Abstract
Full activation of protein kinase B (PKB/Akt) requires phosphorylation on Thr-308 and Ser-473. It is well established that Thr-308 is phosphorylated by 3-phosphoinositide-dependent kinase-1 (PDK1). Ser-473 phosphorylation is mediated by both mammalian target of rapamycin-rictor complex (mTORC2) and DNA-dependent protein kinase (DNA-PK) depending on type of stimulus. However, the physiological role of DNA-PK in the regulation of PKB phosphorylation remains to be established. To address this, we analyzed basal, insulin-induced, and DNA damage-induced PKB Ser-473 phosphorylation in DNA-PK catalytic subunit-null DNA-PKcs-/- mice. Our results revealed that DNA-PK is required for DNA damage-induced phosphorylation but dispensable for insulin- and growth factor-induced PKB Ser-473 phosphorylation. Moreover, DNA-PKcs-/- mice showed a tissue-specific increase in basal PKB phosphorylation. In particular, persistent PKB hyperactivity in the thymus apparently contributed to spontaneous lymphomagenesis in DNA-PKcs-/- mice. Significantly, these tumors could be prevented by deletion of PKBα. These findings reveal stimulus-specific regulation of PKB activation by specific upstream kinases and provide genetic evidence of PKB deregulation in DNA-PKcs-/- mice.
The serine/threonine protein kinase B (PKB),3 also known as Akt, is a downstream effector of phosphatidylinositol 3-kinase (PI3K) and a major regulator of a variety of cellular processes, including metabolism, transcription, survival, proliferation, and growth (1–4). PKB acts on a plethora of substrates, including GSK3β and FoxO transcription factors (3–6). PKB is activated by several stimuli, including hormones, growth factors, cytokines and, as recently reported, also by DNA damage (7–10). Deregulation of PKB is implicated in carcinogenesis and diabetes (1, 11, 12).
Activation of PKB requires phosphorylation at two key regulatory sites as follows: Thr-308 and Ser-473 (of PKBα). Phosphorylation by 3-phosphoinositide-dependent kinase-1 (PDK1) occurs on Thr-308 in the activation loop of PKB. The phosphorylation on Ser-473 within a C-terminal hydrophobic motif leads to full activation of PKB. The mTOR-rictor complex (mTORC2), a member of the PI3K-related kinase (PIKK) family, has been reported to regulate Ser-473 phosphorylation (13–16). Significantly, it was shown that DNA-dependent protein kinase (DNA-PK), a further member of the PIKK family, also regulates PKB Ser-473 phosphorylation (9). In addition, a role for DNA-PK in the activation of PKB by CpG-DNA has been established using bone marrow-derived macrophages (17). Moreover, Bozulic et al. (8) demonstrated recently that DNA-PK phosphorylates PKB Ser-473 upon induction of DNA double strand breaks. However, the regulation of PKB by DNA-PK under physiological conditions remained to be established. This study made use of genetically modified mouse models to investigate PKB regulation by DNA-PK, as phosphorylation of Ser-473 may be stimulus-, signaling pathway-, and/or cell type-specific.
DNA-PK is composed of a 470-kDa catalytic subunit (DNA-PKcs) and the Ku antigen complex (Ku80/Ku70) and involved in V(D)J recombination, repair of DNA double strand breaks by nonhomologous end joining, apoptosis, and transcription regulation (18). Double-stranded DNA ends produced by ionizing radiation or radiomimetic drugs activate DNA-PK, which is a primary sensor of DNA damage (19). DNA-PKcs-/- mice display growth retardation, hypersensitivity to ionizing radiation, and severe immunodeficiency (20–22). Because of defects in V(D)J recombination, the development of T and B cells is blocked at an early progenitor stage in DNA-PKcs-/- mice. Moreover, DNA-PKcs-/- thymus has decreased cellularity and displays hypotrophy. Furthermore, DNA-PKcs-/- mice display an increased rate of T cell lymphoma (23).
Studies of mutant mice carrying a null mutation for each of the three PKB isoforms PKBα, PKBβ, and PKBγ revealed differing phenotypes, some of which are shared by DNA-PKcs-/- mice. Similar to DNA-PKcs-/- mice, PKBα-/- mice display growth retardation and hypersensitivity to DNA damage (24–27). Furthermore, PKBα-/- mice show accumulation of early thymocyte subsets (28–30). PKBβ-/- mice display glucose intolerance and insulin resistance, whereas PKBγ-/- mice have a reduced brain size (31–34). Deletion of PKBβ and PKBγ isoforms revealed that a single functional allele of PKBα is sufficient for mouse development and survival (35).
In this study, we investigated the role of DNA-PK in basal, insulin-induced, and DNA damage-induced phosphorylation of PKB Ser-473 under physiological conditions. We report that DNA-PK phosphorylated PKB on Ser-473 upon DNA damage induced by γ-irradiation in vivo. In contrast, DNA-PK was dispensable for insulin and growth factor-induced PKB activation. Interestingly, analysis of basal PKB Ser-473 phosphorylation in DNA-PKcs-/- mice showed tissue-specific deregulation of the PKB/FoxO pathway. In particular, we provide evidence that persistent PKB hyperactivity in the thymus apparently contributes to spontaneous thymic lymphomagenesis in DNA-PKcs-/- mice. Lymphomagenesis could be prevented by the deletion of PKBα and implies deregulation of PKB in DNA-PKcs-/- thymi.
EXPERIMENTAL PROCEDURES
Mice—DNA-PKcs-/- mice (20) were kindly provided by Prof. Fredrick Alt (Howard Hughes Medical Institute, Harvard Medical School). PKBα-/- mice were described previously (27). For the generation of DNA-PKcs-/-PKBα-/- mice, DNA-PKcs-/- mice were mated with PKBα-/- mice, and the resulting DNA-PKcs+/-PKBα+/- progeny intercrossed. All mice had a C57BL/6;129 mixed background. Mice were housed according to Swiss Animal Protection legislation under specific pathogen-free conditions. All procedures were conducted with the approval of the appropriate authorities.
Cell Culture and Treatments—Mouse embryonic fibroblasts (MEFs) were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, 100 units/ml penicillin, and 100 μg/ml streptomycin. For IGF-1 and serum stimulation, MEFs were starved overnight prior to treatment with IGF-1 (50 ng/ml) or serum (10% fetal calf serum) for the indicated times.
Western Blot Analysis—Protein lysates were prepared by homogenization of various organs in lysis buffer (50 mm Tris-HCl, pH 8.0, 120 mm NaCl, 1% Nonidet P-40, 40 mm β-glycerophosphate, 10% glycerol, 0.05 mm phenylmethylsulfonyl fluoride, 1 mm benzamidine, 50 mm NaF, 1 mm Na3VO4, 1 μm microcystin LR). Homogenates were centrifuged twice (13,000 rpm for 15 min at 4 °C) to remove cell debris. Protein concentrations were determined using the Bradford assay (Bio-Rad). Proteins were separated by 6, 8, or 10% SDS-PAGE and then transferred to Immobilon-P polyvinylidene difluoride membranes (Millipore). Antibodies against phospho-PKB (Ser-473), phospho-PKB (Thr-308), phospho-Foxo4 (Ser-193), phospho-GSK3β (Ser-9), phospho-S6K (Thr-389), and phospho-PKC (Ser-657) were purchased from Cell Signaling. PKCα antibody was from Santa Cruz Biotechnology. Phospho-FoxO3 (Thr-32) and IRS-1 antibodies were from Upstate Biotechnology, Inc./Millipore. DNA-PKcs (Ab-4) and actin (Ab-5) antibodies were from NeoMarkers. PKBα isoform-specific antibody and PKB antibody Ab10 were previously described (27, 36). Anti-TSC2 antibody was generated as described previously (37) and kindly provided by K. Molle (Biozentrum, University of Basel, Switzerland). A rat monoclonal anti-α-tubulin (YL1/2)-producing hybridoma cell line was obtained from American Type Culture Collection. Quantification was performed using ImageQuant TL (Amersham Biosciences) software.
