Abstract
Vibrio vulnificus, a septicemia-causing pathogenic bacterium, acquires resistance against various stresses and expresses virulence factors via an rpoS gene product. In this study, we investigated the transcriptional characteristics of this global regulator. Two distinct transcriptional initiation sites for the rpoS gene, the proximal promoter (Pp) and the distal promoter (Pd), were defined by primer extension experiments. Various rpoS::luxAB transcriptional fusions indicated that Pd is a major promoter of rpoS expression. Western blot analysis showed that RpoS levels were inversely correlated with intracellular levels of 3′,5′-cyclic AMP (cAMP). The expressions of both Pd and Pp were increased in cya and crp mutants. The exogenous addition of cAMP to the cya mutant resulted in repressed expression of rpoS. In addition, rpoS expression was significantly lowered in the cpdA mutant, in which the level of cAMP was elevated because of the absence of 3′,5′-cAMP phosphodiesterase. In vitro transcription assays using the V. vulnificus RNA polymerase showed that the transcripts from both promoters were reduced by addition of the cAMP-cAMP receptor protein (CRP). The cAMP-CRP was shown to bind to two rpoS promoters by electrophoretic mobility shift assays. The alteration of the putative CRP-binding site on each rpoS promoter, via site-directed mutagenesis, abolished the binding of cAMP-CRP as well as regulation by cAMP-CRP. Therefore, this study shows a relationship between the level of intracellular cAMP and the degree of rpoS expression and further demonstrates, for the first time, the direct binding of the cAMP-CRP complex to rpoS upstream regions, which results in repression of rpoS gene expression.
Global and immediate response to diverse environmental stimuli is one of the characteristics of bacterial adaptation and survival. Cellular responses to stressful conditions have been extensively studied in many bacterial species, most notably in Escherichia coli (1). To respond properly to diverse stresses, E. coli requires the rpoS gene product, which is a second principal σ factor, RpoS (σs), to endow it with the ability to mediate changes in bacterial physiology and structure. Vibrio vulnificus, a human pathogen causing a fatal septicemia with rapid pathogenic progression and a high mortality rate (2), has been shown to require RpoS for better survival under nutrient starvation, oxidative stress, UV irradiation, and acidic conditions (3, 4).
RpoS is also required for eliciting phenotypes related to virulence in many pathogenic bacteria belonging to the γ-subdivision of Proteobacteria (5). V. vulnificus has been shown to use RpoS to express its major virulence factors, such as an elastase (6) and other exoproteases (3), and for expression of the fur gene, which encodes a transcriptional regulator of virulence-associated iron-uptake systems (7, 8).
Thus, the intracellular level of RpoS needs to be finely adjusted depending on the environmental conditions, and it is modulated via transcriptional control of the rpoS gene, translational efficiency of rpoS mRNA, proteolysis of the RpoS protein, and the interaction of RpoS with RNA polymerase core subunits (1). Transcriptional regulation of rpoS expression has been shown to be mediated by diverse trans-acting proteins (9). A transcription factor, ArcA, represses rpoS expression by binding to the regulatory regions flanking the major promoter of the E. coli rpoS gene (10). The repressor Fis and the activator PsrA directly bind to the rpoS gene regulatory regions of Salmonella enterica and Pseudomonas spp., respectively (11, 12). In Borrelia burdorferi, another σ factor, RpoN, is involved in the expression of its rpoS gene (13).
The role of cAMP-CRP2 complex as a negative transcriptional regulator in rpoS expression was shown genetically using crp, cya, and crr knock-out mutants of E. coli and other bacterial species (14, 15). However, whether control by cAMP-CRP complex is direct or indirect has not yet been determined, because the effects of cAMP-CRP on rpoS expression were shown only by transcriptional fusion assays, and direct evidence for the binding of cAMP-CRP complex to rpoS promoter regions and the effect of cAMP-CRP on rpoS transcription have not been reported in any bacterial species (1, 16). In addition, the locations of the putative CRP-binding sites of E. coli rpoS were predicted to be the sites centered at –61.5 and +56.5 with respect to the major transcription start site. Because the CRP-binding sites usually overlap with the promoters of the genes repressed by the cAMP-CRP complex, those putative CRP-binding sites are not typical for a negative regulation; one (centered at –61.5) is usually an activating site and the other (centered at +56.5) is located at the downstream region of the rpoS initiation codon (IC) (1, 10). Thus, the molecular basis of how the cAMP-CRP complex modulates the transcription of the rpoS gene has not been completely defined.
We have previously isolated the rpoS homolog from V. vulnificus (17) and characterized the roles of RpoS in bacterial response to various environmental conditions, which bacteria might encounter within a host (4, 6, 7). Although the functions of RpoS are quite conserved in diverse bacteria, the regulatory modes of rpoS gene transcription are distinct in each bacterium (9). Therefore, we are interested in the transcriptional characteristics of V. vulnificus rpoS. In this study, we defined two distinct promoters for the V. vulnificus rpoS gene and demonstrated direct evidence for transcriptional regulation of rpoS via the interaction of the cAMP-CRP complex with these two promoters.
EXPERIMENTAL PROCEDURES
Strains, Plasmids, and Culture Cultivation—Bacterial strains and plasmids used for this study are listed in Table 1. E. coli strains used for plasmid DNA preparation and conjugational transfer were grown in Luria-Bertani (LB) broth or on LB plates containing 1.5% (w/v) agar. V. vulnificus strains were grown in LB medium supplemented with additional 2% (w/v) NaCl (LBS), unless stated otherwise. All medium components were purchased from Difco, and chemicals and antibiotics were purchased from Sigma.
TABLE 1.
Bacterial strains and plasmids used in this study
Primer Extension Experiment—An oligonucleotide, PE+83 (Table 2), complementary to the open reading frame (ORF) of rpoS was end-labeled with [γ-32P]ATP using T4 polynucleotide kinase and then used for cDNA synthesis. RNA was converted to cDNA with SuperScript II reverse transcriptase (Invitrogen). The resultant cDNA products were purified and resolved on a sequencing gel alongside sequencing ladders generated with the same primer used for primer extension. The nucleotide sequence of pINE32 was determined using the dideoxy chain termination method with Top™ DNA polymerase (Bioneer) as described previously (18). The sequencing gels were dried and then visualized upon exposure to a PhosphorImager (Personal Molecular Imager FX, Bio-Rad). Another oligonucleotide, PE-368 (Table 2), was used for synthesis of cDNA to examine rpoS expression derived from the second promoter as described above.
TABLE 2.
Oligonucleotides used in this studya
a Restriction sites are underlined and their usages in cloning experiments are described under “Experimental Procedures.”
b Modified nucleotides for site-directed mutagenesis are italicized and the detailed information is described in the text.
Construction of the ΔcpdA V. vulnificus Strain, HY101—A 762-bp cpdA upstream region was amplified using primers cpd5F and cpd5R (Table 2), which locate at –784 and –27 relative to the cpdA IC, respectively. The resultant PCR product was digested with the appropriate restriction enzymes, SpeI and BglII, and ligated to a suicide vector pDM4 treated with the same enzymes (19) to produce pHY01. A 449-bp DNA fragment was amplified using primers icc3F and icc3R (Table 2), which are located at +302 and +750 relative to the IC of cpdA, respectively. This PCR product was digested with BglII and SphI and cloned into pHY01, resulting in pHY02. Finally, a 1.2-kb kanamycin resistance gene, isolated by digesting pUC4K (GE Healthcare) with BamHI, was inserted into the BglII site of pHY02 to yield pHY03. Plasmid pHY03 in the SM10 λpir (20) strain was mobilized to V. vulnificus AR, a rifampin-resistant derivative strain of V. vulnificus ATCC29307, and the conjugates were selected by plating the mixture of E. coli and V. vulnificus on LBS supplemented with 4 μg/ml chloramphenicol and 50 μg/ml rifampin. A colony with characteristics indicating a double homologous recombination event (resistance to 5% (w/v) sucrose, sensitivity to chloramphenicol, and resistance to kanamycin) was further confirmed by PCR, using primers cpd5F and icc3R, and named HY101.
