Abstract
Signaling through mammalian target of rapamycin complex 1 (mTORC1) is stimulated by amino acids and insulin. Insulin inactivates TSC1/2, the GTPase-activator complex for Rheb, and Rheb·GTP activates mTORC1. It is not clear how amino acids regulate mTORC1. FKBP38 (immunophilin FK506-binding protein, 38 kDa), was recently reported to exert a negative effect on mTORC1 function that is relieved by its binding to Rheb·GTP. We confirm that Rheb binds wild type FKBP38, but inactive Rheb mutants showed contrasting abilities to bind FKBP38. We were unable to observe any regulation of FKBP38/mTOR binding by amino acids or insulin. Furthermore, FKBP38 did not inhibit mTORC1 signaling. The translationally controlled tumor protein (TCTP) in Drosophila was recently reported to act as the guanine nucleotide-exchange factor for Rheb. We have studied the role of TCTP in mammalian TORC1 signaling and its control by amino acids. Reducing TCTP levels did not reproducibly affect mTORC1 signaling in amino acid-replete/insulin-stimulated cells. Moreover, overexpressing TCTP did not rescue mTORC1 signaling in amino acid-starved cells. In addition, we were unable to see any stable interaction between TCTP and Rheb or mTORC1. Accumulation of uncharged tRNA has been previously proposed to be involved in the inhibition of mTORC1 signaling during amino acid starvation. To test this hypothesis, we used a Chinese hamster ovary cell line containing a temperature-sensitive mutation in leucyl-tRNA synthetase. Leucine deprivation markedly inhibited mTORC1 signaling in these cells, but shifting the cells to the nonpermissive temperature for the synthetase did not. These data indicate that uncharged tRNALeu does not switch off mTORC1 signaling and suggest that mTORC1 is controlled by a distinct pathway that senses the availability of amino acids. Our data also indicate that, in the mammalian cell lines tested here, neither TCTP nor FKBP38 regulates mTORC1 signaling.
The current high level of interest in signaling through mTOR3 reflects its ability to integrate multiple signals to control diverse cell functions (1, 2) and its roles in human diseases, including cancer (3, 4). mTOR forms two types of complexes, mTORC1 and mTORC2.
mTORC1 promotes the phosphorylation and activation of the 70-kDa S6 kinases (and thus the phosphorylation of ribosomal protein S6) and the multisite phosphorylation and inactivation of the translational repressors 4E-BP1/2 (1, 5). mTORC1 signaling is promoted by inputs from amino acids, especially leucine, and from hormones such as insulin. Thus, the phosphorylation of S6 requires both amino acids and insulin and is blocked by rapamycin, whereas in 4E-BP1 phosphorylation of Thr-37/46 is induced by amino acids alone and is largely insensitive to rapamycin (6). Nonetheless, extensive data suggest that mTORC1 mediates the phosphorylation of Thr-37/46 in 4E-BP, because this is impaired by inhibitors of the kinase activity of mTOR (other than rapamycin), by the tuberous sclerosis complex (TSC1/2), a negative regulator of Rheb and mTORC1, and by decreasing the cellular levels of mTOR or the mTORC1 component raptor (6, 7).
mTORC1 signaling is activated by the small GTPase Rheb (8) (see scheme in Fig. 1A). Rheb·GTP stimulates the protein kinase activity of mTOR in vitro (9). Insulin and other agents are thought to stimulate mTORC1 by inactivating TSC1/2, the GTPase-activator (GAP) for Rheb (10, 11) (Fig. 1A). However, TSC2 is dispensable for amino acid regulation of mTORC1 signaling (12, 13). Although several other components have been implicated in this, it remains unclear how amino acids activate mTORC1 signaling (13–15). One attractive idea is that amino acids regulate the activity of a GEF that converts Rheb·GDP to active Rheb·GTP.
