Abstract
The enormous macromolecular phycobilisome antenna complex (>4 MDa) in cyanobacteria and red algae undergoes controlled degradation during certain forms of nutrient starvation. The NblA protein (∼6 kDa) has been identified as an essential component in this process. We have used structural, biochemical, and genetic methods to obtain molecular details on the mode of action of the NblA protein. We have determined the three-dimensional structure of the NblA protein from both the thermophilic cyanobacterium Thermosynechococcus vulcanus and the mesophilic cyanobacterium Synechococcus elongatus sp. PCC 7942. The NblA monomer has a helix-loop-helix motif which dimerizes into an open, four-helical bundle, identical to the previously determined NblA structure from Anabaena. Previous studies indicated that mutations to NblA residues near the C terminus impaired its binding to phycobilisome proteins in vitro, whereas the only mutation known to affect NblA function in vivo is located near the protein N terminus. We performed random mutagenesis of the S. elongatus nblA gene which enabled the identification of four additional amino acids crucial for NblA function in vivo. This data shows that essential amino acids are not confined to the protein termini. We also show that expression of the Anabaena nblA gene complements phycobilisome degradation in an S. elongatus NblA-null mutant despite the low homology between NblAs of these cyanobacteria. We propose that the NblA interacts with the phycobilisome via “structural mimicry” due to similarity in structural motifs found in all phycobiliproteins. This suggestion leads to a new model for the mode of NblA action which involves the entire NblA protein.
The phycobilisome (PBS)4 is an extremely large membrane-associated complex in cyanobacteria and red algae that serves as a light-harvesting antenna for the photosynthetic apparatus (1-4). The complex can reach dimensions of about 50 × 30 × 12 nm with an overall molecular mass generally greater than 4 MDa (5). It is composed of pigment-binding proteins collectively called phycobiliproteins (PBPs): phycocyanin (PC), allophycocyanin (APC), and in some cases phycoerythrin (PE) or phycoerythrocyanin (PEC) (3). In all cases three PBP monomers self-assemble into circular disks (trimers). In the case of PC, PEC, and/or PE, the two trimers assemble into a hexamer, and then variable numbers of hexamers assemble into extended cylinders called rods. Rod structures surround a central core, made up of 2, 3, or 5 tightly packed cylinders. Each one of the core cylinders contains four APC trimers, suggested by x-ray crystallography to be more loosely assembled then the rods (1, 3, 5, 6). The internal spaces within the centers of the disks contain a variety of linker proteins whose role is to modify the energy absorption and transfer characteristics as well as to stabilize the PBS structure (7, 8).
The size, composition, and mode of membrane attachment of the PBS are variable, as a function of the environment of the organism, affording optimal energy absorption and transfer. In many cases PBS complexes contain 50% that of the total soluble protein in the cell (2) and, thus, represent a major metabolic expenditure. Under certain conditions of nutrient stress, for example the lack of nitrogen or sulfur, cyanobacteria efficiently utilize the PBS as an internal source of nutrients by degradation processes (9). The onset of PBS degradation leads to a yellowing (bleaching) of the cells, due to the loss of the native PBS pigmentation. The molecular mechanism that leads to the ability to degrade the PBS protein has been studied genetically and biochemically for the past 15 years, and a number of genes have been found to encode for proteins that are required in the bleaching process. The first gene was identified by analysis of a non-bleaching mutant of Synechococcus elongatus sp. 7942 and was, thus, called nblA (10). Additional genes involved in the bleaching process, denoted nblB (11), nblC (12), nblR (13), nblS (14), and ald (15), were subsequently identified. nblB encodes a polypeptide with similarity to α-phycocyanin phycocyanobilin lyase, an enzyme catalyzing the covalent bond formation between the tetrapyrrole chromophore and the α-subunit of the apoprotein. Apparently, NblB is not a lyase as inactivation of such an enzyme is expected to yield a constitutively bleaching rather than a non-bleaching phenotype. Therefore, it was suggested that interaction of NblB with the PBS chromophores is crucial for the degradation process (11).
The nblC, nblR, nblS, and ald genes are required for proper induction of the nblA gene (12-15). In contrast to all the other non-bleaching mutants, which exhibit aberrant PBS degradation under nitrogen and sulfur starvation, inactivation of ald, encoding for alanine dehydrogenase, impairs the PBS degradation, specifically under nitrogen starvation (15).
Only the nblA gene product (the NblA protein) has been shown to physically interact with the PBS (16, 17) and, thus, may single-handedly serve to induce the disassembly of the PBS. The NblA may be the first known member of a novel family of proteins whose function is the disassembly of complexes by physical association, to whom we designate the term “disassembelase.”
A sequence homology comparison of NblA between PBS-containing photosynthetic organisms shows a low degree of homology (<30% identity, Fig. 1 and Ref. 18), which is somewhat surprising as the putative substrates (PE, PC, or APC) are highly homologous (typically 70-90% (19). In addition, NblA has no known homology to any other protein family in the gene banks, suggesting that its function and mode of activity may be unique.
FIGURE 1.
Amino acid alignment of NblA from various organisms. S. elongatus (Syn7942), Anabaena sp. PCC 7120 (Anab7120), Nostoc punctiforme (Nostoc73102), Synechococcus sp. JA-2-3B (SynJA-2-3B), Synechococcus sp. JA-3-3Ab (SynJA-3-3Ab), Synechocystis sp. PCC 6803 (Syn6803), Thermosynechococcus elongatus BP-1 (Thermosyn elo), T. vulcanus (Thermosyn vul), Crocosphaera watsonii WH 8501 (Croco8501), Cyanidioschyzon merolae strain 10D (Cyanidioschyzon), Porphyra purpurea (Porphyra), Cyanidium caldarium (Cyanidium), Gracilaria tenuistipitata var. liui (Gracilaria), Aglaothamnion neglectum (Aglaothamnion), Galdieria sulphuria (Galdieria), and F. diplosiphon (Fremyella). a and b denote chromosomal and plasmid encoded NblAs, respectively. Synechocystis sp. PCC 6803 possesses two chromosomally encoded NblAs. The black background indicates amino acids identical in at least 18 out of the 19 sequences. Similar amino acids that are present in at least 12 sequences are indicated by gray background. Stars indicate the positions of mutations in non-bleaching mutants of S. elongatus described below. Numbering is according to NblA of S. elongatus. Sequences selected for this alignment represent NblA proteins from a verity of organisms including cyanobacteria and red algae as well as mesophilic and thermophilic cyanobacteria.