Construction of Δcya V. vulnificus Strain, KP301—A 3,245-bp DNA fragment containing the cya ORF and its upstream and downstream regions was amplified from the genomic DNA of V. vulnificus ATCC29307 using two primers, cya-F and cya-R (Table 2). The PCR product was digested with ApaI and XbaI and then cloned into pBluescript II SK(+) to produce pVVcya. A mutation in cya was created by deleting the NsiI fragment within the cya coding region in pVVcya, thus yielding pVVΔcya. A 1,973-bp DNA fragment of pVVΔcya digested with ApaI and XbaI was ligated into a suicide vector, pDM4, to generate pDMΔcya. The E. coli SM10 λpir strain carrying pDMΔcya was conjugated to V. vulnificus ATCC29307, and the exconjugants were then selected on thiosulfate citrate bile sucrose medium supplemented with 4 μg/ml chloramphenicol. A colony indicating double homologous recombination was confirmed by PCR, using primers cya-F and cya-R, and named KP301.
Construction of rpoS luxAB Transcriptional Fusions—A set of rpoS::luxAB transcriptional fusions was made by subcloning a series of rpoS promoter DNA fragments into pHK0011 (6). Primer rpoS+88 (Table 2) contained an XbaI restriction site followed by bases corresponding to the 5′-end of the rpoS coding region. Primer rpoS+88 was used in conjunction with one of the following primers to amplify DNA upstream of rpoS: rpoS-105 (for pKP-105), rpoS-368 (for pKP-368), rpoS-732 (for pKP-732), rpoS-891 (for pKP-891), rpoS-1315 (for pKP-1315), and rpoS-1640 (for pKP-1640) (Tables 1 and 2). The PCR products were digested with BamHI and XbaI and then inserted into pHK0011, which had been digested with the same enzymes, to create six different rpoS::luxAB fusions.
Luciferase Assay—The rpoS::luxAB reporters were mobilized into wild type, ΔcpdA, crp, and Δcya V. vulnificus mutants via conjugal transfer. Overnight (16–18 h) cultures of the bacterial cells containing one of these fusions were inoculated into fresh LBS medium containing tetracycline (3 μg/ml) and then grown to the stationary phase. At various time points of bacterial growth, a portion of the samples was taken from each culture and diluted 100-fold with LBS medium. The expression from various lengths of the rpoS promoter was measured by monitoring light production in the presence of 0.006% (v/v) n-decyl aldehyde using a luminometer (TD-20/20; Turners Designs). Light production was expressed in arbitrary relative light units (RLU), and the specific bioluminescence was calculated by normalizing RLU with cell mass (A595), as described previously (7). cAMP at a concentration of 0.5 mm was added exogenously to the culture of Δcya mutant V. vulnificus either with pKP-368 or pKP-1315, and the light emission from these cells was monitored at the various phases of bacterial growth.
Western Blot Analysis—The plasmid pQErpoS was constructed to express the recombinant RpoS protein as a histidine-tagged form, which was then used to produce polyclonal antibodies against V. vulnificus RpoS as described previously (4). Wild type V. vulnificus was harvested at various phases of bacterial growth in LBS broth, and bacterial extracts were then prepared by sonication in TNT buffer (10 mm Tris-HCl, 150 mm NaCl, and 0.05% (v/v) Tween 20, pH 8.0) (21). Forty micrograms of the extracts were fractionated by SDS-PAGE. After a transfer to a Hybond P membrane (Amersham Biosciences), Western blot analysis was performed by serially incubating the filter with anti-V. vulnificus RpoS antibodies (1:5,000) and alkaline phosphatase-conjugated rabbit anti-rat immunoglobulin G (1:5,000; Sigma). The RpoS protein of V. vulnificus was visualized using an nitro blue tetrazolium-5-bromo-4-chloro-3-indolyl phosphate system (Promega). To investigate the role of cAMP-CRP in RpoS formation, cell extracts of wild type, crp, Δcya, and ΔcpdA mutant V. vulnificus were prepared and then examined by Western blot analysis with RpoS-specific antibodies. cAMP was added to the Δcya mutant at a final concentration of 0.5 mm, and the bacterial extract was also prepared for Western blot analysis.
Purification of V. vulnificus RNA Polymerase (RNAP) Coreenzyme and σ Factor 70 (RpoD)—V. vulnificus MO6–24/O was grown in LBS medium at 30 °C for 6 h. Cell pellet was resuspended in the cold grinding buffer (50 mm Tris, 5% (v/v) glycerol, 2 mm EDTA, 233 mm NaCl, 1 mg/ml lysozyme, 1 mm phenylmethylsulfonyl fluoride, 0.5 mm dithiothreitol, and 7 mm β-mercaptoethanol, pH 8.0). After 20 min of incubation on ice, the bacterial cells were disrupted by sonication in the presence of 4% (w/v) sodium deoxycholate. The core RNAP was purified by an immunoaffinity chromatography using the polyol-responsive monoclonal antibody 8RB13 (NeoClone Biotechnology) as described (22, 23). Eluted core RNAP was then dialyzed with the storage buffer (50 mm Tris-HCl, 0.1 mm EDTA, 0.1 mm dithiothreitol, 50% (v/v) glycerol, pH 7.6). The coding region of V. vulnificus rpoD was amplified using oligonucleotide primers, F-rpoDvv and R-rpoDvv (Table 2). The 1.8-kb PCR product was digested with NdeI and XhoI and then ligated to the expression vector, pET28a (Novagen), which was digested with the same enzymes, to produce pSH0505. His-tagged recombinant RpoD protein was expressed in E. coli BL21 in the presence of 1.0 mm isopropyl thio-β-d-galactoside and then purified by a nickel-nitrilotriacetic acid affinity chromatography according to the manufacturer's procedure (Qiagen).
In Vitro Transcription Assay in the Presence of CRP—The plasmids containing the templates for RNAP were constructed by inserting the DNA fragments of rpoS promoter regions to pRLG770 (24). The insert DNA, including rpoS Pd, was amplified using the PCR primers rpoS-IV-1 and rpoS-IV-2. The insert DNA, including rpoS Pp, was amplified using rpoS-IV-3 and rpoS-IV-5 (Table 2). Amplified DNA were digested with EcoRI and HindIII and then ligated to EcoRI/HindIII-treated pRLG770. Purified CRP (0, 72, 144, 216, and 288 nm) and the plasmid (0.25 nm) were mixed in the reaction buffer (50 mm KCl, 20 mm Tris-HCl, 3 mm MgCl2, 0.1 mm dithiothreitol, 0.1 mm EDTA, 0.5 mm cAMP, pH 8.0) and incubated at 37 °C for 40 min. The radiolabeled nucleotide (5 μCi of [α-32P]UTP) and cold nucleotides (25 μm UTP and 500 μm each ATP, CTP, and GTP) were added to the reaction mixture, and the transcription was initiated by adding the V. vulnificus RNAP holoenzyme premixture of 10 nm core RNAP and 10 nm RpoD (supplemental Fig. 1). The in vitro transcription reactions were stopped with the stop buffer (95% formamide, 20 mm EDTA, 0.05% bromphenol blue, and 0.05% xylene cyanol, pH 8.0) after 15 min. The reaction mixtures were electrophoresed on a 6.5% polyacrylamide/bisacrylamide (19:1), 7 m urea denaturing gel. Transcripts were visualized and quantified using Personal Molecular Imager FX and Quantity One software, respectively (Bio-Rad).
