Abstract
In this study, we analyzed the roles for AML1/RUNX1 in the regulation of the c-mpl promoter. Wild-type AML1 activated the c-mpl promoter through the proximal AML-binding site in luciferase assays using 293T and HeLa cells. In accord with this result, electrophoretic mobility shift assay and chromatin immunoprecipitation assays demonstrated that AML1 bound to this site. Next, we analyzed the function of AML1 using a mutant of AML1 lacking the C terminus (AML1dC), which was originally found in a patient with myelodysplastic syndromes. AML1dC dominant-negatively suppressed transcriptional activity of wild-type AML1. However, unexpectedly, AML1dC-transduced murine c-Kit+Sca1+Lineage- cells expressed c-mpl mRNA and c-Mpl protein more abundantly than mock-transduced cells, which led to the enhanced thrombopoietin-mediated proliferation. Moreover, when AML1dC was induced to express during the development of hematopoietic cells from embryonic stem (ES) cells, AML1dC augmented the c-Mpl expression on hematopoietic stem/progenitor cells. Furthermore, we found that early hematopoietic cells that derived from AML1+/- ES cells expressed c-Mpl more intensely than those that developed from wild-type ES cells. In contrast, AML1dC hardly affected c-Mpl expression and maturation of megakaryocytes. As for the mechanism of the different roles of AML1 in the regulation of the c-mpl promoter, we found that AML1 forms a complex with a transcription repressor mSin3A on the c-mpl promoter in hematopoietic stem/progenitor cells, although it forms a complex with a transcription activator p300 on the same promoter in megakaryocytic cells. Together, these data indicate that AML1 can regulate the c-mpl promoter both positively and negatively by changing the binding partner according to cell types.
AML1 (RUNX1) is a family member of heterodimeric transcription factors named core binding factors. AML1 was originally identified at a breakpoint on human chromosome 21 in the t(8;21) translocation and known as the most common targets of chromosomal translocations in human leukemia (1, 2). In addition to chromosomal translocations, recent reports have shown the importance of point mutations of AML1 in hematological malignancies, such as acute myelogenous leukemia (AML)2 and myelodysplastic syndromes (MDS) (3). The Runt domain of AML1 is utilized for DNA binding and heterodimerization with a partner PEBP2β/CBFβ. Although PEBP2β by itself does not bind to the DNA, the association with PEBP2β is necessary for AML1 to elicit its biologic activity (4–6). AML1 can regulate the transcription of the target gene both positively and negatively through the binding to the consensus DNA sequence, TGT/cGGT, possibly dependent on the cellular context and/or its target gene. For example, it positively regulates the expression cytokines and their receptors in myeloid and lymphoid lineage cells (7–12), whereas it negatively regulates CD4 transcription in immature thymocytes (13). Several experiments using conventional and conditional gene targeting in mice demonstrated that AML1 is essential for the early step in definitive hematopoiesis (14). North et al. (15) revealed that AML1 is required for the generation of hematopoietic stem cells (HSCs) from the vitelline and umbilical arteries and from the aorta-gonad-mesonephros (AGM) region. In addition, AML1 is necessary for the transitions from the stage of double-negative (DN)2 to DN3 and DN3 to DN4 in the T-lymphocyte development (9, 17). Furthermore, AML1 plays an important role in the maturation of megakaryocytes and platelet production. AML1 deletion in adult mice led to the impaired polyploidization of megakaryocytes and low platelet production (17, 18), whereas the number of megakaryocyte progenitors was not altered in these mice, suggesting that AML1 is indispensable for the terminal maturation of megakaryocytes. Also, the hereditary loss-of-function mutation of AML1 or PEBP2β causes familial platelet disorder with predisposition to AML (FPD/AML), which is characterized by decreased platelet count and propensity to develop AML (19).
MDS are clonal hematological disorders derived from gene alteration at a level of HSC (20), which are characterized by ineffective hematopoiesis, dysplastic morphology of blood cells, and high possibility to transit to AML. A number of genetic or epigenetic alterations involved in the pathogenesis of MDS have been identified as follows: activating point mutations of signaling molecules such as N-RAS and Flt3 (21, 22); deletion, point mutation, and/or silencing of cell cycle inhibitory molecules such as p15, p16, and p53 (23–25); deletion, point mutation, and generation of chimeric genes for transcriptional factors such as Evi1, IRF-1, AML1 (26–28), and point mutations of the nucleolar protein (Nucleophosmin) (29). Among them, the point mutations of AML1 were found in 15–17% of patients with sporadic MDS/AML (high risk MDS and AML following MDS) (3, 30). Previously, point mutations of AML1 were intensively screened in the N-terminal region, including the Runt domain in patients with AML and MDS, and the researchers found several point mutations, most of which disrupts DNA binding activity of AML1 but not the interaction with PEBP2β (3). In addition, recent reports have revealed that about 50% of point mutations are detected in the C-terminal region in MDS/AML. In addition, a C-terminal AML1 point mutation was also detected FPD/AML (30, 31). Most of the C-terminal mutations of AML1 lead to the premature termination yielding the C-terminally truncated form of AML1, which inhibits transcriptional activity of AML1.
Thrombopoietin (TPO) is a crucial regulator of megakaryopoiesis and platelet production. It stimulates both megakaryocyte progenitor cell growth and subsequent maturation in vitro and in vivo (32). In accord with these data, knock-out mice for TPO or its receptor c-mpl both revealed severely impaired megakaryopoiesis and platelet reduction (about 5% of normal mice) without apparent abnormality in erythropoiesis, granulopoiesis, and lymphopoiesis, suggesting that a physiologic role of the TPO/c-Mpl system is restricted to the megakaryocytic lineage. However, in the later study, the total number of HSCs was found to be reduced in the bone marrow of c-Mpl-/- mice (57). Also, c-Mpl-/- HSCs revealed severely decreased reconstitution activity in transplantation experiments. These results indicate that TPO/c-Mpl-mediated signaling also plays an important role in the growth and survival of HSCs as well as in megakaryopoiesis (33, 34).
Considering the fact that both AML1 and TPO/c-Mpl signaling play crucial roles in the growth and survival of HSCs as well as in megakaryopoiesis, we speculate the transcriptional regulation by AML1 might have some influence on TPO/c-Mpl signaling. So, we here examined the effects of AML1 on the c-Mpl transcription using the promoter analyses. Also, we analyzed the biologic effects of AML1dC on c-Mpl expression in HSC and megakaryocytes and on megakaryocytic differentiation.
EXPERIMENTAL PROCEDURES
Reagents and Antibodies—Recombinant human (h) interleukin-6 (hIL-6), murine (m) IL-3, murine stem cell factor (mSCF), human thrombopoietin (hTPO), human erythropoietin, and an anti-mouse c-Mpl monoclonal antibody were provided by Kirin Brewery Co. (Tokyo, Japan). Human flt3-ligand was purchased from PeproTech (London, UK). Fluorescein isothiocyanate (FITC)-conjugated rat IgG1 and biotinylated rat IgG2b were purchased from Immunotech (Marseilles, France). Biotinylated anti-lineage (Lin) antibodies (Abs) against Gr-1 (RB6–8C5), B220 (RA3–6B2), CD3 (145–2C11), Mac1 (M1/70), and Ter119 (TER119), FITC-labeled anti-Sca-1(D7), phycoerythrin-labeled anti-c-Kit (2B8), phycoerythrin-conjugated anti-Rat Igλ (B46–5), and streptavidin-PerCP-Cy5.5 were purchased from BD Biosciences. The anti-AML-1 Ab (N-20) and normal goat IgG were purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
Plasmid Constructs—The expression vectors for AML1b and AML1-MTG8 (pRCCMV-AML1b and pRCCMV-AML1-MTG8) were kindly provided by Dr. Kitabatashi (National Cancer Center Research Institute, Tokyo, Japan) (35). AML1dC, lacking the C terminus of AML1b (amino acids from 224 to 453), was obtained by the PCR method. Retrovirus expression vectors for AML1b and AML1dC were generated by subcloning these cDNAs into the Mie vector (pMSCV-IRES-EGFP). The expression vector for PEBP2β was provided by Dr. N. A. Speck (Dartmouth Medical School, Hanover, NH) (36).