In Vivo Insulin Stimulation—Insulin stimulation was performed on ∼3-month-old DNA-PKcs-/- mice and wild-type controls (n = 8). Following an overnight fast, a bolus of insulin (1 unit/kg of body weight; human recombinant insulin; Sigma) or saline solution was injected via the inferior vena cava into terminally anesthetized mice. White adipose tissue, liver, skeletal muscle, and heart were collected 20 min after stimulation and immediately snap-frozen. Tissues were homogenized and lysed as described above.
Glucose and Insulin Tolerance Tests—Three-month-old mice were fasted overnight (n = 2 for wild-type; n = 5 for DNA-PKcs-/-). For glucose tolerance tests, glucose (2 g/kg of body weight; d-(+)-glucose anhydrous; Fluka) was given orally, and for insulin tolerance tests, insulin (1 unit /kg; human recombinant insulin, Sigma) was administered by intraperitoneal injection as described previously (35). Blood samples were collected at the indicated times from tail veins, and glucose levels were determined using Glucometer Elite (Bayer).
Total Body γ-Irradiation of Mice—Mice received a single exposure of 1 Gy total body irradiation with x-rays (Asteophysics Research Corp., TORREX 120D x-ray system). The tissues were collected after 30 min and immediately snap-frozen. Tissues were homogenized and lysed as described above.
Flow Cytometry—Thymocytes in suspension were stained at 4 °C for 30 min in FACS buffer (2% fetal calf serum in phosphate-buffered saline) with fluorescein isothiocyanate- or phycoerythrin-conjugated antibodies to CD4 and CD8 cell-surface markers. Antibodies were from Immunotools.
Histological Analysis—Organs were dissected and fixed in 4% paraformaldehyde/phosphate-buffered saline at 4 °C. After dehydration in ethanol, samples were embedded in paraffin. Tissues were cut into 4-μm-thick sections and stored for staining. For hematoxylin-eosin (Sigma) staining, sections were deparaffinized and stained.
Statistics—Data are provided as arithmetic means ± S.E., and n represents the number of independent experiments. Data were tested for significance using one-way analysis of variance. The p values for the Kaplan-Meier survival curve were determined by LogRank tests with the Holm-Sidak multiple comparisons procedure using SigmaStat 3.11 (Systat Software, Inc.) statistics software. Results at p < 0.05 were considered significant.
RESULTS
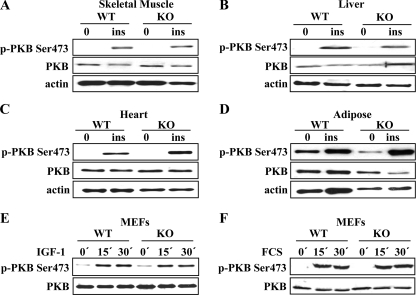
In Vivo, DNA-PK Is Dispensable for PKB Ser-473 Phosphorylation upon Insulin Stimulation—To evaluate the involvement of DNA-PK in the regulation of PKB in response to insulin stimulation, we treated wild-type and DNA-PKcs-/- mice with insulin. The mice were fasted overnight and then injected with a bolus of insulin (1 unit /kg body weight) or a saline control. Twenty minutes after injection, liver, skeletal muscle, adipose, and heart tissues were collected and analyzed by immunoblotting. PKB Ser-473 was robustly (∼20-fold for liver; ∼25-fold for skeletal muscle and heart; ∼10-fold for adipose tissue; data not shown) phosphorylated upon insulin stimulation in all four tissues from both wild-type and DNA-PKcs-/- mice (Fig. 1, A–D). Thus, we concluded that DNA-PK is dispensable for insulin-induced PKB Ser-473 phosphorylation in vivo. Additionally, we treated wild-type and DNA-PKcs-/- mouse embryonic fibroblasts (MEFs) with IGF-1 (Fig. 1E) or serum (Fig. 1F). Neither treatment resulted in impaired PKB Ser-473 phosphorylation in DNA-PKcs-/- MEFs, indicating that DNA-PK is not essential for PKB Ser-473 phosphorylation upon IGF-1 or serum stimulation.
FIGURE 1.
DNA-PK is dispensable for PKB Ser-473 phosphorylation upon insulin and growth factor stimulation. After overnight fasting and insulin (ins) stimulation (1 unit/kg body weight) for 20 min, tissues from 3-month-old (WT) and DNA-PKcs-/- (KO) mice were analyzed for PKB Ser-473 phosphorylation via immunoblotting with phospho-specific antibodies. A, skeletal muscle; B, liver; C, heart; D, adipose tissue. Wild-type and DNA-PKcs-/- MEFs (KO) were stimulated with IGF-1 (50 ng/ml) (E) or serum (10%) (F) for various times, and PKB Ser-473 phosphorylation was analyzed by immunoblotting.
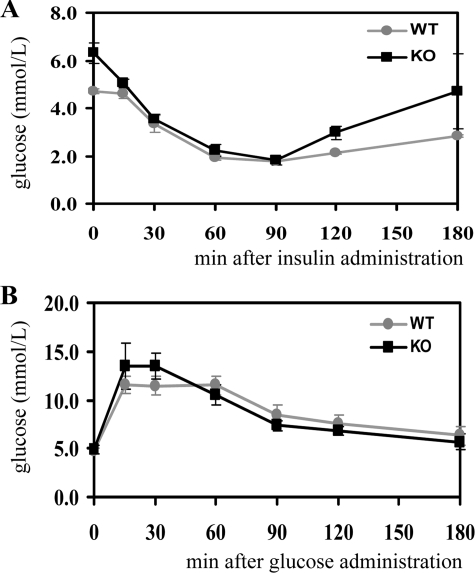
To investigate the role of DNA-PK in the maintenance of glucose metabolism, mice fasted overnight were subjected to an insulin or glucose tolerance test. To evaluate insulin sensitivity, insulin (1 unit/kg) was injected intraperitoneally and blood glucose levels measured at the indicated time points. No significant differences in blood glucose levels were found between DNA-PKcs-/- mice and wild-type controls (Fig. 2A). After oral application of glucose (2 g/kg), blood glucose levels were measured at the indicated times. Wild-type and DNA-PKcs-/- mice displayed a similar response to the glucose treatment (Fig. 2B). Hence, DNA-PKcs-/- mice display neither insulin resistance nor glucose intolerance. Taken together, the results show that DNA-PK is dispensable for PKB Ser-473 phosphorylation in response to insulin and growth factor stimulation and is also not essential for the maintenance of glucose homeostasis.
FIGURE 2.
DNA-PK is dispensable for maintenance of glucose metabolism. For analysis of glucose metabolism, 3-month-old wild-type (WT) and DNA-PKcs-/- (KO) mice after overnight fasting were treated with insulin (1 unit /kg body weight) for insulin tolerance tests (A) or glucose (2 g/kg body weight) for glucose tolerance tests (B). The graphs depict arithmetic means ± S.E. of blood glucose concentrations at the indicated time points following the treatments.