The crystal structure of the NblA protein from the mesophilic cyanobacterium Anabaena sp. PCC 7120 (An-NblA) was recently determined (18). The structure revealed that the monomer consists of a helix-loop-helix motif which dimerizes to form an open four-helical bundle. Although not stated specifically that the NblA dimers were present in solution before crystallization (in fact an earlier report on the NblA from the same organism suggested a trimeric form in solution (20)), it is implicitly stated by these authors that the dimer is the basic structural unit of the NblA. On the basis of short stretches of sequence homology, in vitro analysis of site-directed mutations and in vitro binding assays, these authors also proposed that the NblA protein interacts with the PBPs via the C terminus. Their results were interpreted as showing a strong interaction between the NblA termini and the PBPs, especially with the α-subunit of PE or PC. However, a problem with this suggestion is the large degree of difference in the length of the termini of the NblA of various species for which sequences have been determined. In these sequences the N termini can be significantly longer and the C termini significantly shorter (than the sequence of An-NblA), which would require a somewhat different binding mechanism for each NblA. In addition, it has been shown that the cyanobacterium Synechocystis sp. PCC 6803 contains and requires two forms of the NblA protein to perform its function (16, 21). These two isoforms are also different in the length of their termini. Furthermore, the sequences for two very long NblA proteins have been deposited in the Gen-Bank™: Nostoc sp. 73102 with 114 residues and Synechococcus JA with 90 residues.
Although it can be argued that the NblA of each species may function through a completely different set of interactions, it is also possible that the molecular details on how the NblA functions are quite different from those previously proposed (20). In this study, we present the structures of NblA from the thermophilic cyanobacterium Thermosynechococcus vulcanus (Tv-NblA) and the mesophilic cyanobacterium S. elongatus (Se-NblA). Data obtained from the structural analyses coupled with molecular modeling and examination of in vivo function suggest a very different model for NblA function.
EXPERIMENTAL PROCEDURES
Protein Expression, Purification, and Crystallization—The T. vulcanus and S. elongatus nblA genes encoding for the NblA proteins were expressed, purified, and crystallized as previously described (22). Briefly, both Tv-NblA and Se-NblA recombinant proteins had solubility problems that were alleviated by treatment with low concentrations of urea.
NblA Structure Determination—Native and selenomethionine derivative x-ray diffraction data were collected at the European Synchrotron Radiation Facility (beamlines ID14-1 and BM30, respectively). All data were reduced and scaled using Mosflm (23) or the Denzo/Scalepack package (24), and the final refinement details are tabulated in Table 1. The structure of Tv-NblA determined in the presence of urea (Tv-NblA+urea) was determined by molecular replacement (MR) and single wavelength anomalous dispersion (SAD) (25), with the MR step was performed using Phaser (26) using a single monomer from the 1OJH structure as the search model and with the SAD phases obtained using the positions of the sulfurs from the MR solution and peak anomalous data using CNS (27). Refinement was performed using the MR-SAD phases in CNS, and visualization and manual rebuilding of the structure was performed using XtalView (28) and Coot (29). The Tv-NblA and Se-NblA structures were both obtained by MR using the Tv-NblA+urea structure as the search model. The coordinates and structure factors for the refined Tv-NblA+urea, Tv-NblA, and Se-NblA protein structures have been deposited in the Protein Data Bank with codes 2Q8V, 2QDO, and 3CS5, respectively. For additional details see “Results.”
TABLE 1.
Data collection and refinement statistics
| X-ray data collection | |||||
| PDB code | 2Q8V | 2QDO | 3CS5 | ||
| Organism | T. vulcanus | T. vulcanus | S. elongatus PCC 7942 | ||
| Additive | 2 m urea (present in crystallization liquor) | 2 m urea (present during purification only) | 2 m urea (present during purification only) | ||
| Space group | P3221 | P1 | P4 | ||
| Unit cell parameters | |||||
| a | 43.265 | 42.190 | 78.080 | ||
| b | 43.265 | 42.390 | 78.080 | ||
| c | 149.400 | 50.560 | 70.678 | ||
| α | 90.00 | 69.00 | 90.00 | ||
| β | 90.00 | 83.36 | 90.00 | ||
| γ | 120.00 | 61.25 | 90.00 | ||
| Monomers/Asymmetric Unit |
2
|
4 | 4 | ||
|
Native
|
Se Peak
|
||||
| Wavelength | 0.9792 | 0.97913a | 0.9792 | 0.9792 | |
| Resolution range (Å) | 30-2.5 | 20-3.0 | 20-2.5 | 20-2.2 | |
| Unique reflectionsb | 6108 | 3492 | 9900 | 28717 | |
| <I>/<σ>b | 19.1 (4.4) | 24.2 | 19.5 | 9.75 (3.2) | |
| Rmergeb,c (%) | 7.3 (32.6) | 7.5 | 3.7 (11.3) | 5.7 (26.9) | |
| Completenessb (%) | 99.8 (98.7) | 99.9 | 96.7 (97) | 96.9 (97) | |
| Redundancyb | 6.2 (5.9) | 11.5 | 4.88 (3.62) | 6.8 (3.6) | |
| Refinement | |||||
| R (%)/Rfree (%)d | 24.8/27.9 | 24.5/26.3e | 24.8/34.7f | ||
| Amino acids | 104 | 211 | 196 | ||
| Total number of non-hydrogen atoms | 843 | 1697 | 1644 | ||
| Number of water molecules | 16 | 18 | 27 | ||
| Number of urea molecules | 2 | 0 | 0 | ||
| Average B factor (Å2), protein atoms | 45.6 | 41.8 | 27.5 | ||
| r.m.s.d. | |||||
| Bond length (Å) | 0.012 | 0.010 | 0.022 | ||
| Bond angles (°) | 1.6 | 1.7 | 1.9 | ||
Data set collected at peak wavelength of selenium and used for MR-SAD phasing.
Values in parentheses are for the last shell.
Rsym = ∑hkl∑i|Ii(hkl) - <Ii(hkl)>| /∑hkl∑iIi(hkl).
R/Rfree = ∑hkl||Fobs| - |Fcalc||/∑hkl|Fobs| where R and Rfree are calculated using the working and test reflections respectively. The test reflections (10%) were held aside and not used during the entire refinement process.
Statistics for data with F > 3σ.
Statistics for data with F > 4σ.
Molecular Manipulations of nblA—Inactivation of nblA was described elsewhere (10). Complementation of nblA-inactivated strain (NblAΩ) was done by cloning of nblA (flanked by a kanamycin resistance cassette as a selective marker, Fig. 3C) into a genomic neutral site (12) cloned into Bluescript SK yielding the plasmid pES36. The following primers were used to obtain a PCR product bearing the promoter and the coding region of nblA:5′-CGATCGCAAGTAAAGACC and 3′-CAGCACCTTGAACTCCAT. After transformation of S. elongatus with pES36, homologous recombination allows replacement of the ectopically introduced nblA (together with the antibiotic cassette) with the genomic neutral site. PCR on genomic DNA of transformants verified complete segregation of the modified chromosome. Mutagenesis of nblA was performed by transformation of pES36 into Escherichia coli XL1 Red cells (Stratagene), a strain that introduces random point mutations. The mutagenized nblA library obtained by plasmid preparation from this E. coli strain was used to transform NblAΩ. Transformants were selected for kanamycin resistance, and clones exhibiting high red fluorescence (FL3) were sorted by fluorescence activated cell sorter as previously described (30). A second screen was performed by plating and re-streaking colonies onto nitrogen-depleted solid growth medium.
