Abstract
MicroRNAs (miRs) are short endogenous RNAs that regulate gene expression by incomplete pairing with messenger RNAs. An increasing number of studies show that mammalian microRNAs play fundamental roles in various aspects of cellular function including differentiation, proliferation, and cell death. Recent findings demonstrating the presence of microRNAs in mature neuronal dendrites suggest their possible involvement in controlling local protein translation and synaptic function. HIV-1 Encephalopathy (HIVE) is a manifestation of HIV-1 infection that often results in neuronal damage and dysfunction. While neurons are rarely, if ever, infected by HIV-1, they are exposed to cytotoxic viral and cellular factors including the HIV-1 transactivating factor Tat. In this study, we show that Tat deregulates expression levels of selected microRNAs, including the neuronal mir-128, in primary cortical neurons. We further show that mir-128a inhibits expression of the pre-synaptic protein SNAP25, whereas the anti-mir-128a partially restores Tat/mir-128a-induced downregulation of SNAP25 expression. Altogether, our data provide a novel mechanism by which HIV-Tat perturbs neuronal activity.
MicroRNAs (miRNAs, miRs) are a growing class of short non-coding RNAs that control post-transcriptional gene expression (Ke et al., 2003; Lai, 2003). Mature microRNA derive from longer transcripts (pri-miRs) which are processed to shorter hairpin precursors (pre-miRs) by the action of Drosha enzyme (Lee et al., 2003). The 70-nucleotides precursors are exported to the cytoplasm where they are cleaved by Dicer to produce mature 19–22 nucleotides microRNA, which enter RNA-induced silencing complex (RISC; Hutvagner et al., 2001; Ketting et al., 2001). Translational silencing by the RISC complex appears to regulate a wide variety of cellular and developmental processes (Du and Zamore, 2005; Hwang and Mendell, 2006). An increasing number of studies have shown the presence of microRNAs in the central nervous system (CNS) and their importance for neuronal development (Klein et al., 2005; Cao et al., 2006). As protein synthesis in neurons occurs not only in the cell body but also in axons and dendrites, the enrichment of microRNAs in the neuronal processes (Tai and Schuman, 2006) suggests a possible action of dendritic microRNAs in regulating synaptic function (Kim et al., 2004, 2005), as well as spine development (Schratt et al., 2006).
Synaptic vesicles are critical regulators of pre-synaptic events that are required for proper neurotransmission, including organelle transport, interaction with cytoskeleton, uptake and storage of low molecular weight molecules, and membrane fusion for exo- and endocytosis (reviewed by Burre and Volknandt (2007)). Key regulators of membrane fusion are the soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) proteins (reviewed by Salaun et al. (2004)). Among them, synaptosome associated protein of 25 kDa (SNAP25) is associated with the cell membrane.
Human immunodeficiency virus 1 (HIV-1) infection can lead to HIV-Encephalopathy (HIVE), which is associated with neurological manifestations ranging from mild to severe motor and cognitive dysfunction (Janssen et al., 1989; Sacktor, 2002; McArthur, 2004). HIVE is the histopathological substrate of the clinical entity known as AIDS–dementia complex (Del Valle and Pina-Oviedo, 2006). The viral regulatory protein Tat, which is required for HIV-1 replication, is secreted by HIV-1 infected cells and freely enter surrounding uninfected cells including neurons (Del Valle et al., 2000; Aprea et al., 2006). Exogenous administration of Tat is thought to activate excitatory amino acid receptors increasing the release of intracellular calcium (Haughey et al., 2001).
In this study, we evaluated the effects of Tat-treatment on the expression pattern of microRNAs in rat embryonic cortical neurons. The results show a novel mechanism by which Tat can control expression of specific genes in neurons by modulating expression of selected microRNAs. We focused on one of the microRNAs upregulated by Tat, the neuronal mir-128a, and found that it modulates expression of the synaptic factor SNAP25. Treatment of neuronal cultures with the viral protein Tat also determined a marked downregulation of SNAP25 protein, partially rescued by the antago-mir-128a. Altogether these data show that mir-128a modulates levels of SNAP25 protein in neurons and that Tat-induced downregulation of SNAP25 may result, at least in part, by an increased expression of mir-128a.
Materials and Methods
Primary cells and cell lines
293T cell line was cultured in DMEM containing 10% FBS. Primary cortical neurons were obtained by enzymatic and mechanical treatment from Sprague–Dawley rat embryonic day 17. Cortices were dissected out as previously described (Aprea et al., 2006). Cells were plated on poly-D-lysine coated dishes at a density of 4.5 × 104/cm2 and cultured in Neurobasal medium containing B27 supplement, 0.25 mM Glutamax, and 0.25 mM L-Glutamine (all from Gibco, Invitrogen Corporation, Carlsbad, CA).
Mouse neuronal progenitors were obtained from C57BL/6J mice embryonic day 16. The whole brains were digested for 10 min at 37°C with TrypLE (Gibco) and mechanically dissociated with a fire-polished pasteur pipette in HibernateE processing medium (BrainBits, Springfield, IL). Cells were plated at 3 × 104 cells/cm2 in petri dishes. Neural progenitors were cultured in a serum free medium system composed of Neurobasal medium supplemented with 2 mM glutamine, 2% B27, 1% N2, 20 ng/ml EGF, 20 ng/ml bFGF (all from Gibco, Invitrogen Corporation), and Heparin (1 μg/ml; Sigma-Aldrich, St. Louis, MO). These conditions promote the growth of floating three-dimensional clusters (neurospheres). After two passages, secondary neurospheres were nucleofected using Amaxa electroporator, following the manufacturer’s protocol (Amaxa, Inc., Gaithersburg, MD).
Tat protein and control peptide
For the microRNA microarray, recombinant Tat1-72 protein was a kind gift of Dr. Avindra Nath (Johns Hopkins University, Baltimore, MD), and was used at the concentration of 500 nM. Validation of miR expression by qRT-PCR was performed on the same RNA samples used for the microRNA microarray. For all the other experiments, including dose- and time-dependent miR expression profile, recombinant Tat1-101 was purchased from ImmunoDiagnostics, Inc. (Woburn, MA), and was used at the concentrations of 100 and 500 nM (about 1 and 5 μg/ml, respectively). Arg(9) cell permeable peptide was purchased from AnaSpec (San Jose, CA) and used at the concentration of 500 nM.