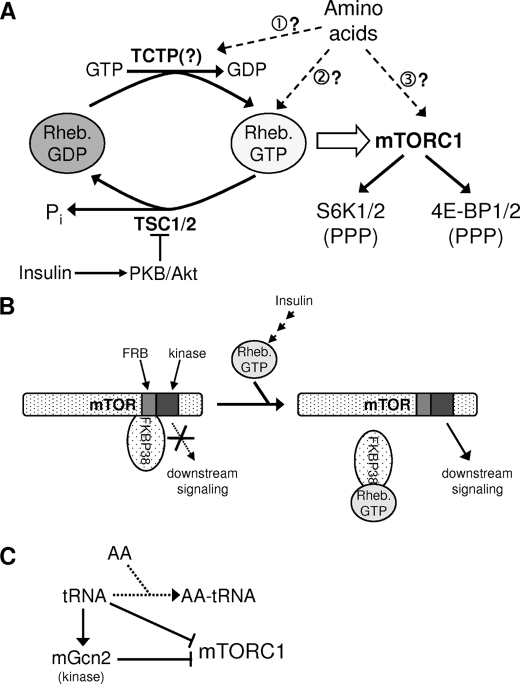
FIGURE 1.
Models for the regulation of Rheb/mTORC1. A, guanine nucleotide-binding status of Rheb is likely controlled by its GAP (TSC1/2, which is inactivated by insulin signaling via Akt) and perhaps by its potential GEF, TCTP. Rheb·GTP activates mTORC1, which regulates the downstream effectors p70 S6K and 4E-BP1; phosphorylation of 4E-BP1 is more complex than shown, as different sites show differential sensitivity to rapamycin. Dashed arrows show ways in which amino acids might promote mTORC1 function, and: numbers refer to points made in the text. B, FKBP38 has been proposed to interact with mTOR/mTORC1 and inhibit its function. Binding of FKBP38 to Rheb·GTP is suggested to result in the release of FKBP38 from mTOR and activation of mTORC1 function. FRB denotes the FKBP12·rapamycin-binding domain of mTOR. C, deficiency of amino acids (AA) may lead to accumulation of uncharged tRNA. Either directly or via the activation of the protein kinase mGcn2, for example, this could lead to inhibition of mTORC1 function.
FKBP38 (also termed FKBP8 (16)) is an immunophilin and belongs to the peptidyl/prolyl cis-trans-isomerase protein family. It has also been shown to bind to and be regulated by calmodulin and by Hsp90 (17–20). FKBP38 is a pro-apoptotic modulator of Bcl-2 (17, 18, 20–22) and has also been shown to regulate cell size (23). A very recent study reported that FKBP38 interacted with mTOR and could inhibit its function, both in vitro and in cells (24). FKBP38 was also reported to bind to Rheb, such that Rheb·GTP induced the release of FKBP38 from mTOR. This would provide a mechanism by which Rheb·GTP could activate mTORC1 signaling (Fig. 1B). It is not clear from the available data whether FKBP38 is involved in the regulation of mTORC1 by amino acids (24).
TCTP (also termed “P23”) is a growth-related and anti-apoptotic protein. Extensive studies have led to numerous functions being ascribed to it (25). We have previously established its microtubule binding/stabilizing activity (26). Knocking out TCTP results in embryonic lethality highlighting its importance in early development (27).
Recent genetic studies in Drosophila implicated Drosophila TCTP (dTCTP) in the control of the dTOR pathway, which controls cell growth and cell number (28). Consistent with this, dTCTP was required for phosphorylation of dS6K. Biochemical evidence suggested that dTCTP acts as a GEF for Rheb (28) (see Fig. 1A). Indeed, Drosophila and human TCTP were shown to mediate GDP/GTP exchange on the corresponding Rheb proteins in vitro (28). The proposed role of TCTP is depicted in Fig. 1A.