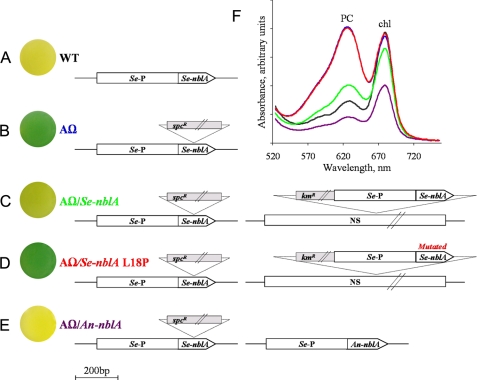
FIGURE 3.
Effects of manipulations of NblA on phycobilisome degradation. A-E, cultures of the relevant strains (round plates at the left of the figure) and schematic presentation of native nblA gene loci and the relevant molecular modifications. A, nblA of S. elongatus (WT). B, a strain possessing insertional inactivated nblA (AΩ). C, AΩ containing native nblA of S. elongatus in a neutral site (NS) in the genome (AΩ/Se-nblA). D, AΩ containing the L18P mutated NblA in a neutral site in the genome. E, AΩ containing the promoter region of nblA of S. elongatus fused to the coding region of the chromosomally located NblA of Anabaena PCC 7120 inserted in a shuttle vector (AΩ/An-nblA). kmR and spcR stand for kanamycin and spectinomycin resistance cassettes, respectively. F, absorbance spectra of cultures of wild type (black), AΩ (blue), AΩ/Se-nblA (green), AΩ/Se-nblA L18P (red), and AΩ/An-nblA (purple). Red and blue curves overlap. Cultures were starved for nitrogen for 48 h before spectral analysis. Absorbance maxima at 620 nm and 680 nm represent PC and chlorophyll a (chl), respectively.
A two-step PCR process was used to fuse the promoter region of Se-nblA to the coding region of An-nblA. The primers used for construction of the fusion product were as follows: 5′-promoter primer, CGATCGCAAGTAAAGACC; 3′-promoter primer (5′ sequence of An-nblA is underlined), GTTCTAATGACAATTCGATTGGTTGGTTCATGGGAGCCTCCGGCAC; 5′-An-nblA primer: (3′ sequence of Se-nblA promoter is underlined), GTGCCGGAGGCTCCCATGAACCAACCAATCGAATTG; 3′-An-nblA primer, CTATGCCGGAGTGGAACC.
The above-designed primers resulted in Se-nblA promoter region and An-nblA coding region, which contained overlapping sequences and, therefore, served as “mega primers” in a third PCR reaction yielding the fusion product. This fusion product was cloned into a shuttle vector and introduced into the NblAΩ.
Molecular Modeling—All molecular models were built using the Swiss-Model server (31) using the coordinates of the Tv-NblA+urea, 1KTP, or 2C7L structures as templates for NblA, PC, and PEC models, respectively. Molecular graphics were performed using Pymol (DeLano, W. L. (2002)). Calculations of all electrostatic potential surfaces were performed using the Pymol algorithm in vacuo, as only a qualitative comparison of the outside surfaces was required. All electrostatic potentials are presented on the same scale.
RESULTS
The remarkably low conservation of sequence homology between members of the NblA family of proteins has been previously noted and remains valid in light of the determination of additional nblA gene sequences (Fig. 1). If the functionality of the NblA protein is obtained through specific pairing of residues between NblA and PBPs, the differences in NblA sequences should also be manifested in comparable changes in the PBPs sequences. However, the sequences of the components of PBPs are actually highly homologous (19). In light of this observation, we decided to determine the structures of additional NblA family members to allow a comparative examination of the structures within this family. Structural analysis could result in additional insight into the mode of NblA action in the mechanism leading eventually to PBS degradation.
Structure Determination of NblA Proteins
The T. vulcanus NblA Structure Crystallized in the Presence of Urea—The expression, isolation, and crystallization of the NblA protein from T. vulcanus (Tv-NblA) and S. elongatus (Se-NblA) have been described elsewhere (22). Briefly, both proteins were obtained by recombinant methods. The resulting proteins were sparingly soluble and required the solubilizing effect of urea to afford the supersaturating conditions needed for crystallization. This method resulted in the growth of crystals of Se-NblA and Tv-NblA that diffracted to better than 2.2 and 2.5 Å, respectively.
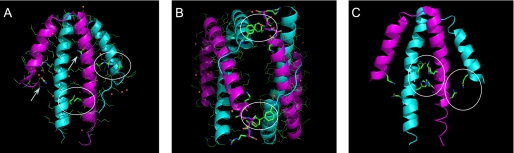
The structure of Tv-NblA in the presence of 2 m urea (Tv-NblA+urea) was determined to 2.5 Å by a combination of MR-SAD. A single An-NblA polypeptide served as the search model (22% sequence identity to Tv-NblA). A single solution was obtained using Phaser (34, 35) which contained a dimer in the asymmetric unit. The positions of the two monomers in the dimer were very near to those found in the Anabaena protein (r.m.s.d. between all Cα atoms in the dimers is 1.8 Å). The positions of 10 sulfur atoms were carved out of the molecular replacement solution, transformed into selenium atoms, and used to obtain SAD phases using diffraction data collected from a selenomethionine derivative Tv-NblA+urea crystal (Table 1). This resulted in excellent electron density maps which were not biased by the original MR model. After density modification, refinement was performed using CNS interspersed with rounds of rebuilding and geometry improvement. The final model includes residues 8-60 and 11-62 in monomers A and B, respectively. Two patches of unoccupied density that were too large to represent water molecules were modeled as urea molecules. The urea molecules form hydrogen bonds with polar side chains and a water molecule. The crystal data and structure refinement statistics are shown in Table 1. As in the previously determined An-NblA structure, the overall structure shows a helix-loop-helix motif with the two monomers associated around an approximate C2 axis (Fig. 2A). The two monomers in the Tv-NblA+urea are very similar (r.m.s.d. of 0.4 Å) and are also similar to the An-NblA monomers (r.m.s.d. ∼1.0 ± 0.3 Å).
FIGURE 2.
NblA crystal structures. Monomers A and B are depicted in cyan and magenta schematics, whereas side chains are colored according to the CPK system. A, Tv-NblA+urea dimer; arrows indicate urea molecules bound to structure, and white circles show dimer stabilizing contacts as described under “Results.” B, Tv-NblA asymmetric unit; circles show dimer-dimer stabilizing interactions. C, dimer of Se-NblA; circles show two areas with polar interactions which stabilize the dimer.
The T. vulcanus NblA Structure in the Absence of Urea—Out of a very large number of trials, only a single crystal of Tv-NblA was obtained in the absence of urea, which proved to be triclinic (Tv-NblA). A molecular replacement solution was obtained using the P3221 Tv-NblA+urea structure and proved to have two dimers in the asymmetric unit. The structure was refined by CNS and includes residues 10-62, 11-62, 9-62, and 11-62 for monomers A-D, respectively (Fig. 2B). The dimers are very similar (r.m.s.d. 0.24 Å), whereas the r.m.s.d. between the Tv-NblA+urea and Tv-NblA dimers is ∼0.3 Å, showing that the presence of urea had little effect on the P3221 structure. This is extremely important because it clearly shows that the use of up to 2 m urea may be used as a method in the preparation of sparingly soluble proteins before performance of crystallization screens. Many such sparingly soluble recombinant proteins are today discarded by structural genomics initiatives as not being able to crystallize. The fact that 2 m urea does not monomerize the NblA dimers indicates that the interactions required to form NblA dimers are much stronger than the spurious interactions that induce precipitation and are strongly likely to have a role in the proteins function in vivo.