Electrophoretic Mobility Shift Assay—V. vulnificus recombinant CRP protein was overexpressed in E. coli BL21 carrying pHK0201 (25), a pRSETA (Invitrogen)-based expression plasmid, and purified by a nickel-nitrilotriacetic acid affinity chromatography according to the manufacturer's procedure (Qiagen). The 394-bp upstream region of the rpoS gene, extending from residues –732 to –339 with respect to the IC of rpoS, was amplified by PCR using 32P-labeled PE-368 and unlabeled rpoS-732 as primers (Table 2). The other rpoS promoter region used for binding assays was made with two primers, PE+83 and rpoS-368 (Table 2), which contained the 456-bp DNA fragment from –373 to +83 with respect to the IC of rpoS. Seven nanomolar of the labeled DNA fragment was incubated with varying concentrations of purified histidine-tagged CRP protein (150–600 nm) for 30 min at 37 °C in a 20-μl reaction mixture containing 1× binding buffer (26), including 500 μm cAMP (Sigma). Following the addition of 3 μl of loading buffer to each reaction, the samples were separated by electrophoresis on a 6% nondenaturing polyacrylamide gel. For competition analyses, the identical but unlabeled rpoS DNA fragment was included as a competitor DNA. Various amounts of competitor DNA (14–70 nm) were added to the reaction mixture containing 7 nm of the labeled DNA prior to the addition of 600 nm CRP. A 378-bp DNA fragment encompassing the promoter region of the gap gene encoding glyceraldehyde-3-phosphate dehydrogenase was amplified from genomic DNA of V. vulnificus using primers gap-F and gap-R (Table 2) and included as nonspecific DNA in the binding assay.
cAMP Assay—V. vulnificus cells grown in LBS were harvested, and the amounts of cAMP in the bacterial lysates were estimated according to the manufacturer's instructions (cAMP Biotrak EIA System, Amersham Biosciences).
Site-directed Mutagenesis of the rpoS Promoters—The putative CRP-binding sites include the sequences homologous to the inverted repeat, TGTGAN6TCACA. Inverted repeats, which are located at –26 to –47 relative to the distal transcription initiation site (CRP-binding site I) and –27 to –48 relative to the proximal transcription initiation site (CRP-binding site II) of the rpoS gene, were mutagenized using primers carrying the substituted nucleotides. To amplify the mutated CRP-binding site I, two sets of primers, rpoS-1315/rpoSM1-R and rpoSM1-F/rpoS+88 (Table 2), were utilized. Two PCR products (an 846-bp PCR product using rpoS-1315 and rpoSM1-R and a 584-bp PCR product using rpoSM1-F and rpoS+88) were used as template DNAs to produce the mutagenized rpoS DNA fragment, encompassing the segment from –1315 and +88 relative to the IC of rpoS, using primers rpoS-1315 and rpoS+88. The resultant mutagenized rpoS promoter DNA was cloned into the pGEM®-T Easy vector (Promega) to produce pGEMT-rpoSmt1. To amplify the mutated CRP-binding site II, two primer sets, rpoS-368/rpoSM2-R and rpoSM2-F/rpoS+88, were used. Two PCR products (a 345-bp PCR product using rpoS-368 and rpoSM2-R, and a 142-bp PCR product using rpoSM2-F and rpoS+88) were used as template DNAs to produce the mutagenized rpoS DNA fragment, encompassing the segment from –368 and +88 relative to the IC of rpoS, using primers rpoS-368 and rpoS+88. The resultant mutagenized rpoS promoter DNA was cloned into the pGEM®-T Easy vector (Promega) to produce pGEMT-rpoSmt2. The mutagenized nucleotide sequences of both pGEMT-rpoSmt1 and pGEMT-rpoSmt2 were confirmed by DNA sequencing. Then the insert DNA fragment of each plasmid, digested with BamHI and XbaI, was ligated to BamHI/XbaI-digested pHK0011, which contained the promoterless luxAB genes. The resultant plasmids, prpoSM1 and prpoSM2, were mobilized into V. vulnificus strains by conjugation, and the exconjugants were selected in thiosulfate citrate bile sucrose medium supplemented with 3 μg/ml tetracycline.
Statistical Analyses—Results were expressed as the means ± S.D. from at least three independent experiments. Statistical analysis was performed using the Student's t test (SYSTAT program, SigmaPlot version 9, Systat Software Inc.). Differences were considered significant at p values <0.01.
Nucleotide Sequence Accession Numbers—The nucleotide sequences of the cpdA and cya genes isolated from V. vulnificus ATCC29307 have been deposited in the GenBank™ data base under the accession numbers AY221025 and AY240931, respectively.
RESULTS
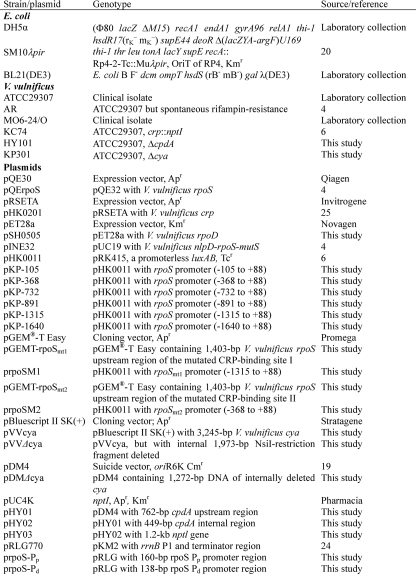
Identification of Two Transcription Start Sites of the V. vulnificus rpoS Gene—RNA prepared from the stationary phase cells was converted to cDNA using primer PE+83, which annealed to the nucleotide sequences between +54 and +83 relative to the IC of rpoS. Upon gel electrophoresis with the sequencing ladder of the rpoS gene with the same primer, a single signal was observed 29 bp upstream of the IC of RpoS (Fig. 1A), which is 52 bp downstream of the stop codon of the adjacent gene, nlpD. Putative –10 (TAAAGT) and –35 (TTGCGA) sequences were discernable, and this promoter was designated as Pp, a promoter proximal to the rpoS ORF (Fig. 1C).
FIGURE 1.
Identification of the transcriptional start sites of the rpoS gene in V. vulnificus. A, primer extension experiment using V. vulnificus RNA and the oligonucleotide primer PE+83 (annealing to the region from +54 to +83 relative to the IC of rpoS). Lanes C, T, A, and G represent the nucleotide sequencing ladders of pINE32. The asterisk indicates the site of transcriptional initiation for rpoS. B, primer extension experiment using the oligonucleotide primer PE-368 (annealing to the region from –368 to –339 relative to IC) as described above. C, two rpoS promoters in the nlpD open reading frame and nlpD-rpoS intergenic space. The promoter Pp is proximal to the IC of rpoS, and the promoter Pd is distal from the IC of rpoS. The promoters, –10 and –35 regions, are underlined, whereas the stop codon of nlpD and the start codon of rpoS are capitalized.
The same RNA was hybridized with another primer PE-368, which contained complementary sequences to nucleotide sequences between –368 and –339, relative to the IC of rpoS. The initiation site of the resultant cDNA was mapped at –473 with respect to the IC of rpoS (Fig. 1B). This putative promoter resided within the ORF of the upstream gene, nlpD, with reasonable promoter sequences (CTGTCA and AATAAC as –35 and –10, respectively), and was designated as Pd, a distal promoter to the rpoS ORF (Fig. 1C).
