elevated ceramide induces the expression of c-jun/c-fos and activated caspase-3, resulting in apoptosis (9).
Spingomyelinases (SMases, EC 3.1.4.12) hydrolyze sphingomyelin, which is a major component of the lipid bilayer of subcellular membranes, to generate ceramide and phosphorylcholine (10). To date, five types of SMases have been described, including the lysosomal acidic SMase (11–13), the cytosolic Zn2+-dependent acidic SMase (14, 15), the membrane neutral magnesium-dependent SMase (16–19), the cytosolic neutral magnesium-independent SMase (20), and the alkaline SMase (21–24). These enzymes differ in their subcellular localization, tissue specificity, and enzymatic properties, especially optimum pH (25). SMases, especially acidic SMase and neutral SMase, are activated in response to growth factors, cytokines, chemotherapeutic agents, irradiation, nutrient removal, and stress (4, 10, 26).
Recent research has identified three distinct neutral SMases in human and mouse; these are designated neutral SMases 1, 2, and 3 (27–29). Although a neutral or acidic SMase is thought to mediate stress-induced ceramide generation and apoptosis, the identity of the stress-induced SMase that mediates apoptosis has not been determined. To understand the molecular mechanisms of stress-induced apoptosis and sphingolipid metabolism, the SMase(s) involved must be cloned and biochemically characterized.
We have focused on zebrafish embryos and cultured cells as experimental models for the characterization of ceramide-induced apoptosis (30–32). Exogenous C2-ceramide induces apoptosis in Japanese flounder embryos (33), and the inhibition of neutral ceramidase in zebrafish embryos induces ceramide generation and apoptosis (34). Recently, we found a zebrafish Mg2+-dependent neutral SMase 1 that produces ceramide and causes thalidomide-induced vascular defects in zebrafish embryos (32). Thus, this enzyme may be the mediator of stress-induced ceramide generation and apoptosis.
In our preliminary studies, zebrafish cells showed high Mg2+-dependent neutral SMase activity. Here, we report the expression cloning and biochemical features of this zebrafish neutral SMase 1 as a key enzyme in stress-induced apoptosis and ceramide generation in zebrafish embryonic (ZE) cells. Because the amino acid sequence of the zebrafish enzyme was expected to have very low sequence homology with the known mammalian neutral SMases, we used an Escherichia coli expression cloning strategy with an SMase assay. Biochemical characterization showed that his enzyme is a mediator of stress-inducible apoptosis in ZE cells.
EXPERIMENTAL PROCEDURES
Materials—Acetyl-Asp-Glu-Val-Asp-4-methylcoumaryl-7-amide (Ac-DEVD-MCA) and benzyloxycarbonyl-Val-Ala-Asp-fluoromethylketone (Z-VAD-fmk) were purchased from the Peptide Institute (Osaka, Japan). C6-7-Nitro-2–1,3-benzoxadiazol-4-yl (C6-NBD) sphingomyelin was obtained from Matreya (Pleasant Gap, PA). [choline-methyl-14C]Sphingomyelin (2.0 GBq/mmol), [choline-methyl-14C]dipalmitoyl-phosphatidylcholine (2.11 GBq/mmol), [1-O-octadecyl-3H]lyso-platelet-activating factor (5.99 TBq/mmol), and l-[U-14C]serine (5.7 GBq/mmol), protein A-Sepharose, polyvinylidene difluoride membranes, the ECL™ Western blotting detection kit, and horseradish peroxidase-labeled secondary antibodies were purchased from GE Healthcare. Mouse monoclonal anti-KDEL antibody and mouse monoclonal anti-58K Golgi protein antibody were purchased from Abcam (Cambridge, MA). Anti-transferrin receptor polyclonal antibody, anti-cadherin polyclonal antibody, and anti-aldolase polyclonal antibody were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-PARP polyclonal antibody, anti-FLAG M2 monoclonal antibody, anti-actin monoclonal antibody, and p3XFLAG-CMV™-14 expression vector were purchased from Sigma-Aldrich. Xpress™ System synthetic oligonucleotides, goat anti-rabbit IgG Alexa Fluor 488-labeled antibody, and anti-mouse IgG Alexa Fluor 594-labeled antibody were purchased from Invitrogen. Premix Taq (Ex Taq™, version 2) and PrimeSTAR™HS DNA polymerase were purchased from Takara Biomedicals (Shiga, Japan).
Cell Culture—Zebrafish embryonic cell line ZE cells and fat-head minnow FHM cells were cultured in Leibovitz's L-15 medium (Invitrogen) supplemented with 2% fetal calf serum (FCS; JRH Biosciences) and 80 μg/ml kanamycin sulfate at 28.5 °C (30, 35). Human leukemia HL-60 cells, human kidney embryonic HEK293 cells, and COS-7 cells were cultured in RPMI 1640 (Invitrogen) supplemented with 10% FCS (JRH Biosciences) and 80 μg/ml kanamycin sulfate at 37 °C in a humidified atmosphere containing 5% CO2.
Heat Shock Treatment—ZE cells were grown at 5 × 105 cells/ml in preheated medium in culture dishes, heat-shocked in an incubator at 37 or 38 °C, and then allowed to recover at 28.5 °C. Cell viability was determined using the trypan blue dye exclusion method (32). After cells were heat-shocked at 37 or 38 °C for 0–11 h, they were stained with 4′,6-diamidino-2-phyenylidole (DAPI (7)), and at least 200 cells were counted under a light microscope. Cells exhibiting nuclear condensation and fragmentation were judged as apoptotic.
Caspase-3 Assay—Caspase-3 activity was measured using a synthetic substrate, Ac-DEVD-MCA, according to the method described previously (30). ZE cells were washed once with PBS and homogenized in lysis buffer (20 mm Hepes, pH 7.5, containing 250 mm sucrose, 50 mm KCl, 2.5 mm MgCl2 and 1 mm dithiothreitol) by passage through a 27-gauge needle. The lysate was centrifuged, and the supernatant was collected. A portion of the supernatant was added to 180 μl of a caspase substrate mixture containing 20 mm Hepes, pH 7.5, 250 mm sucrose, 50 mm KCl, 2.5 mm MgCl2 1 mm dithiothreitol, and 4 μm Ac-DEVD-MCA and then incubated at 37 °C for 60 min. The release of 7-amino-4-methyl-coumarin was measured using a fluorescent spectrophotometer (Millipore) with excitation at 360 nm and emission at 450 nm. Protein concentrations were determined using a Bio-Rad protein assay kit.
Ceramide Measurement—Lipids in the cells were extracted by the methods of Bligh and Dyer (36), and ceramide mass measurements using E. coli diacylglycerol kinase were according to the method described by Okazaki et al. (3). The solvent system used to separate ceramide 1-phosphate was chloroform/acetone/methanol/acetic acid/water (10:4:3:2:1). To calculate the ceramide content, the positive spots on thin-layer chromatography (TLC) plates were measured using a Storm® 860 imaging analyzer (GE Healthcare).
Sphingomyelin Quantification—After heat shock treatment, 1 × 107 cells were harvested, and the lipids were extracted from the cells by the methods of Bligh and Dyer (36) and developed using a solvent system of chloroform/methanol/water (60:35:8, by volume) on a plastic TLC plate with standard sphingomyelin. The spots corresponding to sphingomyelin were scraped, and the lipids were extracted by the method of Bligh and Dyer (36). Inorganic phosphate in the extract was measured by the ammonium molybdate/ascorbic acid method (37) to calculate the sphingomyelin content.
Metabolic Labeling of Ceramide with [14C]Serine—The cells were wash with PBS, seeded at 5 × 105/ml in Leibovitz's L-15 medium supplemented with 370 MBq/ml l-[U-14C]serine and 2% FCS, and incubated at 28.5 °C for 3 days. The labeled cells were heat-shocked at 38 °C for 1 h and then recovered to incubate at 28.5 °C for 3 h. After harvesting the cells, lipids extracted from the cells by the methods of Bligh and Dyer (36) were separated on TLC plates with a solvent system of methyl acetate, propanol-1, chloroform, methanol, and 0.25% KCl (25:25:25: 10:9 by volume). The spots corresponding to ceramide were visualized, and the relative radioactivity was determined by the Storm® 860 analyzer system (GE Healthcare), corrected by the amount of phospholipids in each sample.
Serine Palmitoyltransferase Assay—The cells at 5 × 106/ml were homogenized in a lysis buffer (20 mm Tris-HCl, pH 7.5, containing 5 mm EDTA and 1× Complete® Protease inhibitor mixture (Roche Applied Science)) by passage through a 27-gauge needle. The homogenate was centrifuged at 1300 × g, and the supernatant was then ultracentrifuged at 100,000 × g. The pellets resuspended in 10 mm Tris-HCl buffer, pH 7.4, containing 30% glycerol were used for the enzyme assay as the microsomal fraction. Protein concentration was determined using a Bio-Rad protein assay kit. Serine palmitoyltransferase activity was determined with the microsomal fractions prepared from both the heat-shocked and the control cells. The microsomal fraction containing 80 μg of protein was incubated in 100 mm Hepes buffer, pH 8.3, containing 5 mm dithiothreitol, 50 μm pyridoxal-5′-phosphate, 1 mm 14C-labeled l-serine (370 MBq/mmol), and 200 μm palmitoyl CoA in a total volume of 200 μl at 37 °C for 20 min with shaking. The reactions were stopped by the addition of 1.5 ml of chloroform:methanol (1:2 by volume). Unlabeled sphinganine (25 μl of a 1 mg/ml ethanol solution) was added as a carrier and extracted by phase separation using 1 ml of chloroform and 2 ml of 0.5 m ammonia. After removal of the aqueous phase, the organic phase was washed twice with 2 ml of water to remove unincorporated radiolabeled serine. The resultant organic phase was dried under nitrogen, and the radioactivity of the produced 3-ketodihydrosphingosine was measured by liquid scintillation counting.