The Tv-NblA dimers have limited contact with the N termini of the A and C monomers making contacts with the C termini of the B and D monomers, respectively. The two dimers pack in a V-shape with an angle of about 45° between dimers (not shown). The interface between dimers is very different from the interface between dimers found in the An-NblA structure but is similar to that found in the packing of adjacent dimers in the Tv-NblA-urea structure (data not shown). Indeed, the Tv-NblA+urea and Tv-NblA crystal lattices are quite similar, indicating that the same attractive forces come into play during crystal growth. In both crystal forms, the C-terminal extension (residues 58-63) of the B monomers have a random coil structure which is wedged between two Phe residues that reside on the N-terminal helix of the A monomer on the opposite dimer (Fig. 2B). Both crystal forms are made up of layers of dimers associated by complementary electrostatic interactions (data not shown). The An-NblA crystal lattice also shows a layered structure with tighter packing and larger interaction interfaces (18).
As in the case of the mesophilic An-NblA, the thermophilic Tv-NblA dimeric structure is held together by relatively few interchain polar interactions, indicating a largely hydrophobic interface. However, the Tv-NblA does have more charged residues (four additional basic and two additional acidic residues) which may have the ability to strengthen both the intramolecular α-helical structure as well as the intermolecular structure. Four salt bridges are formed by symmetry-related Glu-17 to Arg-44 and Lys-49 to Asp-50 (Fig. 2A), replacing two charged and two polar interactions between Arg-45 to Glu-45 and Gln-23 to Lys-31 in An-NblA (18). On the other hand, the Tv-NblA lacks the aromatic Tyr-Tyr ring stacking contact in the center of the dimer (18). The two Tyr residues are replaced by Ile-53, which form a tighter (3.15 Å) hydrophobic contact. Thus, as would be expected, the Tv-NblA dimers have the potential to be structurally stable at the elevated growth temperatures of T. vulcanus as shown previously for T. vulcanus phycocyanin (36, 37).
The S. elongatus NblA Structure—Recombinant Se-NblA could only be isolated in the presence of 2 m urea, which was then removed before crystallization. Crystals were obtained that diffracted to better than 2.2 Å. Analysis of the diffraction pattern indicated that the crystals belonged to the P4 tetragonal space group, with slightly less than perfect merohedral twinning (twinning fraction = 0.48) as determined using both CNS and the Merohedral Crystal Twinning Server (46). The Matthews coefficient appeared to indicate the possibility of between 4 and 8 NblA monomers in the asymmetric unit (Vm between 3.85 and 1.92, respectively), perhaps organized as 2-4 dimers. Molecular replacement was performed using the Phaser program within CCP4 (34, 35) using the Tv-NblA+urea structure. The only recognizable solution obtained with reasonable statistics (R < 0.5) contained two dimers in the A.U, with a solvent content of 68%. Refinement of the structures to an acceptable Rfree was not possible with either CNS (27) or Refmac5 in the CCP4 suite (34, 38) due to the high (but not perfect) twinning fraction. We, thus, utilized the refinement protocol implemented in SHELX97 (39), which has previously been shown to be useful in similar cases of near-perfect merohedral twinning (40). After a number of rounds of conjugate gradient least-squares refinement interspersed with manual rebuilding and real-space refinement with Coot (29), the final R and Rfree factors converged to 0.248 and 0.347, respectively (Table 1). Although somewhat high, we are confident that the structure represents a good approximation of both the monomer and dimer structures of Se-NblA. These results strengthen the possibility that the dimeric form of NblA may exist in vivo and may be the active form. However, it is certainly possible that the dimers dissociate in the presence of the PBS (see below). All four Se-NblA monomers include residues Val-12 to Ser-59, which is the terminal residue in S. elongatus NblA, nine residues shorter than An-NblA. The Se-NblA structure shows that the monomer-monomer assembly interface is mostly hydrophobic in nature with three polar interactions (two symmetry-related Gln-43-Lys-44 residues and between symmetry-related His-47-His-47) at the center of the interface (Fig. 2C). The dimer is further stabilized by the two symmetry-related Gln-14 to Asn-49 residues, which contact between the short and long helices. By comparing all three structures, we can see that although the overall monomeric and dimeric structures are conserved, the specific residues that determine the strength of interface bonding power are completely non-conserved.
Elucidation of Amino Acids of NblA Crucial for Its Function
To gain further insight into the role of NblA in phycobilisome degradation we used random mutagenesis to search for amino acids crucial for its function in vivo (Fig. 3). We employed the S. elongatus strain in which the nblA gene was disrupted (NblAΩ, Fig. 3B and Ref. 10). Native Se-nblA was then inserted into a neutral site in the genome of NblAΩ (Fig. 3C). In contrast to NblAΩ, the resulting cyanobacterial strain exhibited phycobilisome degradation, as apparent from the cultures (Fig. 3, B and C). Additionally, absorbance spectra indicated reduced levels of phycocyanin in the nitrogen-starved-complemented strain as compared with the mutant (Fig. 3F, green and blue curves, respectively). For an as yet unknown reason, the complemented strain exhibited slower kinetics of PBS degradation as compared with the wild type.
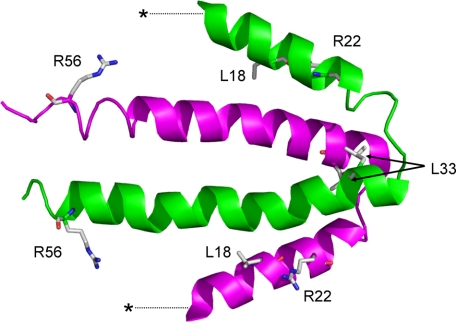
Random mutagenesis of the native nblA was performed (see “Experimental Procedures”), and the library obtained was transformed into NblAΩ. We were interested in transformants that maintained their non-bleaching phenotype, namely those that acquired a non-functional NblA from the library. Isolation of non-bleaching clones by cell-sorting allowed the identification of four point mutations which totally impaired NblA function. For example, replacement of leucine at position 18 with proline (L18P) yielded a non-bleaching phenotype similar to NblAΩ (Fig. 3, D and F (red)). Identical phenotypes were obtained for the following amino acid modifications: R22C, L33I, and R56C (not shown). Mutation of Anabaena NblA at the position equivalent to arginine 56 (K53A) was shown to prohibit interaction with PC in vitro (18). Taken together, the experimental data and the conservation of arginine or lysine at this position in NblA (Fig. 1) indicate the requirement for this specific positive charge for NblA function.
The mutations R22C and L33I identified in this study as well as the S9F mutation (10) highlight positions in NblA that do not exhibit high conservation in terms of the chemical nature of the amino acid but nevertheless are crucial for function. The positions of all mutations identified in this study are depicted on the dimeric structure of Se-NblA (Fig. 4). This illustration indicates that non-functional mutations occur not only at the protein termini (see “Discussion”).