MicroRNA microarray
Eight-day-old rat cortical neurons were subjected to a change of medium in the presence or absence of recombinant Tat1-72 at a concentration of 500 nM. A total of 60 μg RNA was extracted from untreated and Tat-treated neuronal cultures using the mirVana miRNA extraction kit (Ambion, Inc., Austin, TX). One to two micrograms of RNA were subjected to electrophoresis to verify the integrity of the RNAs. RNA processing, microarray fabrication, array hybridization, and data acquisition were performed by a service provider (LC Sciences, Houston, TX). Two different arrays were performed on the RNA extracts; each of them contained the paired samples untreated/Tat-treated. In the first chip Tat-treated 1 sample was labeled with Cy3 and the untreated 1 sample with Cy5. In the second chip Tat-treated 2 was labeled with Cy5 and untreated 2 with Cy3. Each chip used contained 3,200 miRNAs representing multiple rat miRNAs and controls. Each region further comprised a miRNA probe region, which detects miRNA transcripts listed in Sanger miRBase Release 8.0 (http://www.sanger.ac.uk/software/rfam/mirna/).
Quantitative RT-PCR
Levels of representative miRs were evaluated by Real Time RT-PCR using mirVana qRT-PCR miRNA Detection Kit (Ambion, Inc.) following the manufacturer’s protocol. PCR primer pairs for reverse transcription and mature miRs detection were purchased from Ambion, Inc. (rno-mir-100, rno-mir-128a, hsa-mir-374, and control U6). Quantitative real time RT-PCR was performed on at least three different concentrations of template each in triplicate. Relative quantification of the selected microRNAs was calculated using the Comparative Ct method (ΔΔCt), in accordance with Applied Biosystems general guidelines.
MicroRNA gene targets prediction
Gene targets were predicted according to the miRBase Target Database V9.1 (http://microrna.sanger.ac.uk/), as well as TargetScan V3.1 (http://www.targetscan.org). All potential targets were considered.
microRNA functional analysis
To create pSilencer-128a, a genomic fragment (225 bp) bearing the pre-rno-miR-128a was amplified from rat genomic DNA and cloned into the expression vector pSilencer™ 4.1—CMV Expression Vector (Ambion, Inc.). The oligos were: mir-128a Forward: 5′-CGGGATCCTTATGTGATTATATCTTACA AT; mir-128a Reverse: 5′-CCCAAGCTTATGAAGCCAAATGATGCAAAAT. To generate 3′UTR reporter constructs, a 1,220 bp fragment of rat 3′UTR SNAP25 was cloned downstream of the Renilla firefly luciferase cassette in the psiCHECK™ -2 vector (Promega Corporation, Madison, WI). The oligonucleotides for the PCR were: SNAP25-3′UTR Forward: 5′-CCGCTCGAGAGAAGAGAGCTCCTTCATGC, and Reverse: 5′-ATAAGAATGCGGCCGCTACAAAATGTCAAATCAC. The specific mir-128a putative wild type sites in the 3′UTR sequences of SNAP25 were also cloned into the reporter plasmid as double stranded oligos:
SNAP25#1 Top: ccgctcgagTCTTGTAAAACTGTGACATTCCACAGAagatctgcggc cgctaaactat. SNAP25#2 Top: ccgctcgagTATAGATAAACTGTGAGATAAATATCAag atctgcggccgctaaactat. In these sequences, the bases in smaller case represent the XhoI and NotI restriction sites; in italics is the BglII restriction site for the easy screening of positive clones; the nucleotides underlined were replaced with their complementary bases to generate mutated microRNA sites. The oligos for the cloning of mir-128a perfect match (PM) positive control were: 5′-ccgctcgagAAAAGAGA CCGGTTCACTGTGAagatctgcggccgctaaactat, and its complementary sequence.
For miR target validation, 293T cells were plated at the concentration of 8 × 104 cells/well in a 12-well plate. The next day a total amount of 0.6 μg/well of DNA was transfected using FUGENE 6 (Invitrogen Corporation) at the DNA:Fugene 6 ratio of 1:3. pcDNA3 was used to reach the final amount of DNA. Samples were harvested 48 h post-transfection and subjected to the dual luciferase assay system (Promega Corporation) following the manufacturer’s instructions. Relative units represent the ratio between Renilla values and the Luciferase internal control. The experiments were performed in duplicates and repeated at least three times.
Antago-mir-128a was the miRCURY LNA mir-128a probe from Exiqon, Inc. (Woburn, MA). It was used at the concentration of 20 nM.
Western blots
Two-week-old neuronal cultures were incubated over night with Neurobasal medium in the absence of B27 supplement. Next day, Tat101 was added at the concentrations of 100 and 500 nM. Neurons were collected by scraping the plates in the presence of PBS, followed by centrifugation and lysis of the cell pellet in the appropriate volume of RIPA buffer (50 mM Tris pH 7.4, 1% NP-40, 0.25% sodium deoxycholate, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM PMSF, 1 mM sodium orthovanadate, phosphatase inhibitors and protease inhibitor cocktails). Whole cell lysate (15–50 μg) was separated on a 4–15% SDS–PAGE (BioRad, Hercules, CA). SNAP25 antibody was from Abcam, Inc. (Cambridge, MA). Grb2 antibody was from Transduction Laboratories (BD Biosciences, San Jose, CA), anti-alpha-tubulin was from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Experiments were repeated at least three times. Densitometric analysis was performed using Image J software (http://rsb.info.nih.gov/ij/). The intensity of each experimental band was subtracted from the background and normalized for the corresponding values of Grb2.
Immunohistochemistry
Samples from four cases of formalin-fixed, paraffin-embedded HIV-Encephalopathy were collected from the pathology archives of the HIV Manhattan Brain Bank at Mount Sinai Medical School. Sections of 4 μm in thickness were cut using a microtome, and placed on charged glass slides. Immunohistochemistry was then performed according to the manufacturer’s instructions (Vector Laboratories, Burlingame, CA). Our modified protocol includes deparaffination, re-hydration in alcohol up to water, antigen retrieval in Citrate buffer pH 6.0 at 95°C, endogenous peroxidase quenching and blocking in normal horse serum for 2 h at room temperature. SNAP25 (Clone SMI-81, 1:1,000 dilution, Abcam, Inc.) mouse monoclonal primary antibody was incubated overnight in a humidified chamber. After rinsing with PBS, secondary biotinilated antibody was incubated for 1 h (1:200 dilutions), followed by incubation with ABC complexes and developing with Diaminobenzidine (DAB, Boehringer, Ingelheim, Ridgefield, CT), counterstained with hematoxylin and coverslipped.