Luciferase Assays—To construct reporter genes for the c-mpl promoter, various PCR products were subcloned into the luciferase plasmid, PSP72-Luc (37). Luciferase assays were performed with a dual luciferase reporter system (Promega, Madison, WI) as described previously (37). In short, 293T cells (2 × 105 cells) cultured in DMEM containing 10% fetal bovine serum (FBS) were seeded into a 60-mm dish and transfected with the effector genes (2 μg) and reporter gene (2 μg) together with pRL-CMV-Rluc (5 ng), an expression vector for Renilla luciferase, by the calcium phosphate coprecipitation method. After 12 h, the cells were washed and serum-deprived for 24 h. Then the cells were lysed and subjected to the measurement of the firefly and Renilla luciferase activities on a luminometer LB96P (Berthold Japan, Tokyo, Japan). The relative firefly luciferase activities were calculated by normalizing transfection efficiencies according to the Renilla luciferase activities. To perform luciferase assays in HeLa cells, we used a FuGENE 6 (Roche Applied Science) for transfection. The experiments were performed in triplicate, and the similar results were obtained from at least three independent experiments.
Electrophoretic Mobility Shift Assay (EMSA)—EMSA was performed as described previously (38). One probe used as a positive control contained the reported AML1-binding sequence (39). One more probe contained the proximal putative AML1-binding sequence in the human c-mpl promoter (-135/-116, numbered from the first ATG). For competition assays, unlabeled oligonucleotides containing wild-type (WT) (TGTGGT) or mutated (MT) (TGTTAG) AML1-binding site were added to the DNA-binding reaction mixtures. The sequences of the oligonucleotides are as follows: WT AML1, 5′-CGAGTATTGTGGTTAATACG-3′; MT AML1, 5′-CGAGTATTGTTAGTAATACG-3′; c-mpl (-135/-116) WT, 5′-ACCCCAGTGTGGTCTGGATG-3′; and c-mpl (-135/-116) MT, 5′-ACCCCAGTGTTAGCTGGATG-3′.
Chromatin Immunoprecipitation (ChIP) and ReChIP Assays—ChIP assays were performed with a ChIP assay kit (Upstate Biotechnology Inc.). Briefly, 1 × 107 cells were fixed with 1% formaldehyde for 10 min. Cross-links were quenched with 125 mm glycine. After isolation of the nuclear extract, chromatin was sonicated to shear DNA to the length between 200 and 1000 bp. After sonication, AML1-DNA-binding complexes were immunoprecipitated with the anti-AML1 Ab or control goat IgG. The immunoprecipitated DNA was eluted and subjected to the PCRs using AmpliTaq Gold (PerkinElmer Life Sciences), in the following thermal cycling conditions: 94 °C for 10 min, 30 cycles of 94 °C for 30 s, 65 °C for 30 s, and 72 °C for 30 s, followed by 72 °C for 10 min. The sequences of the primer set for the human c-mpl promoter are as follows: sense, 5′-TTTCCCCAGTGTGGTCTGGATGG-3′; antisense, 5′-TTTGCCTTAGCCCATCCTCCCTT-3′. PCR products were electrophoresed on agarose gels and visualized by staining with SYBR Green I (BioWhittaker Molecular Applications, Rockland, ME). In the sequential ChIP (ReChIP) experiments, we performed a first ChIP with the anti-AML1 Ab. Immunoprecipitated complexes were eluted by incubation for 30 min at 37 °C in 50 μl of 10 mmol/liter dithiothreitol. After centrifugation, the supernatant was diluted 20 times with ReChIP buffer (1% Triton X-100, 2 mmol/liter EDTA, 150 mmol/liter NaCl, 20 mmol/liter Tris-HCl, (pH 8.0)) and subjected to the second re-immunoprecipitation and the ChIP procedure. In the ReChIP analysis, PCRs were performed with 35 cycles of amplification (40, 41).
Purification of Murine c-Kit+Sca1+Lin- (KSL) Cells—Bone marrow cells were harvested from 8- to 10-week-old C57BL/6 mice, and mononuclear cells were isolated by density gradient centrifugation. After staining with biotinylated anti-Lin Abs, an FITC-conjugated anti-Sca1 Ab, a phycoerythrin-conjugated anti-c-Kit A, and a streptavidin-PerCP-Cy5.5, KSL cells were sorted on FACS Aria (BD Biosciences).
Preparation of the Conditioned Medium Containing Retrovirus Particles—Conditioned medium containing high tighter retrovirus particles was prepared as reported previously (38). Briefly, retrovirus plasmid DNA was transfected into retrovirus packaging cell line 293gp along with a vesicular stomatitis virus-G envelope expression plasmid by the calcium phosphate coprecipitation method. After 48 h, cultured supernatant was collected and concentrated by 100-fold in volume.
Retrovirus Transfection into Murine Hematopoietic Stem/Progenitor Cells—Purified KSL cells were cultured in DMEM containing 10% FBS, mSCF (100 ng/ml), and hTPO (100 ng/ml). Then the cells were seeded into the culture plates coated with Retronectin (TAKARA BIO, Shiga, Japan) and cultured with conditioned medium containing retrovirus and Polybrene (10 μg/ml) in the presence of mSCF (100 ng/ml) and hTPO (100 ng/ml). After 24 h, cells were washed and cultured in DMEM containing 10% FBS, mSCF (50 ng/ml), human flt3-ligand (30 ng/ml), and hIL-6 (50 ng/ml).
Flow Cytometry—Two days after retrovirus infection, GFP+ cells were sorted by FACS Aria (BD Biosciences). Cell surface marker analyses were performed with FACSCalibur (BD Biosciences). DNA content of cultured cells was examined by staining with propidium iodide and analyzed by the same device. FACS data were analyzed by FlowJo software (TreeStar, Ashland, OR). To analyze TPO-dependent tyrosine phosphorylation of STAT5, we used BD Phosflow™ Technology (BD Biosciences). Two days after retrovirus infection, Mock-, AML1dC-, and AML1b-transduced cells were cultured in DMEM containing 2% FBS without cytokines for 4 h. The cytokine-deprived cells were then stimulated with hTPO (100 ng/ml) for 15 min. Fixed samples were stained with Alexa Fluor®647 STAT5 (pY694) and analyzed by FACS Aria (BD Biosciences).
RT-PCR—For RT-PCR, total RNA was isolated from 7 × 103 GFP+ cells and was reverse-transcribed into cDNA with oligo(dT) primers (Pharmacia, Piscataway, NJ) using SuperScript II reverse transcriptase (Invitrogen). PCR was performed in a total volume of 50 μl using 4 μl of the cDNA product as a template and 1 μl of Advantage cDNA polymerase mix (Clontech). The primer sets to amplify murine c-mpl and β-actin are as follows: c-mpl, 5′-CCTACTGCTGCTAAAGTGGCAAT-3′ and 5′-CAATAGCTTAGTGGTAGGTAGGA-3′; β-actin, 5′-CATCACTATTGGCAACGAGC-3′ and 5′-ACGCAGCTCAGTAACAGTCC-3′. Cycling conditions were 94 °C for 1 min, followed by 22–35 cycles of 94 °C for 30 s and 68 °C for 3 min, followed by 68 °C 5 min. The PCR products were electrophoresed on agarose gels, and their amounts were evaluated by staining with SYBR Green I (BioWhittaker Molecular Applications, Rockland, ME).