FIGURE 4.
Positions of mutations inducing the non-bleaching phenotype on the S. elongatus NblA dimer. Stars indicate the approximate position of the S9F mutation identified previously (10) but could not be identified in the electron density map.
To try and assess the importance of the NblA C-terminal on its activity, wild-type Se-NblA was modified to add a C-terminal FLAG tag (the addition of eight amino acids, DYKDDDDK). This modified NblA was found to be able to complement NblAΩ (not shown), thus reiterating that extension of the C terminus of NblA does not interfere with its function.
Electrostatic Surface Potential of NblA Structures
The previously identified residues important for NblA function in vivo and in vitro (10, 18) and two of the new mutations identified in the screen described above are polar/charged residues. Electrostatic potentials can induce attractive forces with the longest range of any chemical interaction, and then short-range forces (>5 Å) become increasingly important. Structural complementarities are created by the coupling of polar, hydrophobic interactions, and/or hydrogen bonds with geometries that can closely match and form large interaction surfaces.
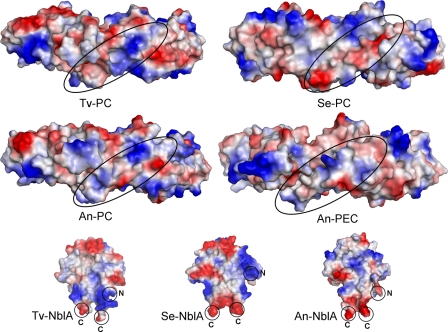
In an effort to identify structural features in the NblA protein that could promote interactions with phycobilisome subunits, we wanted to assess whether electrostatic complementarities are possible. Because structures of PBPs from either S. elongatus or Anabaena have yet to be determined, we built homology-based molecular models of the PC (S. elongatus and Anabaena) and PEC (Anabaena) components based on the high resolution structure of the T. vulcanus PC (1KTP (37)) and Mastigocladus laminosus PEC (2C7L (41)). Fig. 5 shows the electrostatic potential surface of the outer circumference of the Tv-PC structure and the Se-PC, An-PC, and An-PEC models. Upon rod assembly these surfaces form the exterior exposed surface, and due to the 3-fold rotational symmetry of the rod, the surfaces are identical around the entire circumference. The overall distributions of charged patches on these surfaces are somewhat different between the different PBPs. One segment that has significantly different distributions is formed by helices Y-A of the α subunits (Fig. 5, black ovals). On the basis of in vitro interaction analysis, Bienert et al. (18) suggested that the NblA interacts with PEC and PC through residues on these helices. Because in Anabaena rods the potentials of the Y-A helices of PC and PEC are significantly different, it is unlikely that only electrostatic complimentarity could explain the interaction between NblA and the Y-A helices.
FIGURE 5.
Calculated surface electrostatic potentials of PC and PEC monomers and molecular structures of dimers of NblA from three organisms: T. vulcanus (Tv), S. elongatus (Se), and Anabena (An). Surface potentials were calculated using the in vacuo approximation of the Pymol system. The distribution of patches of positive or negative potentials are shown in blue and red, respectively. The Tv-PC (1KTP crystal structure), Se-PC, An-PC, and An-PEC (molecular models) are shown from the faces that point outward from the PBS rod substructure. The black ovals show the position of the α-subunit Y-A helices indicated by Bienert et al. (18) as interacting with the An-NblA protein. The round circles indicate the positions of the N and C termini on the crystal structures of the NblA proteins.
The previously suggested model for NblA-PBS interaction (18) indicates that the NblA associates via its termini. Potentials of N and C termini in all proteins are typically positive and negative, respectively. However adjacent residues may modify the magnitude of the potentials. The electrostatic potentials of the three NblA dimeric structures were, thus, calculated to estimate the magnitude of these potentials (Fig. 5, bottom). The Se-NblA structure is the only one that shows the actual C-terminal potential, as the C termini of both Tv-NblA and An-NblA structures are disordered. For all three proteins the C termini create two strong negative potential patches separated by 12-15 Å. The positions of the NblA N termini are also not definitive due to disorder in all of the crystal structures. The most complete N terminus is that of monomer D in the An-NblA structure (18), which starts at the second residue (Fig. 5, bottom right). However, it appears from both the An-NblA and Tv-NblA structures that the N termini have a random coiled structure that extends down toward the C termini and not outward from the main body of the protein. Fig. 5 (bottom) shows that as expected the potentials of the NblA N termini are indeed positive (only one termini is visible in each dimer as depicted in Fig. 5, bottom) The estimated distances between the N and C termini in the different NblA structures are 18-26 Å, and the two N termini are separated by 25-35 Å. The two monomers in the NblA dimers are related by nearly C2 symmetry. Thus, any electrostatic complementarity between PBPs and NblA should exhibit similar symmetry (although with opposite charges), with distances comparable with the distances between termini. Such symmetric positioning of charged patches, with dimensions similar to those of the NblA charges, is absent on the PBP surface shown in Fig. 5. We can, thus, conclude that it is highly unlikely that the NblA termini associate with the outer rod surface through electrostatic complementarity.
If the termini are not sufficient, the NblA could associate through electrostatics via a more extensive surface interaction. The crystallographic results presented above coupled with the Anabaena structure provide a high degree of confidence that although relatively low in sequence homology, NblA proteins preserve a high degree of structural conservation. On this basis we performed homology-based modeling of additional NblA from diverse species such as Gracilaria, Fremyella diplosiphon, and Cyanidioschyzon. We calculated the surface electrostatic potentials of the different NblAs and compared these surfaces from either crystal structures or models (Fig. 5, bottom and supplemental Fig. 1, respectively). As can be readily seen, the surfaces of the NblA have significantly different distributions of electrostatic potentials. On the other hand, comparison of the two major surfaces within a specific group of PBPs shows almost identical distributions of negative and positive potentials. Such inspection was performed for PC (supplemental Fig. 2, showing both faces of three different PC molecules) and for PEC, PE, and APC (data not shown). Thus, although the overall distribution of potentials of the same PBPs from different organisms are similar, those of the NblAs are different, and there are no clearly identifiable conserved electrostatic complementarities between any of putative pair of proteins (for instance PC and NblA). The lack of similarity in the surface characteristics between different members of the NblA further reduces the likelihood that the NblA associates with the PBS components through electrostatic interactions.
Structural Comparison between NblA and Fragments of PBPs
Because the analysis of the NblA and PBP surfaces shows that electrostatics cannot be sufficient to explain NblA function, a different mode must exist. A possible mode of action of NblA could be based on structural similarities to specific configurations of the PBPs. Such structural resemblance could mimic the native PBS structures, thus undermining the interactions stabilizing the PBS complex and assisting in its disassembly and degradation.