Statistical analysis
All results are represented as mean values ±SD. The significance of differences between means was performed using a two-tailed Student’s t-test. P values ≤0.05 were considered to be statistically significant.
Results
One of the most intriguing properties of the HIV-1 transactivating protein Tat is its ability to enter virtually any cell type, including neurons. To analyze potential effects of Tat on the microRNAs expression profile in neurons, we have isolated RNA suitable for microRNA microarray from E17 rat embryonic cortical neurons cultured for 6 days followed by a 12 h treatment with 500 nM recombinant Tat1-72 (Aprea et al., 2006; Peruzzi, 2006). RNA extracted from untreated cultures served as baseline control.
microRNA expression profile
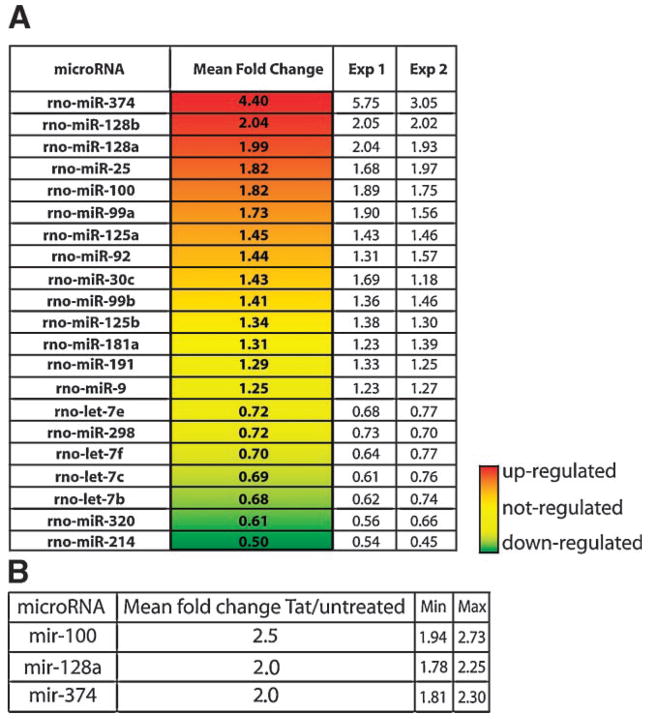
Figure 1A illustrates a collection of microRNAs differentially expressed in rat primary cortical neurons upon Tat treatment in comparison to untreated neurons. microRNAs not statistically significant were excluded from the analysis (see Materials and Methods Section). Among the 21 (~10% of spotted microRNAs) microRNAs present in the list, 15 have been previously shown to be associated with polyribosomes in rat cortical neurons (Kim et al., 2004). These are designated 128a, 128b, 100, 99a, 30b, 30c, let-7c, let-7f, let-7b, let-7e, 125a, 125b, 191, 181a, and 9. A group of six microRNAs (374, 128a, 128b, 100, 25, and 99a) was upregulated in Tat-treated samples (mean fold change from 4.4 to 1.5; P-value <0.01). The presence of Tat in neurons also determined a downregulation of seven microRNAs: let-7e, 298, let-7f, let-7c, let-7b, 320, and 214. The expression pattern of a third group of microRNAs (125a, 92, 30c, 99b, 125b, 181a, 191, and 9) was not changed by Tat-treatment.
Fig. 1.
microRNAs whose expression is modulated (P-value < 0.01) by Tat-treatment in primary neurons. A: The fold change represents microRNA expression level in Tat-treated compared to untreated cells and it is expressed as an average of two experiments shown as Exp 1 and Exp 2. B: Quantitative RT-PCR. The mean fold change represents the amount of microRNA in the Tat-treated sample after normalization with U6 and relative to the untreated sample (detailed in Materials and Methods Section). Minimum and maximum values of the 95% confidence interval are shown.
Validation of microRNAs by qRT-PCR
Expression levels of selected microRNAs (100, 128a, and 374) were evaluated by qRT-PCR as detailed in Materials and Methods Section. Results from quantitative real time PCR are summarized in Figure 1B. Increased expression of microRNAs mir-100, -128a, and -374 after Tat-treatment was confirmed by replicate experiments. Note that the fold change values of the qPCR paralleled with microarray data.
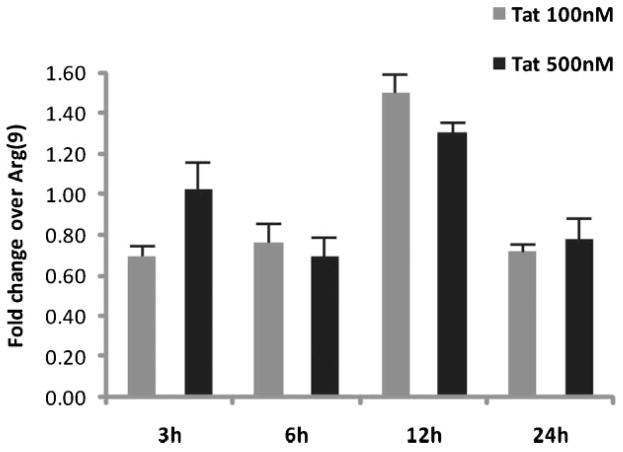
Expression profiles of mir-128a obtained with the first exon Tat1-72 (Fig. 1) was also determined by qRT-PCR analysis in rat cortical neurons treated with recombinant full-length Tat101 at two different concentrations and for various time points (Fig. 2). As in Figure 1B, ΔΔCt (see also Materials and Methods Section) was calculated using U6 RNA levels measured in each experimental sample. As a negative control neurons were treated with a small cell-permeable peptide Arg(9) at the concentration of 500 nM. Results are shown in Figure 2, where the fold change of mir-128a in the Tat-treated samples was normalized for the fold change measured in the Arg(9) peptide-treated neurons. The presence of Tat101 in neurons induces a transient induction of mature mir-128a expression, which peaks at 12 h post-treatment and decreases thereafter.