SMase Assay—The cells were lysed by passage through a 27-gauge needle in lysis buffer (10 mm Tris-HCl buffer, pH 7.5, containing 1 mm EDTA, 0.1% Triton X-100, 1 mm phenylmethylsulfonyl fluoride, 10 μg/ml pepstatin A, 0.15 unit/ml aprotinin, and 50 μg/ml leupeptin). The lysate was centrifuged at 10,000 × g at 4 °C for 10 min. The supernatant was used as an enzyme source. Supernatant protein (60 μg) was mixed in a reaction mixture (100 mm Tris-HCl, pH 7.5, containing 10 mm MgCl2, 5 mm dithiothreitol, 10 μm C6-NBD-sphingomyelin, and 0.1% Triton X-100) and incubated at 37 °C for 1 h. The reaction was stopped by the addition of 900 μl of H2O and 2 ml of chloroform/methanol (2:1 by volume) and then mixed well and centrifuged. The lower organic phase was collected, and the solvent was evaporated. Aliquots were applied to TLC plates. The solvent system used to separate C6-NBD-ceramide and C6-NBD-sphingomyelin was chloroform/methanol/12 mm MgCl2 in water (65:25:4 by volume). C6-NBD-ceramide was visualized by UV irradiation and measured using a TLC scanner with fluorometer (475 nm excitation, 525 nm emission).
Expression Cloning of Neutral SMase cDNA from Zebrafish—Total RNA was isolated from zebrafish embryonic cells using the TRIzol reagent (Invitrogen), and poly(A)+ RNA was purified from total RNA using the Oligotex-dt30 mRNA purification kit (Takara Biomedicals, Shiga, Japan). Double-stranded cDNA was synthesized from poly(A)+ RNA using a Superscript Choice system for cDNA synthesis (Invitrogen). A cDNA library was prepared using the isopropyl-1-thio-β-d-galactopyranoside-inducible expression vector pET-28a (Novagen, Madison, WI) in E. coli BL21(DE3)pLysE cells (Novagen). The library contained 1 × 107 independent clones. Aliquots containing ∼1000 E. coli clones from the library were inoculated in 4 ml of Luria-Bertani (LB) broth and grown overnight at 30 °C in a shaker at 200 rpm. The culture was transferred to 4 ml of fresh LB broth with 100 μg/ml ampicillin, and the above incubation conditions were continued until turbidity at 600 nm reached 0.8. Isopropyl-1-thio-β-d-galactopyranoside was added to the culture to a final concentration of 0.5 mm, and the culture was grown for a further 4 h to induce the expression of the transgene product. Bacterial cells were collected by centrifugation, and neutral SMase activity was assayed. E. coli colonies from the positive pool that had high SMase activity were incubated on LB plates. Each E. coli clone was cultured in 4 ml, and its SMase activity was measured. Finally, a positive cDNA clone encoding neutral SMase from zebrafish was isolated and sequenced. The nucleotide sequence of the isolated zebrafish neutral SMase cDNA was deposited at the DDBJ/GenBank™/EMBL-EBI data bank under accession number AB196165.
Transfection of the Neutral SMase 1 cDNA into ZE Cells—The neutral SMase cDNA from zebrafish containing the complete open reading frame was amplified by PCR using the isolated full-length cDNA as the template, sense primer 5′-TCAGGAGCGGACTGAAGCGGCATCATGGCA-3′, and antisense primer 5′-CCGTCGAGTCCTTTCAAACGGGAGGAATAA-3′. The PCR reaction was carried out with 25 cycles of denaturation at 94 °C for 30 s, annealing at 63 °C for 30 s, and extension at 72 °C for 2 min. The amplified product was subcloned downstream of the human cytomegalovirus promoter of the pTARGET mammalian expression vector (Promega) according to the manufacture's protocol. The nucleotide sequence of the PCR product was confirmed by sequencing. This construct was named pTARGET-ZNSMase.
For the neutral SMase assay, ZE cells were cultured at a density of 1 × 106 cells/60-mm dish in 5 ml of Leibovitz's L-15 medium supplemented with 2% FCS. At ∼90% confluence, each dish of cells was transiently transfected with 5 μg of DNA of pTARGET-ZNSMase vector or mock vector with the FuGENE 6 transfection reagent (Roche Applied Science) according to the manufacturer's protocol. At 24 h after transfection, the cells were washed twice with PBS and homogenized in lysis buffer for neutral SMase assay.
Preparation of Recombinant Neutral SMase—The neutral SMase 1 cDNA was amplified by PCR using sense primer 5′-CATATGGCACCACAGCAGCCCGGCAAACTG-3′ and antisense primer 5′-CATATGTTATTCCTCCCGTTTGAAAGGACT-3′, each containing an NdeI site; the amplimer was subcloned into the pGEM-T Easy plasmid vector by the TA cloning method (Promega). The cloned nucleotide sequence was confirmed by sequencing and subcloned into the NdeI site in the multiple cloning site of the pET-16b vector (Novagen) to fuse a His10 tag sequence to the N terminus of the neutral SMase 1 open reading frame. This construct was named pETZNSMase 1. Neutral SMase 1 was expressed in E. coli BL21(DE3)pLysE cells (Novagen) transformed with pETZNSMase 1. The cells were inoculated in 100 ml of LB broth and grown overnight at 30 °C in a shaker at 200 rpm. The culture was transferred to 1000 ml of fresh LB medium with 100 μg/ml ampicillin in a 5-liter flask, and the above incubation conditions were continued until turbidity at 600 nm reached 0.8. Isopropyl-1-thio-β-d-galactopyranoside was added to a final concentration of 1 mm, and the culture was grown for an additional 4 h to induce the expression of the transgene. Bacterial cells were collected by centrifugation at 4000 × g for 15 min. The N-terminal His-tagged neutral SMase was purified from the bacterial extract by affinity chromatography with HisTrap HP (GE Healthcare) according to the manufacturer's protocol. The bacterial cells were suspended in lysis buffer (50 mm Tris-HCl, pH 7.5, containing 10% glycerol, 1× Complete proteinase inhibitor mixture, 5 mm MgCl2, 60 mm imidazole, 0.1% Triton X-100, 1 mm EDTA, and 1 mg/ml lysozyme) by passage through a 22-gauge needle. All subsequent procedures were carried out at 4 °C. The supernatant was collected by centrifuging the lysate at 10,000 × g for 30 min and then dialyzed against wash buffer (50 mm Tris-HCl, pH 7.5, containing 300 mm sodium chloride, 10% glycerol, 5 mm MgCl2, 60 mm imidazole, 0.1% TritonX-100, and 1 mm EDTA). The dialyzed sample was loaded onto a 5-ml HisTrap HP column (GE Healthcare) that had been equilibrated in wash buffer. After sample loading, the column was washed with 100 ml of wash buffer followed by a 0–100% linear gradient of elution buffer (50 mm Tris-HCl, pH 7.5, containing 300 mm sodium chloride, 10% glycerol, 5 mm MgCl2, 800 mm imidazole, 0.1% TritonX-100, and 1 mm EDTA). The flow rate was 1 ml/min, and 2-ml fractions were collected. The HisTrap HP fractions that had neutral SMase activity were pooled and dialyzed against gel filtration buffer (25 mm Tris-HCl, pH 7.5, containing 150 mm NaCl, 5 mm MgCl2, 0.1% Triton X-100, and 1 mm EDTA) and then loaded onto a Sephacryl S-100 column (HR 16/60, GE Healthcare) that had been equilibrated in gel filtration buffer. The enzyme was eluted at 1 ml/min with 150 ml of gel filtration buffer.
Preparation of Rabbit Polyclonal Antibody against Zebrafish Neutral SMase—To generate a polyclonal antibody against zebrafish neutral SMase 1, the purified recombinant protein was used for immunization in the rabbit. Specific IgG was purified from the serum by an affinity chromatography on a protein A-Sepharose column (GE Healthcare).
