Abstract
GbpC is a large multidomain protein involved in cGMP-mediated chemotaxis in the cellular slime mold Dictyostelium discoideum. GbpC belongs to the Roco family of proteins that often share a central core region, consisting of leucine-rich repeats, a Ras domain (Roc), a Cor domain, and a MAPKKKinase domain. In addition to this core, GbpC contains a RasGEF domain and two cGMP-binding domains. Here, we report on an intramolecular signaling cascade of GbpC. In vitro, the RasGEF domain of GbpC specifically accelerates the GDP/GTP exchange of the Roc domain. Moreover, cGMP binding to GbpC strongly stimulates the binding of GbpC to GTP-agarose, suggesting cGMP-stimulated GDP/GTP exchange at the Roc domain. The function of the protein in vivo was investigated by rescue analysis of the chemotactic defect of gbpC null cells. Mutants that lack a functional guanine exchange factor (GEF), Roc, or kinase domain are inactive in vivo. Together, the results suggest a four-step intramolecular activation mechanism of the Roco protein GbpC: cGMP binding to the cyclic nucleotide-binding domains, activation of the GEF domain, GDP/GTP exchange of Roc, and activation of the MAPKKK domain.
Extracellular cAMP is a chemoattractant for Dictyostelium cells. Upon binding of cAMP to surface receptors, several signaling cascades are activated that cause cells to crawl toward the source of cAMP (1, 2). Some of the signaling molecules involved in chemotaxis are the second messengers phosphatidylinositol 1,4,5-trisphosphate, inositol 1,4,5-trisphosphate, Ca2+, and the product(s) of PLA2,2 as well as the cyclic nucleotides cAMP and cGMP (2-4). These second messengers have important roles in transducing signals that lead to actin polymerization at the front of the cell (5) and phosphorylation of myosin in the back of the cell, which are vital for cells to move (6, 7). One of the second messengers that have been implicated in myosin regulation is cGMP (8, 9). Recently, the proteins that are involved in the formation and degradation of cGMP have been identified and characterized (4, 10). Binding of extracellular cAMP to the surface cAMP receptor cAR1 causes a G-protein-dependent activation of two guanylyl cyclases, soluble guanylyl cyclase (sGC) and membrane-bound guanylyl cyclase (GCA). These two enzymes are responsible for the rapid synthesis of cGMP in cells, whereas three cGMP-degrading enzymes, PDE3, PDE5, and PDE6, cause a subsequent decrease of cGMP in the cells, back to basal levels.
In Dictyostelium, the function of cGMP is transduced by the target protein GbpC. Disruption of the gene coding for GbpC yields a cell line that has impaired regulation of myosin II as well as defects in chemotaxis, comparable with cells that lack both guanylyl cyclases (9). GbpC was shown to contain two cyclic nucleotide-binding domains that are able to bind cGMP with high affinity. Furthermore, this protein has many other domains: leucine-rich repeats and domains that have homology to Ras-GTPases (Roc for Ras of complex proteins), Cor (for C terminus of Roc), MAPKKK, RasGEF, GRAM (a domain in glucosyltransferases, myotubularins, and other putative membrane-associated proteins), and DEP (a domain found in Dishevelled, Egl-10, and Pleckstrin). Based on its domain topology, GbpC was categorized as a member of the Roco family of proteins that are characterized by conserved Roc and Cor domains, often in addition to LRR and MAPKKK (11). GbpC is an unusual member of the Roco family, because it also contains additional RasGEF and cNBDs. The Roco family gained attention recently, when the human Roco protein LRRK2 was found to be involved in Parkinson disease (12).
Ras proteins switch between an inactive GDP-bound state and an active GTP-bound state and can interact with downstream effectors only in the active GTP-bound state. Activation of Ras is mediated by guanine exchange factors (GEFs), which catalyze the exchange of GDP for GTP (13). Among the best characterized downstream effectors of Ras are MAPKKKs (14). For the human Roco protein LRRK2 it has been shown that GTP binding to Roc enhances kinase activity of the MAPKKK domain, which is the presumed output of LRRK2 and the homologous protein LRRK1 (15, 16). So far, it is unclear whether GEFs are necessary to regulate GDP/GTP exchange on the Roc domain of LRRK2. In potential, GbpC has all the domains to provide a complete intramolecular signaling pathway. In this study, the activation mechanism of GbpC was investigated. The results lead to an activation model in which cGMP binding to the cNBDs causes activation of the GEF domain, the subsequent GDP/GTP exchange of the Roc domain, and activation of the MAPKKK domain.
EXPERIMENTAL PROCEDURES
Strains and Cell Culture—We made a new gbpC null cell line in AX3 background, because this facilitates direct comparison with other null mutants in AX3 background. This new cell line was made with the same constructs and strategy as described before for the gbpC null strain in DH1 (9). The clones were examined for correct integration of the knockout construct in the gbpC gene with Southern blotting and PCR (data not shown) and verified in a cGMP binding assay, as described before (17). Wild-type AX3 cells and gbpC null cells were grown in HG5 medium to a maximum density of about 5 × 106 cells/ml. Cells expressing GbpC-derived proteins were grown in the presence of the appropriate selection marker, which was G418 for most constructs but hygromycin for the GbpC-ΔcGMP construct.
Cloning of gbpC Constructs—The complete gbpC cDNA was amplified as domain modules with the following strategy. The 7.9-kb gene was divided in eight DNA parts of approximately equal size, with borders to assure that the various domains of GbpC were completely covered in a specific DNA part. Unique restriction sites were introduced in the PCR primers at the border of two DNA parts (see Fig. 2A). This strategy had several advantages. First, the gene is almost 8 kb long, therefore making it difficult to PCR the complete gbpC cDNA at once. Second, using mutagenesis of one DNA module, it is relatively easy to study the function of specific domains within the complete GbpC protein. Third, another advantage comes from the possibility to express the smaller parts separately, so the various domains of GbpC can be studied by mutagenesis more easily. Primers were designed for each part, which include several sequence elements (see supplemental Table S1 and Fig. 2A). Most forward primers (from 5′to 3′) consisted of a BglII or BamHI site (for cloning in expression plasmids), followed by a Kozak sequence, a start codon, a unique restriction site in the gbpC cDNA (sometimes created by silent mutation), and gbpC cDNA. The reverse primers (from 3′to 5′) generally consisted of gbpC cDNA, a unique restriction site in the gbpC cDNA (sometimes created by silent mutation), a SpeI or XbaI site (for cloning in expression plasmids), and a stop codon. All PCR products were ligated in pGemTeasy (Promega). The separate parts were fused with each other in the correct order with the unique restriction sites. After joining parts 1-5, this 4.9-kb fragment was cut out of pGemTeasy with BamHI/SpeI and ligated into BglII/SpeI-digested MB74-derived plasmid. This resulting plasmid was subsequently digested with NruI/SpeI and joined with the 3-kb fragment of part 6-8 (cut with NruI/XbaI from pGemTeasy), yielding the full gbpC open reading frame. Different MB74-derived plasmids were used to obtain expression plasmids with different selection markers (G418 or hygromycin) or the presence of a C-terminal GFP tag. Mutations were created by site-directed mutagenesis, and the DNA fragments were exchanged with gbpC cDNA using the appropriate unique restrictions enzymes and cloned in a similar way as the original gbpC construct. All PCR products were sequenced to assure mutation-free constructs. Expression and folding of all proteins was monitored using cGMP binding assays (see below), by fluorescent microscopy and Western blotting, using anti-GFP antibodies (Santa Cruz), and by assaying GTP binding, using the GTP-agarose pulldown assay.