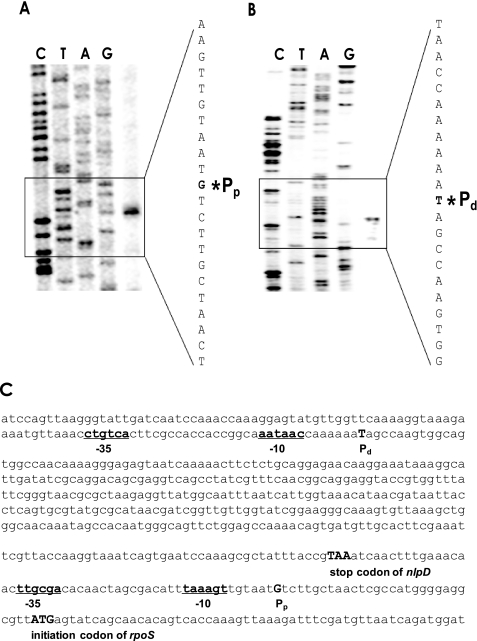
Expressional Analysis of rpoS Promoters Using luxAB-transcriptional Fusions—A series of luxAB-transcriptional fusions were constructed by cloning various lengths of the rpoS promoter regions (Fig. 2A). Two rpoS::luxAB fusions, pKP-105 and pKP-368, contained only the Pp, whereas the remaining four fusions with longer promoter regions had both promoters Pd and Pp. During growth, the expressions of these diverse fusions were determined by measuring their luciferase activities (Fig. 2B). The basal and induced expression levels of pKP-105 were similar to those of pKP-368. The remaining four longer fusions (pKP-732, -891, -1315, and -1640) also showed similar levels of expression compared with each other, but significantly higher expression (over 200-fold) relative to the other two shorter fusions, including only Pp. When bacterial cells entered the stationary phase, the shorter fusions were induced about 3–4-fold, and the longer fusions were induced 12–15-fold. For further analyses, pKP-368 was chosen to represent the expression from Pp only, and pKP-1315 was chosen to represent the expression from both Pd and Pp.
FIGURE 2.
Deletion analysis of the regulatory region of rpoS. A, construction of rpoS::luxAB fusion plasmids. PCR fragments carrying different lengths of the upstream region of rpoS were cloned into pHK0011 to create various transcriptional reporter luxAB fusions. Two promoters, defined by primer extension analyses, are indicated as Pd and Pp. B, expression of rpoS::luxAB fusions. Wild type V. vulnificus carrying one of the six rpoS::luxAB fusions was grown in LBS medium supplemented with 3 μg/ml tetracycline and examined for luminescence as they grew (open circles). Luciferase activities are expressed as normalized values (closed circles): number of relative light units (RLU) divided by the A595 value of each sample.
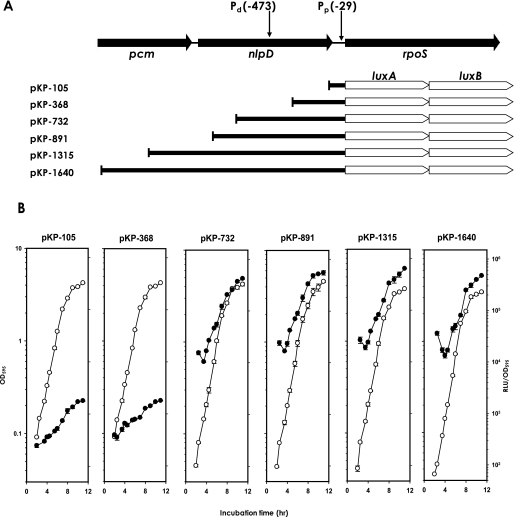
Intracellular Levels of RpoS and cAMP during V. vulnificus Growth—Using polyclonal antibodies specific to V. vulnificus RpoS, intracellular levels of RpoS were measured at various time points along a growth curve (Fig. 3). Upon Western blot analysis with RpoS-specific antibodies, an immunoreactive band of ∼40 kDa was observed in eight different bacterial extracts along the growth curve. The intensity of this band became stronger as the cells entered the stationary phase, as shown by the assay using luxAB-transcriptional fusions. Densitometric quantification of these immunoreactive bands indicates about a 5-fold increase in the RpoS level as bacterial cells entered the stationary phase in LBS medium.
FIGURE 3.
Intracellular levels of cAMP and RpoS at various growth stages of V. vulnificus. V. vulnificus cells grown in LBS were assayed for cAMP using the cAMP Biotrak enzyme immunoassay system (Amersham Biosciences), and presented as femtomoles of cAMP per μg of bacterial protein (circles). At various time points of bacterial growth (designated by numbers from 1 to 8), cells were harvested and then subjected to Western blot analysis (lower panel). The intensities of bands corresponding to RpoS were estimated by densitometry and are also presented in the plot (triangles).
The observation that transcription of the rpoS gene and the amount of RpoS increased at the stationary phase suggests the presence of some intracellular signal(s) that switched on the rpoS expression system. The most probable signals related to the high cell density or stationary phase conditions are quorum-sensing signal molecules and cAMP, as suggested previously (27–29). The luxS mutant V. vulnificus, which is defective in production of the autoinducer-2 (30), showed similar levels of RpoS to the wild type (supplemental Fig. 2). cAMP levels were also estimated from the bacterial cells grown under the same incubation condition used for Western blot analysis (Fig. 3). V. vulnificus grown exponentially in the complex medium, such as salt-enriched LB broth, contained 20–50 fmol of cAMP/μg of bacterial protein. When V. vulnificus entered the stationary phase, its intracellular cAMP contents dramatically decreased to the levels less than 5 fmol of cAMP/μg of bacterial protein. This result suggests an inverse correlation between cAMP concentration and rpoS expression.
Effect of cAMP-CRP on rpoS Expression—The inverse relationship between cAMP contents and rpoS expression implied that cAMP and CRP might repress rpoS gene expression. To test this hypothesis, two representative rpoS::luxAB fusions (pKP-368 and pKP-1315) were transferred to two mutant V. vulnificus strains, deficient in either cya (coding for the cAMP-synthesizing enzyme, adenylate cyclase) or crp (coding for cAMP-receptor protein). Both fusions in crp or Δcya mutant strains showed about a 3-fold increase in their expression over that of the wild type strain (Table 3). Exogenous addition of cAMP (0.5 mm) to the Δcya mutant reduced the expression of both fusions to wild type levels. In addition, another V. vulnificus mutant, which is deficient in the cpdA gene encoding a 3′,5′-cAMP phosphodiesterase, showed an even lower expression of both fusions compared with the wild type strain.
TABLE 3.
Expression of rpoS::luxAB fusions in various genetic backgrounds
|
Genetic background
|
Intracellular cAMP
levela
|
Expression of luxAB-transcriptional fusion
(RLU/OD)b
|
|
|---|---|---|---|
| pKP-368 | pKP-1315 | ||
| fmol/μg protein | |||
| Wild type | 8.4 | 1,320 ± 86 | 267,780 ± 1,450 |
| crp | 421 | 3,430 ± 95 | 739,960 ± 87,700 |
| Δcya | Not detectablec | 3,130 ± 26 | 632,590 ± 17,900 |
| Δcya + cAMP (0.5 mm) | ndd | 1,110 ± 9 | 219,540 ± 9,080 |
| ΔcpdA | 24 | 590 ± 16 | 119,480 ± 4,610 |
cAMP contents were measured from the stationary phase V. vulnificus, and its levels were expressed as moles of cAMP in the unit mass of bacterial cells (determined by protein amount in lysate).
RLU per cell mass determined by A595.
cAMP concentration was below the detection limit of the assay used in this study.
ND means not determined.
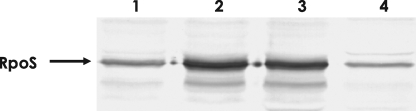
Cellular levels of RpoS proteins in crp and Δcya V. vulnificus mutants were also compared with that of the wild type isogenic strain (Fig. 4). In Western blot analysis using polyclonal antibodies against recombinant RpoS of V. vulnificus, more RpoS proteins were observed in both crp and Δcya strains than in the wild type. The Δcya strain grown in the presence of exogenously added cAMP demonstrated a decreased level of RpoS.
FIGURE 4.
Effect of cAMP and CRP on the intracellular level of RpoS protein. Extracts of various V. vulnificus strains, which were grown to the early stationary phase (A595 of 1.0–1.5), were subjected to Western blot analysis using polyclonal antibodies against V. vulnificus RpoS. Lane 1, wild type; lane 2, crp mutant; lane 3, Δcya mutant; and lane 4, Δcya mutant grown in the presence of exogenously added cAMP (0.5 mm).