OP9 System to Develop Hematopoietic Cells from Murine ES Cells—E14tg2a ES cells and OP9 stromal cells were maintained as described previously (42, 43). To induce differentiation toward hematopoietic cells, ES cells were deprived of leukemia inhibitory factor and seeded onto confluent OP9 cells in 6-well plates at a density 104 cells/well in α-minimum essential medium supplemented with 20% FBS. After 4.5 days, Flk-1+ cells were sorted by FACS or the cells were harvested by 0.25% trypsin/EDTA, and whole cell suspensions were transferred into a new 10-cm dish and incubated in 37 °C for 30 min to remove adherent OP9 cells. The collected floating cells were replated onto OP9 cells at a density 1 × 104 cells/well of 6-well plate or 6 × 104 cells/10-cm dish and cultured under the indicated conditions.
Tetracycline (Tet)-regulated Inducible Expression of AML1dC in ES Cells—To inducibly express AML1dC in ES cells, we utilized a Tet-Off system as reported previously (44, 45), in which transcription of the target mRNA is initiated by the removal of Tet from the culture medium. Briefly, we initially introduced pCAG20-1-tTA and pUHD10-3-puro by electroporation (800 V, 3 microfarads) and selected one clone designated E14 by the culture with 1 μg/ml of Puro and/or 1 μg/ml of Tet, in which the Tet-regulatory system works most effectively. We further transfected pUHD10–3-AML1dC-GFP, which can inducibly express AML1dC and GFP as a single mRNA through the internal ribosome entry site in response to the Tet removal, together with the neomycin-resistant plasmid pcDNA3.1-neo. After the culture with G418 (0.4 mg/ml) in the Tet+ medium, we selected several clones that can inducibly express GFP in response to the Tet deprivation. Subsequently, we examined the Tet-regulated expression of AML1dC in the Tet+ and Tet- medium in these clones, and several clones were subjected to further analyses.
Colony Assays—Two days after retrovirus infection, GFP+ cells (1000 cells/35-mm dish) were cultured in the methylcellulose media M3234 (Stem Cell Technologies, Vancouver, British Columbia, Canada) containing the indicated cytokines. The number of CFU-GM was counted on day 7 and those of BFU-E and CFU-GEMM on day 12.
RESULTS
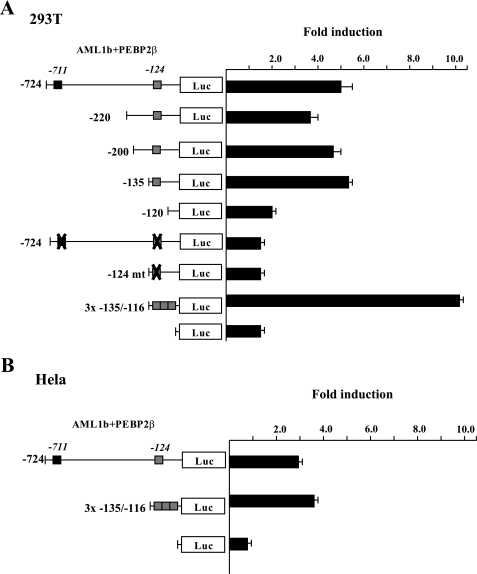
Function of AML1b on c-mpl Promoter Activity—To examine the effect of AML1 on TPO/c-Mpl-mediated signaling, we initially examined whether AML1 transcriptionally regulates the expression of c-mpl. For this purpose, we performed luciferase assays with the -724 construct, a reporter gene containing the proximal 724 bp of the c-mpl promoter using 293T cells. As shown in Fig. 1A, WT AML1b activated the -724 construct about 5.0-fold in the presence of its heterodimerization partner, PEBP2β. As there were two putative AML1-binding sites (TGTGGT) in the -724 construct, we further constructed several deletion mutants. Although deletion of the distal AML1-binding site (-711) and extended deletion up to -135 bp did not influence the c-mpl promoter activation by AML1b, the -120 construct further lacking the proximal AML1-binding site (-124) was scarcely activated by AML1. In addition, AML1b could not activate -124mt construct, in which the proximal AML1-binding site was changed from TGTGGT to TGTTAG. Furthermore, AML1 activated the 3× -135/-116 construct containing three tandem repeats of the proximal AML1-binding site and minimal JunB promoter over 10-fold. Similar results were obtained from luciferase assays using HeLa cells (Fig. 1B). These results suggest that AML1 may regulate the expression of c-mpl through the proximal AML1-binding site in the promoter.
FIGURE 1.
Effects of AML1b on the activity of c-mpl promoter. A, 293T cells were transfected with 2 μg of AML1b, PEBP2β, and 2 μg of indicated reporter gene. The relative firefly luciferase (LUC) activities were calculated by normalizing transfection efficiencies according to the Renilla luciferase activities. The results are shown as the means ± S.D. of triplicate cultures. B, HeLa cells were transfected with 0.5 μg of AML1b and PEBP2β and 1 μg of reporter genes by FuGENE 6.
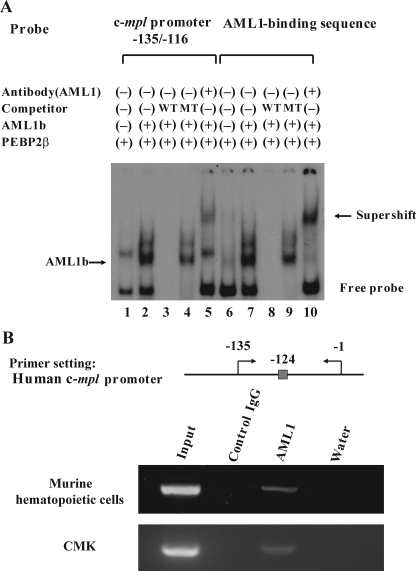
AML1 Transcriptionally Regulates the c-mpl Promoter—To analyze whether AML1 directly binds to the proximal AML1-binding sequence in the c-mpl promoter, we performed EMSA with the corresponding c-mpl (-135/-116) probe. Also, one more probe with the reported AML1-binding sequence (39, 46) was used as a positive control. Nuclear extracts were isolated from 293T cells that were transfected with PEBP2β with or without AML1b. As compared with the nuclear extract from AML1b-untransfected 293T cells (Fig. 2A, lane 1), that from AML1b-transfected cells formed two additional complexes with the c-mpl (-135/-116) probe (lane 2), and their mobilities were almost the same with that detected by the positive control probe (lane 7). These complexes were abolished by the WT competitor (Fig. 2A, lanes 3 and 8) but not by the MT competitor (lanes 4 and 9). Furthermore, these complexes were supershifted by the anti-AML1 Ab (Fig. 2A, lanes 5 and 10). These data indicate that these complexes were formed in a sequence-specific manner and contained AML1. To further test whether endogenous AML1 binds to the c-mpl promoter in vivo, we conducted ChIP assays using the anti-AML1 Ab. To obtain enough numbers of hematopoietic cells, we separated Lin- cells and cultured with mIL-3 and hTPO for 3 days. Nuclear extracts were isolated from 1 × 107 cultured cells. As shown in Fig. 2B, the c-mpl promoter, including the proximal AML1-binding site, was immunoprecipitated by the anti-AML1 Ab but not by control IgG (Fig. 2B). Similar results were also observed using the nuclear extract obtained from a human megakaryocytic cell line CMK (Fig. 2B). Together, these data indicate that endogenous AML1 bind to the proximal AML1-binding site in the c-mpl promoter and suggest that AML1 might regulate its transcription.
FIGURE 2.