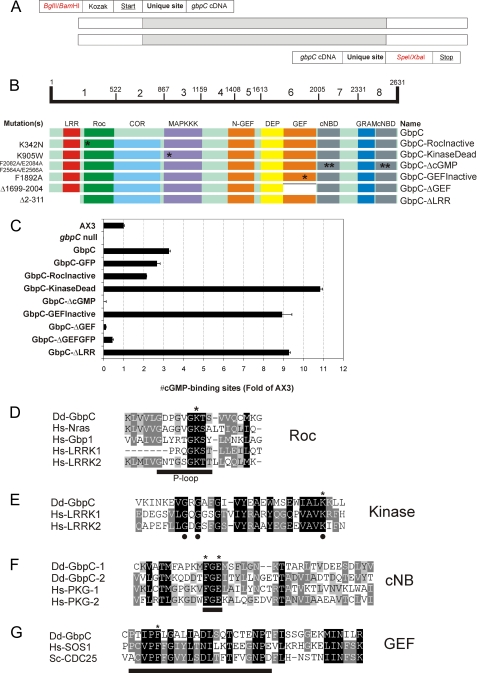
FIGURE 2.
Primer design, GbpC mutants and alignments. The gbpC open reading frame was amplified by PCR from cDNA in eight parts. The used primers consisted of the elements that are shown in A. The forward primer (5′-3′) contains a BglII or BamHI site for cloning in MB74 plasmids (in red), followed by a Kozak-sequence, a start codon (underlined), and a unique restriction site in the gbpC cDNA, which can be used for fusion of various parts. In the reverse primer (3′-5′), a unique site in the gbpC cDNA is followed by a SpeI or XbaI site for cloning in MB74-plasmids (in red) and a stop codon (underlined). B, overview of the GbpC domain structure and the constructs used in this study. The amino acid borders of eight parts of GbpC are indicated by numbers on the bar. The mutations and truncations of used constructs (left) and their corresponding names (right) are shown. The locations of point mutations are highlighted by asterisks. C, expression of GbpC mutants. The cytosols of AX3, gbpC null cells, and gbpC null cells expressing various GbpC-derived proteins were assayed for high affinity cGMP-binding sites, using 10 nm [3H]cGMP. The amount of binding sites is indicative for expression levels and protein stability and presented as a comparison with the amount of binding sites in AX3 cells. cGMP binding was determined in triplicate, and the assay was repeated at least once during the time course of this study to verify stable expression levels throughout the whole study. The average values ± S.D. are presented. D, alignment of the functionally important P-loop in the Roc domain of GbpC with a selection of other GTPases. The P-loop is underlined. A conserved lysine is indicated with an asterisk and was mutated to an asparagine to inactivate the nucleotide-binding abilities of the Roc domain. E, alignment of the first 31 amino acids of the kinase domain of GbpC with those of human LRRK1 and human LRRK2. Residues with dots are thought to be essential for ATP binding. A conserved lysine is indicated with an asterisk and was mutated to a tryptophan. F, alignment of part of the cyclic nucleotide-binding domains of GbpC and human protein kinase G. Underlined is the conserved FGE motif that is thought to be important for cGMP binding. The phenylalanines and glutamic acids of this motif (indicated with asterisks) were mutated to alanines in both cNBDs of GbpC to create a GbpC mutant that was unable to bind cGMP. G, alignment of part of the RasGEF domain of GbpC with human SOS1 and yeast CDC25. Underlined is a helical hairpin that is thought to be important for activity. A conserved phenylalanine is indicated with an asterisk and was mutated to an alanine. Dd, D. discoideum; Hs, H. sapiens.
cGMP Binding Assay—The assay was performed essentially as described before (18). Briefly, the cells were resuspended in lysis buffer (40 mm HEPES/NaOH, pH 7.0, 0.5 mm EDTA, 0.5 mm EGTA, and 10 mg/ml crushed protease inhibitor mixture tablet; mini Complete-tablets, EDTA-free; Roche Applied Science) and lysed using Nuclepore filters. The lysate was precleared by centrifugation for 3 min at 14,000 × g and an additional 5 min at 14,000 × g. Next, 200 μl of lysate was incubated with 200 μl of binding mix (40 mm HEPES/NaOH, pH 7.0, 0.5 mm EGTA, 6 mm MgCl2, and 20 nm [3H]cGMP) for 20 min on ice and subsequently filtered through 0.45 μm of nitrocellulose. The filters were washed twice with 3 ml of ice-cold wash buffer (40 mm HEPES/NaOH, pH 7.0, 0.5 mm EDTA), dried, and analyzed by liquid scintillation counting. Nonspecific binding of [3H]cGMP was determined in the presence of 1 mm cGMP. GbpC-binding sites were determined by subtracting the remaining binding to gbpC null cell lysates from cGMP binding to all other lysates and normalizing the numbers to the amount of binding sites in AX3.
Chemotaxis Assay—Chemotaxis was measured with the small population assay (19). The experiments were performed in the wells of a six-well plate with 1 ml of agar non-nutrient hydrophobic agar (11 mm KH2PO4, 2.8 mm Na2HPO4, 7 g/liter hydrophobic agar) containing 2 μm p-bromophenacyl bromide and 50 μm LY294002 to inhibit PLA2 and phosphatidylinositol 3-kinase, respectively (20). Droplets of about 0.1 μl of 7-h starved cells (6 × 106 cells/ml) were placed on the agar. Chemotaxis toward cAMP was tested after 30 min by placing a second 0.1-μl droplet, with 1 μm cAMP, next to the droplet of cells. The distribution of the cells in the droplet was observed about every 10 min for 90 min and scored positive when at least twice as many cells were pressed against the side of the population closer to higher cAMP concentration as against the other side of the droplet. Recorded is the fraction of droplets scored positive, averaged over three successive observations at and around the moment of the maximal response. The data presented are the means and standard error of the means of at least three independent measurements on different days.