The fact that starving cells for amino acids (especially leucine) leads to inhibition of mTORC1 signaling has led to the suggestion that uncharged tRNA, generated because of insufficiency of amino acids, might act as a proximal negative regulator of mTORC1 (29). In yeast it is well established that uncharged tRNA activates the protein kinase Gcn2p, which phosphorylates eukaryotic initiation factor (eIF) 2 on its α-subunit, leading to inhibition of general protein synthesis (reviewed in Ref.30). Mammalian cells contain an ortholog of Gcn2p (mGcn2) (31, 32). It is thus formally possible that activated mGcn2 can negatively regulate mTORC1 signaling, perhaps by phosphorylating mTORC1 or a regulator of this complex. These proposed tRNA-mediated regulatory inputs to mTORC1 are summarized in Fig. 1C. Given the high level of interest in mTORC1 signaling, it was important to assess the roles of these proposed regulators of mammalian TORC1 signaling, including their possible importance for the control of this protein kinase by amino acids.
EXPERIMENTAL PROCEDURES
Chemicals and Biochemicals—Chemicals and biochemicals were from Sigma unless otherwise indicated. Antibodies for TCTP (26) and 4E-BP1 (6) were described earlier. Antibodies for 4E-BP1 phosphorylated at Thr-37/46 or Thr-70, S6-phosphorylated Ser-235/6 or Ser-240/4, S6, mTOR, and phospho-Akt[Ser-473] were from Cell Signaling Technology. The antibody for 4E-BP1 phosphorylated at Ser-65 was from Santa Cruz Biotechnology. The antibody for 4E-BP1 has been described previously (33). Anti-FLAG and -Myc were from Sigma, and anti-HA was from Roche Applied Science.
Cell Culture, Transfection, and Treatment—HEK293 cells were cultured in high glucose (4.5 g/liter) Dulbecco's modified Eagle's medium with 10% (v/v) fetal bovine serum, 2 mm l-glutamine, 100 μg/ml streptomycin sulfate, and 100 units/ml penicillin G and transfected with plasmids or siRNAs as described earlier (6). Prior to use, cells were starved of serum overnight and in some cases also starved of amino acids by transferring them to Dulbecco's phosphate-buffered saline containing 10 mm d-glucose (for 90 min (6)). As a control, some dishes were transferred to Dulbecco's phosphate-buffered saline containing the same mixture of amino acids as Dulbecco's modified Eagle's medium. Vectors for FLAG-mTOR, myc-Raptor, and FLAG-Rheb have been described before (6, 7). The plasmid pcDNA-3/P23 for expressing murine TCTP (P23) was described earlier (25), and the HA-PRAS40 vector was described in Ref. (7. The vector for HA-FKBP38 uses the same plasmid as used for HA-PRAS40 (pCMV5-HA).
Chinese hamster ovary (CHO) cells were grown as described previously (43) in Dulbecco's modified Eagle's medium/Nutrient Mixture Ham's F-12 (Sigma) supplemented with 9% (v/v) fetal bovine serum, 100 μg/ml streptomycin sulfate, and 100 units/ml penicillin G at 34 °C. The tsH1 line was derived by Thompson et al. (34) and contains a temperature-sensitive leucyl-tRNA synthetase that is active at 34 °C but defective at 39.5 °C. Shifting the cells to the latter temperature mimics the effects of amino acid starvation on protein synthesis (43). The control cells (TR-3) were a single-step temperature revertant of tsH1 and have normal leucyl-tRNA synthetase activity at 39.5 °C (35, 36). Both TR-3 and tsH1 cells were grown in 5% CO2 in a humidified incubator at 34 °C. Where indicated, cells were transferred to 39.5 °C.
CHO cells were starved of amino acids by transferring them to Dulbecco's modified Eagle's medium/Nutrient Mixture Ham's F-12 supplemented with 9% (v/v) dialyzed fetal bovine serum, 100 μg/ml streptomycin sulfate, and 100 units/ml penicillin G but lacking either leucine or glutamine. Amino acid-free serum was prepared by dialysis against cold phosphate-buffered saline. Typically, 100 ml of serum were dialyzed twice against 2 liters for 12 h each time, using a membrane with a cutoff of 3.5 kDa. siRNA—The oligoribonucleotides 5′-AAGGGAGACGACGACGGCGCUAGCUUA-3′ and 5′-AGCUAGCGCCGUCGUCGUCUCCCTT-3′ (Integrated DNA Technologies, Coralville, IA) were used to knock down TCTP, at concentrations of 20–60 nm. Transfection was by the calcium-phosphate method, and cells were left for 40 h before further analysis.