SMase Assay of the Recombinant Enzyme—The SMase activity of the recombinant enzyme was determined using radiolabeled substrate in a mixed micelle assay system as described previously (20). Briefly, [14C]sphingomyelin and bovine brain sphingomyelin were placed in a glass tube and dried under nitrogen. The reaction mixture containing Tris-HCl, pH 7.5, 5 nmol of [14C]sphingomyelin (100,000 dpm), 5 mm dithiothreitol, 0.1% Triton X-100, and 5 mm MgCl2 was prepared by sonication for 5 min. After the addition of 1 μl of enzyme solution to the reaction mixture, the corresponding solution was incubated for 30 min at 37 °C. The reaction was stopped by the addition of 0.2 ml water and 1.5 ml of chloroform/methanol (2:1 by volume). After vortexing and a two-phase separation by centrifugation, 0.2 ml of the upper aqueous phase was removed and added to 2 ml of scintillation solution for radioactivity counting. The reaction was linear with the incubation times up to 3 h. The amount of enzyme added to the reaction mixtures was chosen such that <10% hydrolysis of the substrate occurred. An appropriate blank containing denatured enzyme was run with each reaction and subtracted from the experimental samples. To examine the effect of the magnesium ion on the activity of the purified enzyme, the reaction mixtures were used in the presence or absence of magnesium. For assaying the effects of different pH on the SMase activity, the following buffers were used at final concentration of 100 mm: sodium acetate (pH 4 and 5), PIPES (pH 6 and 7), and Tris (pH 7.5, 8, 8.5, and 9).
Hydrolyzing activity against phosphatidylcholine of the purified enzyme was measured with 10 nmol of [14C]phosphatidylcholine (100,000 dpm) instead of sphingomyelin in the mixed micelle solution for the SMase assay. Hydrolyzing activity against lyso-platelet-activating factor was determined using radiolabeled substrate in a mixed micelle assay system as described previously (38). The recombinant enzyme was added to 100 μl of reaction mixture containing 100 mm Tris-HCl, pH 7.5, 5 mm dithiothreitol, 5 mm MgCl2, and 10 nmol [3H]lyso-PAF (200,000 dpm). The mixture was incubated for 30 min at 37 °C. The lipid was extracted by the method of Bligh and Dyer (36) and separated by TLC in solvent system chloroform/methanol/15 mm CaCl2 in water (60:35:8 by volume). To calculate the monoalkylglycerol content, the TLC plate was exposed to imaging film. The radioactivity of the positive spots scraped from the TLC plate was determined by liquid scintillation counting.
Construction of Neutral SMase Variant—An SMase-FLAG tag fusion construct and an alanine substitution mutant were created by PCR, using zebrafish neutral SMase 1 cDNA as the template and appropriate combinations of the forward and reverse oligonucleotides primers, and introduced into p3XFLAG-CMV™-14 expression vector (Sigma-Aldrich). All of the nucleotide sequences of the vector constructs used for the assays were confirmed by DNA sequencing.
Generation of Zebrafish ZE and Human HEK293 Stable Transfectants—ZE cells were culture in Leibovitz's L-15 medium containing 10% FCS. To obtain stable transfectants, 1 × 106 ZE cells were transfected with 2 μg of DNA of SMase-FLAG tag fusion constructs (wild type) and alanine substitution mutants (H272A mutant) using FuGENE 6 transfection regent (Roche Applied Science) according to the manufacturer's instruction and selected in the presence of 0.4 mg/ml Geneticin (Invitrogen). HEK293 cells were cultured in RPMI 1640 medium containing 10% FCS. 5 × 106 HEK293 cells were transfected with 4 μg of DNA of wild-type vector and H272A mutant vector using FuGENE 6 transfection regent and selected in the presence of 0.8 mg/ml Geneticin. Both ZE and HEK293 cell lines overexpressing the wild type of neutral SMase 1 and its alanine substitution mutant were generated.
Immunofluorescence Microscopy—Transfected ZE and HEK293 cells were cultured on cover glasses, fixed with 4% paraformaldehyde in PBS for 15 min, rinsed with PBS, and permeabilized with 0.1% Triton X-100 in PBS for 3 min at room temperature. After incubation with PBS containing 1% bovine serum albumin and 2% fetal bovine serum for 1 h, the samples were incubated overnight at 4 °C with both anti-zebrafish SMase 1 rabbit IgG (1:2000) and a subcellular marker antibody (anti-KDEL mouse IgG or anti-58K mouse IgG (both from Abcam; 1:200)) in blocking buffer. After three 15-min washes with PBS, the samples were incubated with Alexa Fluor 488-labeled goat anti-rabbit IgG secondary antibody and Alexa Fluor 594-labeled goat anti-mouse IgG secondary antibody for 3 h (1:200; both from Invitrogen). The cells were washed three times with PBS for 15 min, counterstained with DAPI, and observed using a fluorescence microscope (R400; Edge Scientific Instruments, Santa Monica, CA).
For nonpermeation staining of transfected HEK293 cells, the cells were cultured on a cover glass, fixed with 4% paraformaldehyde in PBS for 15 min, and incubated with PBS containing 1% bovine serum albumin and 2% FCS for 1 h. The samples were then incubated with anti-FLAG antibody (1:5000) and anti-cadherin antibody (Santa Cruz Biotechnology; 1:1000) at 4 °C for 1 h. The cells were washed three times with PBS for 15 min and then incubated with secondary antibody as described above. The cells were washed three times with PBS for 15 min, counterstained with DAPI, and imaged by fluorescence microscopy.
Subcellular Fractionation—Subcellular fractionation was performed by the modified method described previously (20). The cells (5 × 106/ml) were homogenized in a lysis buffer (20 mm Tris-HCl, pH 7.5, containing 1 mm EDTA, 1 mm EGTA, 10 mm KCl, 1 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, 10 μg/ml pepstatin A, 0.15 units/ml aprotinin, and 50 μg/ml leupeptin) by passage through a 27-gauge needle. The lysate was centrifuged at 1300 × g for 15 min at 4 °C. The obtained pellet was used as the nuclei fraction. The supernatant was centrifuged at 100,000 × g for 1 h at 4°C. The collected supernatant was used as the cytosolic fraction. The pellet was resuspended in the lysis buffer by homogenization and sonication and then centrifuged at 12,000 × g for 1 h at 4°C. The obtained pellet was used as the microsomal fraction.
Western Blotting—The proteins extracted from the cytosolic and microsomal fractions were separated by SDS-PAGE and electroblotted onto a polyvinylidene difluoride membrane as described by Yabu et al. (30). The anti-zebrafish neutral SMase 1 polyclonal, anti-aldolase polyclonal, anti-actin (control) monoclonal, anti-PARP polyclonal, anti-transferrin polyclonal, and anti-FLAG monoclonal antibodies were used as the primary antibody and signals were detected with the secondary antibody using the ECL™ Western blotting detection kit according to the manufacturer's protocol (GE Healthcare).
Neutral SMase Antisense Oligonucleotides—The phosphorothioate oligonucleotide was synthesized to block the translation of neutral SMase 1: antisense neutral SMase 1, 5′-CGGGCTGCTGTGGTGCCATGATGCC-3′; sense neutral SMase 1, 5′-GGCATCATGGCACCACAGCAGCCCG-3′. The cells were incubated with sense or antisense neutral SMase (0–10 μm) at a concentration 5 × 105 cells/ml in Leibovitz's L-15 medium supplemented with 2% FCS for 48 h before heat shock treatment.
Statistical Analysis—Statistical values are expressed as the mean ± S.D. Differences among groups were analyzed by one-way analysis of variance followed by Bonferroni's post-hoc t test. Comparisons between the two experimental groups were based on two-tailed t tests.
RESULTS
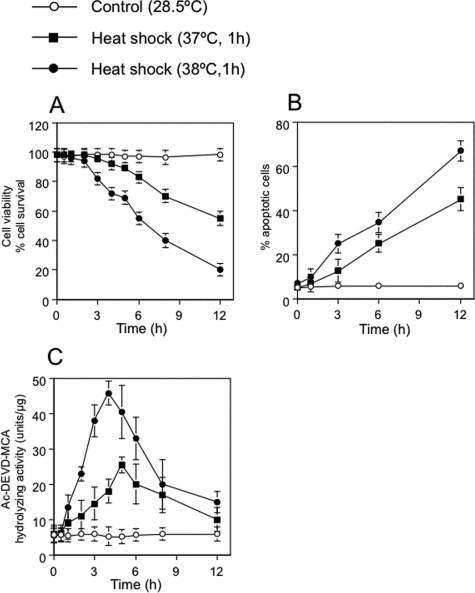
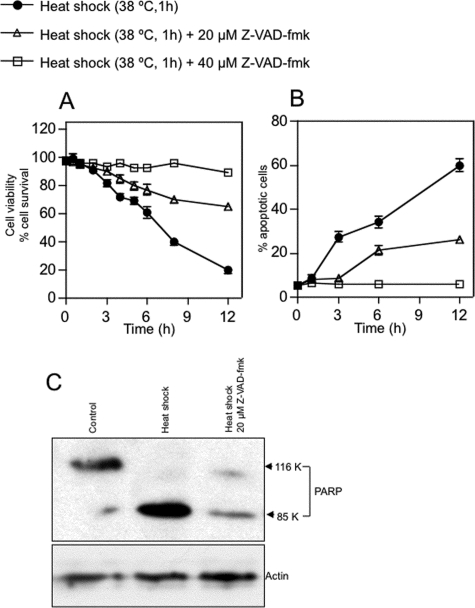
Caspase-3-dependent Apoptosis Induced by Heat Shock—Zebrafish ZE cells and embryos exhibit typical apoptotic features such as membrane blebbing, nuclear and cytoplasmic shrinkage, and nuclear DNA fragmentation (30). We studied whether such apoptotic processes are mediated by ceramide generation and caspase-3 activation under heat stress. When ZE cells were incubated at 37 or 38 °C for 1 h and returned to 28.5 °C, cell survival decreased and apoptotic cell numbers increased in a time- and dose-dependent manner (Fig. 1, A and B). The percentages of cells that showed the morphological changes characteristic of apoptosis (7), as judged by DAPI nuclear staining, were 25 and 35% at 6 h after heat shock treatment for 1 h at 37 and 38 °C, respectively (Fig. 1C). After heat shock at 39 °C for 1 h, all cells died by 1 h in recovery (data not shown). When apoptotic cells were judged by DAPI staining, the percentages of apoptotic cells were similar to those determined by morphological changes. When caspase-3 activity for Ac-DEVD-MCA was assayed after heat shock treatment at 37 or 38 °C for 1 h, a peak of caspase-3 activity was found at 4 h after heat treatment, which then returned to control levels (Fig. 1C). A specific caspase inhibitor, Z-VAD-fmk, which reportedly prevents heat-induced apoptosis (7), actually inhibited heat-induced apoptosis in ZE cells (Fig. 2, A and B). In heat-induced apoptosis, active caspase-3 cleaved PARP (Fig. 2C). Thus, heat-induced apoptosis in ZE cells is mediated by a pathway involving caspase-3 activation.