Purification of Proteins—The Roc-Cor fragment of GbpC (amino acids 326-880) was expressed as glutathione S-transferase fusion protein from the pGEX4T-3 (GE Healthcare) vector in Bl21(DE3)codonplus-RIL Escherichia coli cells (Stratagene). The cells were grown in TB medium (Merck) containing 50 μg/ml ampicillin and 25 μg/ml chloramphenicol, induced at an A600 of 0.8 with 0.1 mm isopropyl-1-thio-β-d-galactopyranoside, and incubated overnight at 25 °C. After protein production, the cells were pelleted (15 min, 4000 × g, and 4 °C), and resuspended in lysis buffer (5 mm dithioerythritol, 50 mm NaCl, 5 mm MgCl2, 50 mm Tris-HCl, pH 7.9, 1 mm GDP). To inhibit protein degradation, 1 mm of phenylmethylsulfonyl fluoride was added. The cells were lysed in a microfluidizer (Microfluidizer Inc.), and 0.1 mg/ml DNase was added. The lysates were cleared by centrifugation (45 min, 100,000 × g at 4 °C), and fusion proteins were purified using a GSH affinity column (GE Healthcare). The glutathione S-transferase tag was cleaved on the column using 200 units of thrombin (SERVA), followed by elution of the protein in lysis buffer and further purification by size exclusion chromatography (Superdex 75 16/20; GE Healthcare). The RasGEF domain was cloned into a pRSETB vector and expressed in Rosetta DE3 bacteria (Merck) as an N-terminal His tag fusion protein. The 63-kDa protein was purified by nickel affinity chromatography, followed by size exclusion chromatography (Superdex 200 16/20; GE Healthcare). The Dictyostelium Ras proteins were purified as previously described (21). Isolated proteins were analyzed using SDS-PAGE, and the protein concentration was determined by the method of Bradford (Bio-Rad).
Guanine Nucleotide Exchange Assays—For measuring GEF activity, the GTP-binding proteins were first incubated at room temperature with a 20-fold excess of the fluorescent GDP analog 2′/3′-O-(N′-methylanthraniloyl)-guanosine-diphosphate (mGDP), in the presence of 10 mm EDTA (22). The mGDP-loaded GTPases were incubated at 25 °C in assay buffer (50 mm Tris-HCl, pH 7.5, 5 mm MgCl2, 50 mm NaCl and 5 mm dithioerythritol), containing 200-fold excess of unlabeled GDP, with the indicated amounts of GEF protein. The stability of the protein was controlled by incubating the mGDP-loaded protein in assay buffer, under the same conditions. The intrinsic exchange activity was measured by incubating the mGDP-loaded G-protein with an excess of unlabeled GDP. The nucleotide exchange was measured in real time as decay in fluorescence using a Spex2 spectrofluorometer (Spex Industries), with excitation and emission wavelengths of 366 and 450 nm, respectively. The obtained data were fitted to a single exponential decay, using the program Grafit (Erithacus Software).
GTP Binding Assay—gbpC null cells expressing GbpC-GFP were washed with lysis buffer (100 mm Tris-HCl, pH 7.4, 50 mm KCl, 1 mm EDTA, 1 mm dithiothreitol, 5 mm MgCl2) and resuspended in lysis buffer, supplemented with 1 mg/ml of crushed protease inhibitor mixture tablets (Roche Applied Science). Cell lysis occurred by passage through a 3.0-μm Nuclepore filter at 4 °C. The lysate was then cleared by a 15-min centrifugation step at 14,000 × g, and the supernatant was used for the GTP-agarose pulldown assay. GTP-agarose beads (40 μl of slurry/reaction; Sigma) were preincubated with bovine serum albumin (100 μg/ml) for 30 min to prevent nonspecific binding, and after washing with lysis buffer, 400 μl of lysate was incubated with the beads in the absence or presence of the indicated amounts of cGMP and GTP for 45 min at 4 °C, under gently turning. Next, the beads were collected by centrifugation and washed two times with lysis buffer. Elution of proteins occurred by incubation for 5 min at 80 °C in SDS loading dye, and the amount of eluted GbpC-GFP was visualized on a Western blot.
Western Blotting—Boiled cells or cell extracts were loaded on 7.5% precast gels (Bio-Rad), separated by SDS-PAGE, and transferred to polyvinylidene difluoride membranes (Invitrogen) that were soaked in transfer buffer (39 mm glycine, 48 mm Tris-HCl, 0.037% SDS, and 20% (v/v) methanol, pH 8.3). After washing three times with TBST (25 mm Tris-HCl, 137 mm NaCl, 5 mm KCl, 0.05% Tween, pH 8.0), the membranes were blocked for 30 min with blocking buffer (5% (w/v) milk powder in TBST), and subsequently incubated with B-2 anti-GFP antibody (Santa-Cruz, 1:500 dilution in blocking buffer) for 2 h at room temperature or overnight at 4 °C. The membranes were washed three times again and incubated with horseradish peroxidase-coupled goat anti-mouse antibody (Santa-Cruz, 1:2000 dilution in blocking buffer) for 1.5 h at room temperature. The bound antibodies were visualized using a chemiluminiscence kit (Lumilight; Roche Applied Science).
RESULTS
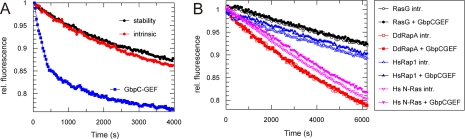
The RasGEF Domain of GbpC Specifically Activates the Roc Domain in Vitro—RasGEFs activate Ras proteins by catalyzing the exchange of GDP to GTP. In Dictyostelium, the RasGEF domain of GbpC is one of the at least 25 RasGEFs that were identified by computer analysis (23). GbpC, also referred to as gef-T, is the only protein in Dictyostelium known so far that possesses both a RasGEF domain and a Ras-like domain called Roc, making it an excellent candidate for intramolecular Ras-GEF-mediated Ras activation. To investigate the potential activation of Roc by the RasGEF domain, we performed in vitro fluorescence nucleotide exchange assays (Fig. 1A). For these experiments, the Roc-Cor fragment of GbpC was expressed in E. coli and isolated as a 63-kDa protein. The isolated Roc-Cor fragment was loaded with the fluorescence GDP analog mGDP and incubated with excess GDP in the presence or absence of a GEF. Nucleotide exchange was measured in real time as the decay of fluorescence, caused by the release of mGDP. As a control, the stability of the protein was measured by incubating the mGDP-loaded protein without GDP, and the subsequent release of mGDP followed, using the same conditions. In the presence of GDP but the absence of GEF, the fluorescence slowly decreases, indicating that the Roc domain has a slow intrinsic exchange activity, like many G-proteins. The RasGEF domain of GbpC, comprising amino acids 1410-2008, was expressed in E. coli and isolated as an N-terminal His-tagged protein. The addition of GbpC-GEF to mGDP-loaded Roc-Cor results in the rapid decrease of fluorescence, demonstrating that the RasGEF domain of GbpC possesses nucleotide exchange activity on the Roc domain. Next, we tested the specificity of this RasGEF/Roc interaction by performing the same assay with several other purified Ras proteins. The addition of GbpC RasGEF to mGDP-loaded Dictyostelium RasG and RapA, and human N-Ras and Rap1 did not result in a decrease in fluorescence (Fig. 1B). GbpC-GEF was also unable to accelerate nucleotide exchange on RasB, RasD, or RasS (data not shown). The Ras and Rap preparations used are activated by CDC25 and C3G, respectively, indicating that purified proteins are stable and functional (21). CDC25 and C3G did not activate nucleotide exchange of mGDP loaded Roc-Cor (data not shown). These results indicate that RasGEF of GbpC specifically activates Roc.