Analysis of the responsive element to AML1b in the c-mpl promoter. A, EMSA was performed with the probe containing putative AML1-binding sequence in the human c-mpl promoter or known AML1-binding sequence (positive control). Nuclear extract was isolated from 293T cells transfected with the indicated genes and subjected to EMSA. In competition assays, a 1000-fold molar excess of unlabeled wild-type or mutant competitor oligonucleotide was added to the binding mixture. B, location of AML1-binding site and the primer set in the c-mpl promoter utilized for the ChIP assay are indicated. The nuclear extract was isolated from primary cultured murine hematopoietic cells and CMK cells, and the chromatin was sonicated. Then AML1-DNA-binding complexes were immunoprecipitated with the anti-AML1 Ab (N-20) or control goat IgG. The immunoprecipitated DNA was eluted and subjected to the PCR analyses. PCR products were electrophoresed on agarose gels and visualized with SYBR Green staining.
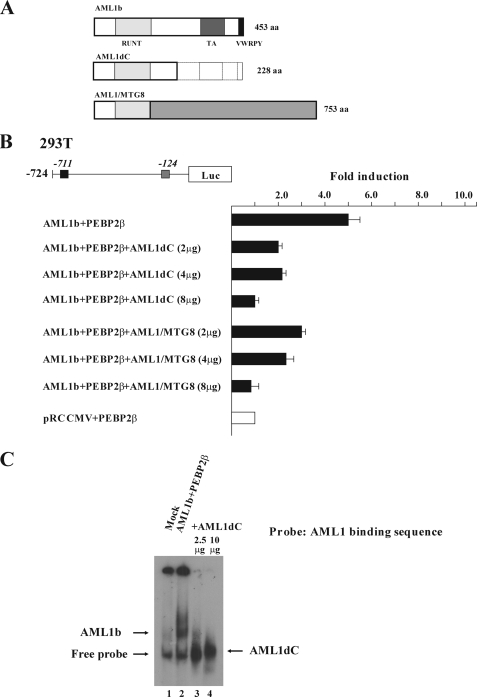
AML1dC Dominant-negatively Suppresses AML1 Function—Next, we examined the effects of a mutant of AML1b, AML1dC, on the c-mpl promoter activity. AML1dC is a C-terminal deletion mutant of AML1b (Fig. 3A), which was originally identified in a patient with MDS (30). Harada et al. (30) reported that this mutant suppressed transactivation activity of AML1 for macrophage colony-stimulating factor receptor. As was the case with the macrophage colony-stimulating factor receptor, AML1dC dose-dependently suppressed AML1b/PEBP2β-induced c-mpl promoter activity in 293T cells as efficiently as AML1-MTG8, which is known to act as a dominant-negative repressor of AML1 (Fig. 3B). To clarify how AML1dC inhibits AML1 activity, we performed EMSA with the c-mpl (-135/-116) probe. As shown in Fig. 3C, only 2.5 μg of cotransfected AML1dC was able to effectively cancel the DNA-binding complex formed by 10 μg of transfected wild-type AML1b, which was more prominent when 10 μg of AML1dC was cotransfected (Fig. 3C, lanes 3 and 4). These results suggest that AML1dC dominant-negatively suppresses the function of AML1 by inhibiting its DNA binding activity.
FIGURE 3.
AML1dC dominant-negatively suppresses AML1 function. A, horizontal bars show WT AML1b (453 amino acids (aa)), C-terminal deletion mutant of AML1b (288 amino acids), and AML1/MTG8 (753 amino acids). In the case of AML1dC, the insertion of ACCGT into 669–670 causes frameshift mutation and results in truncation of WT AML1b. RUNT indicates the runt domain; TA indicates the transactivation domain; VWRPY indicates the VWRPY motif. B, 293T cells were transfected with 2 μg of AML1b, PEBP2β, and indicated doses of AML1dC or AML1/MTG8. The results are shown as the means ± S.D. of triplicate cultures. C, EMSA was performed with the probe containing AML1-binding sequence. Nuclear extracts were isolated from 293T cells transfected with empty vector (10 μg) or AML1b (10 μg), PEBP2β (10 μg), and indicated doses of AML1dC.
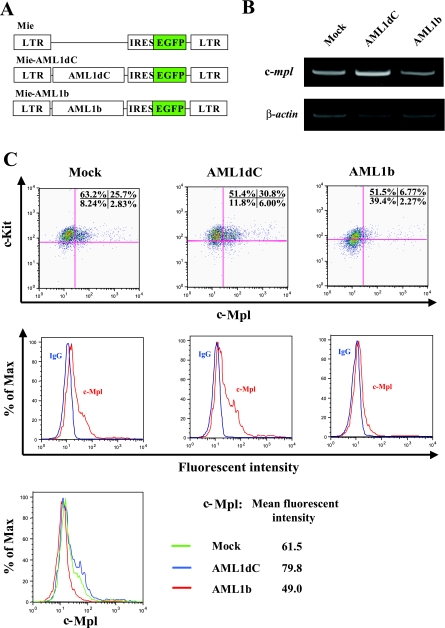
AML1dC Enhances c-Mpl Expression in Hematopoietic Stem/Progenitor Cells—Because TPO/c-Mpl signaling plays an important role in the proliferation and survival of hematopoietic stem/progenitor cells (33, 34), we next examined the function of AML1 in the c-Mpl regulation in hematopoietic stem/progenitor cells. For this purpose, we transduced AML1dC and AML1b into murine KSL cells with a retrovirus system (Fig. 4A). Two days after retrovirus infection, we sorted retrovirus-transduced cells, which are detected by the GFP expression, and performed RT-PCR analysis. Unexpectedly, in contrast to the result of luciferase assays in 293T and HeLa cells that suggests AML1 positively regulates the c-mpl expression, AML1dC-transduced cells expressed c-mpl mRNA more abundantly than mock-transduced cells in several repeated experiments (representative result is shown in Fig. 4B). In accord with this result, FACS analysis showed that the c-Mpl was more intensely expressed in AML1dC-transduced cells than in mock-transduced cells (mean fluorescent intensity, AMLdC 79.8 versus Mock 61.5) (Fig. 4C). On the other hand, c-mpl mRNA and cell-surface c-Mpl expression were suppressed in AML1b-transduced cells than those in Mock-transduced cells (mean fluorescent intensity: AMLb 49.0 versus Mock 61.5) (Fig. 4, B and C).
FIGURE 4.
Analysis of the c-Mpl expression in murine hematopoietic stem/progenitor cells. A, structure of Mie (Mock), Mie-AML1dC, and Mie-AML1b retroviruses. B, 2 days after retroviral transfection, GFP+ cells were sorted and subjected to RT-PCR to examine the expression of c-mpl and β-actin mRNA. C, at the same point, the surface phenotype of GFP+ fraction of Mie, Mie-AML1dC, and Mie-AML1b-transduced cells was examined by FACS. Dot plots of cell-surface expressions of c-Kit and c-Mpl (upper panels) are shown. Histogram plots and mean fluorescent intensities of c-Mpl expression (middle and lower panels) are shown. LTR, long terminal repeat; IRES, internal ribosome entry site.