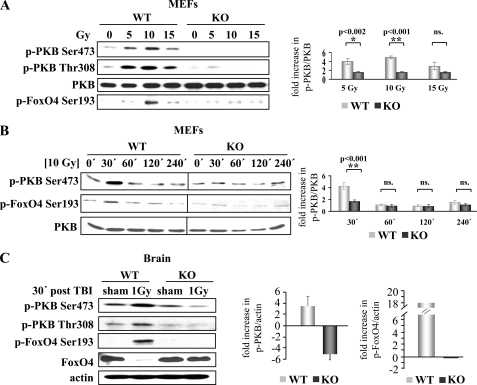
DNA-PK Is the Physiological PKB Ser-473 Kinase upon γ-Irradiation-induced DNA Damage—DNA-PK is activated by DNA double strand breaks induced by γ-irradiation and radiomimetic drugs (18, 19). To investigate the role of DNA-PK in PKB Ser-473 phosphorylation induced by γ-irradiation, we analyzed the PKB response to DNA damage in MEFs. Phosphorylation of PKB was promoted in wild-type MEFs in a dose-dependent manner (∼4-fold for 5 Gy and 5-fold for 10 Gy), whereas this response was significantly impaired (p = 0.0017 for 5 Gy and p < 0.001 for 10 Gy) in DNA-PKcs-/- MEFs (Fig. 3A). Moreover, the levels of phosphorylated FoxO4 Ser-193 also increased in a dose-dependent manner, reaching a peak at 10 Gy (Fig. 3A). Further increase in DNA damage at 15 Gy irradiation led to compromised PKB and FoxO4 phosphorylation (Fig. 3A), which is reminiscent of the dependence of PKB activation on the extent of DNA damage. Further analysis showed that the phosphorylation of PKB Ser-473 and FoxO4 Ser-193 remained impaired in DNA-PKcs-/- MEFs compared with WT MEFs at additional time points analyzed (Fig. 3B). Subsequently, we investigated DNA-PK-dependent PKB activation induced by γ-radiation in vivo. Wild-type and DNA-PKcs-/- mice were subjected to 1 Gy total body irradiation, and tissues were collected after 30 min. PKB was phosphorylated on both Thr-308 and Ser-473 (∼3.5-fold) in the brains of wild-type animals upon γ-irradiation. Strikingly, this activation was compromised in the DNA-PKcs-/- brain (Fig. 3B). Likewise, levels of phosphorylated FoxO4 increased in wild-type irradiated brain (∼18-fold), whereas there was no induction of FoxO4 phosphorylation in DNA-PKcs-/--irradiated brain (Fig. 3B). Immunoblot analysis with an antibody that preferentially recognizes the unphosphorylated form (because of the fact that the peptides used for production of the antibodies against phosphoprotein and total protein were derived from the same sequence) showed a decrease in total FoxO4 protein upon irradiation. This indicates a robust phosphorylation of FoxO4 Ser-193 upon DNA damage (Fig. 3B). In summary, in vivo and ex vivo results both indicate that PKB activation is promoted and that DNA-PK is required for phosphorylation of PKB Ser-473 in response to DNA damage induced by γ-irradiation. Overall, this implies stimulus-specific regulation of PKB activation by DNA-PK.
FIGURE 3.
DNA-PK is the physiological PKB Ser-473 kinase in the response to γ-irradiation. PKB and FoxO4 phosphorylation were analyzed by phospho-specific antibodies in wild-type (WT) and DNA-PKcs-/- (KO) MEF lysates 30 min after the indicated doses ofγ-irradiation (A) or at indicated time points after 10 Gy irradiation (B). Right panels, quantification of fold increase in PKB Ser-473 phosphorylation after γ-irradiation determined by phospho-PKB (p-PKB)Ser-473/PKB ratio with respect to nonirradiated WT and KO controls. Data were tested for significance using one-way analysis of variance with the Holm-Sidak multiple comparisons procedure. ns, not significant. C, analysis of PKB and FoxO4 phosphorylation in WT and DNA-PKcs-/- (KO) brain 30 min after 1 Gy total body γ-irradiation or sham irradiation. Right panel, quantification of fold increase in p-PKB Ser-473/actin ratio and phospho-FoxO4 (p-FoxO4) Ser-193/actin ratio after γ-irradiation with respect to sham-irradiated WT and KO controls.
PKB Is Hyperactivated in DNA-PKcs-/- Thymus—To investigate how loss of DNA-PK is reflected in basal PKB Ser-473 phosphorylation, we analyzed a panel of tissues from wild-type and DNA-PKcs-/- mice. No differences were observed in skeletal muscle, liver, spleen, and brain (supplemental Fig. 1A), and a mild increase was found in adipose and brown fat (∼2- and 2.5-fold, respectively) (Fig. 4, A and B). However, a robust 8-fold increase in PKB Ser-473 phosphorylation was found in DNA-PKcs-/- thymus compared with wild-type (Fig. 4C). p70 ribosomal protein S6 kinase (S6K), acting downstream of PKB, has been shown to repress upstream signaling by induction of insulin receptor substrate-1 (IRS-1) degradation (38–40). In contrast, neither the phosphorylation of S6K1 on the hydrophobic motif residue Thr-389 nor the IRS-1 protein levels were significantly different in DNA-PKcs-/- and wild-type thymus and adipose (Fig. 4C and supplemental Fig. 1B). Hence, we concluded that the increased PKB activity in the DNA-PKcs null background was not because of defective S6K/IRS-1-mediated feedback regulation in these tissues. Interestingly, in some cases we observed increase of both S6K Thr-389 phosphorylation as well as IRS-1 protein levels in DNA-PKcs-/- brown fat (supplemental Fig. 1B). The marked increase in PKB phosphorylation upon loss of an upstream kinase prompted us to further investigate components of the PKB signaling pathway in DNA-PKcs-/- thymus. Further analysis of PKB downstream targets revealed that there was no significant difference in GSK3β Ser-9 phosphorylation, whereas the FoxO4 transcription factor was strongly phosphorylated in DNA-PKcs-/- thymus compared with the wild type. This suggests that the PKB/FoxO pathway is deregulated in DNA-PKcs-/- thymus (Fig. 4C).
FIGURE 4.
Analysis of basal PKB phosphorylation in DNA-PKcs-/- tissues. Tissues were collected from 2-month-old random-fed WT and DNA-PKcs-/- mice and analyzed by immunoblotting. A, top panel, PKB phosphorylation in WT and DNA-PKcs-/- (KO) adipose tissue. Bottom panel, quantification of PKB Ser-473 phosphorylation determined by p-PKB Ser-473/PKB ratio relative to wild-type ratio. B, top panel, PKB phosphorylation in WT and DNA-PKcs-/- (KO) brown fat. Bottom panel, quantification of PKB Ser-473 phosphorylation determined by p-PKB Ser-473/PKB ratio relative to wild-type ratio. C, top panel, PKB Ser-473, PKB Thr-308, GSK3β Ser-9, FoxO3a Thr-32, and S6K Thr-389 phosphorylation and total IRS-1 protein levels in WT and DNA-PKcs-/- (KO) thymi. Bottom panel, quantification of PKB Ser-473 phosphorylation determined by p-PKB Ser-473/PKB ratio relative to wild-type ratio.