Immunoprecipitation, Western Blotting, and Related Methods—These procedures were performed as described in Ref. 7. Generally, cells were lysed using the buffer given in Ref. 7. For all experiments involving immunoprecipitation, the lysis buffer described in Ref. 37 was used for lysis and for all subsequent steps. In other experiments we used our standard lysis buffer containing 1% (v/v) Triton X-100 (38).
RESULTS
FKBP38 Binds to Rheb Independently of Amino Acids or Insulin—A recent study (24) proposed that FKBP38 acts as a negative regulator of mTORC1 by binding to mTOR. That report also suggested that Rheb·GTP bound to FKBP38, releasing the latter protein from mTOR and thereby eliciting the activation of mTORC1. We began our study by testing whether FKBP38 can indeed interact with Rheb. As the available antisera for FKBP38 and Rheb fail to detect the endogenous proteins (data not shown), we transfected HEK293 cells with vectors encoding HA-FKBP38 and FLAG-Rheb and then immunoprecipitated each protein separately using the antibody for the corresponding epitope. Immunoprecipitates were analyzed by SDS-PAGE followed by Western blot. As shown in Fig. 2A, HA-FKBP38 and FLAG-Rheb did co-immunoprecipitate, confirming a mutual interaction (but not proving that it is direct). To explore further the interaction between FKBP38 and Rheb, we made use of two different types of mutant, which have been shown to be unable to activate mTORC1 signaling.
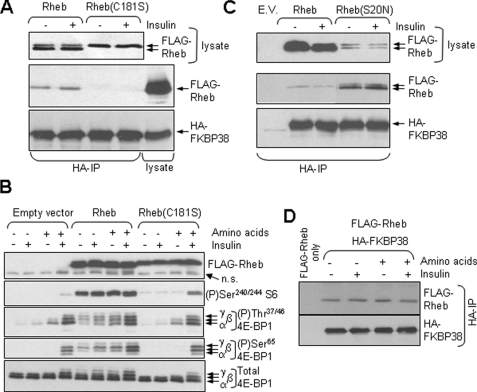
FIGURE 2.
Wild type Rheb, but not the inactive C181S mutant, binds FKBP38 in a constitutive manner. A, HEK293 cells were transfected with vectors encoding FLAG-tagged Rheb or its C181S mutant and HA-tagged FKBP38. Cells were starved of serum overnight and, where indicated, treated with insulin (100 nm, 30 min). Cells were then lysed, and samples were either subjected to direct Western blot analysis (top section; using anti-FLAG) or to immunoprecipitation (IP) with anti-HA prior to SDS-PAGE/Western blot with anti-FLAG (middle section) and anti-HA (bottom section). Samples of cell lysate were analyzed in parallel in the middle and bottom sections. In the top section, the arrows indicate the slower migrating (nonfarnesylated) and faster migrating (farnesylated) forms of Rheb. B. HEK293 cells were transfected with vectors for wild type Rheb, the C181S mutant, or the empty vector. Twenty four hours later, cells were starved overnight for serum and, where shown (–), of amino acids for 60 min. Where indicated, cells were treated with insulin (100 nm for a further 30 min). Samples of cell lysate were analyzed by SDS-PAGE and Western blot using the indicated antisera. n.s. denotes a nonspecific band detected by anti-FLAG. The differentially phosphorylated forms of 4E-BP1 are indicated (α-γ). C, as A, but using wild type Rheb and the S20N mutant.E.V., empty vector. D, cells were transfected (with vectors for HA-FKBP38 and also FLAG-Rheb where indicated) and treated as in B. Samples of lysate were subjected to immunoprecipitation (IP) using anti-HA, and samples were analyzed by SDS-PAGE and Western blot using anti-FLAG and -HA, as indicated.