FIGURE 1.
Temperature- and time-dependent induction of apoptosis by heat. A, effect of heat shock on cell survival in ZE cells. Control cells were maintained in Leibovitz's L-15 medium supplemented with 2% FCS at 28.5 °C. Cells (at 1 × 106 cells/ml) were heat-shocked at 37 or 38 °C for 1 h, allowed to recover at 28.5 °C for 0 to 11 h, and harvested. Viable cell numbers were determined by using trypan blue dye exclusion. B, DAPI staining assessment of cell survival. The apoptotic effects of heat stress were temperature- and time-dependent. C, heat shock-induced activation of caspase-3 was assessed by measuring the hydrolysis of Ac-DEVD-MCA before and after heat shock treatment. The values shown represent the means of three independent experiments; error bars represent standard deviations.
FIGURE 2.
Effect of Z-VAD-fmk on heat stress-induced apoptosis in ZE cells. A, the caspase inhibitor Z-VAD-fmk inhibits heat shock-induced cell death. ZE cells were treated with 0, 20, or 40 μm Z-VAD-fmk for 1 h, heat-shocked at 38 °C for 1 h (at 1 × 106 cells/ml), allowed to recover at 28.5 °C for 0 to 11 h, and harvested. B, DAPI staining assessment of nuclear fragmentation indicated suppressive effects of Z-VAD-fmk on heat-induced apoptosis. Values shown represent the means of three independent experiments; error bars represent standard deviations. C, suppression of PARP cleavage by caspase inhibition. Cells were treated with 0 or 20 μm Z-VAD-fmk for 1 h, heat-shocked, and harvested after 4 h. Levels of PARP and actin were determined by Western blotting as described under “Experimental Procedures.”
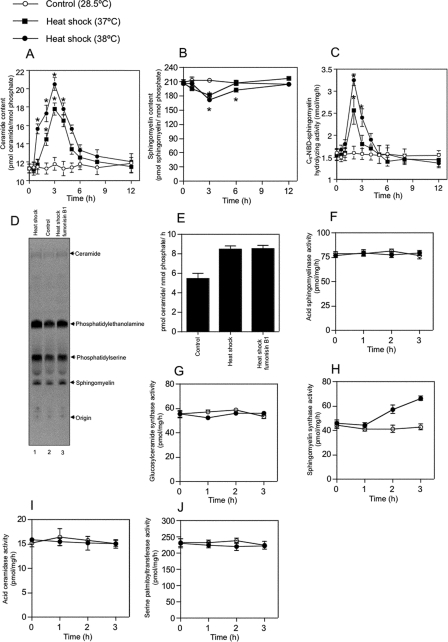
When the intercellular ceramide content was measured using a diacylglycerol kinase assay, heat shock transiently generated ceramide. A peak of approximately twice the basal ceramide levels was induced after heat shock for 3 h (Fig. 3A). A decrease in cellular sphingomyelin levels almost paralleled the increase in cellular ceramide levels after heat shock for 3 h (Fig. 3B).
FIGURE 3.
Heat stress increases ceramide content, decreases sphingomyelin content, and activates Mg2+-dependent neutral SMase in ZE cells. ZE cells were heat shocked at 37 or 38 °C for 1 h and allowed to recover at 28.5 °C for 0 to 11 h. A and B, cellular lipids were extracted at the indicated times. Changes in ceramide levels (A) were quantified using a diacylglycerol kinase-based assay. Changes in sphingomyelin levels (B) were quantified by phosphate measurement after TLC separation. C, changes in the activity of Mg2+-dependent neutral SMase were measured using C6-NBD-sphingomyelin as a substrate. D and E, effect of fumonisin B1 on heat shock-induced ceramide production. ZE cells were radiolabeled with 370 MBq/ml l-[U-14C]serine at 28.5 °C for 48 h, incubated at 28.5 °C for 1 h (lane 2), heat-shocked at 38 °C for 1 h, and allowed to recover at 28.5 °C for 2 h in the presence of 100 μm fumonisin B1 (lane 1) or in the absence of fumonisin B1 (lane 3). The lipids were extracted, separated on a TLC plate, and then detected using a Storm 860 imaging analyzer as described under “Experimental Procedures.” ZE cells were heat-shocked at the indicated temperatures for 1 h and allowed to recover at 28.5 °C for 0, 1 2, or 3 h. F–J, changes were measured in the activities of the ceramide-metabolizing enzymes acidic SMase (F), glucosylceramide synthase (G), sphingomyelin synthase (H), acidic ceramidase (I), and serine palmitoyltransferase (J). ZE cells were heat-shocked at the indicated temperatures for 1 h and allowed to recover at 28.5 °C for 0, 1 2, or 3 h. A and B, each value shown represents the mean of three independent experiments; error bars represent standard deviations. *, p < .05 versus control.
The biochemical mechanism of ceramide generation by de novo biosynthesis under heat stress condition was investigated in ZE cells. First, we examined whether ceramide biosynthesis was inhibited by an inhibitor of ceramide synthase, i .e. fumonisin B1. The cells were radiolabeled with [14C]serine following pretreatment with fumonisin B1; the attempt was unsuccessful in the case of both ceramide and sphingomyelin under normal condition (data not shown), indicating that fumonisin B1 inhibited ceramide generation by ceramide synthase in ZE cells. On the contrary, when ZE cells were radiolabeled with [14C]serine, the labeled ceramide was induced by heat stress, but the treatment of fumonisin B1 did not repress heat-induced ceramide generation (Fig. 3, D and E). These findings revealed that heat-induced ceramide generation is not mediated by ceramide synthase. Thus, heat shock affects the sphingomyelin cycle by increasing cellular ceramide levels and decreasing cellular sphingomyelin levels.
Activation of Neutral SMase in Heat-induced Apoptosis—To confirm the importance of SMase-dependent ceramide generation in heat-induced apoptosis, we measured the activities of ceramide-related enzymes such as Mg2+-dependent neutral SMase, acid SMase, sphingomyelin synthase, glucosylceramide synthase, acid ceramidase, and serine palmitoyltransferase. Heat shock treatment induced the activity of Mg2+-dependent neutral SMase in a time- and dose-dependent manner (Fig. 3C). Heat shock treatment also induced the activity of sphingomyelin synthase (Fig. 3G). In contrast, the activities of acid SMase, sphingomyelin synthase, glucosylceramide synthase, acid ceramidase, and serine palmitoyltransferase in ZE cells were unaffected by heat shock (Fig. 3, F–H and J). In addition, Mg2+-independent neutral SMase activity was not detected in ZE cells. Mg2+-dependent neutral SMase activity was increased from 1.58 to 2.56 nmol/mg/h or 3.25 nmol/mg/h by heat shock for 1 h at 37 or 38 °C, respectively.
Heat shock induced Mg2+-dependent neutral SMase activity in ZE cells as early as 1 h after exposure and reached a peak of activity 2 h after heat shock (Fig. 3C). Sphingomyelin synthase activity was slowly increased from 45.5 to 66.3 pmol/mg/h by heat shock for 1 h at 38 °C (Fig. 3H) Thus, ceramide is generated via sphingomyelin hydrolysis and catalyzed mainly by activated Mg2+-dependent neutral SMase as an early event in the induction of apoptosis after heat shock.