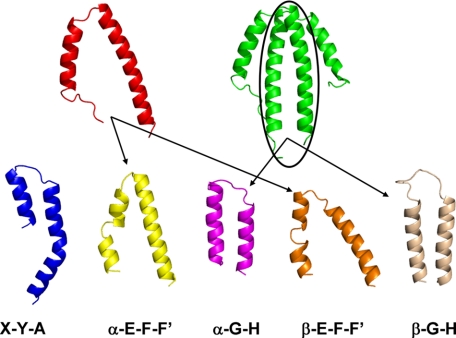
As shown by structure determination, the NblA monomer has a helix-loop-helix structure which dimerizes into a flat four-helical bundle in solution. The putative substrates are the PBPs, all of which are architecturally built up by the folding of a series of α-helices, separated by turns of different lengths and angles. We used the 1KTP crystal structure of PC to try and find possible structural homologies with the Tv-NblA crystal structure. Fig. 6 shows a series of helix-loop-helix motifs, carved out of the 1KTP structure, and shows them in orientations that are similar to that of the NblA monomer or dimer. Different PBPs exhibit a high degree of structural preservation (5), and thus, almost identical motifs can be carved out of APC, PEC, or PE structures. Fragments made up of helices E-F-F′ from both the α and β subunits, respectively, appear to be the most like the NblA monomer, with a similar angle between the helices. The major form of the NblA in vivo may be the dimer seen in all crystal structures. Upon association, each of the NblA monomers contributes a long helix (residues 32-58 in Tv-NblA) that is nearly parallel (with rather even spacing of 6.5-8.5 Å between the two backbones) and that together form a structure that is similar in its dimensions to the A-B-E (not shown) and G-H helices of both α and β-PC (or PE) subunits (Fig. 6). The A-B-E pair is involved in trimer-trimer association (required to enable hexamer assembly). On the basis of crystal structures of isolated components (5) the G-H helices of the β subunit are required for the hexamer-hexamer assembly needed for rod formation by PC and/or PE (see “Discussion”).5
FIGURE 6.
Comparison between Nbla and PC helix-loop-helix fragments. NblA is shown as both monomer (red) and dimer (green). The Tv-PC structure (1KTP) was carved to show five structures which show the same motif; helices X-Y-A (blue), helices G-H (α-subunit in magenta, and β-subunit in wheat), helices E-F-F′ (α-subunit in yellow, and β-subunit in orange). Notice that the two long helices of the NblA dimer form a non-covalently bonded helix-loop-helix motif as well (black circle).
NblA of Anabaena Complements the Non-bleaching Phenotype of NblAΩ of S. elongatus
If the mode of NblA function is structure-dependent as we suggest, it is possible that although NblAs of various organisms share limited primary sequence homology (Fig. 1), they may be exchangeable. To test this hypothesis we cloned the coding region of the chromosomally located NblA of Anabaena PCC 7120 (An-NblA) under the regulatory region of nblA of S. elongatus. This resulted in phycobilisome degradation as evident by culture pigmentation (Fig. 3, E and F (purple)) despite the fact that these two protein sequences share only 26% amino acid similarity (Fig. 1). This supports our proposal that the common helix-loop-helix structure and perhaps the propensity to dimerize as shown by structure determination is crucial for NblA function.
Intriguingly, S. elongatus bearing the An-NblA possesses 70% of the PC amount of the wild type strain when comparing cultures grown in nutrient-replete media (not shown). The reason for this difference remains to be examined; it may stem from the different nature of the foreign NblA or a modified regulation of An-NblA due to the dissimilar cellular context. It is noteworthy that complementation of NblAΩ with Se-NblA in a shuttle vector or in the neutral site exhibited similar PC degradation indicating that the phenotype obtained by the An-NblA expressing strain is not due to its plasmid location.
The Cellular Level of the NblA Protein in S. elongatus Remains Low during Nutrient Starvation
A large variety of methods was employed in an attempt to detect NblA in cellular extracts of S. elongatus including affinity purification and immunoblot analysis of a strain bearing FLAG-tagged NblA (capable of complementation of NblAΩ) and analysis by mass spectrometry of all cellular proteins smaller than 10 kDa. Additionally, isolation of PBSs in the course of starvation followed by mass spectrometry revealed all components of the phycobilisome (including small linker proteins) but not NblA (data not shown). It may be possible that the NblA protein of S. elongatus is rapidly degraded together with the PBS and, therefore, is not accumulated to a detectable level. Previous studies, however, indicated the presence of NblA from Tolypothrix PCC 7601 and Synechocystis PCC 6803 (16, 42, 43); thus, it may be suggested that NblA stability differs between various cyanobacterial strains (see “Discussion”).
DISCUSSION
Proposed Model for NblA Function—The present study has been designed to identify the mechanism by which functional NblA protein disrupts the structure of the PBS during nutrient stress.
Previous studies based on in vitro interaction of NblA with the PBS suggested that the protein interacts directly with rod components (16, 42, 43). The most detailed study was performed by Bienert et al. (18) who identified interactions in vitro with PC and PEC and assigned critical roles for Leu-51 and Lys-53 (numbering according to the Anabaena chromosomal NblA sequence), located at the C terminus of the second helix. Coupled with the S9F mutation identified in S. elongatus (10), these authors proposed that the NblA dimer interacts with the components of the PBS rods via the four protein termini. In the present study we have shown that four additional mutations (Figs. 3 and 4), located on both α-helices, afford the non-bleaching phenotype. Three of the mutations (L18P, R22C, and L33I) are not at the protein termini, thus highlighting the functional importance of NblA residues, which are not confined to the protein termini. None of the newly identified mutations appear to be at a position that would affect the stability of the NblA dimer. Our mutational approach, revealing amino acids crucial for function along the two helices as well as the demonstration that An-NblA is functional in S. elongatus despite the low primary sequence similarity, support the idea that the helix-loop-helix structural motif is essential for function.
As detailed extensively under “Results” (Fig. 5, supplemental Figs. 1 and 2), the electrostatic potentials of both NblA and PBPs are not distributed in a complementary manner. We, thus, propose that the NblA requires a larger interaction surface for complex formation. The NblA is made up of a structural motif very similar to that of the PBPs, the helix-loop-helix (Fig. 6). This similarity could enable structural mimicry, by which the NblA weakens the interactions between PBPs by mimicking the interactions required for rod stability.