Fig. 2.
Modulation of mir-128a expression by Tat in primary neurons. Quantitative RT-PCR shows a time and dose-dependent expression of mir-128a in Tat-treated neuronal cultures. The diagram represents results from three independent experiments each performed in triplicate. The fold change is calculated over the control (neurons treated with the poly-Arginine, Arg(9), peptide) previous normalization of each sample with the U6 internal control (see also Materials and Methods Section). Standard deviation is shown.
Next, we analyzed possible gene targets for mir-128a according to the miRBase Target Database V8.0 (http://microrna.sanger.ac.uk/).
Results from prediction algorithms suggested that mir-128a could target molecules of the soluble N-ethylmaleimide-sensitive factor attached protein (SNAP) receptor (SNARE) complex, including SNAP25 and synaptic vesicle glycoprotein 2A (SV2A).
Mir-128a functional analysis
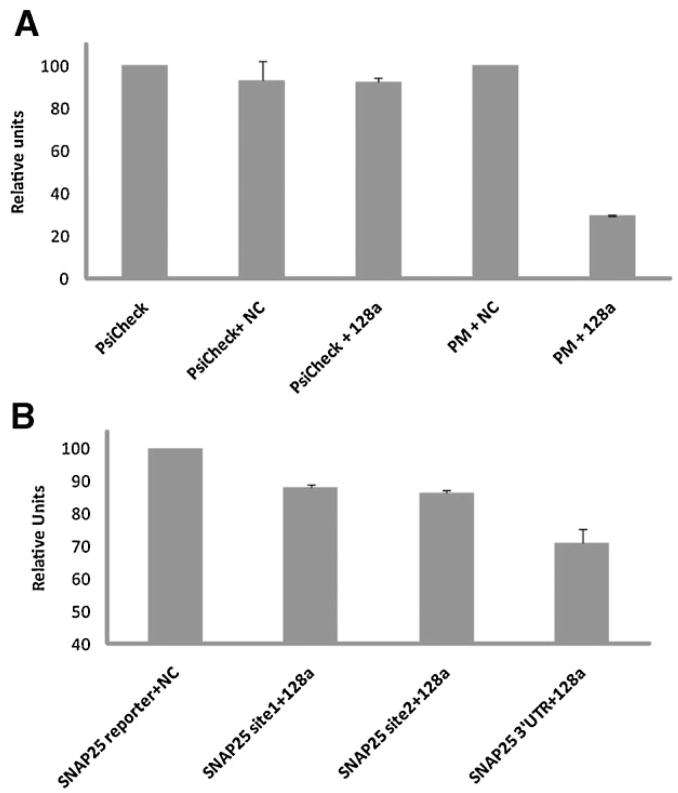
To test the possibility that mir-128a could modulate the expression of pre-synaptic proteins we performed a microRNA functional assay using SNAP25 3′UTR gene target. For this analysis, a genomic sequence corresponding to the rat pre-mir-128a was cloned into the pSilencer expression vector (pSil-mir-128a). Expression of mature mir-128a in 293T cells was verified by qRT-PCR at 24 and 48 h post-transfection (data not shown). Renilla/luciferase reporter constructs contained the entire 3′UTR sequence of SNAP25, as well as the single mir-binding sites downstream to the firefly renilla gene (psiCHECK2 plasmid, see Materials and Methods Section). For luciferase assays, the reporter vectors were co-expressed together with mir-128a or the control non-targeting sequence (NC) into 293T cells and assayed 48 h post-transfection. Results are show in Figure 3 as percent of the basal values. We first tested the efficiency of mir-128a to target its own sequence in co-transfection experiments (Fig. 3A). The ratio Renilla/Luciferase (R/L) in the control empty vector (PsiCheck) was scaled to 100%. A small, non-specific, 10% decreased ratio was observed when PsiCheck was co-transfected with pSil-NC (non-targeting sequence) or with pSil-mir-128a. The R/L ratio relative to the reporter plasmid carrying the mir-128a perfect match sequence (PM) did not change with the addition of NC control plasmid (PM + NC). By converse, co-transfection of PM with mir-128a resulted in a reproducible 75% reduction of R/L ratio, confirming that mir-128a is overexpressed and functional in this cellular system. Next, we determined the inhibitory effect of mir-128a on the 3′UTR of SNAP25 (Fig. 3B). The inhibitory effect of mir-128a on the SNAP25 single sites 1 and 2 was about 15%, increasing to nearly 30% for the entire 3′UTR sequence.
Fig. 3.
Inhibitory effect of mir-128a on SNAP25 predicted gene target. Luciferase and Renilla values were determined 24 h (A) or 48 h (B) post-transfection. A: Diagram relative to the inhibitory effect of mir-128a on the control perfect match sequence. B: Inhibitory effect of mir-128a on SNAP25 3′UTR and single mir-128a sites 1 and 2. Relative units represent the ratio between Renilla values and the Luciferase internal control. The experiments were performed in duplicates and repeated at least three times.
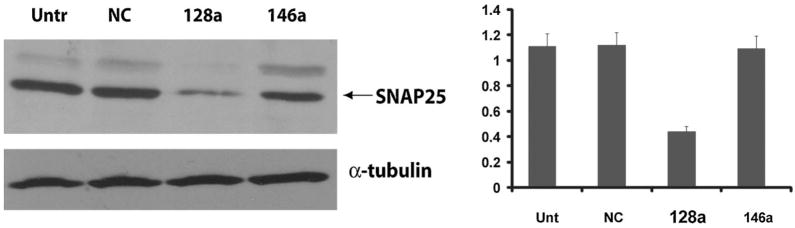
The next step in determining the inhibitory effect of mir-128a activity on SNAP25 3′UTR was to evaluate expression of SNAP25 protein upon forced expression of mir-128a. For these experiments we utilized primary cultures of mouse neuronal progenitors (Neuronal Stem Cells, NSC), which can be efficiently nucleofected. Basal expression levels of mir-128 were determined by qRT-PCR on RNA extracts from mouse NSC (data not shown). The activity of mir-128a on SNAP25 protein was determined 24 h after nucleofection (see Materials and Methods Section) with pSIL-mir-128a expression vector. Figure 4 shows downregulation of SNAP25 protein levels in mir-128a transfected cells but not in cells transfected with a control plasmid (pSIL-NC) or in untransfected cells. Expression of SNAP25 protein did not change in cells transfected with a non-relevant microRNA, mir-146a, confirming the specificity of mir-128a on its target sequence.