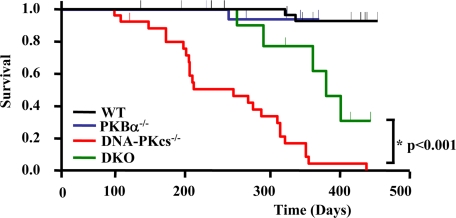
Constitutive PKB Activity Contributes to the Development of Spontaneous Thymic Lymphomas in DNA-PKcs-/- Mice and Can Be Prevented by Deletion of PKBα—PKB deregulation has been implicated in various types of cancer, including thymic lymphoma (41). Transgenic mice expressing a constitutively active PKB in T cells develop T cell lymphomas (42–44). Consistent with previous results (23), we observed an increased frequency of thymic lymphomas in DNA-PKcs-/- mice (Fig. 5A). Thymic tumors already appeared in 3-month-old animals and were 30-fold larger than nonmalignant DNA-PKcs-/- thymi (0.010 ± 0.001 g versus 0.313 ± 0.106 g; p = 0.03) (Table 1). PKBα is highly expressed in thymocytes particularly at the early stages of development (28–30), where DNA-PKcs-/- T cells are arrested (20–22). Reasoning that persistent PKB activity in DNA-PKcs-/- thymus might contribute to thymic lymphoma formation, we investigated whether deletion of PKBα, prevented formation of thymic tumors in DNA-PKcs-/- mice. About 27% (6/22) of DNA-PKcs-/- mice aged 3–7 months displayed thymic tumors, as against zero in wild-type mice (0/14) (Fig. 5A). Significantly, none of the DNA-PKcs-/-PKBα-/- double knock-out (DKO) mice analyzed at 2–15 months (0/6) exhibited thymic tumors (Fig. 5A). The analysis of DNA-PKcs-/-PKBα-/- DKO mice over a wide age range ruled out delayed progression of thymic tumors. Further immunoblot analysis of DNA-PKcs-/-PKBα-/- DKO thymi revealed that elevated PKB Ser-473 and FoxO phosphorylation in DNA-PKcs-/- thymi were restored to wild-type levels with deletion of PKBα (Fig. 5B). This suggests that increased PKB and FoxO phosphorylation in DNA-PKcs-/- thymi is because of deregulation of PKBα. In line with previous reports (23), we observed reduced viability correlated with the occurrence of thymic tumors in DNA-PKcs-/- mice (Fig. 6). Hence, we investigated whether deletion of PKBα gene improved survival of mice lacking DNA-PKcs. The survival of DNA-PKcs-/-PKBα-/- DKO mice significantly increased compared with DNA-PKcs-/- mice (p < 0.001). The longevity of mice increased from ∼238 days for DNA-PKcs-/- to 375 days for DNA-PKcs-/- PKBα-/- DKO supporting the involvement of PKB deregulation in the reduced life span of DNA-PK-deficient mice because of tumorigenesis (Fig. 6).
FIGURE 5.
Deletion of PKBα prevents spontaneous development of thymic tumors. A, incidence of thymic tumors from wild-type (WT), DNA-PKcs-/- (KO) analyzed between 3 and 7 months age and DNA-PKcs-/- PKBα-/- double knock-out (DKO) mice analyzed between 2 and 15 months age. B, PKB Ser-473, FoxO4 Ser-193, and FoxO3a Thr-32 phosphorylation in WT thymus, DNA-PKcs-/- tumor, and DNA-PKcs-/-PKBα-/- (DKO) thymi.
TABLE 1.
Thymic weights of wild-type and DNA-PKcs–/– mice (nonmalignant control and tumor) n indicates number of mice analyzed.
| Tissue | Thymic weight (±S.E.) | Thymic/body weight (±S.E.) | n |
|---|---|---|---|
| g | % | ||
| Wild-type thymus | 0.045 (±0.004) | 0.181 (±0.017) | n = 8 |
| DNA-PKcs–/– nonmalignant thymus | 0.010 (±0.001)a | 0.038 (±0.005)a | n = 8 |
| DNA-PKcs–/– thymic tumor | 0.313 (±0.106)b | 1.231 (±0.450)c | n = 5 |
p < 0.001 DNA-PKcs–/– nonmalignant thymus versus wild-type thymus
p = 0.003 DNA-PKcs–/– thymic tumor versus DNA-PKcs–/– nonmalignant thymus
p = 0.005 DNA-PKcs–/– thymic tumor versus DNA-PKcs–/– nonmalignant thymus
FIGURE 6.
Deletion of PKBα improves survival of DNA-PKcs-/- mice. Kaplan-Meier survival curves for wild-type (WT), DNA-PKcs-/-, PKBα-/-, and DNA-PKcs-/-PKBα-/- double knock-out (DKO) mice analyzed by LogRank analysis and the Holm-Sidak multiple comparisons procedure. The survival of DNA-PKcs-/-PKBα-/-DKO mice significantly increased compared with DNA-PKcs-/- mice (p < 0.001). Mice that survived longer than 2 months were included in the analysis. Numbers of mice analyzed are as follows: WT 52; DNA-PKcs-/- 21; PKBα-/- 26; and DNA-PKcs-/-PKBα-/- (DKO) 13.
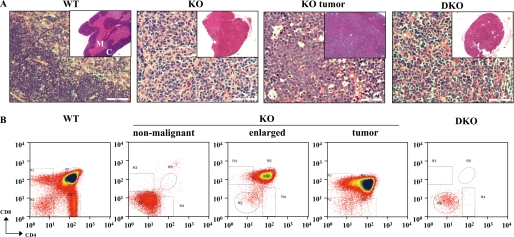
Histological analysis of thymi by hematoxylin-eosin and Ki67 staining showed tumors from DNA-PKcs-/- mice to consist of atypical cells with a high proliferative index (Fig. 7A and data not shown). DNA-PKcs-/-PKBα-/- DKO thymi showed disruption of the cortico-medullary boundary similar to DNA-PKcs-/- thymi (see insets in Fig. 7A). To further investigate the role of PKB in DNA-PKcs-/- tumors, thymocytes isolated from wild-type, DNA-PKcs-/-, and DNA-PKcs-/-PKBα-/- DKO mice were analyzed by flow cytometry using antibodies against CD4 and CD8 cell-surface markers (Fig. 7B). In DNA-PKcs-/- thymus, development of T cells was blocked at the CD4-CD8- double-negative (DN) stage; about 80% of DNA-PKcs-/- T cells were CD4-CD8- (DN) cells compared with about 2% DN stage cells in the wild type (Fig. 5B and Table 2). T cells escaped from this developmental block in tumors from DNA-PKcs-/- mice, where there was a marked increase in CD4+CD8+ double-positive (DP) cells (80%). Interestingly, flow cytometric analysis of T cells from the two thymus lobes of a DNA-PKcs-/- animal displayed differing profiles; one was totally blocked at the DN stage and the other closely resembled the profile of an advanced tumor (Fig. 5B). Consistent with this, the latter was significantly enlarged (0.008 g versus 0.027 g). In contrast, DNA-PKcs-/-PKBα-/- DKO thymocytes were almost exclusively blocked DN stage cells (Fig. 5B and Table 2), which suggests that PKBα has a role in neoplastic expansion of T cells in the DNA-PKcs-/- thymus.
FIGURE 7.
Deletion of PKBα prevents neoplastic expansion of DNA-PKcs-/- thymocytes. A, histological analysis of thymi from wild-type (WT), DNA-PKcs-/- (KO), and DNA-PKcs-/-PKBα-/- double knock-out (DKO) mice and thymic tumors from DNA-PKcs-/- mice. Thymi were fixed with 4% paraformaldehyde, processed with paraffin, sectioned, and stained with hematoxylin-eosin (×400 magnification; scale bar indicates 50 μm). The insets show ×40 magnification. M, medulla; C, cortex. B, flow cytometric analysis of thymocytes from WT, DNA-PKcs-/- (KO), and DNA-PKcs-/-PKBα-/- (DKO) mice for CD4 and CD8 markers.