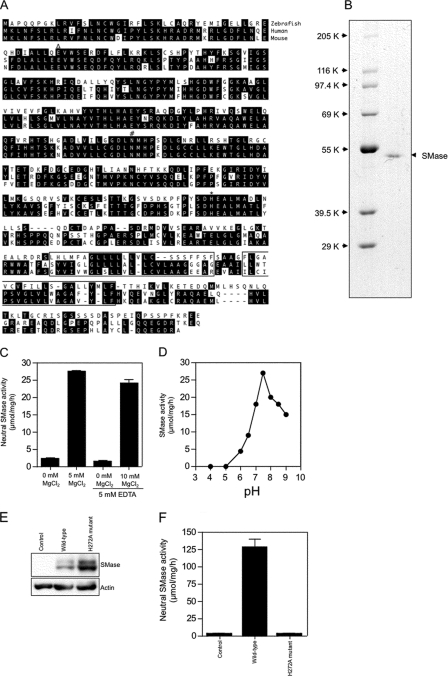
Expression Cloning of cDNA Encoding Mg2+-dependent Neutral SMase—ZE cells had ∼14-fold higher Mg2+-dependent neutral SMase activity than other fish and mammalian cell lines (Table 1). The ZE cell line has been studied previously for apoptotic responses under stresses such as UV irradiation, heat shock, cold shock, and starvation (35). We used ZE cells to characterize the neutral SMase. To isolate cDNA encoding neutral SMase, we screened bacterial clones that showed a neutral SMase activity against C6-7-nitro-2–1,3-benzoxadiazol-4-yl-sphingomyelin as a substrate from a cDNA library from ZE cells in an E. coli pET-28a expression vector. A single cDNA clone showing neutral SMase activity was isolated. This clone had a 1260-bp open reading frame that encoded a protein of 420 amino acids, with a putative molecular weight of 46,900 (Fig. 4A). A FASTA search of the Swiss-Prot/PIR protein sequence data base revealed that zebrafish neutral SMase possessed significant homology with the human neutral SMase 1 (29). The amino acid sequence identity of the neutral SMase 1 with human and mouse homologs was 44 and 46%, respectively (Fig. 4A). According to Tomiuk et al. (29), conserved amino acid residues in neutral SMase 1 were identified as the putative magnesium-binding glutamine residue (Gln-49 (39), the substrate-binding asparagine residue (Asn-180 (40), and the histidine residue (His-272) of the active site, deduced from studies of the Bacillus cereus IAM 1208 enzyme (Ref. 41; Fig. 4A). Secondary structure analysis performed using the SMART program (42) predicted that the C-terminal region of neutral SMase 1 contains two transmembrane domains, suggesting that this protein binds to cellular membranes (Fig. 4A).
TABLE 1.
Comparison of Mg2+-dependent neutral SMase activity in various cell types
| Cell type | Neutral SMase activitya |
|---|---|
| pmol/mg/h | |
| ZE | 1585.5 ± 19 |
| FHM | 1345.2 ± 11 |
| HEK293 | 134.7 ± 8 |
| HL-60 | 110.2 ± 5 |
Mean ± S.D. (n = 3)
FIGURE 4.
Structure and Mg2+-dependent neutral SMase activity of the cloned zebrafish enzyme. A, predicted amino acid sequence of zebrafish Mg2+-dependent neutral SMase 1. The two putative transmembrane domains identified by the SMART program are boxed. The putative Mg2+-complexing glutamine residue (▵), the asparagine residue involved in substrate binding (#), and the histidine residue that acts as the catalytic base (*) are shown. Human (NM_009213) and mouse (NM_009213) homologs of zebrafish Mg2+-dependent neutral SMase 1 (GenBank™ accession number AB196165) were identified using a FASTA search of the GenBank™ data base, and the deduced amino acid sequences were aligned. Amino acid residues conserved in at least two of the proteins are shaded in black. B, the purified recombinant enzyme was subjected to reducing 10% SDS-PAGE and visualized by staining with Coomassie Brilliant Blue R-250. C, effect of Mg2+ ions on the activity of recombinant neutral SMase. Measurements with Mg2+ ions and EDTA were performed in the presence of 10 mm EDTA. D, effect of pH on neutral SMase activity. To determine the optimum pH, the activity of the purified recombinant enzyme was measured at 37 °C for 30 min in various buffers (100 mm): acetate (pH 4 and 5), PIPES (pH 6, 6.5, and 7), and Tris (pH 7.5, 8, 8.5, and 9). Each value shown represents the mean of three independent experiments at each pH. E, the neutral SMase activity in lysates of ZE cell lines stably transfected with FLAG-tagged wild-type or H272A mutant Mg2+-dependent neutral SMase constructs was assayed. Lane 1, control; lane 2, wild type; lane 3, H272A mutant. The expressed proteins in the cell lines were detected with anti-FLAG or anti-actin antibodies by Western blotting as described under “Experimental Procedures.” F, neutral SMase activity against in each line is shown. Column 1, control; column 2, wild type; column 3, H272A mutant. Each value represents the mean of three independent experiments; error bars represent standard deviations.
To determine the enzyme activity of the cloned Mg2+-dependent neutral SMase, a cDNA expression construct under the control of the human cytomegalovirus promoter (pTRAGET-ZSMase) was introduced into COS-7 and ZE cells, and SMase activity in the cell lysates was assayed. The transfectants showed 10.9- and 7.8-fold higher sphingomyelin hydrolyzing activity than did those of the empty vector in COS-7 and ZE cells, respectively (Table 2). We showed that overexpression of the cloned enzyme in ZE cells expressed transgene products (Fig. 4E). The transfectant of wild-type SMase had a higher activity of Mg2+-dependent neutral SMase than transfectants with the H272A mutant or nontransfectants (control) (Fig. 4F).
TABLE 2.
Transient overexpression of cloned recombinant zebrafish neutral SMase in COS-7 and ZE cells
| Cell type | Neutral SMase activitya |
|---|---|
| pmol/mg/h | |
| COS-7 | |
| Untransfected | 120.2 ± 2.3 |
| Mock-transfected | 123.3 ± 2.4 |
| pTARGET-ZNSMase | 1,321.7 ± 4.5 |
| ZE | |
| Untransfected | 1,585.5 ± 19 |
| Mock-transfected | 1,567.2 ± 23 |
| pTARGET-ZNSMase | 12,491.2 ± 24.6 |
Mean ± S.D. (n = 3)
A recombinant protein with the His tag sequence at the N terminus was produced, using the expression vector pET-16b carrying the neutral SMase cDNA. The affinity purification by His tag chromatography of the recombinant protein yielded a high purity protein, as indicated by a single protein band with a molecular weight of 49,400 on SDS-PAGE (Fig. 4B). The purified recombinant Mg2+-dependent neutral SMase showed high activity toward the substrate [choline-methyl-14C]sphingomyelin, whereas the enzyme had no activity against the phospholipid [choline-methyl-14C]phosphatidylcholine and very little activity against [1-O-octadecyl-9,10-3H]lyso-platelet-activating factor (Table 3). The activity was absolutely dependent on magnesium ions (Fig. 4C), optimally at pH 7.5 (Fig. 4D). The membrane-bound Mg2+-dependent neutral SMase encoded by our isolated clone was designated “neutral SMase 1:SMPD2” (29, 43).
TABLE 3.
Substrate specificity of the cloned recombinant neutral SMase
| Substrate | Activitya |
|---|---|
| pmol/mg/h | |
| [14C]Sphingomyelin | 27 ± 0.76b |
| [14C]Phosphatidylcholine | 0b |
| [3H]Lyso-PAF | 1.5 ± 0.8c |
Mean ± S.D. (n = 3)
Activity was determined in the presence of 0.1% Triton X-100
Activity was determined in the absence of Triton X-100
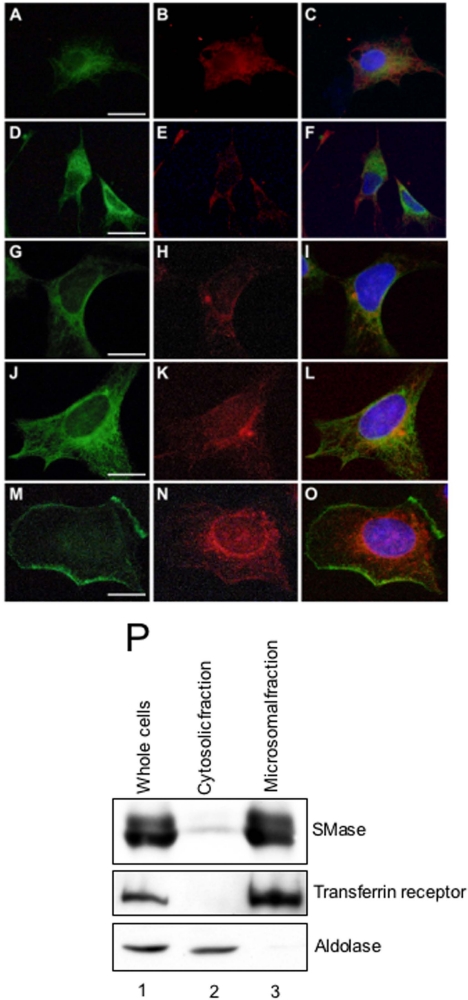
Subcellular Localization of Neutral SMase 1—To examine the subcellular localization of the neutral SMase 1, we constructed an SMase-FLAG fusion and established its stable transfectants in both the ZE and HEK293 cell lines. The transfectants overexpressing wild-type SMase 1 were double-stained with anti-zebrafish neutral SMase 1 antibody together with anti-58K protein antibody (Golgi marker) or anti-KDEL protein antibody (ER marker) by immunofluorescence microscopy. Subcellular localization of SMase 1 with that of the Golgi marker (Fig. 5, A and B) showed that the SMase 1 exhibited major signals in the Golgi (Fig. 5C). Consequently, staining with ER (Fig. 5, D and E), SMase 1 was not colocalized in the ER (Fig. 5F). In the case of HEK293 transfectants, the zebrafish SMase 1 was also colocalized in both the Golgi and the ER (Fig. 5, G–L). When we used a non-permeation treatment without 0.1% Triton-X 100 in the staining steps to detect the SMase 1 in the cell membrane, the enzyme was localized differentially from the cell membrane cadherin, revealing the disappearance of the SMase 1 in the cell membrane (Fig. 5, M–O). The subcellular localization of neutral SMase 1 in ZE cells was also examined by centrifugal fractionation. The activities of neutral SMase in the microsomal fraction in both the ZE cells and the SMase 1-overexpressed transfectant were higher than those of the nuclear and cytosolic fractions (Table 4). Western blotting showed that SMase 1 was detected in the microsomal fraction but not in the cytosolic fraction (Fig. 5P). Therefore, neutral SMase 1 was present in the microsomal fraction.