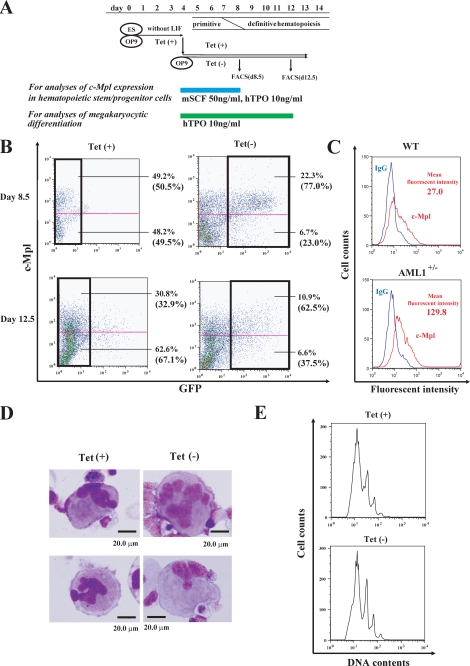
AML1dC Enhances c-Mpl Expression in Hematopoietic Cells That Derived from Murine ES Cells—To further explore the effects of AML1dC on the c-Mpl expression during the development of hematopoietic stem/progenitor cells, we utilized the Tet-Off system in the OP9 system. In the OP9 system, after deprivation of leukemia inhibitory factor from the culture medium, Flk-1+ hemangioblasts that have both the potential to develop into hematopoietic cells and endothelial cells develop from ES cells after 4.5 days, and definitive hematopoietic stem/progenitor cells appear after 8.5 days (Fig. 5A) (42, 43). After sorting Flk-1+ cells on day 4.5, we inducibly expressed AML1dC by depriving Tet from the culture medium, cultured for 4 days, and performed FACS analysis on day 8.5. After the culture with Tet, 50.5% of AML1dC-transduced ES cells were positive for c-Mpl in the GFP-negative fraction (Fig. 5B, upper left panel). In contrast, after the culture without Tet, 77.0% of AML1dC-transduced ES cells were positive for c-Mpl in the GFP-positive fraction (Fig. 5B, upper right panel). Similar results were obtained in other clones of AML1dC (data not shown). Tet deprivation by itself did not influence c-Mpl expression in mock-transduced ES cells (data not shown). Together with the results obtained from AML1dC- and AML1b-transduced KSL cells, these results indicate that wild-type AML1 is a negative regulator of the c-mpl transcription in hematopoietic stem/progenitor cells.
FIGURE 5.
Effects of AML1dC on c-Mpl expression on hematopoietic progenitor cells and megakaryocytes and megakaryopoiesis. A, experimental design using the OP9 system. ES cells were deprived of leukemia inhibitory factor and cultured on OP9 cells for 4.5 days. Then Flk-1+ cell were sorted, replated onto OP9 cells, and cultured with mSCF and hTPO (for the analysis of c-Mpl expression in hematopoietic stem/progenitor cells) or only hTPO (for the analysis of megakaryocytic differentiation) with or without Tet for the time indicated. B, c-Mpl expression of nonadherent cells was examined by the direct immunofluorescence method on day 8.5 and day 12.5. The percentage of each fraction is indicated. The relative frequency of GFP- fraction in cultured cells with Tet and the relative frequency of GFP+ fraction in cultured cells without Tet were shown in parentheses. C, c-Mpl expression on early hematopoietic cells that derived from WT and AML1+/- ES cells on day 8.5. D and E, after 12.5-day cultures with TPO, megakaryocytic cells, which derived from ES cells expressing AMLdC, were subjected to morphological analysis (D), and DNA content analysis by propidium iodide staining (E).
Haploinsufficiency of AML1 Also Enhances c-Mpl Expression in ES-derived Hematopoietic Cells—AML1dC might influence the expression of c-Mpl on hematopoietic stem/progenitor cells not only as a dominant-negative mutant but also through the unknown mechanisms. So, it is important to examine the expression of c-Mpl on hematopoietic stem/progenitor cells, in which the expression of AML1 was simply reduced. For this purpose, we developed hematopoietic stem/progenitor cells from murine AML1+/- ES cells and examined the c-Mpl expression on these cells, because AML1-null-ES cells cannot differentiate into definitive hematopoietic cells (47). As a result, we found that early hematopoietic cells that derived from AML1+/- ES cells expressed c-Mpl more intensively than those that developed from WT ES cells (mean fluorescent intensity, AML1+/- 129.8 versus WT 27.0) (Fig. 5C). This result again suggests that AML1 is a negative regulator of c-Mpl expression in hematopoietic stem/progenitor cells.
AML1dC Does Not Influence the c-Mpl Expression in Megakaryocytes or Their Maturation—Except for immature hematopoietic cells, c-Mpl is exclusively expressed on megakaryocytic cells and plays essential roles in megakaryopoiesis and subsequent platelet production. So we next analyzed the effects of AML1dC on megakaryocytic maturation and the c-Mpl expression on mature megakaryocytes. For this purpose, we cultured AML1dC-transduced ES clones in the presence of TPO for 12.5 days (Fig. 5D). On day 12.5, morphologic analysis showed that AML1dC-transduced ES cells were able to possess polyploid nucleus, which is characteristic of mature megakaryocytes, regardless of the presence or absence of Tet (Fig. 5D). Also, FACS analysis on day 12.5 showed that Tet deprivation neither inhibited polyploidization of megakaryocytes (Fig. 5E) nor their c-Mpl expression (Fig. 5B, lower panels) in AML1dC-transduced ES cells.
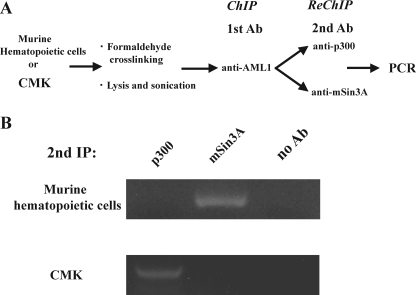
AML1 Forms Different Transcriptional Complex on the c-mpl Promoter in Hematopoietic Stem/Progenitor Cells and Megakaryocytes—Our findings suggested AML1 differentially regulates the c-Mpl expression in hematopoietic stem/progenitor cells and megakaryocytic cells. Because AML1 forms a transcriptional complex with various molecules, we hypothesized that AML1 may change the binding partner included in the transcriptional complex, thereby regulating the c-mpl promoter either positively or negatively according to cell types. To assess this hypothesis, we performed ChIP-ReChIP assays. Nuclear extracts were prepared from 3-day-cultured Lin-Sca1+ cells and CMK cells. After the first ChIP with the AML1 Ab, the eluted samples, including the transcriptional complex of AML1, were re-immunoprecipitated by the anti-p300 Ab and anti-mSin3A Ab respectively (Fig. 6A). As shown in Fig. 6B, upper panel, the anti-mSin3A Ab but not the anti-p300 Ab immunoprecipitated the c-mpl promoter from the transcriptional complex of AML1 obtained from Lin-Sca1+ cells. In contrast, the c-mpl promoter was immunoprecipitated by the anti-p300 Ab but not by the anti-mSin3A Ab from the transcriptional complex of AML1 isolated from CMK cells (Fig. 6B). These results suggest that AML1 represses the c-mpl promoter by forming a complex with a transcriptional corepressor mSin3A in hematopoietic stem/progenitor cells, although it activates the c-mpl promoter by forming a complex with a transcriptional activator p300 in megakaryocytic CMK cells.
FIGURE 6.
AML1 forms different transcriptional complexes on the c-mpl promoter in hematopoietic stem/progenitor cells and megakaryocytes. A, experimental design of ChIP-ReChIP assays. Murine Lin-Sca1+ cells were cultured for 3 days with mSCF (50 ng/ml), mIL-3 (10 ng/ml), and hTPO (50 ng/ml). These cultured cells and CMK cells were cross-linked and subjected to the ChIP-ReChIP assays. B, after the second immunoprecipitation (IP), PCR analyses were performed using the primer set shown in Fig. 2 with the eluted DNA as a template.