Rheb undergoes farnesylation (because of the presence of a CAAX (where AA is aliphatic amino acid) box at its C terminus (39)). Mutation of the Cys residue to Ser (C181S mutant) prevents farnesylation (as is evidenced by altered mobility shown in Fig. 2A) and results in drastically reduced GTP loading (see e.g. Ref. 40). The C181S mutant is also defective in restoring mTORC1 signaling (phosphorylation of Ser-65 in 4E-BP1 or Ser-240/244 in S6) in amino acid-starved cells (Fig. 2B; consistent with earlier data (39, 41)). (Note that 4E-BP1 runs as three distinct bands on SDS-PAGE (Fig. 2B) termed α-γ in order of increasing phosphorylation.) The C181S mutant is also incapable of binding to FKBP38 (Fig. 2A).
Mutation of the final Met of Rheb to Leu should create a site for a different lipid modification, geranylgeranylation, which might substitute for farnesylation. We therefore created and tested a C181S/M184L double mutant. Its mobility was similar to that of the C181S mutant (data not shown), and like this variant it neither supported mTORC1 signaling nor bound to Rheb (not shown).
The mutation of Ser-20 to Asn (S20N) yields a Rheb mutant that cannot bind guanine nucleotides and fails to activate mTORC1 signaling (9, 40). The Rheb(S20N) mutant expressed at lower levels than the wild type protein in HEK293 cells, but relatively much more of the mutant was recovered in HA-FKBP38 immunoprecipitates (Fig. 2C). This shows the following: (i) the ability to bind GTP or GDP is not required for Rheb to interact with FKBP38; and (ii) the inability of the Rheb[C181S] mutant to bind FKBP38 arises from an effect that is distinct from its (in)ability to bind GTP. The reason for its inability to bind FKBP38 remains to be determined.
Our data do confirm the finding of the earlier study (24) that Rheb can bind FKBP38. However, taken together, the data for the two mutants tested here are not consistent with the idea that FKBP38 preferentially binds to Rheb·GTP as suggested by the earlier study (24).
Furthermore, in multiple experiments we were unable to see any consistent difference in the amount of wild type FKBP38 that co-immunoprecipitated with Rheb under conditions where mTORC1 signaling is impaired (amino acid starvation) or activated (treatment of amino acid-replete cells with insulin; Fig. 2D). This finding suggests that the association of FKBP38 with Rheb is independent of amino acids or insulin; it is not consistent with the idea (24) that agents that activate mTORC1 signaling do so by increasing the association of Rheb with FKBP38, thereby alleviating the proposed inhibitory effect of FKBP38 on mTORC1.
FKBP38 Also Binds mTOR but Does Not Inhibit mTORC1 Signaling in HEK293 Cells—To study whether FKBP38 also bound to mTOR, we co-expressed HA-FKBP38 and FLAG-mTOR in HEK293 cells. HA immunoprecipitation of FKBP38 consistently brought down mTOR, as shown in Fig. 3A. This is in agreement with earlier data (24). However, the levels of mTOR associated with FKBP38 were the same whether the cells had been starved of serum or of amino acids, or treated with insulin, prior to lysis (Fig. 3A), even though these conditions do have marked effects on mTORC1 signaling (as shown in Fig. 2B).
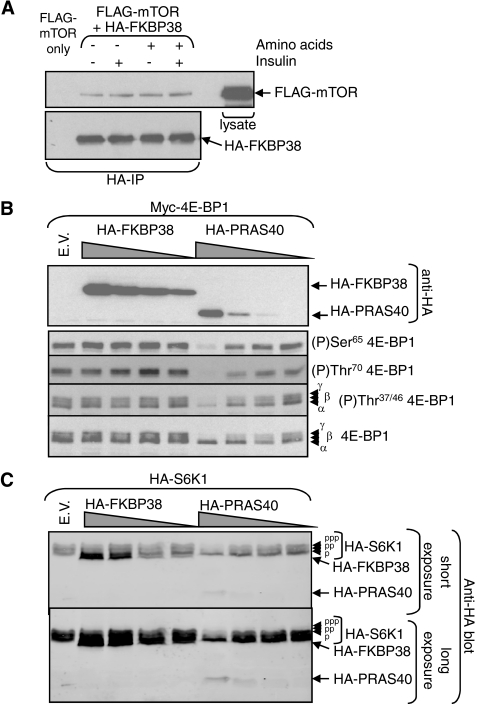
FIGURE 3.