FIGURE 5.
Subcellular localization of neutral SMase 1. Stably transfected ZE cells (A–F) and HEK293 cells (G–O) overexpressing FLAG-tagged SMase were fixed and permeabilized with 0.1% Triton X-100. The cells were double-stained with rabbit anti-zebrafish neutral SMase 1 antibody and an antibody against either 58K protein (a Golgi marker) (A–C, G–I) or KDEL protein (an ER marker) (D–F, J–L) and then stained with fluorescent secondary antibodies. The cells overexpressing FLAG-tagged SMase were fixed and stained without permeation treatment (L–O). The cells were double-stained with mouse anti-FLAG antibody and an antibody against rabbit cadherin (a cell membrane marker) and then stained with fluorescent secondary antibodies. Signals for SMase 1 (A, D, G, J, and N) and signals for subcellular makers such as Golgi (B and H), ER (E and K), and cell membrane (M), were observed. The overlay images (C, F, I, L, and O) indicate that SMase 1 and the subcellular marker were colocalized either in the same place or adjacent to one another. Bar = 10 μm. P, whole lysate of ZE cells (lane 1) were fractionated into cytosolic (lane 2) and microsomal (lane 3) fractions by ultracentrifugation. These fractions were analyzed by Western blotting using antibodies against zebrafish neutral SMase 1, aldolase (a cytosolic marker), and transferrin receptor (a cell membrane marker).
TABLE 4.
Neutral SMase activity in the subcellular fractions of SMase-overexpressing and non-overexpressing ZE cells
|
Subcellular fraction
|
Neutral SMase
activitya
|
|
|---|---|---|
| Control | pTARGET-ZNSMase-transfected | |
| pmol/mg/h | ||
| Whole cells | 1,590 ± 2 | 12,500 ± 25 |
| Nuclear fraction | 551 ± 1 | 1,008 ± 12 |
| Cytosolic fraction | 0 | 0 |
| Microsomal fraction | 33,800 ± 2 | 58,300 ± 100 |
Mean ± S.D. (n = 3)
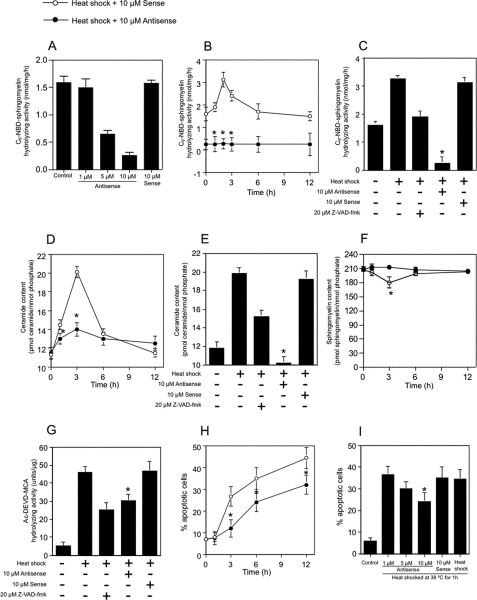
Knockdown Experiment with an Antisense Oligonucleotides—To examine whether ceramide generation by Mg2+-dependent neutral SMase was involved in heat-induced apoptosis, the phosphorothioate oligonucleotide for neutral SMase 1 was used to repress the levels of this protein. The activity of Mg2+-dependent neutral SMase after 48 h of antisense oligonucleotide treatment was decreased in an oligonucleotide-dependent manner (Fig. 6A). Treatment with an antisense oligonucleotide for Mg2+-dependent neutral SMase reduced not only the basal activity of Mg2+-dependent neutral SMase from 1.58 to 0.25 nmol/mg/h but also heat shock-activated neutral SMase from 3.12 to 0.25 nmol/mg/h (Fig. 6, B and C). The treatment with the antisense oligonucleotide repressed induced endogenous ceramide under heat shock (Fig. 6, D and E) but did not affect the basal level of endogenous ceramide (Fig. 6D). Antisense oligonucleotide treatment repressed the decrease in cellular sphingomyelin content (Fig. 6F), the increase in caspase-3 activity (Fig. 6G), and the induction of apoptosis under heat stress (Fig. 6, H and I), whereas treatment with the corresponding sense oligonucleotides had no such effect. Thus, the Mg2+-dependent neutral SMase deficiency induced by antisense oligonucleotide treatment repressed heat-induced apoptosis, suggesting that heat shock normally activates the enzyme to process sphingomyelin into ceramide.
FIGURE 6.
Antisense inhibition of zebrafish Mg2+-dependent neutral SMase 1 blocks heat shock-mediated apoptosis and ceramide generation. A, effect of antisense oligo treatment on Mg2+-dependent neutral SMase activity. ZE cells were pretreated with an antisense (0–10 μm) or sense (10 μm) oligonucleotide targeting neutral SMase 1 mRNA for 48 h; heat-shocked at 38 °C for 1 h; allowed to recover at 28.5 °C; and harvested after 0, 1, 2, 3, 6, or 12 h. Some cells were also treated with 20 μm Z-VAD-fmk for 1 h before being heat-shocked. The neutral SMase activity in the cells was measured as described under “Experimental Procedures.” B and C, Mg2+-dependent neutral SMase activity. D and E, ceramide content. F, sphingomyelin content. G, caspase-3 activity. H, DAPI assay. I, cell viability. Values and bars indicate the means and standard deviations, respectively, of three independent experiments. *, p < .05 versus sense oligonucleotide-treated cells.
DISCUSSION
Ceramide is a lipid signaling molecule in stress-induced apoptosis. It is generated by the hydrolysis of sphingomyelin through the action of an SMase that is activated by stresses such as heat, UV and γ-irradiation, Fas ligand and tumor necrosis factor-α, serum withdrawal, and hypoxia (4, 26, 44, 45). At least three distinct neutral SMases have been identified in human and mouse: neutral SMases 1, 2, and 3 (27–29). However, the neutral SMase(s) responsible for stress-induced ceramide generation and apoptosis has not been identified. To characterize the molecular mechanism of stress-induced ceramide generation and apoptosis, we cloned a membrane-bound Mg2+-dependent neutral SMase 1 from zebrafish ZE cells and showed that this enzyme was activated in response to heat shock, generating ceramide and inducing apoptosis. The isolated neutral SMase 1 showed enzymatic characteristics similar to those of the known mammalian neutral SMase 1. The zebrafish and mammalian enzymes share common structural features such as a magnesium-binding site, a substrate-binding site, and an active center His residue. They also have two transmembrane domains in the C-terminal region, indicating that the neutral SMase 1 is membrane-bound.
Furthermore, the activity and substrate specificity of the enzyme are similar to those of the mammalian neutral SMase 1 (43). The recombinant zebrafish neutral SMase 1 had specific hydrolyzing activity against sphingomyelin and produced ceramide. In addition, the enzyme was responsible for heat-inducible ceramide generation and apoptosis. The human enzyme also possesses Mg2+-dependent neutral SMase activity (43). However, the mechanism responsible for ceramide generation and induction of apoptosis has not been well characterized.
The overexpression of the human neutral SMase 1 has no effect on sphingomyelin metabolism but decreases levels of lyso-PAF, suggesting that it has a lyso-PAF phospholipase C activity (38). In knock-out mice, the loss of function of neutral SMase 1 results in neither lipid storage abnormalities nor deficient sphingomyelin metabolism (46). The overexpression of human neutral SMase 1 in human Jurkat cells results in no effect on CD95/Fas receptor-induced ceramide production or apoptosis, suggesting that it is not a candidate enzyme for the execution phase of death receptor-induced apoptosis (47). On the other hand, zebrafish neutral SMase 1 showed specific hydrolyzing activity against sphingomyelin and only low lyso-PAF phospholipase C activity, indicating that SMase 1 primarily regulates ceramide generation on sphingomyelin metabolism.