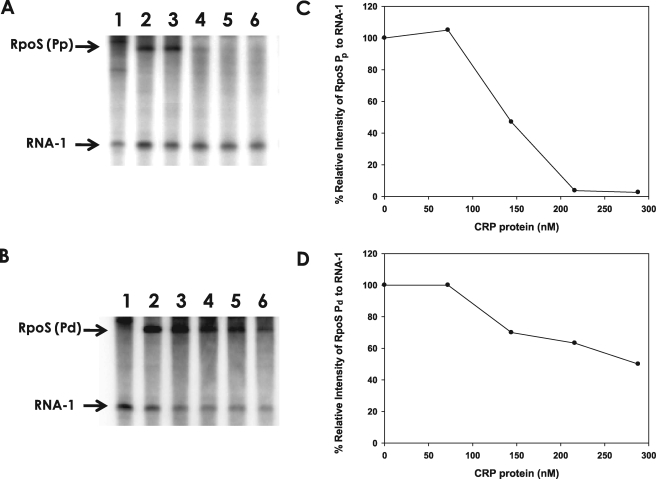
We then reconstituted the CRP-dependent repression of two rpoS promoters in vitro. The upstream regions of two rpoS promoters, Pd and Pp, were cloned into pRLG770 to produce prpoS-Pd and prpoS-Pp, respectively. In vitro transcription reaction with each plasmid DNA in the presence of core RNAP and RpoD of V. vulnificus produced two discrete transcripts, the plasmid-encoded RNA-1 (108 nucleotides long) and the longer transcript (188 nucleotides long RNA from prpoS-Pd or 210 nucleotides long RNA from prpoS-Pp) (Fig. 5). The results of in vitro transcription assays showed that cAMP-CRP has little or no effect on the synthesis of RNA-1 but repressed syntheses of the transcripts from the Pd and Pp promoters (Fig. 5, A and B). Effect of cAMP-CRP complex on transcription repression of Pp was more pronounced than that of Pd, because the slope of the decrease in relative intensities of rpoS Pp transcript was steeper than that of rpoS Pd transcript over the CRP concentration range used (Fig. 5, C and D). rpoS Pp transcript was not produced in the presence of more than 200 nm CRP. On the other hand, production of rpoS Pd transcript was less sensitive to CRP concentration. About 50% of the rpoS Pd transcript production was repressed in the presence of >200 nm CRP.
FIGURE 5.
Repression of rpoS transcription by the cAMP-CRP complex. A, in vitro transcription at the rpoS promoter Pp in the presence of cAMP-CRP. The pRLG770 (24)-based plasmid carrying the rpoS Pp (prpoS-Pp) was used for a template for core RNAP, RpoD, and CRP derived from V. vulnificus (supplemental Fig. 1). Produced 32P-labeled transcripts were separated on 6.5% polyacrylamide/bisacrylamide (19:1), 7 m urea denaturing gel showing two RNA bands (the control RNA of 108-nucleotide-long RNA-1 and the 210-nucleotide-long rpoS RNA) on each lane. Lane 1, pRLG770 (no rpoS promoter); lanes 2–6, prpoS-Pp incubated with 0, 72, 144, 216, or 288 nm of CRP. B, in vitro transcription at the rpoS promoter Pd in the presence of cAMP-CRP. The pRLG770 (24)-based plasmid carrying the rpoS Pd (prpoS-Pd) was used for an assay, which resulted in production of RNA-1 and 188-nucleotide-long rpoS RNA. Lane 1, pRLG770; lanes 2–6, prpoS-Pd incubated with 0, 72, 144, 216, or 288 nm of CRP. C and D, plots illustrating the relative intensity of rpoS transcript. The intensities of two RNA bands on each lane, the 108-base RNA-1 transcript, and the longer transcript starting from one of the rpoS promoters were estimated by densitometric reading (the plot C is from A, and the plot D is from the B), and then each rpoS transcript was normalized to RNA-1 on the same lane. Relative intensity of the normalized rpoS transcript was indicated as the percentage of that in the absence of CRP.
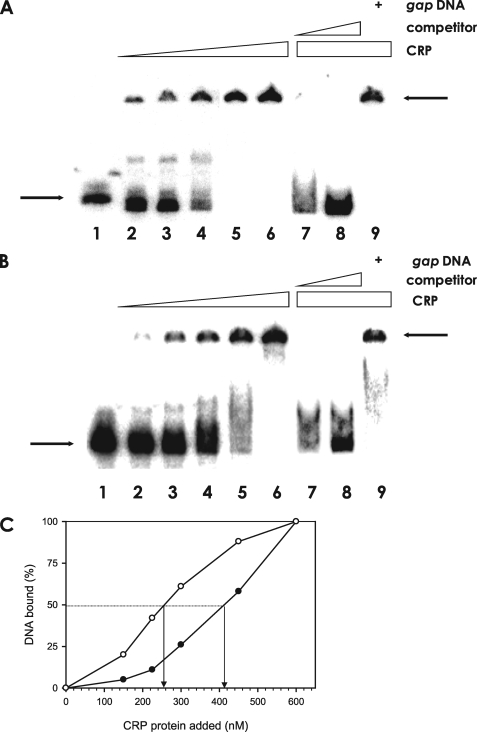
Specific Binding of cAMP-CRP Complex to the rpoS Promoters—This study clearly demonstrated that cAMP and CRP negatively affect the expression of rpoS. To determine whether the cAMP-CRP complex acts directly by binding to the rpoS promoter regions, we performed an electrophoretic mobility shift assay (EMSA) using V. vulnificus CRP protein and two different DNA fragments containing Pd or Pp. As shown in Fig. 6A, the addition of CRP and cAMP resulted in a shift of the 456-bp Pp DNA fragments to a band with slower mobility. Binding of cAMP-CRP to the Pp DNA was specific, because formation of the slower moving band was abolished by including excess unlabeled Pp DNA in the reaction mixture, but it retained its retarded mobility in the presence of a DNA fragment unrelated to the rpoS promoter sequence, such as the V. vulnficus gap promoter.
FIGURE 6.
Specific binding of the cAMP-CRP complex to the rpoS promoters. A, binding assay of CRP to the rpoS promoter carrying Pp. The 456-bp DNA fragment of the rpoS upstream region (Pp) was radiolabeled and incubated, with increasing amounts of CRP up to 600 nm. The identical but unlabeled competitor DNA fragment was included in the reaction mixtures in various amounts. Lane 1, probe DNA alone; lanes 2–6, probe DNA incubated with 150, 225, 300, 450, or 600 nm CRP, respectively; lane 7, probe DNA incubated with 600 nm CRP and 14 nm of the identical but unlabeled DNA; lane 8, probe DNA incubated with 600 nm CRP and 70 nm of the identical but unlabeled DNA; and lane 9, probe DNA incubated with 600 nm CRP and 70 nm gap promoter DNA. The arrows on the left side indicate the unbound DNA probe, whereas the arrows on the right side indicate DNA bound to CRP. B, binding assay of CRP to the rpoS promoter carrying Pd. Labeled Pd DNA was incubated with increasing amounts of CRP. For competition analysis, the identical but unlabeled 394-bp DNA fragment was included in the binding reactions containing 600 nm CRP. Lane 1, probe DNA alone; lanes 2–6, probe DNA incubated with 150, 225, 300, 450, or 600 nm CRP, respectively; lane 7, probe DNA incubated with 600 nm CRP and 14 nm of the identical but unlabeled DNA; lane 8, probe DNA incubated with 600 nm CRP and 70 nm of the identical but unlabeled DNA; and lane 9, probe DNA incubated with 600 nm CRP and 70 nm nonspecific gap promoter DNA. The arrows on the left side indicate unbound DNA probe, whereas the arrows on the right side indicate the DNA bound to CRP. C, plot showing the affinity of cAMP-CRP to each promoter of rpoS. The intensities of bound DNA fragments, Pp (open circle) and Pd (closed circle), were estimated by densitometer and plotted against the CRP concentrations. Arrows indicate the concentrations of CRP causing half-maximal binding corresponding to the Kd.