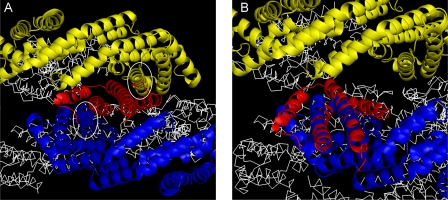
Can the NblA interact with an already formed PBS substructure (rod or core)? If the crystal structures of isolated PBP components represent the assembly of the PBS substructures (1, 5), PC and PE (αβ)3 trimers assemble into (αβ)6 hexamers which associate further into rods. All of the α-helices of the PBP are perpendicular to the direction of the rod, i.e. the surface “seen” by the approaching NblA is that of a rather even cylinder (Fig. 5 shows PC monomers from the side facing outwards from the rod). We have recently determined the structure of a complete PC rod (including the linker proteins) from T. vulcanus which shows that the organization of the PC subunits in the rod is identical to that seen in the high resolution structures obtained by the crystallization of trimers.5 There is, however, one rather large hole in the interface formed by two (αβ)6 hexamers. The interface is formed by four β-subunits that assemble back-to-back, leaving a significant gap into the internal space of the rod (Fig. 7). This is the only orifice large enough to allow the penetration of either NblA monomers or dimers. The NblA dimer is V-shaped, and the proposed model shows penetration of the narrow part of the NblA into the gap. Steric hindrance (between the wider end of the NblA and the rod proteins) only occurs if the NblA penetrates farther into the internal space. In this potential arrangement (using the Tv-PC structure as a template), an NblA dimer (red) has penetrated into the orifice (Fig. 7A) and aligned its two long helices with helices H (residues 155-174) of the β-subunits of two PC associated monomers (blue and yellow), thus interacting with the X-Y loops and perhaps disrupting the interface. Notice that if the hexamer is disrupted, the two long helices of a NblA dimer (residues 32-58) can shift in position and mimic the interaction between helices G-H of the lower PC monomer with helices X-Y of the upper PC-monomer (Fig. 7B). Formation of interactions between the PC and NblA proteins in this mode could lower rod stability and initiate PBS disassembly. Molecular modeling of extended structures of PE, based on the crystallization of hexamers, show an identical hole (data not shown). Thus, the NblA protein could interact with all of the rod components.
FIGURE 7.
Hypothetical models for the mode of NblA-PBS interaction. A rod structure was built using the Tv-PC 1KTP crystal structure and its symmetry-related monomers. The interface between two (αβ)6 hexamers is shown, with an orifice formed between two PC monomers (blue and yellow schematic representation). Other PC monomers are shown in white ribbon for clarity. Panel A, a NblA dimer (red) has wedged itself into the orifice parallel to helix H of both PC monomers (white circles). The rod assembly and the NblA dimer were treated as rigid bodies. Panel B shows how the two long helices of the NblA dimer can mimic the G-H helices of a PC monomer, with the small helices disrupting the interface contacts.
The center of the rod structure is occupied by different linker proteins (7); however, the exact positions of the linker proteins is unknown (7). The single crystal structure that contains a linker protein is of the small core linker (LC7.8), which is entirely enclosed with the trimeric APC hole. There are no apparent appendages that could jut out toward the surface and interact with the NblA. Attempts to visualize the position of the rod linkers by crystallography in PE (44, 45) or by modeling (32) have not provided evidence that the linkers protrude out of the sides of the rod or penetrate into the hole described above. In our aforementioned unpublished rod structure, there is no electron density within the holes formed by association of hexamers, and thus, there is no evidence for linker-NblA interactions.
The proposed model in which the NblA and PBS interact through the structural mimicry of PBP motifs by the NblA is further strengthened by the complementation achieved by the An-NblA protein in the S. elongatus NblA-null mutant. The two proteins have little sequence homology (Fig. 1), and the surface electrostatic potentials are quite different (Fig. 5 and supplemental Fig. 1). However, the structures are almost identical, affording similar possibilities for association of helix-loop-helix motifs.
Two of the mutations identified in this study appear to affect the structural motif critical for function. The L18P mutation could potentially insert a kink in the middle of the N-terminal α-helix, thus substantially modifying the structure required for function. The L33I mutation is at a position that could be critical for proper alignment of the two long α-helices in the dimer. It is not yet clear how the two arginine-to-cysteine mutations affect NblA function. However, we believe on the basis of the distribution of the mutations that the entire NblA protein is involved in PBS disassembly. It is not improbable that certain NblA residues could have additional functional roles beyond association with the PBPs, for example recruitment of NblB or of a protease.
As described above, the level of NblA protein in S. elongatus was below the levels of detection of the methods employed by us. A number of studies have shown that the level of nblA mRNA increases under stress (16, 21, 42). The presence of elevated levels of mRNA does not necessarily indicate the concomitant elevation of levels of protein. Although the PBS complexes (the NblA target) can comprise up to 50% of the total soluble protein in cyanobacterial cells, the amount of NblA protein has not been reported to increase to a level identifiable by Coomassie Blue staining of SDS-PAGE. In Synechocystis sp. PCC 6803, a cyanobacterium with two forms of NblA, low levels of protein were identified by immunoblotting (33). In the filamentous cyanobacterium Tolypothrix PCC 7601, NblA monomers could only be identified immunologically in cells grown in red light (42, 43). In green light-grown cells, a protein of twice the molecular weight of the NblA was labeled by the polyclonal antibodies, and its molecular identity could only be suggested as an SDS resistant dimer. Because there appear to be only small and relatively constant amounts of NblA during the disassembly process, it can be proposed that the NblA protein is rapidly degraded during its activity, and the elevated levels of mRNA provide sufficient levels of newly synthesized NblA to compensate for the rate of its degradation. As indicated above, in S. elongatus, we could not identify the presence of NblA protein by affinity purification, immunoblotting, or mass spectrometry, and we, thus, suggest that in this organism the rate of NblA degradation may be very fast. Low NblA protein levels coupled with its rapid degradation can permit the cells to quickly resume normal growth upon cessation of nutrient starvation.
Previously described experiments performed on S. elongatus cells, in which the ratios of PC to APC and of various linkers were measured after the onset of nutrient starvation (Refs. 2 and 9 and references therein), show that PBS disassembly occurs in an ordered fashion, with the loss of rod hexamers before degradation of the core. Because the circumferences of the core cylinders are covered by the rods, the NblA protein would have to initially induce the removal of the rods to access the core components. At this point the entire PBS could be degraded. The model we present here is consistent with the observation suggesting a sequential degradation (rods before core); however, it does not explain the directional progression of degradation (from rod tips down toward the core). Further biochemical experiments will be required to elucidate these details.
Supplementary Material
Acknowledgments
We gratefully thank the staff of the European Synchrotron Radiation Facility for provision of synchrotron radiation facilities and assistance (beamlines BM30 and ID14-1).
The atomic coordinates and structure factors (codes 2Q8V, 2QDO, and 3CS5) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
This work was supported by the Israel Science Foundation founded by the Israel Academy of Sciences and Humanities (Grants 1045/06 and 683/03-17.1 (to N. A. and R. S., respectively) and German Israel Foundation Grant I-729-27.9/2002 (to R. S.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
Footnotes
The abbreviations used are: PBS, phycobilisome; PBP, phycobiliprotein; APC, allophycocyanin; PC, phycocyanin; PE, phycoerythrin; PEC, phycoerythrocyanin; r.m.s.d., root mean square deviation; Se, S. elongatus PCC 7942; Tv, T. vulcanus; An, Anabaena sp. PCC 7120; r.m.s.d., root mean square deviation; MR, molecular replacement; SAD, single wavelength anomalous dispersion.