The specific inactivation of neutral SMase 1 by antisense RNA is repressed in ceramide-mediated apoptosis triggered by T cell receptor ligation (48). The loss of function of neutral SMase 1 represses amyloid-β peptide-induced ceramide generation and apoptosis in rat oligodendrocytes (49). Thus, the known mammalian neutral SMase 1 have biochemical characteristics that are closely related to the zebrafish enzyme, and their biological significance in ceramide metabolism is suggested. Because zebrafish neutral SMase 1 hydrolyzes sphingomyelin and induces apoptosis in vivo and in vitro, such functions in mammalian neutral SMase 1 should be examined with regard to ceramide metabolism and the induction of apoptosis.
Several lines of evidence support the induction of ceramide generation and stress-induced apoptosis in ZE cells under heat stress at 37–38 °C, similar to heat-shocked human HL-60 cells (7). Heat shock and recovery following at 28.5 °C caused apoptosis in a dose- and time-dependent manner in ZE cells. This apoptotic process showed enhanced caspase-3 activity during recovery after heat shock; the apoptosis could be rescued by the presence of the caspase inhibitor Z-VAD-fmk, indicating that the induced apoptosis in ZE cells was regulated by a caspase-3-dependent mechanism. Heat shock (8), Fas ligand, and tumor necrosis factor-α have been shown to activate neutral and/or acid SMase in generating ceramide (4, 26, 50). In our experiments, heat-induced apoptosis with ceramide generation occurred in a temperature- and time-dependent manner in parallel with SMase activation and a decrease in sphingomyelin content in ZE cells (Fig. 3). The temporally produced ceramide may be the proapoptotic signal for apoptosis in stressed cells, and ceramide was reduced to the normal physiological levels by the activation of sphingomyelin synthase during the recovery after heat shock (Fig. 3H). In contrast, the activities of other ceramide-related enzymes, such as acid SMase, glucosylceramide synthase, acid ceramidase, and serine palmitoyltransferase, were unaffected in ZE cells by heat shock (Fig. 3, F, G, I, and J). These findings support an important function of the neutral SMase in heat shock-induced ceramide generation. Unlike our present findings, de novo biosynthesis for ceramide generation has been reported in bacteria and yeast, whereas heat shock induced ceramide generation from dihydroshingosine by the action of ceramide synthase and serine palmitoyltransferase (51). In zebrafish cells, ceramide generation was not affected by fumonisin B1 under the prescribed heat shock conditions (Fig. 3, D and E), indicating that ceramide synthase is not responsible for heat-induced ceramide generation in the fish cells.
Ceramide has been considered to act as a signaling molecule upstream of Bcl-2 and caspase-3 and -9 (7, 52–55). In our study, caspase-3 activation by heat shock was followed by ceramide generation. However, the mechanism of caspase-3 activation must not involve directly ceramide. Other proapoptotic signaling molecules, such as Apaf-1 and unknown ceramide-binding protein, may induce the caspase cascade.
Hostetler and Yazaki (17) reported that the activity of a microsomal neutral SMase in rat liver is different from the enzyme in the cell membrane fraction. The Mg2+-dependent neutral SMase may be localized at the cytosolic side of the cell membrane during apoptosis (56–58). The subcellular localization of human and mouse neutral SMase 1 was also shown in the ER membranes (29, 43). In our immunocytochemical experiments, the zebrafish enzyme was localized primarily in the Golgi, and subcellular fractionation indicated that it was distributed in the microsomal fraction (Fig. 5). Because zebrafish neutral SMase 1 is predicted to possess two transmembrane domains in its C-terminal region and has an intracellular catalytic domain (Fig. 4A), the enzyme may bind primarily to the Golgi membranes.
The localization of neutral SMase 1 in the Golgi supports the cellular functions of the enzyme in ceramide generation for vesicle-membrane fusion and endocytosis. SMase has been reported to induce aggregation and fusion by ceramide release (59) and to disturb the lipid bilayer structure in favor of a nonlamellar and micellar phase (60). Thus, SMase activation and ceramide generation may be induced by local vesicle formation and fusion under heat shock conditions. Sphingomyelin and phosphatidylcholine are asymmetrically distributed in the outer leaflet, and the aminophospholipids are in the inner leaflet of the lipid bilayer (61, 62). As the neutral SMase is suggested to bind to cellular membranes, heat shock may induce the temporal loss of the asymmetric phospholipids distribution and the SMase activation. In addition, the neutral SMase 1 activity is regulated by glutathione levels (63). The enzyme activities are inhibited by glutathione at 1 mm in the ZE cells in vitro and in vivo.3 Therefore, the depletion of glutathione may be an important mechanism in activation of the SMase.
Neutral SMase 1 plays a critical role in zebrafish development. Knockdown of the SMase 1 gene showed blockage of epiboly formation and neurogenesis, hence generation of abnormal phenotype. Such phenotypic arrest was rescued by microinjection with exogenous ceramide to the normal phenotype. Thus, SMase 1 and its production of ceramide are essential for programmed cell death and differentiation.3 The enzyme expressed in the notochord at segmentation stages and depletion of ceramide by the SMase knockdown result in thalidomide-induced ceramide generation and vascular defect (32). In contrast to ceramide, sphingosine 1-phosphate inhibits SMase-dependent ceramide generation and restores thalidomide-induced vascular defect with an increase of expression of vascular endothelial growth factors (VEGF) receptors. Therefore, neutral SMase 1 is an essential mediator of normal development and embryogenesis as well as a proapoptotic signal in stress-induced apoptosis.
Acknowledgments
We thank Dr. Anwar Hossain for critical reading of the manuscript.
This work was supported by grants from the Japan Science and Technology Corporation, the Japan Society for the Promotion of Science, and the Ministry of Agriculture, Forestry, and Fisheries of Japan. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: C2-ceramide, N-acetylsphingosine (a ceramide analog); MCA, 4-methylcoumaryl-7-amide; C6-NBD-sphingomyelin, C6-7-nitro-2-1,3-benzoxadiazol-4-yl-sphingomyelin; [14C]phosphatidylcholine, l-3-phosphatidyl[N-methyl-14C]choline,1,2-dipalmitoyl; [14C]sphingomyelin, [N-methyl-14C]sphingomyelin; DAPI, 4′,6-diamidino-2-phenylidole; ER, endoplasmic reticulum; FCS, fetal calf serum; HEK, human embryonic kidney; PAF, [1-O-octadecyl-3H]lyso-platelet-activating factor; PBS, phosphate-buffered saline; SMase, sphingomyelinase; LB, Luria-Bertani broth; PARP, poly(ADP-ribose) polymerase; PIPES, 1,4-piperazinediethanesulfonic acid; ZE, zebrafish embryonic; Z-VAD-fmk, benzyloxycarbonyl-Val-Ala-Asp-fluoromethylketone.