TABLE 2.
Percentages of T cell subsets in wild type, DNA-PKcs–/–, and DNA-PKcs–/–PKBα–/– DKO mice T cell subsets that are double negative (DN), double positive (DP), CD4+CD8– (CD4+), and CD4–CD8+ (CD8+) for CD4 and CD8 cell surface markers. Data provided as % mean (±S.E.)
| T cell | WT | DNA-PKcs–/– | DNA-PKcs–/– | DNA-PKcs–/– | DNAPKcs–/–PKBα–/– |
|---|---|---|---|---|---|
| Subsets | non-malignant | enlarged | Tumor | DKO | |
| DN | 1.66 (±0.14) | 78.72 (±2.12) | 1.84 | 0.7 | 83.71 (±1.18) |
| DP | 79.99 (±1.80) | 0.55 (±0.20) | 82.47 | 79.66 | 0.1 (±0.1) |
| CD4+ | 6.46 (±0.47) | 2.50 (±0.62) | 0.41 | 0.27 | 1.84 (±0) |
| CD8+ | 1.67 (±0.16) | 0.64 (±0.24) | 2.55 | 4.41 | 0.46 (±0.14) |
DISCUSSION
In this study, we evaluated the in vivo role of DNA-PK in basal PKB Ser-473 phosphorylation as well as in the responses to various stimuli. We found that DNA-PK is dispensable for insulin- and growth factor-induced PKB activation. Furthermore, DNA-PK is not essential for the maintenance of glucose metabolism. In contrast, our in vivo and ex vivo results revealed that DNA-PK is required for phosphorylation of PKB Ser-473 upon DNA damage induced by γ-irradiation. Taken together, this implies stimulus-specific regulation of PKB Ser-473 phosphorylation by specific upstream kinases. The impaired phosphorylations of both PKB Ser-473 and Thr-308 residues observed in DNA-PKcs-/- cells as well as in PDK1-/- cells (8) treated with γ-irradiation suggest the requirement of phosphorylation of these two sites for full activation of PKB and imply that two phosphorylation steps are tightly connected and inter-dependent (8, 45). Hence, both Thr-308 by PDK1 and Ser-473 phosphorylations by DNA-PK appear to be essential for activation of PKB in response to DNA damage induction (8). The fact that increased doses of γ-irradiation led to compromised PKB phosphorylation suggests the dependence of PKB activation upon the extent of DNA damage, which is consistent with data obtained with human umbilical vein endothelial cells (8). In addition, the PKB response to γ-irradiation includes phosphorylation of the downstream target FoxO4 (this study) as well as regulation of p21 (8) and GSK3 (8, 46) placing PKB as an important mediator of DNA damage signaling. In addition to its response to irradiation-induced double strand DNA breaks, PKB is activated by various other DNA damage inducers. Numerous studies have demonstrated the importance of functional PKB signaling for survival after DNA damage. Doxorubicin promotes PKB activation (Ser-473 and Thr-308 phosphorylation) in mouse embryonic fibroblasts (8). In addition, doxorubicin-induced PKB activation was shown in several breast cancer cell lines (7) as well as in vivo by which elevated myocardial PKB signaling ameliorates doxorubicin-induced congestive heart failure and promotes heart growth in mice (47). Furthermore, a recent study showed that doxorubicin leads to phosphorylation of PKB and concominantly to PKB-dependent inactivation and nuclear exclusion of FoxO4 in human colon carcinoma cell line (48). Moreover, PKB activation is promoted by etoposide (49), cisplatin (50–52), and UV light (26, 53) in several different cell types. Notably, PKBα is important for survival after UV irradiation, as MEFs lacking PKBα undergo irradiation-induced apoptosis to a much larger extent than the wild-type MEFs (26). These studies place PKB as an important mediator of DNA damage signaling.
Our results revealed deregulation of PKB phosphorylation in DNA-PKcs-/- mice in a tissue-specific manner. The marked increase in basal PKB Ser-473 phosphorylation in the absence of an upstream kinase is unexpected. Elevated PKB Ser-473 phosphorylation in DNA-PK-/- tissues could most likely be mediated by mTORC2 and reminiscent of cross-talk between the two PKB Ser-473 kinases, DNA-PK and mTOR. This regulation might take place at multiple levels of the pathway as our results showed that elevated PKB Ser-473 phosphorylation could also be observed without the accompanying changes in S6K Thr-389 phosphorylation. Therefore, although we cannot completely rule out the contribution of an S6K-mediated mechanism, our data suggest the existence of an alternative mechanism of regulation where deletion of DNA-PK could lead to its disruption and result in deregulation of PKB.
It has been suggested that mTORC2 is necessary for modulation of PKCα Ser-657 phosphorylation (54), and it was shown that overexpression of mTORC2 components leads to enhanced PKCα activity. Knockdown of rictor or mTOR leads to impaired PKCα phosphorylation (54, 55). Furthermore, ablation of rictor or mLST8 was shown to severely compromise PKCα activity (14). We therefore examined whether PKCα phosphorylation was altered in DNA-PKcs-/- thymus. We observed increased phosphorylation of PKCα Ser-657 (supplemental Fig. 2A) reminiscent of increased mTORC2 activity in DNA-PKcs-/- thymus. Furthermore, a recent study suggested that TSC1-TSC2 complex positively regulates mTORC2 independent of its effects on mTORC1 (56). Interestingly, we observed increased total TSC2 levels in DNA-PKcs-/- thymus (supplemental Fig. 2B). Although circumstantial evidence on whether this has an effect on increased PKB Ser-473 phosphorylation via positively regulating mTORC2 in DNA-PKcs-/- thymus remains to be established, this result supports our hypothesis that hyperphosphorylation of PKB Ser-473 could be mediated by mTORC2. On the other hand, the possibility that a third Ser-473 kinase leading to increased PKB phosphorylation in DNA-PKcs-/- thymus cannot be completely excluded. For instance, another PIKK family member, ataxia telangiectasia mutated has also been proposed to function as PKB Ser-473 kinase (57).
Recent studies proposed a role for Tel2 to function as a coordinator among PIKKs and suggested the existence of cross-talk between different PIKKs where alteration of one PIKK may influence the other (58–61). It has been shown that Tel2 directly interacts with and stabilizes the protein levels of PIKKs, including DNA-PK and mTOR (59, 61). It has also been proposed that Tel2 may serve as a scaffold protein that mediates signal transduction from PIKKs to their target proteins (60). However, further study is required to understand the mechanistic role of Tel2 as a mediator of PIKK functions. It will be intriguing to see whether Tel2 could have a role mediator role affecting the downstream signaling of PIKKs, in particular within the context of mTOR and DNA-PK.
We have found that the PKB/FoxO pathway is deregulated in DNA-PKcs-/- thymus and shown that increased PKB activity in a DNA-PKcs null background contributes to spontaneous formation of thymic tumors by allowing neoplastic expansion of thymocytes. Significantly, these tumors could be prevented by deletion of PKBα that is highly expressed in thymocytes, particularly in DN and DP stages (28–30). Furthermore, subsequent deletion of the PKBα gene improves the survival of DNA-PKcs-/- mice. However, the survival of DNA-PKcs-/- PKBα-/- DKO mice was not restored to that of PKBα-/- mice, which is not surprising given that DNA-PK has a wide variety of functions, some of which are independent of PKBα. Alternatively, the loss of DNA-PK in a PKBα-/- background could lead to deterioration of DNA-PK-dependent PKB functions in tissues other than the thymus. Moreover, deregulation of other isoforms of PKB in DNA-PK-/- mice could possibly affect overall organismal fitness and survival. Finally, loss of DNA-PK-independent PKBα functions could lead to additive effects in DNA-PKcs-/-PKBα-/- DKO mice. Further studies using tissue-specific DNA-PK-/-PKBα-/- double knock-out mice will be necessary to evaluate these possibilities.