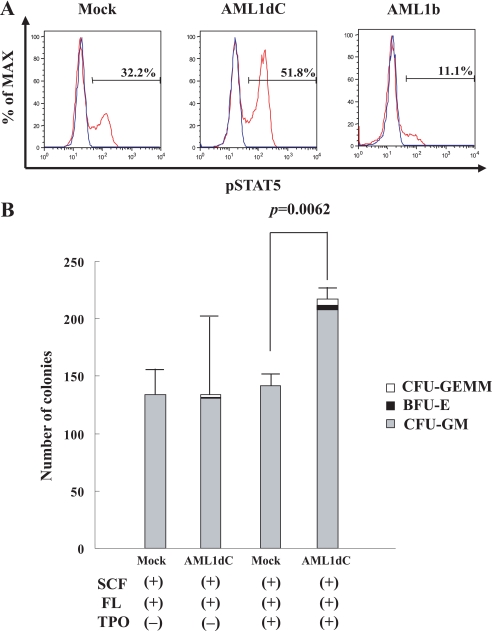
AML1dC Enhances TPO Signaling and TPO-dependent Colony Forming Activity—To assess the biologic significance of the AML1dC-enhanced c-Mpl expression in hematopoietic stem/progenitor cells, we initially compared TPO-induced tyrosine phosphorylation of STAT5 between AML1dC- and Mock-transduced KSL cells by flow cytometry. As a result, we found that the stimulation with TPO for 15 min activated STAT5 more effectively in AML1dC-transduced cells than in Mock-transduced cells (% of activated cells: AML1dC 51.8% versus Mock 32.2%) (Fig. 7A). Meanwhile, TPO-induced STAT5 activation in AML1b-transduced cells was distinctly attenuated compared with Mock-transduced cells (% of activated cells, AML1b 11.1% versus Mock 32.2%). Also, we performed colony assays using these cells under several conditions with or without TPO. As shown in Fig. 7B, although AML1dC- and Mock-transduced KSL cells developed almost equivalent numbers of hematopoietic colonies in the absence of TPO, AML1dC-transduced KSL cells yielded more and larger colonies than Mock-transduced KSL cells in the presence of TPO. In particular, CFU-GEMM was formed from AML1dC-transduced KSL cells but not from Mock-transduced KSL cells. These results suggest that the augmented c-Mpl expression by AML1dC led to the enhanced proliferation (in part, self-renewal) and survival of KSLs through the TPO/c-Mpl signaling (Fig. 7B).
FIGURE 7.
AML1dC enhances TPO signaling and TPO-dependent colony forming activity. A, after 15-min of TPO stimulation, tyrosine phosphorylation of STAT5 was examined by FACS in Mock-, AML1dC-, and AML1b-transduced cells, which are gated as a GFP+ fraction. Red line, with TPO stimulation. Blue line, without TPO stimulation. B, 2 days after retrovirus infection, GFP+ cells (1000 cells/dish) were seeded into the methylcellulose media with indicated cytokines. The number of CFU-GM was counted on day 7 and those of BFU-E and CFU-GEMM on day 12. The results are shown as mean ± S.D. of triplicate experiments. CFU-GEMM (open square); BFU-E (closed square); CFU-GM (gray square).
DISCUSSION
Because both AML1 and TPO/c-Mpl signaling play important roles in the growth of HSCs and megakaryopoiesis, we assumed that AML1 might regulate TPO/c-Mpl signaling. Also, in a recent paper, Heller et al. (31) reported that platelet surface c-Mpl expression was decreased in FPD/AML patients, suggesting that AML1 would augment c-Mpl expression in megakaryocytes. To clarify this relationship, in this study we performed luciferase assays, EMSA, and ChIP assays with the c-mpl promoter. As a result, we found that AML1 directly binds to the proximal AML-binding sequence between -137 and -122 bp of the c-mpl promoter, thereby regulating its activity. In agreement with the suggestive data by Heller et al. (31), AML1 activated the c-mpl promoter in luciferase assays using 293T cells and HeLa cells. However, the enforced expression of a dominant-negative form of AML1, AML1dC, in KSL cells by the retrovirus system enhanced c-Mpl expression in hematopoietic stem/progenitor cells and exogenous AML1b transduction into KSL cells and attenuated c-Mpl expression and TPO-induced STAT5 activation. Also, the induced expression of AML1dC during the development of hematopoietic cells in the OP9 system enhanced c-Mpl expression on hematopoietic stem/progenitor cells. Furthermore, early hematopoietic cells that derived from AML1+/- ES cells expressed c-Mpl more intensively than those that developed from WT ES cells. These results suggest that AML1 is a negative regulator of c-Mpl expression in these cells, which is opposite to its role in megakaryocytes. As for this inconsistent result observed in hematopoietic stem/progenitor cells, we speculate that AML1 would be able to regulate the c-mpl promoter both positively and negatively according to cell types. In fact, AML1 and its heterodimeric partner, PEBP2β, have been reported to form a transcriptional complex with various molecules and to change its function dependently on cell types. When AML1 forms a complex with p300/CBP and MOZ, this complex strongly stimulates AML1-mediated transcription (48). On the other hand, when AML1 combines with mSin3A, this complex works as a transcriptional repressor (49). In agreement with this speculation, we found that AML1 forms a complex with mSin3A on the c-mpl promoter in hematopoietic stem/progenitor cells, whereas it formed a complex with p300 on the same promoter in megakaryocytic cells (Fig. 6B). These results suggest that AML1 plays distinct roles in the regulation of the c-mpl promoter dependent on cell types by changing the binding partner.
In a previous paper, Ichikawa et al. (17) reported that conditional targeting of AML1 in adult mice led to the impaired polyploidization of megakaryocytes, resulting in the low platelet production. Also, patients with hereditary FPD/AML, which was caused by the heterozygous point mutations of the AML1 gene or the PEBP2β gene revealed low platelet numbers in the peripheral blood (19). To clarify the roles of AML1 in megakaryocytic maturation, several studies have been performed. Consequently, Bernardin-Fried et al. (50) found that AML1 directly activates the cyclin D3 promoter, thereby enhancing DNA synthesis required for polyploidization. Also, Goldfarb and co-workers (51, 52) showed that AML1 binds to and activates the promoter of megakaryocyte-specific genes, αIIb integrin, and glycoprotein Ibα, in combination with a transcription factor specific for the erythroid/megakaryocyte lineage, GATA-1, thereby promoting phenotypic maturation of megakaryocytes (51, 52). Most AML1 mutations observed in FPD/AML and AML are clustered within the Runt domain in the N-terminal region (19, 53–55). Heterozygous Runt domain mutations show haploinsufficient phenotype because of their reduced DNA binding and PEBP2β binding (30, 53). On the other hand, because C-terminal deletion mutants of AML1 have enhanced DNA-binding potential, they strongly suppress wild-type AML1 function through the blocking of its DNA binding in a dominant-negative manner (30). In line with this result, we also found that AMLdC lacking the C-terminal amino acids 224–453 dominant-negatively suppressed DNA binding of WT AML1 in EMSA using nuclear extracts of 293T cells. However, in this study, we found that AML1dC scarcely influenced the morphology or polyploidization of megakaryocytes. This result seems to be at variance with previous reports indicating the importance of AML1 in megakaryopoiesis (as described above). However, because an apparent abnormality was not detected in megakaryopoiesis and platelet production in AML1 heterozygous knock-out mice (14), it was speculated that pure haploinsufficiency of AML1 would not impair maturation of megakaryocytes or platelet production. Although genuine haploinsufficiency of AML1 was observed in some cases, it was also speculated that a greater majority of mutant AML1 proteins are assumed to act in a dominant-negative manner (56). So, at present, it remains unknown to what degree AML1 activity must be suppressed to cause the defect in megakaryopoiesis. In addition, there is a possibility that although AML1dC was found to act as a dominant-negative suppressor of AML1 in 293T cells in this study, AML1dC protein might be far more labile than wild-type AML1 in megakaryocytes. Alternatively, it is also possible that AMLdC might reveal some unknown biologic effect on megakaryopoiesis in combination with other transcriptional regulators such as GATA-1. Although the precise mechanism remains to be clarified, our data suggest that AMLdC lacking the C-terminal amino acids 224–453 by itself would not be responsible for the impaired megakaryopoiesis in the original MDS patient. Further studies using several C-terminal mutants are required to clarify the roles of mutants of AML1 in impaired megakaryopoiesis in MDS patients.