Fig. 4.
Inhibitory effect of mir-128a on SNAP25 protein. Representative Western blot showing levels of expression of SNAP25 in mouse neuronal progenitors upon transfection of pSIL-mir-128a and controls. Alpha-tubulin was used as loading control. The diagrams on the right show densitometric analysis (average of three experiments) of the SNAP25 protein levels after normalization with alpha-tubulin.
Decreased expression of SNAP25 in Tat-treated neuronal cultures involves activity of mir-128a
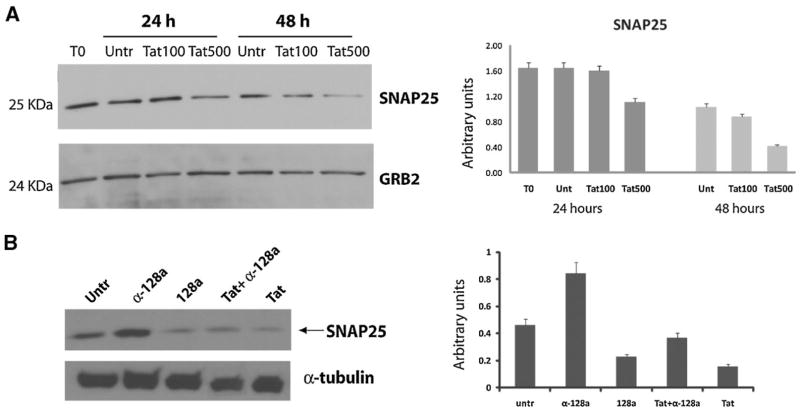
We determined expression levels of SNAP25 in rat cortical neurons treated with recombinant Tat (Tat101) at two different concentrations of 100 and 500 nM and at two time points of 24 and 48 h. Results shown in Figure 5A clearly indicate a dose- and time-dependent reduction of SNAP25 levels in Tat-treated compared to untreated neurons, with the maximal effect at 48 h and higher dose of Tat.
Fig. 5.
Tat-induced down regulation ofSNAP25 involves the activity of mir-128a. A: Western blot analysis performed on cellular lysates obtained at 24 and 48 h from Tat-treated (at 100 and 500 nM) rat cortical neurons and control. Grb2 antibody was used to show equal loading of protein lysates. Densitometric analysis relative to SNAP25 expression after treatment with Tat101 was normalized for Grb2 levels and calculated from three independent experiments. B: Western blot analysis performed on neuronal progenitors lysates showing the contribution of mir-128a in the Tat-mediated downregulation of SNAP25. Treatment of Tat-transfected cells with antago-mir-128a partially prevents Tat-induced reduction of SNAP25 levels. Alpha-tubulin was used as loading control. The diagram on the right shows densitometric analysis (average of three independent experiments) of the SNAP25 protein levels after normalization with alpha-tubulin.
Next, we utilized mouse neuronal progenitors to determine the contribution of mir-128a in the Tat-induced downregulation of SNAP25 expression. The results are shown in Figure 5B where mouse NSC were nucleofected with pEYFP-Tat101 plasmid alone or in the presence of antago-mir-128a (α-128a). Levels of SNAP25 decreased upon expression of either Tat or mir-128a when compared to untransfected cells. By converse, treatment of cells with antago-mir-128a resulted in increased levels of SNAP25 in untransfected and in Tat-transfected cells. These data suggest an involvement of mir-128a activity in the Tat-mediated downregulation of SNAP25 protein.
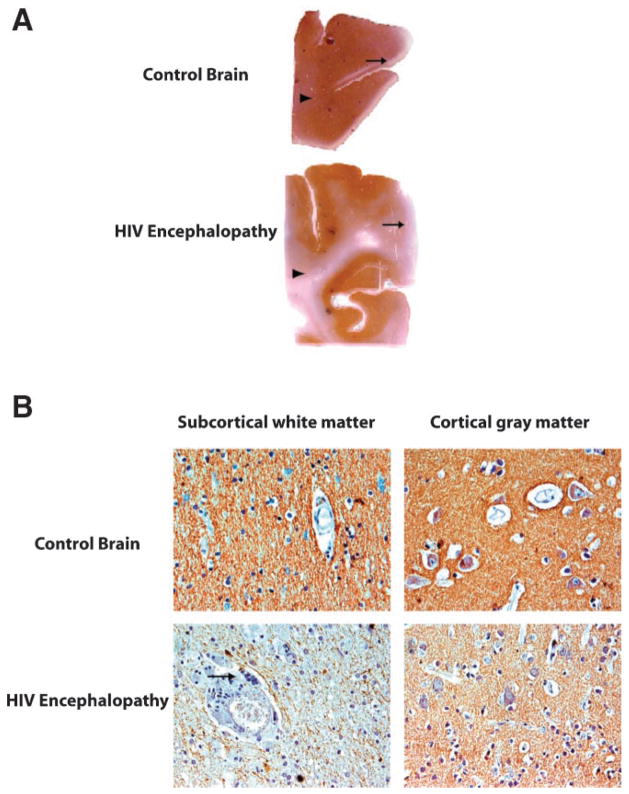
Finally, to correlate the clinical significance of these findings, we performed immunohistochemical experiments in a collection of samples from HIVE cases and normal control brains, using a specific antibody for SNAP25. Figure 6A shows montages of these cases demonstrating a significant reduction of SNAP25 in the HIV-Encephalopathy cases compared to the normal brain. Examination at higher magnification revealed a strong reduction of SNAP25 expression in the sub-cortical white matter and a moderate reduction in the cortex (Fig. 6B).
Fig. 6.
Decreased expression of SNAP25 protein in HIV-Encephalopathy. A: Full montages of the brain from normal and HIVE cases demonstrate that while SNAP25 is present throughout the cortex and white matter in the normal brain, its levels are significantly decreased in both the cortex (arrow) and the white matter (arrowhead). B: Expression of SNAP25 is also significantly reduced in the sub-cortical white matter in an area of inflammation where a giant multinucleated cell can be seen (arrow), and also decreased in the cortex on an HIVE affected area.