FIGURE 1.
The Roc domain of GbpC is activated by the GbpC-RasGEF domain. A, the purified Roc-Cor fragment of GbpC was loaded with the fluorescent GDP analog mGDP, and the decay of fluorescence was followed over time in the presence of excess GDP. The stability of the protein was measured by incubating the mGDP-loaded protein in assay buffer without further additions. The intrinsic exchange activity was measured by incubating the mGDP-loaded protein with an excess of unlabeled GDP. The addition of the purified RasGEF domain of GbpC causes a dramatic increase in the exchange rate, meaning that GbpC uses its own RasGEF to activate the Roc domain. B, same assay as in A, but the Roc-Cor fragment of GbpC was replaced by several other GTPases. Intrinsic exchange rates and exchange rates in the presence of GbpC-RasGEF are shown. No differences were found upon the addition of the GbpC-RasGEF domain, confirming specificity of GbpC-RasGEF for its own Roc. All of the exchange assays were repeated at least twice on different days, yielding similar results. Dd, Dictyostelium discoideum; Hs, Homo sapiens.
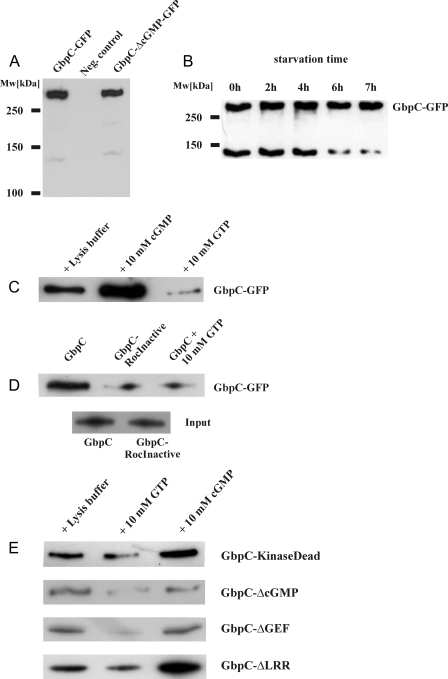
Expression of GbpC Mutants—To characterize the activation mechanism of GbpC in vitro and in vivo, we cloned the complete 7.9-kb gbpC cDNA in MB74-derived Dictyostelium expression plasmids. Furthermore, we expressed several mutated and truncated GbpC-derived proteins in gbpC null cells (Fig. 2B). All of the strains expressing mutant GbpC proteins were tested for the presence of high affinity binding sites in a radio nucleotide binding assay, using 10 nm [3H]cGMP (Fig. 2C). Because GbpC is the only high affinity cGMP-target in Dictyostelium, comparison of the amount of these cGMP-binding sites between cell lines gives an accurate indication about the expression level of GbpC. Moreover, cGMP binding assures correct folding of at least the cNBDs of the protein. Expression and correct protein folding was further monitored with GFP fluorescence, Western blotting, and binding to GTP-agarose. Expression of GbpC in gbpC null cells results in a [3H]cGMP binding activity that is about 3-fold higher compared with wild-type AX3 cells (Fig. 2C). Expression of GbpC, fused to a C-terminal GFP tag, can also be monitored on a Western blot with anti-GFP antibodies: a ∼320-kDa band is visible, as well as a smaller band of ∼140 kDa, probably representing a degradation product (Fig. 3A). At extended starvation times, the presence of this product gradually decreases (Fig. 3B). The functional activity of GbpC and GbpC mutants was tested in vitro for their ability to bind GTP and in vivo to restore the chemotactic defects of gbpC null cells.
FIGURE 3.
Roc-mediated GTP-binding by GbpC is specific and is stimulated by cGMP. A and B, full-length GbpC-GFP (∼320 kDa) and GbpC-ΔcGMP-GFP, expressed in gbpC null cells, can be visualized by Western blotting. A truncated protein (∼140 kDa) is also visible, and its presence is developmentally regulated. Equal amounts of cells were taken at several time points during starvation. The cells were boiled in SDS loading buffer and resolved by SDS-PAGE, and GbpC-GFP expression was visualized with Western blotting, using anti-GFP antibodies. C, GbpC-GFP was expressed in gbpC null cells and pulled down from lysates with GTP-agarose beads in the presence of equal volumes of lysis buffer, cGMP, or GTP, respectively. Bound protein was visualized on a Western blot, using anti-GFP antibodies. D, GbpC and the GbpC-RocInactive mutant with an inactive Roc domain were pulled down with GTP-agarose. The results show very little binding of GbpC-RocInactive to GTP-agarose. The inputs are shown as a control, indicating equal expression levels of both constructs. The proteins were visualized on a Western blot, using anti-GFP antibodies. E, cGMP stimulates GTP binding in mutants with abolished kinase or LRR domains but does not stimulate GTP binding in a mutant that is unable to bind cGMP or in a mutant with a deletion of the catalytic part of the RasGEF domain. GFP-fused GbpC mutants were pulled down with GTP-agarose as in A and visualized with anti-GFP antibodies on a Western blot. Representatives of at least three independent experiments on different days are presented.
cGMP Binding to GbpC Activates the Roc Domain—We hypothesized that cGMP binding to the cyclic nucleotide-binding domains (cNBDs) of GbpC could lead to the activation of GbpC. This activation could occur through direct activation of the kinase domain or via an intramolecular signaling cascade, involving Roc activation through RasGEF. In the latter case, this would mean that the GDP/GTP exchange activity of Roc should increase upon cGMP binding, resulting in an increased amount of Roc in the active GTP-bound state. To investigate the role of cGMP binding in protein function, we incubated lysates of gbpC null cells expressing GbpC-GFP with GTP-agarose beads. The amount of GbpC-GFP bound to these beads is a marker for the activation state of the Roc domain (16, 24). After incubation of the lysate with GTP-agarose beads, the beads were extensively washed, and bound GbpC-GFP was eluted and detected by Western blot (Fig. 3C). Similarly to LRRK1 and LRRK2, we observed that small but significant amounts of GbpC-GFP could be affinity-purified with GTP-agarose beads, but not in the presence of an excess GTP, suggesting a specific GTP-interaction. Interestingly, the addition of cGMP to the incubation mixture causes a 2-3-fold increase of GbpC binding to the GTP beads, suggesting that cGMP binding to GbpC-GFP causes activation of the Roc domain (Fig. 3C).