FKBP38 interacts with mTOR but does not inhibit mTORC1 signaling. A, HEK293 cells were transfected with vectors encoding FLAG-mTOR and HA-FKBP38 (or where indicated only with FLAG-mTOR). Twenty four hours later, cells were starved overnight for serum and, where shown (–) of amino acids for 60 min. Where indicated, cells were treated with insulin (for a further 30 min). Samples of lysate were subjected to immunoprecipitation (IP) with anti-HA, and the immunoprecipitates were analyzed by SDS-PAGE/Western blot with the indicated antibodies. A sample of lysate was run as a control on the anti-FLAG blot. B, HEK293 cells were transfected with a vector encoding Myc-tagged 4E-BP1 and varying amounts of vectors encoding HA-FKBP38 or HA-PRAS40 (or where indicated, with the empty vector, E.V.). Samples of cell lysate were analyzed by SDS-PAGE/Western blot with the indicated antibodies. The positions of the different species of 4E-BP1 are shown. C, HEK293 cells were transfected with a vector for HA-S6K1 and varying amounts of vectors encoding HA-FKBP38 or HA-PRAS40 (or where indicated, with the empty vector). Two differentially phosphorylated forms of p70 S6K1 (p, pp) are resolved on this gel system (the faster moving (less phosphorylated) of which runs just above HA-FKBP38, upper section). The lower section shows a longer exposure on which the signal for HA-PRAS40 is evident.
It is conceivable that the lack of effect of amino acids or insulin on the association of this proposed inhibitor with mTORC1 (Fig. 3A) may be related to the fact that FKBP38 is overexpressed in these experiments. However, whatever the reason for this lack of regulation of FKBP38/mTOR binding, the model proposed in Ref. 24 predicts that this “constitutive” association should impair the regulation of mTORC1 signaling (24), because it proposes that FKBP38 is a negative regulator of mTORC1. We therefore expressed HA-FKBP38 in HEK293 cells (where we can achieve high levels of overexpression) along with vectors for either of two targets of mTORC1, 4E-BP1 and p70 S6K1. As a control, we also co-transfected cells with a vector encoding HA-PRAS40, which we have previously shown to interfere with mTORC1 signaling (7).
Cells were starved of serum and subsequently treated with insulin. In the case of 4E-BP1, expressing FKBP38 did not impair the phosphorylation of any site in 4E-BP1 that we studied even when FKBP38 was expressed at high levels (Fig. 3B). In contrast, ectopic expression of PRAS40 did inhibit mTORC1 signaling, and did so even when it was expressed at much lower levels than FKBP38 as judged by immunoblot using an antibody to their common HA tag (Fig. 3B).
To study whether FKBP38 affected the activation state of p70 S6K1, samples of cell lysate were analyzed using gels in which the multisite phosphorylation of p70 S6K1 that is associated with its activation causes a retarded mobility. (Note: HA-FKBP38 migrates just below the least phosphorylated and fastest moving species of p70 S6K1.) Expression of FKBP38 had no detectable effect on the mobility of p70 S6K1 (Fig. 3C), whereas PRAS40, expressed at far lower levels, did cause the loss of the slower migrating, hyperphosphorylated, form(s). Thus, even when expressed at relatively high levels, FKBP38 had no effect on the phosphorylation of either of two well established targets of mTORC1, 4E-BP1 and p70 S6K1.