N. Adir, unpublished data.
References
- 1.Adir, N., Dines, M., Klartag, M., McGregor, A., and Melamed-Frank, M. (2006) in Microbiology Monographs: Inclusions in Prokaryotes (Shively, J. M., ed) pp. 47-77, Springer-Verlag, Berlin
- 2.Grossman, A. R., Schaefer, M. R., Chiang, G. G., and Collier, J. L. (1993) Microbiol. Rev. 57 725-749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.MacColl, R. (1998) J. Struct. Biol. 124 311-334 [DOI] [PubMed] [Google Scholar]
- 4.Glazer, A. N. (1989) J. Biol. Chem. 264 1-4 [PubMed] [Google Scholar]
- 5.Adir, N. (2005) Photosynth. Res. 85 15-32 [DOI] [PubMed] [Google Scholar]
- 6.Brejc, K., Ficner, R., Huber, R., and Steinbacher, S. (1995) J. Mol. Biol. 249 424-440 [DOI] [PubMed] [Google Scholar]
- 7.Liu, L. N., Chen, X. L., Zhang, Y. Z., and Zhou, B. C. (2005) Biochim. Biophys. Acta 1708 133-142 [DOI] [PubMed] [Google Scholar]
- 8.Reuter, W., Wiegand, G., Huber, R., and Than, M. E. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 1363-1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collier, J. L., and Grossman, A. R. (1992) J. Bacteriol. 174 4718-4726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collier, J. L., and Grossman, A. R. (1994) EMBO J. 13 1039-1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dolganov, N., and Grossman, A. R. (1999) J. Bacteriol. 181 610-617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sendersky, E., Lahmi, R., Shaltiel, J., Perelman, A., and Schwarz, R. (2005) Mol. Microbiol. 58 659-668 [DOI] [PubMed] [Google Scholar]
- 13.Schwarz, R., and Grossman, A. R. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 11008-11013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Waasbergen, L. G., Dolganov, N., and Grossman, A. R. (2002) J. Bacteriol. 184 2481-2490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lahmi, R., Sendersky, E., Perelman, A., Hagemann, M., Forchhammer, K., and Schwarz, R. (2006) J. Bacteriol. 188 5258-5265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li, H., and Sherman, L. A. (2002) Arch. Microbiol. 178 256-266 [DOI] [PubMed] [Google Scholar]
- 17.Baier, K., Lehmann, H., Stephan, D. P., and Lockau, W. (2004) Microbiology 150 2739-2749 [DOI] [PubMed] [Google Scholar]
- 18.Bienert, R., Baier, K., Volkmer, R., Lockau, W., and Heinemann, U. (2006) J. Biol. Chem. 281 5216-5223 [DOI] [PubMed] [Google Scholar]
- 19.Apt, K. E., Collier, J. L., and Grossman, A. R. (1995) J. Mol. Biol. 248 79-96 [DOI] [PubMed] [Google Scholar]
- 20.Strauss, H., Misselwitz, R., Labudde, D., Nicklisch, S., and Baier, K. (2002) Eur. J. Biochem. 269 4617-4624 [DOI] [PubMed] [Google Scholar]
- 21.Richaud, C., Zabulon, G., Joder, A., and Thomas, J. C. (2001) J. Bacteriol. 183 2989-2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dines, M., Sendersky, E., Schwarz, R., and Adir, N. (2007) J. Struct. Biol. 158 116-121 [DOI] [PubMed] [Google Scholar]
- 23.Leslie, A. G. W. (1992) Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography No. 26.
- 24.Otwinowski, Z. (1993) in Data Collection and Processing (Sawyer, L., Isaacs, L., and Baily, S., eds) SERC Daresbury Laboratory, Daresbury, UK
- 25.Schuermann, J. P., and Tanner, J. J. (2003) Acta Crystallogr. D Biol. Crystallogr. 59 1731-1736 [DOI] [PubMed] [Google Scholar]
- 26.McCoy, A. J., Grosse-Kunstleve, R. W., Storoni, L. C., and Read, R. J. (2005) Acta Crystallogr. D Biol. Crystallogr. 61 458-464 [DOI] [PubMed] [Google Scholar]
- 27.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T., and Warren, G. L. (1998) Acta Crystallogr. D. Biol. Crystallogr. 54 905-921 [DOI] [PubMed] [Google Scholar]
- 28.McRee, D. E., and David, P. R. (1999) Practical Protein Chemistry, 2nd Ed., Academic Press, Inc., New York
- 29.Emsley, P., and Cowtan, K. (2004) Acta Crystallogr. D. Biol. Crystallogr. 60 2126-2132 [DOI] [PubMed] [Google Scholar]
- 30.Perelman, A., Shaltiel, J., Sendersky, E., and Schwarz, R. (2004) Biotechniques 36 948-950 and 952 [DOI] [PubMed] [Google Scholar]
- 31.Schwede, T., Kopp, J., Guex, N., and Peitsch, M. C. (2003) Nucleic Acids Res. 31 3381-3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parbel, A., and Scheer, H. (2000) Int. J. Photoenergy 2 31-40 [Google Scholar]
- 33.Baier, K., Nicklisch, S., Grundner, C., Reinecke, J., and Lockau, W. (2001) FEMS Microbiol. Lett. 195 35-39 [DOI] [PubMed] [Google Scholar]
- 34.CCP4 Acta Crystallogr. D Biol. Crystallogr. (1994) 50 760-763 [DOI] [PubMed] [Google Scholar]
- 35.McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C., and Read, R. J. (2007) J. Appl. Crystallogr. 40 658-674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adir, N., Dobrovetsky, Y., and Lerner, N. (2001) J. Mol. Biol. 313 71-81 [DOI] [PubMed] [Google Scholar]
- 37.Adir, N., Vainer, R., and Lerner, N. (2002) Biochim. Biophys. Acta 1556 168-174 [DOI] [PubMed] [Google Scholar]
- 38.Murshudov, G. N., Vagin, A. A., and Dodson, E. J. (1997) Acta Crystallogr. Sect. D 53 240-255 [DOI] [PubMed] [Google Scholar]
- 39.Sheldrick, G. M. (1998) in Direct Methods for Solving Macromolecular Structures (Fortier, S., ed) pp. 131-142, Kluwer Academic Publishers, Dordrecht, The Netherlands
- 40.Contreras-Martel, C., Martinez-Oyanedel, J., Bunster, M., Legrand, P., Piras, C., Vernede, X., and Fontecilla-Camps, J. C. (2001) Acta Crystallogr. D Biol. Crystallogr. 57 52-60 [DOI] [PubMed] [Google Scholar]
- 41.Schmidt, M., Krasselt, A., and Reuter, W. (2006) Biochim. Biophys. Acta 1764 55-62 [DOI] [PubMed] [Google Scholar]
- 42.Luque, I., Ochoa De Alda, J. A., Richaud, C., Zabulon, G., Thomas, J. C., and Houmard, J. (2003) Mol. Microbiol. 50 1043-1054 [DOI] [PubMed] [Google Scholar]
- 43.de Alda, J. A., Lichtle, C., Thomas, J. C., and Houmard, J. (2004) Microbiology 150 1377-1384 [DOI] [PubMed] [Google Scholar]
- 44.Ficner, R., Lobeck, K., Schmidt, G., and Huber, R. (1992) J. Mol. Biol. 228 935-950 [DOI] [PubMed] [Google Scholar]
- 45.Ritter, S., Hiller, R. G., Wrench, P. M., Welte, W., and Diederichs, K. (1999) J. Struct. Biol. 126 86-97 [DOI] [PubMed] [Google Scholar]
- 46.Yeates, T. O. (1997) Methods Enzymol. 276 344-358 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.