GTP Binding to the Roc Domain Is Required for GbpC Activity—To assess the importance of the Roc domain in the activity of GbpC, we expressed in gbpC null cells a GbpC mutant that was predicted to be unable to bind GDP and GTP. In LRRK1, mutation of Lys650 caused a dramatic decrease in nucleotide affinity by the Roc domain, thus making the domain inactive (16). Other GTPases with the same mutation were also shown to have the same defect, which is explained by the inactivation of the essential P-loop that is located in the nucleotide-binding pocket (25). An alignment of the Roc domain of GbpC with these domains shows that the same conserved lysine is present at position 342, thus serving as a good candidate to inactivate the Roc domain (Fig. 2D). Therefore, we created a K342N mutation in GbpC and called this mutant GbpC-RocInactive. To verify the loss of nucleotide binding capacity, a Roc domain with this mutation was purified from E. coli, analogous to the purification of the Roc domain that we used for the exchange assay in Fig. 1. Contrary to the wild-type Roc domain, it was impossible to load the mutated Roc domain with GDP or GTP, confirming that the mutation indeed abolished the capacity of the Roc domain to bind these nucleotides (data not shown). To investigate the role of the Roc domain for binding to GTP beads, we performed the GTP-agarose pulldown assay with GbpC-RocInactive (Fig. 3D). Both GbpC-GFP and GbpC-Roc-InactiveGFP were expressed at similar levels in gbpC null cells. In contrast to GbpC-GFP, it was not possible to specifically pull down GbpC-RocInactiveGFP, confirming that GTP binding is mediated by the Roc domain.
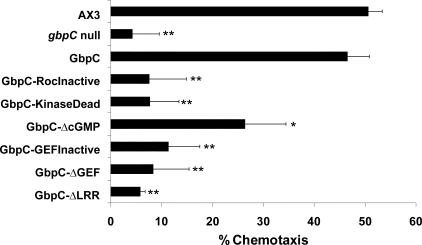
GbpC plays a critical role in cAMP-induced chemotaxis of Dictyostelium cells, which was measured by a small population/drop assay (19). In this assay, starved cells are placed on NN-agar in small drops, close to drops containing cAMP, and observed for the ability to chemotax toward these cAMP drops. Chemotaxis of 7 h starved Dictyostelium cells is mediated by at least three signaling pathways: phosphatidylinositol 3-kinase, PLA2, and a cGMP pathway (20). Wild-type cells show around 50% chemotaxis, when assayed with 10-6 m cAMP, in the presence of the PI3K inhibitor LY and the PLA2 inhibitor p-bromophenacyl bromide. Recently, it was found that this remaining chemotaxis is abolished in cells that have disruptions in genes that encode guanylyl cyclases or GbpC (20). Here, we performed a rescue analysis by expressing GbpC or GbpC mutants in gbpC null cells (Fig. 4). Expression of GbpC in gbpC null cells restores chemotaxis to approximately wild-type levels. In contrast, expression of the GbpC-RocInactive mutant in gbpC null cells was unable to rescue the chemotaxis defect of gbpC null cells (Fig. 4), although the mutant was expressed at a similar level as GbpC (Fig. 2C). This suggests that a functional Roc domain is essential for a functional GbpC protein in vivo.
FIGURE 4.
Chemotaxis data of GbpC mutants. Seven-hour starved cells were analyzed in a small population assay and scored for their ability to chemotaxis toward drops with 10-6 m cAMP in the presence of the PI3K inhibitor LY and the PLA2 inhibitor p-bromophenacyl bromide. GbpC mutants were expressed in gbpC null cells and compared with the chemotaxis data of AX3 and gbpC null cells. The data presented are the means and standard error of the means of at least three independent measurements on different days. The data were analyzed by Student's t test; **, p < 0.001 versus GbpC and not significantly different from gbpC null at p > 0.05; *, significantly less than GbpC at p < 0.05 and significantly above gbpC null at p < 0.01.
A Functional Kinase Domain Is Essential for GbpC Activity—Active kinase domains have a fully conserved lysine in their activation loops. Mutating this lysine into a tryptophan results in a kinase domain that is unable to bind ATP, thus making it inactive (16, 26, 27). The corresponding mutation in GbpC is K905W (Fig. 2E). Proper expression of this kinase dead mutant in gbpC null cells was verified with a cGMP binding assay, showing an expression level that was about 3-fold more compared with expression of GbpC in gbpC null cells (Fig. 2C). In chemotaxis assays, similar results were found as for the Roc-Inactive mutant; the cells were unable to chemotax, suggesting that the kinase domain is also essential for GbpC to be functional in vivo (Fig. 4). To test whether the kinase domain lies upstream or downstream of the Roc domain in a signaling cascade through GbpC, a GTP-agarose pulldown assay was performed, showing that cGMP-stimulated Roc activation was still present in this mutant, as was seen for the wild-type protein (Fig. 3E). This shows that the Roc domain lies upstream of the kinase domain.