For another set of EMSA with cAMP-CRP, a 394-bp DNA containing Pd was used as a probe (Fig. 6B). When the labeled Pd DNA was incubated with cAMP-CRP, formation of the cAMP-CRP-DNA complex was detected as a slower moving DNA band. The specificity of cAMP-CRP binding to this DNA was also confirmed by competition experiments, in which excess unlabeled 394-bp Pd DNA competed out the binding of cAMP-CRP to the 32P-labeled Pd DNA in a dose-dependent manner. In contrast, inclusion of nonspecific gap promoter DNA in the binding assays did not disrupt the CRP-Pd DNA interaction. The apparent affinity of cAMP-CRP complex to each rpoS promoter was compared by extrapolating the CRP concentration required for 50% binding of the labeled DNA (Fig. 6C). The dissociation binding constants (Kd) for CRP to Pp and Pd were ∼250 and 410 nm, respectively. Thus, the results from both the in vitro transcription assays (Fig. 5) and EMSA (Fig. 6) suggest that the promoter Pp has higher affinity to cAMP-CRP complex.
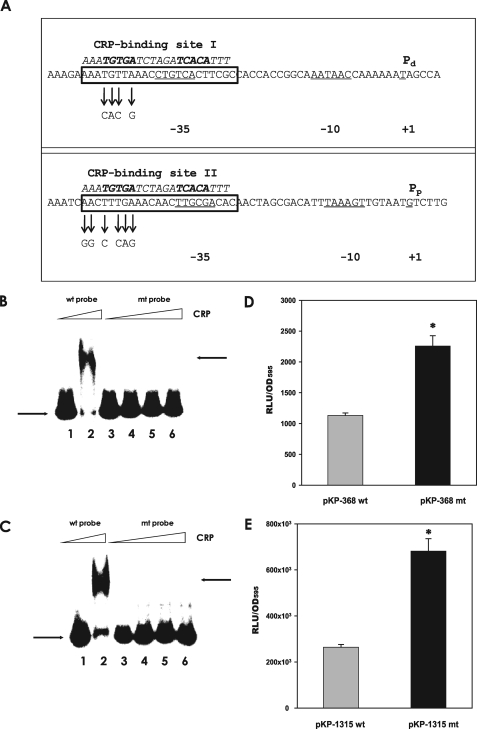
Effect of Mutation of the Putative CRP-binding Sites on Binding of CRP and Expression of rpoS—The nucleotide sequences of the two rpoS promoters were analyzed to determine whether they include the putative cAMP-CRP binding sequence. Nucleotide sequences of both promoters showed considerable homology to the consensus sequence for the E. coli cAMP-CRP-binding site (AAATGTGATCTAGATCACATTT; see Ref. 31) at the upstream regions including their –35 regions (Fig. 7A). To verify if cAMP-CRP binding to these putative sites occurs and to determine whether these interactions are important in rpoS transcription from Pd and Pp, the putative binding sites were modified by site-directed mutagenesis. The DNA fragments containing the mutagenized nucleotide sequences (mt probes) were then used for electrophoretic mobility shift assays. No binding of cAMP-CRP to mt probes was observed (Fig. 7, B and C, lanes 3–6), although the amount of CRP added to the binding assay was two times (up to 765 nm CRP) more than that of the original nucleotide sequences (wild type probes) (Fig. 7, B and C, lane 2).
FIGURE 7.
Effect of mutation of the putative CRP-binding sites on CRP binding and rpoS regulation. A, presence of the putative CRP-binding sites in each promoter region of the rpoS gene. Two promoters for the rpoS gene are indicated as Pp and Pd along with their –10 and –35 regions (CRP-binding site I in the Pd promoter and CRP-binding site II in the Pp promoter). Putative CRP-binding sites are presented in the boxes implied from the conserved nucleotide sequences for CRP binding indicated by italicized letters above the V. vulnificus DNA sequence. Site-directed mutagenized nucleotides in the mutant rpoS promoters are indicated with arrows. B, binding assay of CRP to the rpoS promoter carrying Pd and mutagenized Pd. Labeled Pd (wild type probe) and mutagenized Pd DNA (mt probe) were incubated with CRP, as described in Fig. 5. Lane 1, Pd DNA without CRP; lane 2, Pd with 340 nm CRP; lane 3, mutagenized Pd without CRP; lanes 4–6, mutagenized Pd with 340, 510, and 765 nm CRP, respectively. The arrows on the left side indicate the unbound DNA probe, whereas the arrows on the right side indicate the DNA bound to CRP. C, binding assay of CRP to the rpoS promoter carrying Pp and mutagenized Pp. Labeled Pp (wild type probe) and mutagenized Pp DNA (mt probe) were incubated with increasing amounts of CRP. Lane 1, Pp DNA without CRP; lane 2, Pp with 340 nm CRP; lane 3, mutagenized Pp without CRP; lanes 4–6, mutagenized Pp with 340, 510, and 765 nm, CRP, respectively. The arrows on the left side indicate unbound DNA probe, whereas the arrows on the right side indicate the DNA bound to CRP. D and E, effect of mutation on the expression of rpoS::luxAB transcriptional fusions. Wild type V. vulnificus carrying pKP-368 or pKP-368mt (pKP-368 including the mutated CRP-binding site II) and wild type carrying pKP-1315 or pKP-1315mt (pKP-1315, including the mutated CRP-binding site I) were grown in LBS medium supplemented with 3μg/ml tetracycline, and their luciferase activities were measured. Luciferase activities are expressed as normalized values: the number of RLU divided by the A595 value of each sample. Data with p values of <0.01 are indicated with an asterisk.
In addition, the mutagenized DNAs were used to construct the luxAB -transcriptional fusions, pKP-368mt and pKP-1315mt, which are basically the same as pKP-368 and pKP-1315, but include the mutated CRP-binding sites. These mutant fusions in the wild type strain showed derepressed levels of expression (Fig. 7, D and E), which were similar to the expression levels of pKP-368 and pKP-1315 in the crp and Δcya strains. These results suggest that rpoS expression from Pd and Pp is repressed by direct binding of cAMP-CRP to the regions of –26 to –47 relative to Pd (CRP-binding site I) and –27 to –48 relative to Pp (CRP-binding site II) (Fig. 7A).
DISCUSSION
V. vulnificus is a normal inhabitant of marine estuarine environments and can be delivered into humans via ingestion of seafood or contact with seawater, whereupon it may cause fatal septicemia or gastroenteritis (2). Thus, this bacterium is expected to use efficient survival strategies to sense fluctuations in its surrounding conditions and to express the necessary defense elements against given stresses. In a previous investigation, we identified RpoS as a key regulator mediating the survival of V. vulnificus (4), as found in other bacteria. Variation of the intracellular levels of this global transcription factor, under certain conditions, is critical for modulating the expression of target genes in a finely coordinated manner.
Modulation of the amount of RpoS, which has been extensively investigated, is achieved by several levels of controls, including the regulation of transcription, translation, and proteolysis (1). In E. coli, one of the major regulatory mechanisms modulating the level of RpoS operates at the post-translational level via an RpoS-specific chaperone/protease system, ClpXP (32). Under glucose-starved conditions or in the exponential stage, a recognition factor, RssB, binds to the RpoS protein and enhances RpoS degradation by recruiting ClpXP to the RssB-RpoS complex (33). Translation of E. coli rpoS mRNA is regulated by small noncoding RNAs (34). A specific secondary structure in the rpoS mRNA, which inhibits efficient translational progress, is disrupted by being paired with DsrA RNA with the help of an RNA chaperon, Hfq, resulting in increased translation of rpoS mRNA (35). Similarly, V. vulnificus also utilizes some of the above regulatory mechanisms, both at the post-transcriptional and the post-translational levels via Hfq and ClpX, respectively. For example, the levels of RpoS, determined by Western blot analysis, were significantly reduced in the Δhfq V. vulnificus mutant (36) and highly elevated in the ΔclpX V. vulnificus mutant (supplemental Fig. 3).