PKB has been shown to play an important role in DN to DP stage transition and to be essential for thymocyte survival and differentiation (28–30). Therefore, persistent PKB activity in the DNA-PKcs-/- thymus, where DN thymocytes predominate, could contribute to malignant transition. It was reported recently that conditional and simultaneous disruption of FoxO1, FoxO3, and FoxO4 genes in mice leads to the development of thymic lymphomas and that thymocytes from these mice show increased proliferation (62, 63). Moreover, disruption of FoxO function was shown to accelerate Myc-driven lymphomagenesis (64). Therefore, deregulation of PKB/FoxO downstream events affecting proliferation or apoptosis may play a role in thymic tumor induction in DNA-PKcs-/- mice.
Given the importance of PKB activity in T cell development and the role of PKB in lymphomagenesis, the signaling components leading to PKB activation in DNA-PK-deficient thymus need to be established. Pre-T cell receptor (pre-TCR), Notch, and interleukin-7 (IL-7) signaling provide critical signals for the survival, proliferation, and differentiation of T cells (30, 65–68). Activation of the PI3K/PKB pathway is common to all three receptors and points to the PI3K/PKB pathway as a central player that translates these signals into multiple functional outcomes. As pre-TCR signaling requires a functionally rearranged TCRβ, which is lacking in DNA-PKcs-/- thymocytes as a result of defective V(D)J recombination, it is unlikely that PKB activity increases in response to pre-TCR signaling. Hence, deregulation of Notch and/or IL-7 signaling and their role in PKB activation need to be investigated in DNA-PKcs-/- thymus. Frequent mutational activation of Notch has been identified in human T cell acute lymphoblastic leukemia (T-ALL) and subsequently in mouse thymic lymphomas, including those from severe combined immunodeficient mice, a DNA-PK deficient mouse model (66, 69, 70). Recent studies provided evidence that Notch-1 up-regulates the PI3K/PKB pathway in T-ALL cells bearing Notch-1 mutations and showed synergistic suppression of growth in cells treated with small molecule inhibitor of Notch and rapamycin (71, 72). In other studies of T-ALL cells, rapamycin inhibited IL-7-mediated cell cycle progression and cellular proliferation (73). The rapamycin derivative, CCI-779, was shown to reduce mTORC2 signaling and inhibit PKB activation in hematopoietic malignancies, although another study reported that PKB activation remained unchanged using a different rapamycin derivative, RAD001 (74, 75). Rapamycin treatment was shown to inhibit PKB phosphorylation in a tissue-dependent manner, particularly in thymus (76). It is plausible that PKB hyperactivity in the thymus in the absence of DNA-PK could be mediated by mTORC2, and tissue-specific disruption of the PKB/FoxO pathway could render DNA-PK-deficient thymic lymphomas susceptible to rapamycin treatment. A number of small molecule inhibitors of DNA-PK have been developed, and the inhibition of DNA-PK sensitizes cells to DNA-damaging agents (77, 78). However, the use of DNA-PK inhibitors needs to be evaluated cautiously in the treatment of thymic lymphomas as loss of DNA-PK activity could lead to increased PKB signaling. Taken together, these results provide genetic evidence that PKB is deregulated in DNA-PKcs-/- thymi and demonstrate a key role for PKBα in thymic tumor progression in DNA-PKcs-/- mice. In general, the results indicate elaborate regulation of PKB phosphorylation by upstream kinases and its deregulation leading to malignant transformation.
Supplementary Material
Acknowledgments
We thank P. A. Jeggo (University of Sussex, East Sussex, UK) for providing DNA-PKcs-/- MEFs and F. W. Alt (Howard Hughes Medical Institute, Harvard Medical School, Boston) for DNA-PKcs-/- mice. We also thank H. Kohler (FMI FACS), S. Bichet, A. Bogucki, and M. Wrobel (FMI Molecular Histology) for help with experiments and P. King, E. Fayard, and A. Hergovich for critical reading of the manuscript.
This work was supported in part by Oncosuisse Grant OCS-01667-02-2005. Friedrich Miescher Institute is part of the Novartis Research Foundation. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
Footnotes
The abbreviations used are: PKB, protein kinase B; WT, wild type; KO, knockout; DKO, double knock-out; Gy, gray; MEF, mouse embryonic fibroblast; S6K, S6 kinase; DN, double-negative; DP, double-positive; PI3K, phosphatidylinositol 3-kinase; PIKK, PI3K-related kinase; DNA-PK, DNA-dependent protein kinase; mTOR, mammalian target of rapamycin; IGF-1, insulin-like growth factor 1; TCR, T cell receptor; IL-7, interleukin-7; T-ALL, T cell acute lymphoblastic leukemia.
References
- 1.Brazil, D. P., and Hemmings, B. A. (2001) Trends Biochem. Sci. 26 657-664 [DOI] [PubMed] [Google Scholar]
- 2.Fayard, E., Tintignac, L. A., Baudry, A., and Hemmings, B. A. (2005) J. Cell Sci. 118 5675-5678 [DOI] [PubMed] [Google Scholar]
- 3.Manning, B. D., and Cantley, L. C. (2007) Cell 129 1261-1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parcellier, A., Tintignac, L. A., Zhuravleva, E., and Hemmings, B. A. (2008) Cell. Signal. 20 21-30 [DOI] [PubMed] [Google Scholar]
- 5.Greer, E. L., and Brunet, A. (2005) Oncogene 24 7410-7425 [DOI] [PubMed] [Google Scholar]
- 6.van der Horst, A., and Burgering, B. M. (2007) Nat. Rev. Mol. Cell Biol. 8 440-450 [DOI] [PubMed] [Google Scholar]
- 7.Li, X., Lu, Y., Liang, K., Liu, B., and Fan, Z. (2005) Breast Cancer Res. 7 R589-R597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bozulic, L., Surucu, B., Hynx, D., and Hemmings, B. A. (2008) Mol. Cell 30 203-213 [DOI] [PubMed] [Google Scholar]
- 9.Feng, J., Park, J., Cron, P., Hess, D., and Hemmings, B. A. (2004) J. Biol. Chem. 279 41189-41196 [DOI] [PubMed] [Google Scholar]
- 10.Hambardzumyan, D., Becher, O. J., Rosenblum, M. K., Pandolfi, P. P., Manova-Todorova, K., and Holland, E. C. (2008) Genes Dev. 22 436-448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mitsiades, C. S., Mitsiades, N., and Koutsilieris, M. (2004) Curr. Cancer Drug Targets 4 235-256 [DOI] [PubMed] [Google Scholar]