T. Yabu, unpublished results.
References
- 1.Obeid, L. M., Linardic, C. M., Karolak, L. A., and Hannun, Y. A. (1993) Science 259 1769-1771 [DOI] [PubMed] [Google Scholar]
- 2.Pena, L. A., Fuks, Z., and Kolesnick, R. (1997) Biochem. Pharmacol. 53 615-621 [DOI] [PubMed] [Google Scholar]
- 3.Okazaki, T., Bell, R. M., and Hannun, Y. A. (1989) J. Biol. Chem. 264 19076-19080 [PubMed] [Google Scholar]
- 4.Hannun, Y. A., and Luberto, C. (2000) Trends Cell Biol. 10 73-80 [DOI] [PubMed] [Google Scholar]
- 5.Obeid, L. M., and Hannun, Y. A. (2003) Sci. Aging Knowledge Environ. 2003 27. [DOI] [PubMed] [Google Scholar]
- 6.Verheij, M., Bose, R., Lin, X. H., Yao, B., Jarvis, W. D., Grant, S., Birrer, M. J., Szabo, E., Zon, L. I., Kyriakis, J. M., Haimovitz-Friedman, A., Fuks, Z., and Kolesnick, R. N. (1996) Nature 380 75-79 [DOI] [PubMed] [Google Scholar]
- 7.Kondo, T., Matsuda, T., Kitano, T., Takahashi, A., Tashima, M., Ishikura, H., Umehara, H., Domae, N., Uchiyama, T., and Okazaki, T. (2000) J. Biol. Chem. 275 7668-7676 [DOI] [PubMed] [Google Scholar]
- 8.Chang, Y., Abe, A., and Shayman, J. A. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 12275-12279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kondo, T., Matsuda, T., Tashima, M., Umehara, H., Domae, N., Yokoyama, K., Uchiyama, T., and Okazaki, T. (2000) J. Biol. Chem. 275 8872-8879 [DOI] [PubMed] [Google Scholar]
- 10.Liu, B., Obeid, L. M., and Hannun, Y. A. (1997) Semin. Cell Dev. Biol. 8 311-322 [DOI] [PubMed] [Google Scholar]
- 11.Ferlinz, K., Hurwitz, R., Vielhaber, G., Suzuki, K., and Sandhoff, K. (1994) Biochem. J. 301 855-862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heller, M., and Shapiro, B. (1966) Biochem. J. 98 763-769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kanfer, J. N., Young, O. M., Shapiro, D., and Brady, R. O. (1966) J. Biol. Chem. 241 1081-1084 [PubMed] [Google Scholar]
- 14.Schissel, S. L., Schuchman, E. H., Williams, K. J., and Tabas, I. (1996) J. Biol. Chem. 271 18431-18436 [DOI] [PubMed] [Google Scholar]
- 15.Spence, M. W., Byers, D. M., Palmer, F. B., and Cook, H. W. (1989) J. Biol. Chem. 264 5358-5363 [PubMed] [Google Scholar]
- 16.Gatt, S. (1976) Biochem. Biophys. Res. Commun. 68 235-241 [DOI] [PubMed] [Google Scholar]
- 17.Hostetler, K. Y., and Yazaki, P. J. (1979) J. Lipid Res. 20 456-463 [PubMed] [Google Scholar]
- 18.Nilsson, A. (1969) Biochim. Biophys. Acta 176 339-347 [DOI] [PubMed] [Google Scholar]
- 19.Rao, B. G., and Spence, M. W. (1976) J. Lipid Res. 17 506-515 [PubMed] [Google Scholar]
- 20.Okazaki, T., Bielawska, A., Domae, N., Bell, R. M., and Hannun, Y. A. (1994) J. Biol. Chem. 269 4070-4077 [PubMed] [Google Scholar]
- 21.Duan, R. D., Bergman, T., Xu, N., Wu, J., Cheng, Y., Duan, J., Nelander, S., Palmberg, C., and Nilsson, A. (2003) J. Biol. Chem. 278 38528-38536 [DOI] [PubMed] [Google Scholar]
- 22.Duan, R. D., Cheng, Y., Hansen, G., Hertervig, E., Liu, J. J., Syk, I., Sjostrom, H., and Nilsson, A. (2003) J. Lipid Res. 44 1241-1250 [DOI] [PubMed] [Google Scholar]
- 23.Duan, R. D., Nyberg, L., and Nilsson, A. (1995) Biochim. Biophys. Acta 1259 49-55 [DOI] [PubMed] [Google Scholar]
- 24.Nyberg, L., Duan, R. D., Axelson, J., and Nilsson, A. (1996) Biochim. Biophys. Acta 1300 42-48 [DOI] [PubMed] [Google Scholar]
- 25.Marchesini, N., and Hannun, Y. A. (2004) Biochem. Cell Biol. 82 27-44 [DOI] [PubMed] [Google Scholar]
- 26.Hannun, Y. A. (1996) Science 274 1855-1859 [DOI] [PubMed] [Google Scholar]
- 27.Hofmann, K., Tomiuk, S., Wolff, G., and Stoffel, W. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 5895-5900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krut, O., Wiegmann, K., Kashkar, H., Yazdanpanah, B., and Kronke, M. (2006) J. Biol. Chem. 281 13784-13793 [DOI] [PubMed] [Google Scholar]
- 29.Tomiuk, S., Hofmann, K., Nix, M., Zumbansen, M., and Stoffel, W. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 3638-3643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yabu, T., Kishi, S., Okazaki, T., and Yamashita, M. (2001) Biochem. J. 360 39-47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yabu, T., Todoriki, S., and Yamashita, M. (2001) Fish. Sci. 67 333-340 [Google Scholar]
- 32.Yabu, T., Tomimoto, H., Taguchi, Y., Yamaoka, S., Igarashi, Y., and Okazaki, T. (2005) Blood 106 125-134 [DOI] [PubMed] [Google Scholar]
- 33.Yabu, T., Ishibashi, Y., and Yamashita, M. (2003) Fish. Sci. 69 1218-1223 [Google Scholar]
- 34.Yoshimura, Y., Tani, M., Okino, N., Iida, H., and Ito, M. (2004) J. Biol. Chem. 279 44012-44022 [DOI] [PubMed] [Google Scholar]
- 35.Imamura, S., Ojima, N., and Yamashita, M. (2003) FEBS Lett. 549 14-20 [DOI] [PubMed] [Google Scholar]
- 36.Bligh, E. G., and Dyer, W. J. (1959) Can. J. Biochem. Physiol. 37 911-917 [DOI] [PubMed] [Google Scholar]
- 37.Sawai, H., Okazaki, T., Yamamoto, H., Okano, H., Takeda, Y., Tashima, M., Sawada, H., Okuma, M., Ishikura, H., Umehara, H., and Domae, N. (1995) J. Biol. Chem. 270 27326-27331 [DOI] [PubMed] [Google Scholar]
- 38.Sawai, H., Domae, N., Nagan, N., and Hannun, Y. A. (1999) J. Biol. Chem. 274 38131-38139 [DOI] [PubMed] [Google Scholar]
- 39.Fujii, S., Ogata, K., Inoue, B., Inoue, S., Murakami, M., Iwama, S., Katsumura, S., Tomita, M., Tamura, H., Tsukamoto, K., Ikezawa, H., and Ikeda, K. (1999) J. Biochem. 126 90-97 [DOI] [PubMed] [Google Scholar]
- 40.Fujii, S., Inoue, B., Yamamoto, H., Ogata, K., Shinki, T., Inoue, S., Tomita, M., Tamura, H., Tsukamoto, K., Ikezawa, H., and Ikeda, K. (1998) J. Biochem. 124 1178-1187 [DOI] [PubMed] [Google Scholar]
- 41.Matsuo, Y., Yamada, A., Tsukamoto, K., Tamura, H., Ikezawa, H., Nakamura, H., and Nishikawa, K. (1996) Protein Sci. 5 2459-2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Letunic, I., Copley, R. R., Pils, B., Pinkert, S., Schultz, J., and Bork, P. (2006) Nucleic Acids Res. 34 257-260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tomiuk, S., Zumbansen, M., and Stoffel, W. (2000) J. Biol. Chem. 275 5710-5717 [DOI] [PubMed] [Google Scholar]
- 44.Kronke, M. (1999) Chem. Phys. Lipids 102 157-166 [DOI] [PubMed] [Google Scholar]
- 45.Stoffel, W. (1999) Chem. Phys. Lipids 102 107-121 [DOI] [PubMed] [Google Scholar]
- 46.Zumbansen, M., and Stoffel, W. (2002) Mol. Cell. Biol. 22 3633-3638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tepper, A. D., Ruurs, P., Borst, J., and van Blitterswijk, W. J. (2001) Biochem. Biophys. Res. Commun. 280 634-639 [DOI] [PubMed] [Google Scholar]
- 48.Tonnetti, L., Veri, M. C., Bonvini, E., and D'Adamio, L. (1999) J. Exp. Med. 189 1581-1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee, J. T., Xu, J., Lee, J. M., Ku, G., Han, X., Yang, D. I., Chen, S., and Hsu, C. Y. (2004) J. Cell. Biol. 164 123-131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sawai, H., and Hannun, Y. A. (1999) Chem. Phys. Lipids 102 141-147 [DOI] [PubMed] [Google Scholar]
- 51.Jenkins, G. M., Cowart, L. A., Signorelli, P., Pettus, B. J., Chalfant, C. E., and Hannun, Y. A. (2002) J. Biol. Chem. 277 42572-42578 [DOI] [PubMed] [Google Scholar]
- 52.Tepper, A. D., de Vries, E., van Blitterswijk, W. J., and Borst, J. (1999) J. Clin. Investig. 103 971-978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tepper, A. D., Cock, J. G., de Vries, E., Borst, J., and van Blitterswijk, W. J. (1997) J. Biol. Chem. 272 24308-24312 [DOI] [PubMed] [Google Scholar]
- 54.Sawada, M., Nakashima, S., Banno, Y., Yamakawa, H., Hayashi, K., Takenaka, K., Nishimura, Y., Sakai, N., and Nozawa, Y. (2000) Cell Death Differ. 7 761-772 [DOI] [PubMed] [Google Scholar]
- 55.Gamen, S., Marzo, I., Anel, A., Pineiro, A., and Naval, J. (1996) FEBS Lett. 390 232-237 [DOI] [PubMed] [Google Scholar]
- 56.Linardic, C. M., and Hannun, Y. A. (1994) J. Biol. Chem. 269 23530-23537 [PubMed] [Google Scholar]
- 57.Andrieu-Abadie, N., and Levade, T. (2002) Biochim. Biophys. Acta 1585 126-134 [DOI] [PubMed] [Google Scholar]
- 58.Andrieu, N., Salvayre, R., and Levade, T. (1996) Eur. J. Biochem. 236 738-745 [DOI] [PubMed] [Google Scholar]
- 59.Oorni, K., Hakala, J. K., Annila, A., Ala-Korpela, M., and Kovanen, P. T. (1998) J. Biol. Chem. 273 29127-29134 [DOI] [PubMed] [Google Scholar]
- 60.Veiga, M. P., Arrondo, J. L., Goni, F. M., and Alonso, A. (1999) Biophys. J. 76 342-350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zwaal, R. F., and Schroit, A. J. (1997) Blood 89 1121-1132 [PubMed] [Google Scholar]
- 62.Bevers, E. M., Comfurius, P., Dekkers, D. W., Harmsma, M., and Zwaal, R. F. (1998) Lupus 7 126-131 [DOI] [PubMed] [Google Scholar]
- 63.Martin, S. F., Sawai, H., Villalba, J. M., and Hannun, Y. A. (2007) Arch. Biochem. Biophys. 459 295-300 [DOI] [PubMed] [Google Scholar]