Conditional deletion of AML1 in adult mice leads to the expansion of the HSC compartment and reduction of common lymphoid progenitors (CLPs) as well as impaired megakaryopoiesis (17, 18), suggesting that AML1 enhances differentiation of HSC toward the CLP compartment. Meanwhile, c-mpl mRNA is expressed on HSCs and common myeloid progenitors but not on CLPs (16). However, considering our result that AML1-deficient hematopoietic stem/progenitor cells are hyper-responsive to TPO because of the enhanced c-Mpl expression, these reports may simply indicate that AML1-deficient HSCs and common myeloid progenitors would overgrow as compared with CLPs in response to the TPO stimulation. Also, it was speculated that MDS stem cells harboring AML1dC might be hyper-responsive to TPO, leading to the accumulation of oxidative stress that causes second genetic abnormalities.
In conclusion, we show here that AML1 acts as a negative regulator of c-Mpl expression in hematopoietic stem/progenitor cells which is opposite to its role in megakaryocytes. Also, we found that hematopoietic stem/progenitor cells harboring AML1dC were hyperproliferative in response to TPO. Further studies focusing on the roles of various types of AML1 mutants would be useful to clarify the physiologic roles for AML1 and to understand the pathophysiology of MDS.
Acknowledgments
We thank Dr. Iwama for providing a vesicular stomatitis virus-G expression plasmid and technical advice.
This work was supported by grants from the Ministry of Education, Science, Sports, and Culture and Technology of Japan and the Sankyo Foundation of Life Science. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: AML, acute myelogenous leukemia; HSC, hematopoietic stem cells; EMSA, electrophoretic mobility shift assay; ES, embryonic stem cell; TPO, thrombopoietin; ChIP, chromatin immunoprecipitation; FACS, fluorescence-activated cell sorter; DMEM, Dulbecco's modified Eagle's medium; FBS, fetal bovine serum; Ab, antibody; MDS, myelodysplastic syndrome; FITC, fluorescein isothiocyanate; CLP, common lymphoid progenitor; GFP, green fluorescent protein; RT, reverse transcription; DN, double-negative; h, human; m, murine; IL, interleukin; WT, wild type; MT, mutant; Tet, tetracycline; SCF, stem cell factor; BFU-E, burst-forming unit-erythroid; CFU-GM, colony-forming unit-granulocyte macrophage; CFU-GEMM, colony-forming unit-granulocyte erythrocyte monocyte macrophage.
References
- 1.Miyoshi, H., Shimizu, H., Kozu, T., Maseki, N., Kaneko, Y., and Ohki, M. (1991) Proc. Natl. Acad. Sci. U. S. A. 88 10431-10434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Golub, T. R., Barker, G. F., Bohlander, S. K., Hebert, S. W., Ward, D. C., Bray-Ward, P., Morgan, E., Raimondi, S. C., Rowley, J. D., and Gilliland, D. G. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 4917-4921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Osato, M. (2004) Oncogene 23 4284-4296 [DOI] [PubMed] [Google Scholar]
- 4.Sasaki, K., Yagi, H., Bronson, R. T., Tominaga, K., Matsunashi, T., Deguchi, K., Tani, Y., Kishimoto, T., and Komori, T. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 12359-12363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang, Q., Stacy, T., Miller, J. D., Lewis, A. F., Gu, T. L., Huang, X., Bushweller, J. H., Bories, J. C., Alt, F. W., Ryan G., Liu, P. P., Wynshaw-Boris, A., Binder, M., Marín-Padilla, M., Sharpe, A. H., and Speck, N. A. (1996) Cell 87 697-708 [DOI] [PubMed] [Google Scholar]
- 6.Niki, M., Okada, H., Takano, H., Kuno, J., Tani, K., Hibino, H., Asano, S., Ito, Y., Satake, M., and Noda, T. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 5697-5702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cameron, S., Taylor, D. S., TePas, E. C., Speck, N. A., and Mathey-Prevot, B. (1994) Blood 83 2851-2859 [PubMed] [Google Scholar]
- 8.Otto, F., Lübbert, M., and Stock, M. (2003) J. Cell. Biochem. 89 9-18 [DOI] [PubMed] [Google Scholar]
- 9.Taniuchi, I., Osato, M., Egawa, T., Sunshine, M. J., Bae, S. C., Komori, T., Ito, Y., and Littman, D. R. (2002) Cell 111 621-633 [DOI] [PubMed] [Google Scholar]
- 10.Woolf, E., Xiao, C., Fainaru, O., Lotem, J., Rosen, D., Negreanu, V., Bernstein, Y., Goldenberg, D., Brenner, O., Berke, G., Levanon, D., and Groner, Y. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 7731-7736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takahashi, A., Satake, M., Yamaguchi-Iwai, Y., Bae, S. C., Lu, J., Maruyama, M., Zhang, Y. W., Oka, H., Arai, N., Arai, K., and Ito, Y. (1995) Blood 86 607-616 [PubMed] [Google Scholar]
- 12.Zhang, D. E., Fujioka, K., Hetherington, C. J., Shapiro, L. H., Chen, H. M., Look, A. T., and Tenen, D. G. (1994) Mol. Cell. Biol. 14 8085-8095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang, H., Zhang, F., Kurozu, T., and Peterlin, B. M. (2005) Mol. Cell. Biol. 25 10675-10683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Okuda, T., van Deursen, J., Hiebert, S. W., Grosveld, G., and Downing, J. R. (1996) Cell 84 321-330 [DOI] [PubMed] [Google Scholar]
- 15.North, T. E., de Bruijn, M. F., Stacy, T., Talebian, L., Lind, E., Robin, C., Binder, M., Dzierzak, E., and Speck, N. A. (2002) Immunity 16 661-672 [DOI] [PubMed] [Google Scholar]
- 16.Akashi, K., Traver, D., Miyamoto, T., and Weissman, I. L. (2000) Nature 404 193-197 [DOI] [PubMed] [Google Scholar]
- 17.Ichikawa, M., Asai, T., Chiba, S., Kurokawa, M., and Ogawa, S. (2004) Nat. Med. 10 299-304 [DOI] [PubMed] [Google Scholar]
- 18.Growney, J. D., Shigematsu, H., Li, Z., Lee, B. H., Adelsperger, J., Rowan, R., Curley, D. P., Kutok, J. L., Akashi, K., Williams, I. R., Speck, N. A., and Gilliland, D. G. (2005) Blood 106 494-504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Song, W. J., Sullivan, M. G., Legare, R. D., Hutchings, S., Tan, X., Kufrin, D., Ratajczak, J., Resende, I. C., Haworth, C., Hock, R., Loh, M., Felix, C., Roy, D. C., Busque, L., Kurnit, D., Willman, C., Gewirtz, A. M., Speck, N. A., Bushweller, J. H., Li, F. P., Gardiner, K., Poncz, M., Maris, J. M., and Gilliland D. G. (1999) Nat. Genet. 23 166-175 [DOI] [PubMed] [Google Scholar]
- 20.Hirai, H. (2003) Jpn. J. Clin. Oncol. 33 153-160 [DOI] [PubMed] [Google Scholar]