TCTP Does Not Associate Stably with mTOR or Rheb—It has recently been proposed that TCTP may act as the GEF for Rheb (28), converting inactive Rheb·GDP to the GTP-bound form that can activate mTORC1 (9). Some GEFs bind stably to their cognate partner in its nucleotide-free state,: e.g. we have shown that eIF2B co-purifies with eIF2 (42). Indeed, TCTP, the proposed GEF for Rheb, has been reported to bind Rheb when both are overexpressed in human cells (28). We therefore tested whether TCTP co-immunoprecipitated with FLAG-Rheb. Although FLAG-Rheb expressed well (Fig. 4A, 3rd to 6th lanes), no co-immunoprecipitation with TCTP was observed, even when TCTP was also overexpressed (Fig. 4A, 5th and 6th lanes). This differs from the recently published data for dTCTP (28).
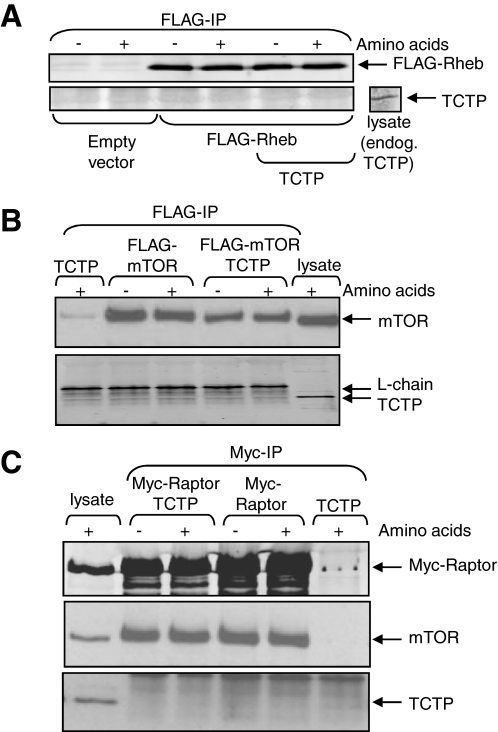
FIGURE 4.
Probing the potential interactions of TCTP with Rheb, mTOR, or Raptor. A, HEK293 cells were transfected with vectors for untagged wild type TCTP and/or FLAG-Rheb (or empty vector). Forty hours later, the cells were lysed, and samples were immunoprecipitated with anti-FLAG (for Rheb). Immunoprecipitates were analyzed by Western blot using the indicated antibodies. Cell lysate was run as a positive control for the anti-TCTP antibody (right). B and C, HEK293 cells were transfected with vectors for TCTP and/or FLAG-tagged mTOR (B) or Myc-tagged raptor (C), as indicated. Forty hours later, cell lysates were prepared and subjected to immunoprecipitation (IP) with anti-FLAG (B) or anti-Myc (C). Samples were analyzed by SDS-PAGE and immunoblot using the indicated antisera. Samples of lysate were analyzed in parallel as positive controls.
The Rheb-GAP complex TSC1/2 interacts with mTORC1 (9, 12), and it was therefore possible that TCTP, the proposed Rheb-GEF, also bound mTORC1. To study this, we expressed either FLAG-tagged mTOR (Fig. 4B) or the myc-raptor (Fig. 4C) in HEK293 cells, in some cases with TCTP. Although a band was apparent at the approximate size of TCTP in the FLAG-mTOR immunoprecipitates, this was only one of several bands detected by the anti-TCTP antiserum. Furthermore, its intensity was not increased when TCTP was overexpressed, as one would have expected (Fig. 4B). Myc-raptor co-immunoprecipitated with endogenous mTOR, indicating that mTORC1 complexes were pulled down, but no clear band was seen for TCTP (Fig. 4C). We therefore have no evidence that TCTP interacts stably with Rheb, mTOR, or raptor. It of course remains possible that such interactions do occur but are not stable enough to allow co-immunoprecipitation, e.g. of Rheb and TCTP.