The regulation of E. coli rpoS expression at the transcriptional level includes complex mechanisms. Trans-acting factors involved in rpoS transcription respond to intracellular signals related to the cessation of growth caused by environmental stresses. The involvement of the intracellular molecule, guanosine 3′,5′-bispyrophosphate (ppGpp), as a positive signal of rpoS expression has been reported (37). The barA gene product, first identified as a regulator of ompR (38), has been demonstrated to induce rpoS transcription (39). A two-component regulatory system for anaerobiosis, ArcA/ArcB, is involved in modulation of rpoS expression (10). The phospho-ArcA represses transcription of the rpoS gene by directly binding to the rpoS promoter region.
Although the importance of CRP in the regulation of rpoS genes has been reported in E. coli and S. enterica serovar Typhimurium (14, 11), the regulatory mechanism of rpoS transcription by the cAMP-CRP complex requires further study. The role of cAMP-CRP complex as an important negative transcriptional regulator was proven only genetically via rpoS-fusion assays using knock-out mutants unable to produce CRP or cAMP (14, 16). Repression of rpoS transcription by cAMP-CRP was also confirmed by the phenotype of a crr knock-out mutant (15). EIIAglc, which is a crr gene product and a component of the glucose uptake system, acts indirectly on the repression of rpoS expression by modulating the activity of adenylate cyclase. However, the molecular basis of transcriptional modulation of the rpoS gene by cAMP-CRP has not yet been identified in any bacterial system.
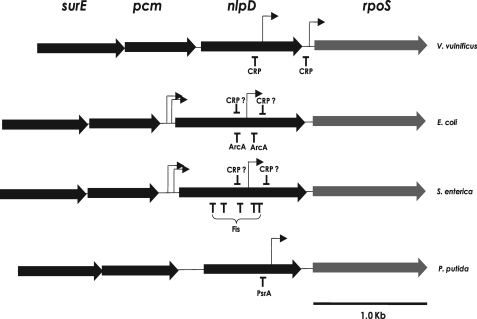
Because rpoS transcription is quite different among bacterial species (9), we examined the expression of the rpoS gene in V. vulnificus at the level of transcription. We preliminarily analyzed the transcription units of the rpoS gene and found two transcripts for rpoS,3 as occurred in E. coli (40). However, the sizes of the V. vulnificus rpoS transcripts were distinct from those of E. coli and other bacterial species (Fig. 8). In E. coli and S. enterica, the rpoS gene is expressed as a polycistronic nlpD-rpoS mRNA and an ∼2.0-kb-long rpoS monocistronic mRNA. Polycistronic nlpD-rpoS mRNA originates from two closely spaced promoters upstream of the nlpD gene. The second promoter responsible for monocistronic rpoS mRNA is located within the nlpD gene, which had been identified as a major rpoS promoter (41). In E. coli growing in LB medium, the polycistronic nlpD-rpoS mRNA was constitutively expressed, but the monocistronic rpoS mRNA was induced only at the stationary phase (14). In the case of P. putida rpoS, a single promoter located within the nlpD ORF was identified (Fig. 8).
FIGURE 8.
Genetic organization and expression of rpoS genes in some Gram-negative bacteria. The genetic organization of rpoS and its upstream genes (nlpD, pcm, and surE) of V. vulnificus were compared with those of E. coli (14), S. enterica serovar Typhimurium (11), and P. putida (12). The transcription start sites for each rpoS gene are designated with arrows, and the binding site locations for transcription factors are designated by vertical lines with the name of each transcription factor. In the case of E. coli and S. enterica, CRP-binding sites are putative and are thus designated with question marks.
Primer extension experiments identified a transcription initiation site 29 bp upstream of the rpoS ORF and another initiation site for a larger transcript at –473 with respect to the IC of V. vulnificus rpoS (Fig. 1, A and B). Thus, a shorter rpoS transcript of 1,021 bp was detected in addition to a 1,518-bp monocistronic rpoS mRNA in V. vulnificus. The series of rpoS::luxAB fusions indicated that the promoter located within the nlpD gene (Pd) is a major promoter, as shown in other bacteria (Fig. 2). In contrast, the promoter located just upstream of the rpoS (Pp) is a minor factor for rpoS expression. Expression of both promoters was induced as the cells entered the stationary phase in LBS medium. However, the affinity of each promoter to the cAMP-CRP complex was quite different (Fig. 5, C and D, and Fig. 6C), which might suggest each promoter has a differential role in V. vulnificus in response to specific stresses. Therefore, further investigation of the roles of each promoter is required to understand its relative contribution to rpoS gene expression under various conditions. Unlike E. coli, the upstream region of nlpD is not involved in rpoS expression, because luciferase activities of two fusions containing the upstream regions of nlpD (pKP-1315 and pKP-1640) were similar to those of two fusions without the nlpD upstream region (pKP-732 and pKP-891) (Fig. 2). This further implies that nlpD transcription in V. vulnificus may start at the further upstream region of the pcm or surE genes.
RpoS levels of V. vulnificus were inversely correlated to the intracellular concentrations of cAMP; for example, stationary phase cells grown in complex media, such as LBS medium, contained less cAMP (<5 fmol of cAMP/μg of bacterial protein) than exponential phase cells (∼50 fmol of cAMP/μg of bacterial protein) (Fig. 3). This pattern of intracellular cAMP fluctuation was similar to that shown in E. coli cells grown in LB medium (42). The role of cAMP-CRP in rpoS transcription was confirmed in various V. vulnificus strains, in which CRP was knocked out or the synthesis/degradation of cAMP was altered (Table 3). Expression of rpoS fusions was decreased in the ΔcpdA mutant that has an intracellular cAMP level approximately twice that estimated in its isogenic wild type strain. rpoS expression was highly increased in the crp and Δcya mutants but decreased in the Δcya mutant in the presence of exogenously added cAMP (Table 3 and Fig. 4). Therefore, rpoS expression is tightly dependent upon the cAMP-CRP complex at the transcriptional level in V. vulnificus.
V. vulnificus growing in a glucose-based minimal medium, such as artificial seawater supplemented with glucose as a sole carbon source, contained significant cAMP levels when cells entered the stationary phase because of carbon source depletion.4 In addition, E. coli cells grown in a minimal medium, such as M9 supplemented with glucose, did not show induction of rpoS transcription during the stationary phase; cellular cAMP levels were also highly increased during this phase (14). Thus, rpoS transcription is highly repressed by cAMP-CRP complex under this condition, and thus regulation at the post-transcriptional level is important in the stationary phase induction of RpoS.
In this study, we further confirmed the role of the cAMP-CRP complex in rpoS expression by demonstrating the repression of rpoS expression by addition of CRP to the in vitro transcription reaction (Fig. 5) and the direct interaction of both rpoS promoters with recombinant CRP protein (Fig. 6). The interactions between the rpoS promoters and cAMP-CRP complex are mediated by DNA sequences homologous to the CRP-binding consensus sequence (Fig. 7A). Alteration of these putative CRP-binding sites resulted in the disappearance of specific interaction with the cAMP-CRP complex in vitro (Fig. 7, B and C) and abolishment of transcriptional repression by the cAMP-CRP complex in vivo (Fig. 7, D and E). This is the first study showing the direct interaction of the cAMP-CRP complex with the rpoS promoters.
The data presented in this study do not exclude the possibility that other transcriptional factors are involved in rpoS expression in V. vulnificus. The transcriptional fusions of rpoS in Δcya and crp mutants still showed the induction of expression at the stationary phase (data not shown). This suggests that the induction of rpoS transcription is partly controlled by another factor, which is independent of cAMP and CRP. In silico screening of the V. vulnificus genome suggests the absence of a gene homologous to the transcriptional activator PsrA found in Pseudomonas putida (12). Therefore, there must be other transcriptional factors in V. vulnificus that remain to be elucidated in future studies.