Discussion
In neurons, a distinct subset of mRNAs is localized in dendrites at relatively high concentrations (Steward and Schuman, 2001; Eberwine et al., 2002). Transported mRNAs are subject to translational control including silencing and subsequent activation (or derepression), ensuring spatio-temporal expression of specific proteins (Mohr and Richter, 2001). Among the molecular mechanisms involved in translational control, regulation of translation by microRNAs has received special attention (Gebauer and Hentze, 2004). Several studies have suggested that microRNAs are present in dendrites (Steward and Schuman, 2001; Kim et al., 2004; Schratt et al., 2006; Tai and Schuman, 2006; Hengst and Jaffrey, 2007; Kye et al., 2007) in complex with Dicer, Argonaute, and Fragile X mental retardation protein (FMRP; Lugli et al., 2005).
The HIV-1 regulatory factor, Tat, exerts its transactivating activity by binding to the transactivating response (TAR) element of the HIV-1 LTR. Interestingly, the host TAR RNA-binding protein (TRBP), a cofactor in Tat-TAR interactions, is a key component of Dicer-mediated dsRNA processing (Haase et al., 2005). It has been reported that Tat binds to and inhibits the activity of Dicer in HIV-infected or Tat-overexpressing HeLa cells (Bennasser et al., 2005). Suppression of microRNAs synthesis in HIV-1 infected cells would facilitate viral replication (Bennasser et al., 2005). However, there are no reports about a similar activity of Tat in cells that are not infected by HIV-1, including neurons. Of interest, Lukiw et al. have recently found that induction of reactive oxygen species (ROS) by metals results in increased levels of a population of microRNAs, including mir-128, in human brain cells. Since Tat is known to induce ROS production (Kruman et al., 1998), one could speculate that, among other effects, Tat-induced ROS production could, in turn, determine a change in microRNA expression.
Although quantification of Tat in the brain of HIV-1 infected individuals remains unknown, its detection in neurons of HIVE tissue samples in proximity of microglial nodules (Hudson et al., 2000; Aprea et al., 2006), may suggest a significant presence of this viral protein, at least locally. In this study, we evaluated whether Tat could affect microRNA expression profile in primary neurons. To ensure maximal activity of this viral protein within a short period of time we treated embryonic cortical neurons with Tat72 at a concentration of 500 nM for 12 h. While the fold change of expression of microRNAs in Tat-treated versus untreated samples (Fig. 1A) may appear modest, we have reproducibly found the same results by using qRT-PCR in at least five different preparations of primary neurons, treated with different batches of recombinant Tat, with two different isoforms of Tat, 72 and 101 aa, and each of them repeated in triplicate. On the other hand, since the relationship of cause-effect for microRNAs does not follow a linear pattern, a small variation in microRNAs expression might result in a stronger effect by targeting multiple 3′UTR sequences, and microRNAs might be responsible for the fine-tuning of cellular events (Hornstein and Shomron, 2006; Cao et al., 2007).
We further monitored changes in the expression of mir-128a at various time points after treatment of cortical neurons with two different concentrations of Tat. For this study we utilized the full-length Tat101 at the doses of 100 and 500 nM. Similarly to the effects observed in neurons treated with Tat72 at 500 nM, quantitative RT-PCR shows an increased expression of mature mir-128a in Tat101 treated cells (Fig. 2).
We have identified 21 microRNAs that were regulated upon Tat treatment (Fig. 1A). Interestingly, more than 60% of them have been previously shown to be associated with polyribosomes in rat cortical neurons (Kim et al., 2004). In addition, among the six microRNAs that showed increased expression upon Tat-treatment (mir-374, -128a, -128b, -25, -100, and -99a), mir-128a has been found to be enriched in the brain (Sempere et al., 2004), and preferentially expressed in mature neurons (Smirnova et al., 2005). Recent reports also show altered expression of mir-128 in pathological conditions, such as acute lymphoblastic leukemia (Mi et al., 2007), glioblastomas (Ciafre et al., 2005), and Alzheimer disease (Lukiw, 2007). However, the function of mir-128 in the neurons remains to be elucidated.
To validate gene target prediction we started the analysis of one putative target of mir-128a, SNAP25 3′UTR, within the cluster of synaptic proteins and performed microRNA-functional assays (Fig. 3). Our data show that SNAP25 could be indeed a true target for mir-128a (Figs. 3–5). Interestingly, the observed Tat-mediated downregulation of SNAP25 was partially inhibited by the activity of anti-mir-128 (Fig. 5). Finally, we observed a clear reduction of SNAP25 labeling in the sub-cortical white matter and, to a lesser extent, in the gray matter of brain tissue from HIVE patients compared to control samples (Fig. 6). This is consistent with previous studies showing downregulation of genes involved in synaptic plasticity and its importance in the pathology of HIV-associated neurocognitive dysfunction (Masliah et al., 1997, 2004; Everall et al., 1999; Moore et al., 2006).
There are several mechanisms involved in the Tat-induced molecular effects and we have previously showed the participation of the proteasome-ubiquitin pathway in promoting downregulation of microtubule-associated protein 2, a key dendritic protein (Aprea et al., 2006). Here, we have explored a novel pathway by which Tat can interfere with synaptic function by altering expression of microRNAs, endogenous mRNA translational inhibitors. Intuitively, decreased expression of pre-synaptic proteins like SNAP25 by the action of microRNAs may compromise proper synaptic activity. Whether this is an effect due to a decreased dendritic arborization observed in HIVE tissues or an early event remains to be determined. Our in vitro study using recombinant Tat shows increased microRNA expression (within 12 h from the treatment of primary neurons) and decreased levels of synaptic proteins as early as 24 h in the absence of a clear reduction of dendritic branches, suggesting that increased upregulation of specific microRNAs could precede neuronal damage. Indeed, selective regional loss of exocytotic pre-synaptic vesicle proteins in Alzheimer’s disease brains has been correlated with loss of cognitive function (Shimohama et al., 1997; Sze et al., 2000). Although data presented here show an inhibitory effect of mir-128a on SNAP25 3′UTR gene target and protein, it remains to be determined the role of mir-128a in regulating synaptic activity in normal and in neurodegenerative disorders including HIVE.
Acknowledgments
Contract grant sponsor: National Institute of Mental Health;
Contract grant numbers: MH071162, MH079751.
Contract grant sponsor: National Institute of Health;
Contract grant numbers: P01NS030916, P01NS43980.