- 12.Whiteman, E. L., Cho, H., and Birnbaum, M. J. (2002) Trends Endocrinol. Metab. 13 444-451 [DOI] [PubMed] [Google Scholar]
- 13.Sarbassov, D. D., Guertin, D. A., Ali, S. M., and Sabatini, D. M. (2005) Science 307 1098-1101 [DOI] [PubMed] [Google Scholar]
- 14.Guertin, D. A., Stevens, D. M., Thoreen, C. C., Burds, A. A., Kalaany, N. Y., Moffat, J., Brown, M., Fitzgerald, K. J., and Sabatini, D. M. (2006) Dev. Cell 11 859-871 [DOI] [PubMed] [Google Scholar]
- 15.Shiota, C., Woo, J. T., Lindner, J., Shelton, K. D., and Magnuson, M. A. (2006) Dev. Cell 11 583-589 [DOI] [PubMed] [Google Scholar]
- 16.Yang, Q., Inoki, K., Ikenoue, T., and Guan, K. L. (2006) Genes Dev. 20 2820-2832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dragoi, A. M., Fu, X., Ivanov, S., Zhang, P., Sheng, L., Wu, D., Li, G. C., and Chu, W. M. (2005) EMBO J. 24 779-789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith, G. C., and Jackson, S. P. (1999) Genes Dev. 13 916-934 [DOI] [PubMed] [Google Scholar]
- 19.Burma, S., and Chen, D. J. (2004) DNA Repair 3 909-918 [DOI] [PubMed] [Google Scholar]
- 20.Gao, Y., Chaudhuri, J., Zhu, C., Davidson, L., Weaver, D. T., and Alt, F. W. (1998) Immunity 9 367-376 [DOI] [PubMed] [Google Scholar]
- 21.Kurimasa, A., Ouyang, H., Dong, L. J., Wang, S., Li, X., Cordon-Cardo, C., Chen, D. J., and Li, G. C. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 1403-1408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taccioli, G. E., Amatucci, A. G., Beamish, H. J., Gell, D., Xiang, X. H., Torres Arzayus, M. I., Priestley, A., Jackson, S. P., Marshak Rothstein, A., Jeggo, P. A., and Herrera, V. L. (1998) Immunity 9 355-366 [DOI] [PubMed] [Google Scholar]
- 23.Espejel, S., Martin, M., Klatt, P., Martin-Caballero, J., Flores, J. M., and Blasco, M. A. (2004) EMBO Rep. 5 503-509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen, W. S., Xu, P. Z., Gottlob, K., Chen, M. L., Sokol, K., Shiyanova, T., Roninson, I., Weng, W., Suzuki, R., Tobe, K., Kadowaki, T., and Hay, N. (2001) Genes Dev. 15 2203-2208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cho, H., Thorvaldsen, J. L., Chu, Q., Feng, F., and Birnbaum, M. J. (2001) J. Biol. Chem. 276 38349-38352 [DOI] [PubMed] [Google Scholar]
- 26.Feng, J., Tamaskovic, R., Yang, Z., Brazil, D. P., Merlo, A., Hess, D., and Hemmings, B. A. (2004) J. Biol. Chem. 279 35510-35517 [DOI] [PubMed] [Google Scholar]
- 27.Yang, Z. Z., Tschopp, O., Hemmings-Mieszczak, M., Feng, J., Brodbeck, D., Perentes, E., and Hemmings, B. A. (2003) J. Biol. Chem. 278 32124-32131 [DOI] [PubMed] [Google Scholar]
- 28.Fayard, E., Gill, J., Paolino, M., Hynx, D., Hollander, G. A., and Hemmings, B. A. (2007) PLoS ONE 2 e992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Juntilla, M. M., Wofford, J. A., Birnbaum, M. J., Rathmell, J. C., and Koretzky, G. A. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 12105-12110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mao, C., Tili, E. G., Dose, M., Haks, M. C., Bear, S. E., Maroulakou, I., Horie, K., Gaitanaris, G. A., Fidanza, V., Ludwig, T., Wiest, D. L., Gounari, F., and Tsichlis, P. N. (2007) J. Immunol. 178 5443-5453 [DOI] [PubMed] [Google Scholar]
- 31.Cho, H., Mu, J., Kim, J. K., Thorvaldsen, J. L., Chu, Q., Crenshaw, E. B., III, Kaestner, K. H., Bartolomei, M. S., Shulman, G. I., and Birnbaum, M. J. (2001) Science 292 1728-1731 [DOI] [PubMed] [Google Scholar]
- 32.Easton, R. M., Cho, H., Roovers, K., Shineman, D. W., Mizrahi, M., Forman, M. S., Lee, V. M., Szabolcs, M., de Jong, R., Oltersdorf, T., Ludwig, T., Efstratiadis, A., and Birnbaum, M. J. (2005) Mol. Cell. Biol. 25 1869-1878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garofalo, R. S., Orena, S. J., Rafidi, K., Torchia, A. J., Stock, J. L., Hildebrandt, A. L., Coskran, T., Black, S. C., Brees, D. J., Wicks, J. R., McNeish, J. D., and Coleman, K. G. (2003) J. Clin. Investig. 112 197-208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tschopp, O., Yang, Z. Z., Brodbeck, D., Dummler, B. A., Hemmings-Mieszczak, M., Watanabe, T., Michaelis, T., Frahm, J., and Hemmings, B. A. (2005) Development (Camb.) 132 2943-2954 [DOI] [PubMed] [Google Scholar]
- 35.Dummler, B., Tschopp, O., Hynx, D., Yang, Z. Z., Dirnhofer, S., and Hemmings, B. A. (2006) Mol. Cell. Biol. 26 8042-8051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andjelkovic, M., Alessi, D. R., Meier, R., Fernandez, A., Lamb, N. J., Frech, M., Cron, P., Cohen, P., Lucocq, J. M., and Hemmings, B. A. (1997) J. Biol. Chem. 272 31515-31524 [DOI] [PubMed] [Google Scholar]
- 37.van Slegtenhorst, M., Nellist, M., Nagelkerken, B., Cheadle, J., Snell, R., van den Ouweland, A., Reuser, A., Sampson, J., Halley, D., and van der Sluijs, P. (1998) Hum. Mol. Genet. 7 1053-1057 [DOI] [PubMed] [Google Scholar]
- 38.Harrington, L. S., Findlay, G. M., Gray, A., Tolkacheva, T., Wigfield, S., Rebholz, H., Barnett, J., Leslie, N. R., Cheng, S., Shepherd, P. R., Gout, I., Downes, C. P., and Lamb, R. F. (2004) J. Cell Biol. 166 213-223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shah, O. J., Wang, Z., and Hunter, T. (2004) Curr. Biol. 14 1650-1656 [DOI] [PubMed] [Google Scholar]
- 40.Um, S. H., Frigerio, F., Watanabe, M., Picard, F., Joaquin, M., Sticker, M., Fumagalli, S., Allegrini, P. R., Kozma, S. C., Auwerx, J., and Thomas, G. (2004) Nature 431 200-205 [DOI] [PubMed] [Google Scholar]
- 41.Yang, Z. Z., Tschopp, O., Baudry, A., Dummler, B., Hynx, D., and Hemmings, B. A. (2004) Biochem. Soc. Trans. 32 350-354 [DOI] [PubMed] [Google Scholar]
- 42.Jones, R. G., Parsons, M., Bonnard, M., Chan, V. S., Yeh, W. C., Woodgett, J. R., and Ohashi, P. S. (2000) J. Exp. Med. 191 1721-1734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Malstrom, S., Tili, E., Kappes, D., Ceci, J. D., and Tsichlis, P. N. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 14967-14972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rathmell, J. C., Elstrom, R. L., Cinalli, R. M., and Thompson, C. B. (2003) Eur. J. Immunol. 33 2223-2232 [DOI] [PubMed] [Google Scholar]