The RasGEF Domain Is Essential for GbpC Activity and Mediates cGMP-stimulated Roc Activation—To investigate the role of the Ras-GEF domain of GbpC, we inactivated the GEF domain in GbpC by deleting the catalytic part of the GEF domain (Fig. 2B), thus assuring that no GEF-mediated GDP/GTP exchange on the Roc domain could occur. This mutant, called GbpC-ΔGEF, was still able to bind GTP specifically, confirming that GTP binding by the Roc domain can occur independently of the RasGEF domain (Fig. 3E). However, stimulated Roc activation by cGMP could not be found for this mutant, strongly suggesting that cGMP binding to the cNBDs causes the RasGEF domain to catalyze GDP/GTP exchange at the Roc domain. The importance of the RasGEF domain is underlined in the observation that the GbpC-ΔGEF mutant is inactive in vivo in the chemotaxis assay (Fig. 4). We observed that cells expressing GbpC-ΔGEFGFP exhibit good fluorescence intensity comparable with cells expressing wild-type GbpC-GFP but that cGMP binding to the lysates of this mutant was lower compared with the wild-type protein (Fig. 2C and data not shown), which may suggest that the mutant protein has a somewhat lower affinity for cGMP or that some parts of the protein are not folded correctly. Therefore, to verify the chemotaxis results of the GbpC-ΔGEF mutant, a less severe mutation was introduced in the catalytic part of the GEF domain, analogous to a mutation that was described before in the RasGEF protein SOS1 (28). In that study, a mutated phenylalanine resulted in a RasGEF domain with a 50-fold decreased affinity for its Ras domain. This residue is located in a helical hairpin that is thought to be important in the activity of GEF domains. The corresponding phenylalanine is conserved in GbpC, and we mutated it to an alanine (F1892A), yielding the GbpC-GEFInactive mutant (Fig. 2G). Cells expressing the mutant protein exhibit very good cGMP binding activity, suggesting good expression and correct folding (Fig. 2C). The GbpC-GEFInactive mutant did not restore the chemotactic defect of gbpC null cells, confirming the results of the GbpC-ΔGEF mutant, meaning that an intact GEF domain is essential for the function of GbpC in vivo (Fig. 4).
Inactive cNBDs Do Not Completely Inactivate GbpC—GbpC contains two cyclic nucleotide-binding domains that specifically bind cGMP with high affinity (9). From the GTP-agarose pulldown assay, we concluded that cGMP binding causes activation of the Roc domain. A mutant that is unable to bind cGMP would lead to an inactive protein, if Roc activation is fully dependent on cGMP binding. Both cGMP-binding sites of GbpC possess FGE motifs, which are thought to be important for cGMP binding (29). We abolished the motifs of both domains by mutating them into an AGA motif (F2082A, E2084A, F2564A, and E2566A; Fig. 2F). Abolishing either one of the FGE motifs created GbpC mutants with diminished but still substantial cGMP binding (data not shown). As expected, abolishing both motifs yields a mutant protein that was unable to bind cGMP (from now on called GbpC-ΔcGMP) (Fig. 2C). To confirm proper expression, we made a C-terminal GFP construct of this mutant. The fluorescence intensity of the cells expressing this protein was approximately similar to cells expressing GbpC-GFP, which was also confirmed by Western blot (Fig. 3A). Next, we tested this mutant in the GTP-agarose pulldown assay. GbpC-ΔcGMP was able to specifically bind GTP, but the stimulation by cGMP, as seen for the wild-type protein, was abolished (Fig. 3E). This result also confirms that the cGMP-stimulated Roc activation is caused by binding of cGMP to the cyclic nucleotide-binding domains of GbpC. Expression of GbpC-ΔcGMP in gbpC null cells partly restored chemotaxis; statistically, the chemotactic activity of this mutant is significantly more than chemotaxis of gbpC null cells but less than chemotaxis of gbpC null cells expressing wild-type GbpC (Fig. 4). This partial rescue suggests that (an) additional GbpC-activating factor(s), next to cGMP, is present in cells.
The LRR Are Essential for Chemotactic Activity in Vivo but Not for cGMP-stimulated Roc Activation in Vitro—Leucine-rich repeats are thought to be important in protein-protein interactions (30). A truncated GbpC protein was created that lacks the first 311 amino acids, comprising these LRR repeats (Fig. 2B). The expression of this GbpC-ΔLRR protein was comparable with the kinase dead mutant as indicated by the cGMP binding activity (Fig. 2C). Moreover, cGMP-stimulated Roc activation was also present in this mutant (Fig. 3E), excluding an important function for the LRR in cGMP binding, GEF activity, and Roc activation. Interestingly, when this mutant was tested in the chemotaxis assay, no rescue whatsoever of the gbpC null phenotype could be observed (Fig. 4), suggesting that the LRR are essential for GbpC to function in vivo, similar to the mammalian Roco protein LRRK2 (31). Similar results were obtained for larger N-terminal deletions of GbpC; proteins that lack more N-terminal amino acids in addition to the LRR are also consistently inactive in vivo (data not shown). Most importantly, expression of the kinase domain alone in gbpC null cells does not contribute to chemotaxis, whereas expression in wild-type cells has no effect on chemotaxis, suggesting that the kinase domain needs to be in the context of the rest of GbpC to be active in the cell.
DISCUSSION
GbpC is a complex multi-domain protein that belongs to the Roco family of proteins. These proteins share a conserved Roc-Cor domain module, often in addition to LRR and MAPKKK domains. Additional regulatory domains are present in some Roco proteins (11). GbpC contains a C-terminal extension with a RasGEF and two cNBDs that bind cGMP with high affinity. This domain extension is homologous to the entire sequence of the Rap-GEF GbpD, suggesting that GbpC arose from a fusion of an ancestor Roco gene with the ancestor GbpD gene. It should be noted that GbpD, despite having two cNBDs, does not bind cyclic nucleotides, and GbpC is the only cGMP-target protein in Dictyostelium identified (9). We have shown here that the RasGEF domain of GbpC specifically induces GDP/GTP exchange of the Roc domain of GbpC. The RasGEF domain of GbpC does not activate any other member of the Ras superfamily, and vice versa, the Roc domain of GbpC is not activated by any other GEF, including C3G and CDC25 that activate Rap and Ras GTPases and GbpD (21).
The presence of GEF, Ras, and MAPKKK domains in one protein strongly suggests that GbpC possesses a complete Ras signaling pathway in one protein. To investigate this pathway, we developed two assays: cGMP-stimulated binding of GbpC to GTP-coated beads in vitro and rescue of the chemotactic defect of gbpC null cells in vivo. During the time course of this study, it was found to be technically difficult to develop a suitable kinase assay for GbpC, because the protein is very large, and no suitable phosphorylation targets of the protein are known at present. This prompted us to assay the final response of the pathway, which is the chemotactic effects in vivo, rather than examining direct kinase activity of the protein. GbpC plays an important role in Dictyostelium chemotaxis, together with phosphatidylinositol 3-kinase and PLA2 (20, 32). The recognition that these three parallel pathways mediate the transduction of chemotactic cAMP signals allowed us to develop an assay to analyze the activity of GbpC in vivo. In this assay, chemotaxis is measured in the presence of chemical inhibitors of phosphatidylinositol 3-kinase and PLA2, by which the chemotactic response becomes critically dependent on guanylyl cyclases and GbpC. Using the GTP binding and chemotaxis assays, we reach four major conclusions on the regulation of GbpC. First, binding of GbpC to GTP-agarose beads is abolished upon mutational inactivation of the Roc domain but not by inactivation of the GEF or cGMP-binding domains. Furthermore we observed that the wild-type Roc-Cor domain, purified from E. coli, is in a GDP-bound state, which becomes free of nucleotides upon mutation of Lys342 in the P-loop of the Roc domain. Together, these observations strongly suggest that the Roc domain is responsible for binding of GbpC to GTP-agarose beads. Second, cGMP stimulates GbpC binding to GTP-agarose beads, which no longer occurs upon inactivation of the GEF or cNBDs. Because cGMP binding to GbpC is specifically lost upon inactivation of the cNBDs, we conclude that cGMP-mediated activation of Roc occurs through cGMP binding to cNBDs and subsequent activation of the GEF domain. Third, a mutant defective in the MAPKKK domain still has cGMP-stimulated GTP binding to the Roc domain, suggesting that the kinase domain lies downstream of cGMP-stimulated Roc activation, thereby probably representing the output of the protein. Fourth, the GEF, Roc, MAPKKK, and LRR domains are absolutely required in vivo to rescue the chemotactic defect of gbpC null cells, whereas cGMP binding to the cNBD is partially required.