- 21.Paquette, R. L., Landaw, E. M., Pierre, R. V., Kahan, J., Lübbert, M., Lazcano, O., Isaac, G., McCormick, F., and Koeffler, H. P (1993) Blood 82 590-599 [PubMed] [Google Scholar]
- 22.Yokota, S., Kiyoi, H., Nakao, M., Iwai, T., Misawa, S., Okuda, T., Sonoda, Y., Abe, T., Kahsima, K., Matsuo, Y., and Naoe, T. (1997) Leukemia (Basingstoke) 11 1605-1609 [DOI] [PubMed] [Google Scholar]
- 23.Uchida, T., Kinoshita, T., Nagai, H., Nakahara, Y., Saito, H., Hotta, T., and Murate, T. (1997) Blood 90 1403-1409 [PubMed] [Google Scholar]
- 24.Quesnel, B., Guillerm, G., Vereecque, R., Wattel, E., Preudhomme, C., Bauters, F., Vanrumbeke, M., and Fenaux, P. (1998) Blood 91 2985-2990 [PubMed] [Google Scholar]
- 25.Sugimoto, K., Hirano, H., Toyoshima, H., Chiba, S., Mano, H., Takaku, F., Yazaki, Y., and Hirai, H (1993) Blood 81 3022-3026 [PubMed] [Google Scholar]
- 26.Russell, M., List, A., Greenberg, P., Woodward, S., Glinsmann, B., Parganas, E., Ihle, J., and Taetle, R. (1994) Blood 84 1243-1248 [PubMed] [Google Scholar]
- 27.Willman, C. L., Sever, C. E., Pallavicini, M. G., Harada, H., Tanaka, N., Slovak, M. L., Yamamoto, H., Harada, K., Meeker, T. C., List, A. F., et al. (1993) Science 259 968-971 [DOI] [PubMed] [Google Scholar]
- 28.Harada, H., Harada, Y., Tanaka, H., Kimura, A., and Inaba, T. (2003) Blood 101 673-680 [DOI] [PubMed] [Google Scholar]
- 29.Yoneda-Kato, N., Look, A. T., Kirstein, M. N., Valentine, M. B., Raimondi, S. C., Cohen, K. J., Carroll A. J., and Morris, S. W. (1996) Oncogene 12 265-275 [PubMed] [Google Scholar]
- 30.Harada, H., Harada, Y., Niimi, H., Kyo, T., Kimura, A., and Inaba, T. (2004) Blood 103 2316-2324 [DOI] [PubMed] [Google Scholar]
- 31.Heller, P. G., Glembotsky, A. C., Gandhi, M. J., Cummings, C. L., Pirola, C. J., Marta, R. F., Kornblihtt, L. I., Drachman, J. G., and Molinas, F. C. (2005) Blood 105 4664-4670 [DOI] [PubMed] [Google Scholar]
- 32.Kaushansky, K., Lok, S., Holly, R. D., Broudy, V. C., Lin, N., Bailey, M. C., Forstrom, J. W., Buddle, M. M., Oort, P. J., Hagen F. S., Roth, G. J., Papayannopoulou, T., and Foster, D. C. (1994) Nature 369 568-571 [DOI] [PubMed] [Google Scholar]
- 33.Solar, G. P., Kerr, W. G., Zeigler, F. C., Hess, D., Donahue, C., de Sauvage, F. J., and Eaton, D. L (1998) Blood 92 4-10 [PubMed] [Google Scholar]
- 34.Fox, N., Priestley, G., Papayannopoulou, T., and Kaushansky, K (2002) J. Clin. Investig. 110 389-394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shimizu, K., Kitabayashi, I., Kamada, N., Abe, T., Maseki, N., Suzukawa, K., and Ohki, M. (2000) Blood 96 288-296 [PubMed] [Google Scholar]
- 36.Gu, T. L., Goetz, T. L., Graves, B. J., and Speck, N. A. (2000) Mol. Cell. Biol. 20 91-103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matsumura, I., Kitamura, T., Wakao, H., Tanaka, H., Hashimoto, K., Albanese, C., Downward, J., Pestell, R. G., and Kanakura, Y. (1999) EMBO J. 18 1367-1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Satoh, Y., Matsumura, I., Tanaka, H., Ezoe, S., Sugahara, H., Mizuki, M., Shibayama, H., Ishiko, E., Ishiko, J., Nakajima, K., and Kanakura, Y. (2004) J. Biol. Chem. 279 24986-24993 [DOI] [PubMed] [Google Scholar]
- 39.Meyers, S., Downing, J. R., and Hiebert, S. W. (1993) Mol. Cell. Biol. 13 6336-6345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jalvy, S., Renault, M. A., Lam Shang Leen, L., Belloc, I., Reynaud, A., Gadeau, A. P., and Desgranges, C. (2007) Circ. Res. 100 1292-1299 [DOI] [PubMed] [Google Scholar]
- 41.Wei, X., Xu, H., and Kufe, D. (2007) Cancer Res. 67 1853-1858 [DOI] [PubMed] [Google Scholar]
- 42.Nakano, T., Kodama, H., and Honjo, T. (1994) Science 265 1098-1101 [DOI] [PubMed] [Google Scholar]
- 43.Niwa, H., Burdon, T., Chambers, I., and Smith, A. (1998) Genes Dev. 12 2048-2060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Era, T., and Witte, O. N. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 1737-1742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakata, S., Matsumura, I., Tanaka, H., Ezoe, S., Satoh, Y., Ishikawa, J., Era, T., and Kanakura, Y. (2004) J. Biol. Chem. 279 55578-55586 [DOI] [PubMed] [Google Scholar]
- 46.Vangala, R. K., Heiss-Neumann, M. S., Rangatia, J. S., Singh, S. M., Schoch, C., Tenen, D. G., Hiddemann, W., and Behre, G. (2003) Blood 101 270-277 [DOI] [PubMed] [Google Scholar]
- 47.Okuda, T., Takeda, K., Fujita, Y., Nishimura, M., Yagyu, S., Yoshida, M., Akira, S., Downing, J. R., and Abe, T. (2000) Mol. Cell. Biol. 20 319-328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kitabayashi, I., Aikawa, Y., Nguyen, L. A., Yokoyama, A., and Ohki, M. (2001) EMBO J. 20 7184-7196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lutterback, B., Westendorf, J. J., Linggi, B., Isaac, S., Seto, E., and Hiebert, S. W. (2000) J. Biol. Chem. 275 651-656 [DOI] [PubMed] [Google Scholar]
- 50.Bernardin-Fried, F., Kummalue, T., Leijen, S., Collector, M. I., Ravid, K., and Friedman, A. D. (2004) J. Biol. Chem. 279 15678-15687 [DOI] [PubMed] [Google Scholar]
- 51.Elagib, K. E., and Goldfarb, A. N. (2007) Crit. Rev. Eukaryotic Gene Expression 17 271-280 [DOI] [PubMed] [Google Scholar]
- 52.Elagib, K. E., Racke, F. K., Mogass, M., Khetawat, R., Delehanty, L. L., and Goldfarb, A. N. (2003) Blood 101 4333-4341 [DOI] [PubMed] [Google Scholar]
- 53.Michaud, J., Wu, F., Osato, M., Cottles, G. M., Yanagida, M., Asou, N., Shigesada, K., Ito, Y., Benson, K. F., Raskind, W. H., Rossier, C., Antonarakis, S. E., Israels, S., McNicol, A., Weiss, H., Horwitz, M., and Scott, H. S. (2002) Blood 99 1364-1372 [DOI] [PubMed] [Google Scholar]
- 54.Osato, M., Asou, N., Abdalla, E., Hoshino, K., Yamasaki, H., Okubo, T., Suzushima, H., Takatsuki, K., Kanno, T., Shigesada, K., and Ito, Y. (1999) Blood 93 1817-1824 [PubMed] [Google Scholar]
- 55.Preudhomme, C., Warot-Loze, D., Roumier, C., Grardel-Duflos, N., Garand, R., Lai, J. L., Dastugue, N., Macintyre, E., Denis, C., Bauters, F., Kerckaert, J. P., Cosson, A., and Fenaux, P. (2000) Blood 96 2862-2869 [PubMed] [Google Scholar]
- 56.Osato, M., Yanagida, M., Shigesada, K., and Ito, Y. (2001) Int. J. Hematol. 74 245-251 [DOI] [PubMed] [Google Scholar]
- 57.Kimura, S., Roberts, A. W., Metcalf, D., and Alexander, W. S. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 1195-1200 [DOI] [PMC free article] [PubMed] [Google Scholar]