- 45.Scheid, M. P., Marignani, P. A., and Woodgett, J. R. (2002) Mol. Cell. Biol. 22 6247-6260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boehme, K. A., Kulikov, R., and Blattner, C. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 7785-7790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taniyama, Y., and Walsh, K. (2002) J. Mol. Cell. Cardiol. 34 1241-1247 [DOI] [PubMed] [Google Scholar]
- 48.Lupertz, R., Chovolou, Y., Unfried, K., Kampkotter, A., Watjen, W., and Kahl, R. (August 6, 2008) Carcinogenesis 10.1093/carcin/bgn184 [DOI] [PubMed]
- 49.Yu, H. G., Ai, Y. W., Yu, L. L., Zhou, X. D., Liu, J., Li, J. H., Xu, X. M., Liu, S., Chen, J., Liu, F., Qi, Y. L., Deng, Q., Cao, J., Liu, S. Q., Luo, H. S., and Yu, J. P. (2008) Int. J. Cancer 122 433-443 [DOI] [PubMed] [Google Scholar]
- 50.Winograd-Katz, S. E., and Levitzki, A. (2006) Oncogene 25 7381-7390 [DOI] [PubMed] [Google Scholar]
- 51.Yang, X., Fraser, M., Abedini, M. R., Bai, T., and Tsang, B. K. (2008) Br. J. Cancer 98 803-808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Belyanskaya, L. L., Hopkins-Donaldson, S., Kurtz, S., Simoes-Wust, A. P., Yousefi, S., Simon, H. U., Stahel, R., and Zangemeister-Wittke, U. (2005) Int. J. Cancer 117 755-763 [DOI] [PubMed] [Google Scholar]
- 53.Kim, M. S., Oh, Y. J., Lee, S., Kim, J. E., Kim, K. H., and Chung, J. H. (2006) Photochem. Photobiol. 82 645-650 [DOI] [PubMed] [Google Scholar]
- 54.Sarbassov, D. D., Ali, S. M., Kim, D. H., Guertin, D. A., Latek, R. R., Erdjument-Bromage, H., Tempst, P., and Sabatini, D. M. (2004) Curr. Biol. 14 1296-1302 [DOI] [PubMed] [Google Scholar]
- 55.Masri, J., Bernath, A., Martin, J., Jo, O. D., Vartanian, R., Funk, A., and Gera, J. (2007) Cancer Res. 67 11712-11720 [DOI] [PubMed] [Google Scholar]
- 56.Huang, J., Dibble, C. C., Matsuzaki, M., and Manning, B. D. (2008) Mol. Cell. Biol. 28 4104-4115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Viniegra, J. G., Martinez, N., Modirassari, P., Losa, J. H., Parada Cobo, C., Lobo, V. J., Luquero, C. I., Alvarez-Vallina, L., Ramony Cajal, S., Rojas, J. M., and Sanchez-Prieto, R. (2005) J. Biol. Chem. 280 4029-4036 [DOI] [PubMed] [Google Scholar]
- 58.Anderson, C. M., Korkin, D., Smith, D. L., Makovets, S., Seidel, J. J., Sali, A., and Blackburn, E. H. (2008) Genes Dev. 22 854-859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chang, M., and Lingner, J. (2008) Science 320 60-61 [DOI] [PubMed] [Google Scholar]
- 60.Kanoh, J., and Yanagida, M. (2007) Genes Cells 12 1301-1304 [DOI] [PubMed] [Google Scholar]
- 61.Takai, H., Wang, R. C., Takai, K. K., Yang, H., and de Lange, T. (2007) Cell 131 1248-1259 [DOI] [PubMed] [Google Scholar]
- 62.Paik, J. H., Kollipara, R., Chu, G., Ji, H., Xiao, Y., Ding, Z., Miao, L., Tothova, Z., Horner, J. W., Carrasco, D. R., Jiang, S., Gilliland, D. G., Chin, L., Wong, W. H., Castrillon, D. H., and DePinho, R. A. (2007) Cell 128 309-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tothova, Z., Kollipara, R., Huntly, B. J., Lee, B. H., Castrillon, D. H., Cullen, D. E., McDowell, E. P., Lazo-Kallanian, S., Williams, I. R., Sears, C., Armstrong, S. A., Passegue, E., DePinho, R. A., and Gilliland, D. G. (2007) Cell 128 325-339 [DOI] [PubMed] [Google Scholar]
- 64.Bouchard, C., Lee, S., Paulus-Hock, V., Loddenkemper, C., Eilers, M., and Schmitt, C. A. (2007) Genes Dev. 21 2775-2787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Juntilla, M. M., and Koretzky, G. A. (2008) Immunol. Lett. 116 104-110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Aster, J. C., Pear, W. S., and Blacklow, S. C. (2008) Annu. Rev. Pathol. 3 587-613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pallard, C., Stegmann, A. P., van Kleffens, T., Smart, F., Venkitaraman, A., and Spits, H. (1999) Immunity 10 525-535 [DOI] [PubMed] [Google Scholar]
- 68.Vella, A., Teague, T. K., Ihle, J., Kappler, J., and Marrack, P. (1997) J. Exp. Med. 186 325-330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tsuji, H., Ishii-Ohba, H., Katsube, T., Ukai, H., Aizawa, S., Doi, M., Hioki, K., and Ogiu, T. (2004) Cancer Res. 64 8882-8890 [DOI] [PubMed] [Google Scholar]
- 70.Tsuji, H., Ishii-Ohba, H., Ukai, H., Katsube, T., and Ogiu, T. (2003) Carcinogenesis 24 1257-1268 [DOI] [PubMed] [Google Scholar]
- 71.Chan, S. M., Weng, A. P., Tibshirani, R., Aster, J. C., and Utz, P. J. (2007) Blood 110 278-286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Palomero, T., Sulis, M. L., Cortina, M., Real, P. J., Barnes, K., Ciofani, M., Caparros, E., Buteau, J., Brown, K., Perkins, S. L., Bhagat, G., Agarwal, A. M., Basso, G., Castillo, M., Nagase, S., Cordon-Cardo, C., Parsons, R., Zuniga-Pflucker, J. C., Dominguez, M., and Ferrando, A. A. (2007) Nat. Med. 13 1203-1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Barata, J. T., Cardoso, A. A., Nadler, L. M., and Boussiotis, V. A. (2001) Blood 98 1524-1531 [DOI] [PubMed] [Google Scholar]
- 74.Zeng, Z., Sarbassov dos, D., Samudio, I. J., Yee, K. W., Munsell, M. F., Ellen Jackson, C., Giles, F. J., Sabatini, D. M., Andreeff, M., and Konopleva, M. (2007) Blood 109 3509-3512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tamburini, J., Chapuis, N., Bardet, V., Park, S., Sujobert, P., Willems, L., Ifrah, N., Dreyfus, F., Mayeux, P., Lacombe, C., and Bouscary, D. (2008) Blood 111 379-382 [DOI] [PubMed] [Google Scholar]
- 76.Sarbassov, D. D., Ali, S. M., Sengupta, S., Sheen, J. H., Hsu, P. P., Bagley, A. F., Markhard, A. L., and Sabatini, D. M. (2006) Mol. Cell 22 159-168 [DOI] [PubMed] [Google Scholar]
- 77.Nutley, B. P., Smith, N. F., Hayes, A., Kelland, L. R., Brunton, L., Golding, B. T., Smith, G. C., Martin, N. M., Workman, P., and Raynaud, F. I. (2005) Br. J. Cancer 93 1011-1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhao, Y., Thomas, H. D., Batey, M. A., Cowell, I. G., Richardson, C. J., Griffin, R. J., Calvert, A. H., Newell, D. R., Smith, G. C., and Curtin, N. J. (2006) Cancer Res. 66 5354-5362 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.