Supplementary Material
Acknowledgments
We thank Kyung-Je Park for the initial investigation of the rpoS fusions; Sung-Min Kim for the construction of ΔcpdA mutant and the measurement of cAMP contents; and Mi-Ae Lee for the careful proofreading of this manuscript.
This work was supported by Grant R01-2007-000-10159-0 (to K.-H. L.) from KOSEF and a grant from the Extreme Molecular Genomics Research Program of the Ministry of Marine Affairs and Fisheries (to K.-H. L.), Republic of Korea. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBank™/EBI Data Bank with accession number(s) AY221025 and AY240931.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3.
Footnotes
The abbreviations used are: CRP, cAMP receptor protein; IC, initiation codon; ORF, open reading frame; RNAP, RNA polymerase; EMSA, electrophoretic mobility shift assay; RLU, relative light unit; mt, mutagenized.
K.-J. Park and K.-H. Lee, unpublished data.
S.-M. Kim and K.-H. Lee, unpublished data.
References
- 1.Hengge-Aronis, R. (2002) Microbiol. Mol. Biol. Rev. 66 373–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Strom, M. S., and Paranjpye, R. N. (2000) Microbes Infect. 2 177–188 [DOI] [PubMed] [Google Scholar]
- 3.Hülsmann, A., Rosche, T. M., Kong, I.-S., Hassan, H. M., Beam, D. M., and Oliver, J. D. (2003) Appl. Environ. Microbiol. 69 6114–6120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Park, K.-J., Kang, M.-J., Kim, S. H., Lee, H.-J., Lim, J.-K., Choi, S. H., Park, S.-J., and Lee, K.-H. (2004) J. Bacteriol. 186 3304–3312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hengge-Aronis, R. (2000) in The General Stress Response in Escherichia coli (Storz, G., and R. Hengge-Aronis, R., eds) pp. 161–178, ASM Press, Washington, D. C.
- 6.Jeong, H. S., Jeong, K. C., Choi, H. K., Park, K.-J., Lee, K.-H., Rhee, J. H., and Choi, S. H. (2001) J. Biol. Chem. 278 45072–45081 [DOI] [PubMed] [Google Scholar]
- 7.Lee, H.-J., Park, K.-J., Lee, A. Y., Park, S. G., Park, B. C., Lee, K.-H., and Park, S.-J. (2003) J. Bacteriol. 185 5891–5896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Litwin, C. M., and Quackenbush, J. (2001) Microb. Pathog. 31 295–307 [DOI] [PubMed] [Google Scholar]
- 9.Venturi, V. (2003) Mol. Microbiol. 49 1–9 [DOI] [PubMed] [Google Scholar]
- 10.Mika, F., and Hengge, R. (2005) Genes Dev. 19 2770–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hirsch, M., and Elliott, T. (2005) J. Bacteriol. 187 1568–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kojic, M., Aguilar, C., and Venturi, V. (2002) J. Bacteriol. 184 2324–2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burtnick, M. N., Downey, J. S., Brett, P. J., Boylan, J. A., Frye, J. G., Hoover, T. R., and Gherardini, F. C. (2007) Mol. Microbiol. 65 277–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lange, R., and Hengge-Aronis, R. (1994) Genes Dev. 8 1600–1612 [DOI] [PubMed] [Google Scholar]
- 15.Ueguchi, C., Misonou, N., and Mizuno, T. (2001) J. Bacteriol. 183 520–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McCann, M. P., Fraley, C. D., and Matin, A. (1993) J. Bacteriol. 175 2143–2149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park, K.-J., Kim, S. H., Kang, M.-J., Chung, D. H., Ha, S.-D., Kim, K.-S., Jahng, D., and Lee, K.-H. (2004) J. Microbiol. Biotechnol. 12 1063–1066 [Google Scholar]
- 18.Roh, J.-B., Lee, M.-A., Lee, H.-J., Kim, S.-M., Cho, Y., Kim, Y.-J., Seok, Y.-J., Park, S.-J., and Lee, K.-H. (2006) J. Biol. Chem. 281 34775–34784 [DOI] [PubMed] [Google Scholar]
- 19.Milton, D. L., O'Toole, R., Hörstedt, P., and Wolf-Watz, H. (1996) J. Bacteriol. 178 1310–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller, V. L., and Mekalanos, J. J. (1988) J. Bacteriol. 170 2575–2583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sambrook, J., and Russell, D. W. (2001) Molecular Cloning: A Laboratory Manual, 3rd Ed., pp. 17.1–17.17, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- 22.Bergendahl, V., Thompson, N. E., Foley, K. M., Olson, B. M., and Burgess, R. L. (2003) Protein Expression Purif. 31 155–160 [DOI] [PubMed] [Google Scholar]
- 23.Thompson, N. E., Hanger, D. A., and Burgess, R. R. (1992) Biochemistry 31 7003–7008 [DOI] [PubMed] [Google Scholar]
- 24.Newlands, J. T., Gaal, T., Mecsas, J., and Gourse, R. L. (1993) J. Bacteriol. 175 661–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jeong, H. S., Lee, M. H., Lee, K.-H., Park, S.-J., and Choi, S. H. (2003) J. Biol. Chem. 276 13875–13880 [DOI] [PubMed] [Google Scholar]
- 26.González-Gil, G., Kahmann, R., and Muskhelishvili, G. (1998) EMBO J. 17 2877–2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Milton, D. L. (2006) Int. J. Med. Microbiol. 296 61–71 [DOI] [PubMed] [Google Scholar]
- 28.Botsford, J. L. (1981) Microbiol. Rev. 45 620–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Latifi, A., Foglino, M., Tanaka, K., Williams, P., and Lazdunski, A. (1996) Mol. Microbiol. 21 1137–1146 [DOI] [PubMed] [Google Scholar]
- 30.Kim, Y. R., and Rhee, R. H. (2003) Biochem. Biophys. Res. Commun. 304 405–410 [DOI] [PubMed] [Google Scholar]
- 31.Busby, S., and Ebright, R. H. (1999) J. Mol. Biol. 293 199–213 [DOI] [PubMed] [Google Scholar]
- 32.Schweder, T., Lee, K.-H., Lomovskaya, O., and Matin, A. (1996) J. Bacteriol. 178 470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Studemann, A., Noirclerc-Savoye, M., Klauck, E., Becker, G., Schneider, D., and Hengge-Aronis, R. (2003) EMBO J. 22 4111–4120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gottesman, S. (2004) Annu. Rev. Microbiol. 58 303–328 [DOI] [PubMed] [Google Scholar]
- 35.Majdalani, N., Cunning, C., Sledjeski, D., Elliott, T., and Gottesman, S. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 12462–12467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee, H.-J., Bang, S. H., Lee, K.-H., and Park, S.-J. (2007) J. Bacteriol. 189 2629–2636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gentry, D. R., Hernandez, V. J., Nguyen, L. H., Jensen, D. B., and Cashel, M. (1993) J. Bacteriol. 175 7982–7989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nagasawa, S., Tokishiba, S., Aiba, H., and Mizno, T. (1992) Mol. Microbiol. 6 799–807 [DOI] [PubMed] [Google Scholar]
- 39.Mukhopadhyay, S., Audia, J. P., Roy, R. N., and Schellhorn, H. E. (2000) Mol. Microbiol. 37 371–381 [DOI] [PubMed] [Google Scholar]
- 40.Arnqvist, A., Olsén, A., and Normark, S. (1994) Mol. Microbiol. 13 1021–1032 [DOI] [PubMed] [Google Scholar]
- 41.Lange, R., Fischer, D., and Hengge-Aronis, R. (1995) J. Bacteriol. 177 4676–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ma, Z., Richard, H., and Foster, J. W. (2003) J. Bacteriol. 185 6852–6859 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.