These conclusions suggest a model for the activation of GbpC (Fig. 5); binding of cGMP to the cNBDs leads to an activation of the RasGEF domain, most likely through a conformational change in the protein. Roc is active in the GTP-bound state. In the absence of cGMP or GEF activity, a small fraction of GbpC is in the active GTP-bound state; the GTP-bound fraction is strongly enhanced by cGMP-stimulated GEF activity. Finally, GTP-activated Roc stimulates the activity of the MAPKKK domain, which very likely acts as the signaling output of GbpC by phosphorylating downstream proteins. It has been shown for the homologous mammalian Roco proteins LRRK1 and LRRK2 that their kinase activities are strongly dependent on the Roc domain; activation of this domain by GDP/GTP exchange causes an increase in kinase activity in vitro (15, 16). It is very likely that GbpC works in a similar way; the phosphorylation and activation of myosin light chain kinase A is activated in a cGMP- and GbpC-dependent process (8). In addition, the phosphorylation states of the stress-activated protein kinase SAPKα and the transcription factor StatC have been shown to be regulated by cGMP in Dictyostelium (33, 34). Recently, the crystal structure of the Roc domain of LRRK2 was solved, and it was suggested that the protein is activated in cis by forming a dimer (35). Disturbance of dimer formation leads to decreased GTPase activity, and the result is an overactive Roc and Kinase domain. At present, it is unclear whether GbpC signaling occurs in cis or in trans. Attempts to show dimer formation have been unsuccessful so far.3
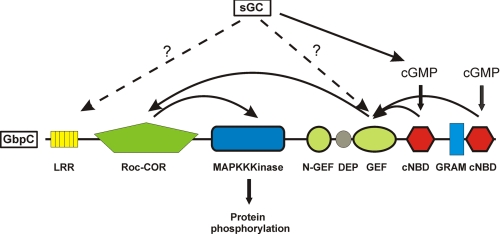
FIGURE 5.
Model for the activation mechanism of GbpC. The essential core of GbpC consists of at least the LRR, GEF, Roc, and kinase domain. Inactivation or deletion of one of these domains abolishes all GbpC activity in vivo. cGMP is produced by sGC, and upon binding of cGMP to the cNBDs, GDP/GTP exchange on the Roc domain is stimulated by the RasGEF domain, which is exposed during cGMP binding. GTP binding to the Roc domain causes activation of the kinase domain, which is the output of the protein, analogous to other Roco proteins like LRRK1 and LRRK2. Abolished cGMP binding to GbpC does not fully inactivate its activity, and therefore a cGMP-independent activation mechanism also exists, hypothetically via a similar mechanism as the sGC protein.
The role of cGMP and LRR for activation of GbpC requires further attention. The GbpC-ΔcGMP mutant is still capable of partially rescuing the gbpC null chemotaxis defect. Strikingly, this result resembles the outcome of a previous study, in which a catalytically inactive sGC was also capable of partially rescuing the chemotactic defect of gc null cells, which cannot produce cGMP anymore (20). In that study, two functions for sGC in chemotaxis were identified: one function as an enzyme producing cGMP, thereby regulating the rear of the cell, and a second function as a protein that translocates to the actin cytoskeleton at the front of the cell. The similarities with our current results for GbpC of cGMP-dependent and cGMP-independent contribution of chemotaxis make us hypothesize that sGC and GbpC can communicate with each other through two mechanisms: first, through cGMP, because sGC produces a pool of cGMP which stimulates GbpC activity, and second, sGC protein may interact with GbpC protein that may function as a second input signal for the activation of GbpC in vivo. In this respect, it is interesting that a cGMP-binding protein has been shown to regulate guanylyl cyclase activity in Dictyostelium (36). Because GbpC is the only high affinity cGMP-binding protein in Dictyostelium, and sGC was the only guanylyl cyclase activity present in those experiments, this hints to a close interaction between GbpC and sGC. The notion that the LRR domain of GbpC is essential for its function in vivo, but not for cGMP-regulated activation of the Roc domain in vitro, suggests that the LRR of GbpC provides a second input signal for GbpC to function in the cell. In mammalian LRRK2, the LRR domain may play a similar role, because also in this Roco protein, the absence of the LRR domain does not affect kinase activity in vitro but does interfere with toxic effects of wild-type LRRK2 in vivo (31).
Supplementary Material
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1.
Footnotes
The abbreviations used are: PLA2, phospholipase A2; GEF, guanine exchange factor; sGC, soluble guanylyl cyclase; MAPKKK, mitogen-activated protein kinase kinase kinase; GFP, green fluorescent protein; cNBD, cyclic nucleotide-binding domain; LRR, leucine-rich repeat.