We thank J. Otte and L. Hodge for technical support, and Thersa Sweet for editorial assistance. The work was supported by grants from National Institute of Mental Health awarded to FP: MH071162 and MH079751. The immunohistochemistry analysis was performed by the Neuropathology core facility and supported by grants from National Institute of Health to LDV: P01NS030916; and S. Amini: P01NS43980.
Literature Cited
- Aprea S, Del Valle L, Mameli G, Sawaya BE, Khalili K, Peruzzi F. Tubulin-mediated binding of human immunodeficiency virus-1 Tat to the cytoskeleton causes proteasomal-dependent degradation of microtubule-associated protein 2 and neuronal damage. J Neurosci. 2006;26:4054–4062. doi: 10.1523/JNEUROSCI.0603-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennasser Y, Le SY, Benkirane M, Jeang KT. Evidence that HIV-1 encodes an siRNA and a suppressor of RNA silencing. Immunity. 2005;22:607–619. doi: 10.1016/j.immuni.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Burre J, Volknandt W. The synaptic vesicle proteome. J Neurochem. 2007;101:1448–1462. doi: 10.1111/j.1471-4159.2007.04453.x. [DOI] [PubMed] [Google Scholar]
- Cao X, Yeo G, Muotri AR, Kuwabara T, Gage FH. Noncoding Rnas in the mammalian central nervous system. Annu Rev Neurosci. 2006;29:77–103. doi: 10.1146/annurev.neuro.29.051605.112839. [DOI] [PubMed] [Google Scholar]
- Cao X, Pfaff SL, Gage FH. A functional study of miR-124 in the developing neural tube. Genes Dev. 2007;21:531–536. doi: 10.1101/gad.1519207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciafre SA, Galardi S, Mangiola A, Ferracin M, Liu CG, Sabatino G, Negrini M, Maira G, Croce CM, Farace MG. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem Biophys Res Commun. 2005;334:1351–1358. doi: 10.1016/j.bbrc.2005.07.030. [DOI] [PubMed] [Google Scholar]
- Del Valle L, Pina-Oviedo S. HIV disorders of the brain: Pathology and pathogenesis. Front Biosci. 2006;11:718–732. doi: 10.2741/1830. [DOI] [PubMed] [Google Scholar]
- Del Valle L, Croul S, Morgello S, Amini S, Rappaport J, Khalili K. Detection of HIV-1 Tat and JCV capsid protein, VP1, in AIDS brain with progressive multifocal leukoencephalopathy. J Neurovirol. 2000;6:221–228. doi: 10.3109/13550280009015824. [DOI] [PubMed] [Google Scholar]
- Du T, Zamore PD. microPrimer: The biogenesis and function of microRNA. Development. 2005;132:4645–4652. doi: 10.1242/dev.02070. [DOI] [PubMed] [Google Scholar]
- Eberwine J, Belt B, Kacharmina JE, Miyashiro K. Analysis of subcellularly localized mRNAs using in situ hybridization, mRNA amplification, and expression profiling. Neurochem Res. 2002;27:1065–1077. doi: 10.1023/a:1020956805307. [DOI] [PubMed] [Google Scholar]
- Everall IP, Heaton RK, Marcotte TD, Ellis RJ, McCutchan JA, Atkinson JH, Grant I, Mallory M, Masliah E. Cortical synaptic density is reduced in mild to moderate human immunodeficiency virus neurocognitive disorder. HNRC Group. HIV Neurobehavioral Research Center. Brain Pathol. 1999;9:209–217. doi: 10.1111/j.1750-3639.1999.tb00219.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebauer F, Hentze MW. Molecular mechanisms of translational control. Nat Rev Mol Cell Biol. 2004;5:827–835. doi: 10.1038/nrm1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase AD, Jaskiewicz L, Zhang H, Laine S, Sack R, Gatignol A, Filipowicz W. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep. 2005;6:961–967. doi: 10.1038/sj.embor.7400509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haughey NJ, Nath A, Mattson MP, Slevin JT, Geiger JD. HIV-1 Tat through phosphorylation of NMDA receptors potentiates glutamate excitotoxicity. J Neurochem. 2001;78:457–467. doi: 10.1046/j.1471-4159.2001.00396.x. [DOI] [PubMed] [Google Scholar]
- Hengst U, Jaffrey SR. Function and translational regulation of mRNA in developing axons. Semin Cell Dev Biol. 2007;18:209–215. doi: 10.1016/j.semcdb.2007.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornstein E, Shomron N. Canalization of development by microRNAs. Nat Genet. 2006;38:S20–S24. doi: 10.1038/ng1803. [DOI] [PubMed] [Google Scholar]
- Hudson L, Liu J, Nath A, Jones M, Raghavan R, Narayan O, Male D, Everall I. Detection of the human immunodeficiency virus regulatory protein tat in CNS tissues. J Neurovirol. 2000;6:145–155. doi: 10.3109/13550280009013158. [DOI] [PubMed] [Google Scholar]
- Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- Hwang HW, Mendell JT. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer. 2006;94:776–780. doi: 10.1038/sj.bjc.6603023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen RS, Saykin AJ, Cannon L, Campbell J, Pinsky PF, Hessol NA, O’Malley PM, Lifson AR, Doll LS, Rutherford GW, et al. Neurological and neuropsychological manifestations of HIV-1 infection: Association with AIDS-related complex but not asymptomatic HIV-1 infection. Ann Neurol. 1989;26:592–600. doi: 10.1002/ana.410260503. [DOI] [PubMed] [Google Scholar]
- Ke XS, Liu CM, Liu DP, Liang CC. MicroRNAs: Key participants in gene regulatory networks. Curr Opin Chem Biol. 2003;7:516–523. doi: 10.1016/s1367-5931(03)00075-9. [DOI] [PubMed] [Google Scholar]
- Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001;15:2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Krichevsky A, Grad Y, Hayes GD, Kosik KS, Church GM, Ruvkun G. Identification of many microRNAs that copurify with polyribosomes in mammalian neurons. Proc Natl Acad Sci USA. 2004;101:360–365. doi: 10.1073/pnas.2333854100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HK, Kim YB, Kim EG, Schuman E. Measurement of dendritic mRNA transport using ribosomal markers. Biochem Biophys Res Commun. 2005;328:895–900. doi: 10.1016/j.bbrc.2005.01.041. [DOI] [PubMed] [Google Scholar]
- Klein ME, Impey S, Goodman RH. Role reversal: The regulation of neuronal gene expression by microRNAs. Curr Opin Neurobiol. 2005;15:507–513. doi: 10.1016/j.conb.2005.08.011. [DOI] [PubMed] [Google Scholar]
- Kruman II, Nath A, Mattson MP. HIV-1 protein Tat induces apoptosis of hippocampal neurons by a mechanism involving caspase activation, calcium overload, and oxidative stress. Exp Neurol. 1998;154:276–288. doi: 10.1006/exnr.1998.6958. [DOI] [PubMed] [Google Scholar]
- Kye MJ, Liu T, Levy SF, Xu NL, Groves BB, Bonneau R, Lao K, Kosik KS. Somatodendritic microRNAs identified by laser capture and multiplex RT-PCR. RNA (New York, NY) 2007;13:1224–1234. doi: 10.1261/rna.480407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai EC. microRNAs: Runts of the genome assert themselves. Curr Biol. 2003;13:R925–R936. doi: 10.1016/j.cub.2003.11.017. [DOI] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Lugli G, Larson J, Martone ME, Jones Y, Smalheiser NR. Dicer and eIF2c are enriched at postsynaptic densities in adult mouse brain and are modified by neuronal activity in a calpain-dependent manner. J Neurochem. 2005;94:896–905. doi: 10.1111/j.1471-4159.2005.03224.x. [DOI] [PubMed] [Google Scholar]
- Lukiw WJ. Micro-RNA speciation in fetal, adult and Alzheimer’s disease hippocampus. Neuroreport. 2007;18:297–300. doi: 10.1097/WNR.0b013e3280148e8b. [DOI] [PubMed] [Google Scholar]
- Masliah E, Heaton RK, Marcotte TD, Ellis RJ, Wiley CA, Mallory M, Achim CL, McCutchan JA, Nelson JA, Atkinson JH, Grant I. Dendritic injury is a pathological substrate for human immunodeficiency virus-related cognitive disorders. HNRC Group. The HIV Neurobehavioral Research Center. Ann Neurol. 1997;42:963–972. doi: 10.1002/ana.410420618. [DOI] [PubMed] [Google Scholar]
- Masliah E, Roberts ES, Langford D, Everall I, Crews L, Adame A, Rockenstein E, Fox HS. Patterns of gene dysregulation in the frontal cortex of patients with HIV encephalitis. J Neuroimmunol. 2004;157:163–175. doi: 10.1016/j.jneuroim.2004.08.026. [DOI] [PubMed] [Google Scholar]
- McArthur JC. HIV dementia: An evolving disease. J Neuroimmunol. 2004;157:3–10. doi: 10.1016/j.jneuroim.2004.08.042. [DOI] [PubMed] [Google Scholar]
- Mi S, Lu J, Sun M, Li Z, Zhang H, Neilly MB, Wang Y, Qian Z, Jin J, Zhang Y, Bohlander SK, Le Beau MM, Larson RA, Golub TR, Rowley JD, Chen J. MicroRNA expression signatures accurately discriminate acute lymphoblastic leukemia from acute myeloid leukemia. Proc Natl Acad Sci USA. 2007;104:19971–19976. doi: 10.1073/pnas.0709313104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr E, Richter D. Messenger RNA on the move: Implications for cell polarity. Int J Biochem Cell Biol. 2001;33:669–679. doi: 10.1016/s1357-2725(01)00047-4. [DOI] [PubMed] [Google Scholar]
- Moore DJ, Masliah E, Rippeth JD, Gonzalez R, Carey CL, Cherner M, Ellis RJ, Achim CL, Marcotte TD, Heaton RK, Grant I. Cortical and subcortical neurodegeneration is associated with HIV neurocognitive impairment. AIDS. 2006;20:879–887. doi: 10.1097/01.aids.0000218552.69834.00. [DOI] [PubMed] [Google Scholar]
- Peruzzi F. The multiple functions of HIV-1 Tat: Proliferation versus apoptosis. Front Biosci. 2006;11:708–717. doi: 10.2741/1829. [DOI] [PubMed] [Google Scholar]
- Sacktor N. The epidemiology of human immunodeficiency virus-associated neurological disease in the era of highly active antiretroviral therapy. J Neurovirol. 2002;8:115–121. doi: 10.1080/13550280290101094. [DOI] [PubMed] [Google Scholar]
- Salaun C, James DJ, Greaves J, Chamberlain LH. Plasma membrane targeting of exocytic SNARE proteins. Biochim Biophys Acta. 2004;1693:81–89. doi: 10.1016/j.bbamcr.2004.05.008. [DOI] [PubMed] [Google Scholar]
- Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, Greenberg ME. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5:R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimohama S, Kamiya S, Taniguchi T, Akagawa K, Kimura J. Differential involvement of synaptic vesicle and presynaptic plasma membrane proteins in Alzheimer’s disease. Biochem Biophys Res Commun. 1997;236:239–242. doi: 10.1006/bbrc.1997.6940. [DOI] [PubMed] [Google Scholar]
- Smirnova L, Grafe A, Seiler A, Schumacher S, Nitsch R, Wulczyn FG. Regulation of miRNA expression during neural cell specification. Eur J Neurosci. 2005;21:1469–1477. doi: 10.1111/j.1460-9568.2005.03978.x. [DOI] [PubMed] [Google Scholar]
- Steward O, Schuman EM. Protein synthesis at synaptic sites on dendrites. Annu Rev Neurosci. 2001;24:299–325. doi: 10.1146/annurev.neuro.24.1.299. [DOI] [PubMed] [Google Scholar]
- Sze CI, Bi H, Kleinschmidt-DeMasters BK, Filley CM, Martin LJ. Selective regional loss of exocytotic presynaptic vesicle proteins in Alzheimer’s disease brains. J Neurol Sci. 2000;175:81–90. doi: 10.1016/s0022-510x(00)00285-9. [DOI] [PubMed] [Google Scholar]
- Tai HC, Schuman EM. MicroRNA: MicroRNAs reach out into dendrites. Curr Biol. 2006;16:R121–R123. doi: 10.1016/j.cub.2006.02.006. [DOI] [PubMed] [Google Scholar]