W. N. van Egmond and A. Kortholt, unpublished observations.
References
- 1.Jin, T., and Hereld, D. (2006) Eur. J. Cell Biol. 85 905-913 [DOI] [PubMed] [Google Scholar]
- 2.Varez-Curto, E., Weening, K. E., and Schaap, P. (2007) Biochem. J. 401 309-316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Insall, R., and Andrew, N. (2007) Curr. Opin. Microbiol. 10 578-581 [DOI] [PubMed] [Google Scholar]
- 4.Veltman, D. M., Bosgraaf, L., and Van Haastert, P. J. (2004) Vitam. Horm. 69 95-115 [DOI] [PubMed] [Google Scholar]
- 5.Sasaki, A. T., and Firtel, R. A. (2006) Eur. J. Cell Biol. 85 873-895 [DOI] [PubMed] [Google Scholar]
- 6.de la Roche, M. A., Smith, J. L., Betapudi, V., Egelhoff, T. T., and Cote, G. P. (2002) J. Muscle Res. Cell Motil. 23 703-718 [DOI] [PubMed] [Google Scholar]
- 7.Bosgraaf, L., and van Haastert, P. J. (2006) Eur. J. Cell Biol. 85 969-979 [DOI] [PubMed] [Google Scholar]
- 8.Goldberg, J. M., Wolpin, E. S., Bosgraaf, L., Clarkson, B. K., Van Haastert, P. J., and Smith, J. L. (2006) FEBS Lett. 580 2059-2064 [DOI] [PubMed] [Google Scholar]
- 9.Bosgraaf, L., Russcher, H., Smith, J. L., Wessels, D., Soll, D. R., and van Haastert, P. J. (2002) EMBO J. 21 4560-4570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bader, S., Kortholt, A., and Van Haastert, P. J. (2007) Biochem. J. 402 153-161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bosgraaf, L., and van Haastert, P. J. (2003) Biochim. Biophys. Acta 1643 5-10 [DOI] [PubMed] [Google Scholar]
- 12.Zimprich, A., Biskup, S., Leitner, P., Lichtner, P., Farrer, M., Lincoln, S., Kachergus, J., Hulihan, M., Uitti, R. J., Calne, D. B., Stoessl, A. J., Pfeiffer, R. F., Patenge, N., Carbajal, I. C., Vieregge, P., Asmus, F., Muller-Myhsok, B., Dickson, D. W., Meitinger, T., Strom, T. M., Wszolek, Z. K., and Gasser, T. (2004) Neuron 44 601-607 [DOI] [PubMed] [Google Scholar]
- 13.Bos, J. L., Rehmann, H., and Wittinghofer, A. (2007) Cell 129 865-877 [DOI] [PubMed] [Google Scholar]
- 14.Chong, H., Vikis, H. G., and Guan, K. L. (2003) Cell Signal. 15 463-469 [DOI] [PubMed] [Google Scholar]
- 15.Guo, L., Gandhi, P. N., Wang, W., Petersen, R. B., Wilson-Delfosse, A. L., and Chen, S. G. (2007) Exp. Cell Res. 313 3658-3670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Korr, D., Toschi, L., Donner, P., Pohlenz, H. D., Kreft, B., and Weiss, B. (2006) Cell Signal. 18 910-920 [DOI] [PubMed] [Google Scholar]
- 17.Goldberg, J. M., Bosgraaf, L., van Haastert, P. J., and Smith, J. L. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 6749-6754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bosgraaf, L., Waijer, A., Engel, R., Visser, A. J., Wessels, D., Soll, D., and van Haastert, P. J. (2005) J. Cell Sci. 118 1899-1910 [DOI] [PubMed] [Google Scholar]
- 19.Konijn, T. M. (1970) Experientia 26 367-369 [DOI] [PubMed] [Google Scholar]
- 20.Veltman, D. M., Keizer-Gunnik, I., and Van Haastert, P. J. (2008) J. Cell Biol. 180 747-753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kortholt, A., Rehmann, H., Kae, H., Bosgraaf, L., Keizer-Gunnink, I., Weeks, G., Wittinghofer, A., and van Haastert, P. J. (2006) J. Biol. Chem. 281 23367-23376 [DOI] [PubMed] [Google Scholar]
- 22.Lenzen, C., Cool, R. H., and Wittinghofer, A. (1995) Methods Enzymol. 255 95-109 [DOI] [PubMed] [Google Scholar]
- 23.Wilkins, A., Szafranski, K., Fraser, D. J., Bakthavatsalam, D., Muller, R., Fisher, P. R., Glockner, G., Eichinger, L., Noegel, A. A., and Insall, R. H. (2005) Genome Biol. 6 R68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greggio, E., Lewis, P. A., van der Brug, M. P., Ahmad, R., Kaganovich, A., Ding, J., Beilina, A., Baker, A. K., and Cookson, M. R. (2007) J. Neurochem. 102 93-102 [DOI] [PubMed] [Google Scholar]
- 25.Praefcke, G. J., Kloep, S., Benscheid, U., Lilie, H., Prakash, B., and Herrmann, C. (2004) J. Mol. Biol. 344 257-269 [DOI] [PubMed] [Google Scholar]
- 26.Huse, M., and Kuriyan, J. (2002) Cell 109 275-282 [DOI] [PubMed] [Google Scholar]
- 27.Greggio, E., Jain, S., Kingsbury, A., Bandopadhyay, R., Lewis, P., Kaganovich, A., van der Brug, M. P., Beilina, A., Blackinton, J., Thomas, K. J., Ahmad, R., Miller, D. W., Kesavapany, S., Singleton, A., Lees, A., Harvey, R. J., Harvey, K., and Cookson, M. R. (2006) Neurobiol. Dis. 23 329-341 [DOI] [PubMed] [Google Scholar]
- 28.Hall, B. E., Yang, S. S., Boriack-Sjodin, P. A., Kuriyan, J., and Bar-Sagi, D. (2001) J. Biol. Chem. 276 27629-27637 [DOI] [PubMed] [Google Scholar]
- 29.Diller, T. C., Madhusudan, Xuong, N. H., and Taylor, S. S. (2001) Structure 9 73-82 [DOI] [PubMed] [Google Scholar]
- 30.Kobe, B., and Deisenhofer, J. (1994) Trends Biochem. Sci. 19 415-421 [DOI] [PubMed] [Google Scholar]
- 31.Iaccarino, C., Crosio, C., Vitale, C., Sanna, G., Carri, M. T., and Barone, P. (2007) Hum. Mol. Genet. 16 1319-1326 [DOI] [PubMed] [Google Scholar]
- 32.Chen, L., Iijima, M., Tang, M., Landree, M. A., Huang, Y. E., Xiong, Y., Iglesias, P. A., and Devreotes, P. N. (2007) Dev. Cell 12 603-614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun, B., Ma, H., and Firtel, R. A. (2003) Mol. Biol. Cell 14 4526-4540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Araki, T., Tsujioka, M., Abe, T., Fukuzawa, M., Meima, M., Schaap, P., Morio, T., Urushihara, H., Katoh, M., Maeda, M., Tanaka, Y., Takeuchi, I., and Williams, J. G. (2003) J. Cell Sci. 116 2907-2915 [DOI] [PubMed] [Google Scholar]
- 35.Deng, J., Lewis, P. A., Greggio, E., Sluch, E., Beilina, A., and Cookson, M. R. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 1499-1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuwayama, H., and Van Haastert, P. J. (1996) J. Biol. Chem. 271 23718-23724 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.