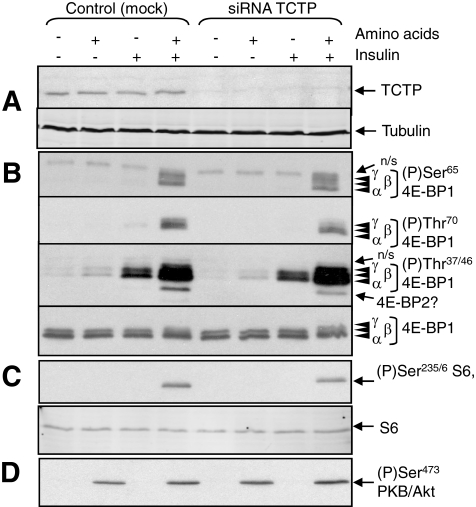
TCTP Knockdown Does Not Impair mTORC1 Signaling—To test the role of TCTP in controlling mTORC1 signaling in human cells, we asked whether knocking down its expression affected the phosphorylation of proteins that are regulated by mTORC1, such as 4E-BP1 and ribosomal protein S6. If TCTP does act as a Rheb-GEF (28), then depleting TCTP would be expected to impair their phosphorylation. After 40 h treatment of HEK293 cells with siRNAs directed against TCTP mRNA, the levels of TCTP protein consistently decreased to almost undetectable levels (Fig. 5A). It is conceivable that a small amount of TCTP may remain, especially as it is an abundant protein (43). Total levels of the mTORC1 targets, ribosomal protein S6 and 4E-BP1, were identical in knockdown and control cells (Fig. 5, B and C).
FIGURE 5.
Knockdown of TCTP does not consistently affect the phosphorylation of S6 and 4E-BPs. HEK293 cells were transfected with siRNAs targeting TCTP or mock-transfected, as indicated. Cells were starved of serum and, where indicated, amino acids. In some cases, cells were treated with insulin (100 nm, 30 min). Cells were then lysed and, after normalizing protein content, samples were analyzed by Western blot using the indicated antibodies. A, immunoblots for TCTP and tubulin, as loading control.B, immunoblots for total 4E-BP1 and for specific phosphorylation sites, as indicated. The Thr(P)-37/46 phosphospecific antibody also appears to recognize the corresponding site(s) in 4E-BP2 (6). 4E-BP1 runs as three distinct species (α-γ), with γ being most highly phosphorylated. n.s. indicates nonspecific cross-reactions seen with these batches of antibodies. C, blots for S6 and for S6 phosphorylated at Ser-235/6. D, immunoblots for Akt phosphorylated at Ser-473.
Insulin activates mTORC1 signaling and enhances the phosphorylation of its target proteins, p70 S6K1 and 4E-BP1. Insulin appears to inactivate TSC2, the Rheb-GAP. It was therefore important to study the effect of knocking down TCTP in the absence of insulin, where TSC2 is active and the requirement for a Rheb-GEF would thus be greater (as depicted in Fig. 1A).
Thr-37/46 in 4E-BP1 are partially phosphorylated in the presence of amino acids without insulin (Fig. 5B). However, both amino acids and insulin are required for phosphorylation of Thr-70 and Ser-65 (see Fig. 5B) (6). In multiple experiments, we observed no consistent effect of knocking down TCTP on the phosphorylation of 4E-BP1 at any of these sites. In most cases, no effect was observed, as shown in Fig. 5B. In some experiments, knocking down TCTP did slightly increase the proportion of 4E-BP1 migrating in the least phosphorylated form (α) and decreased the overall levels of phosphorylation of Thr-37/46. The band just below 4E-BP1 seen on longer exposure of immunoblots for Thr(P)-37/46 is probably 4E-BP2, because Thr-46 in 4E-BP2 lies in a similar sequence context to Thr-37/46 in 4E-BP1 and likely cross-reacts with the anti-4E-BP1[Thr(P)-37/46] antibody (6). In some experiments, its strength was decreased on blots from TCTP knockdown cells (not shown). This variability did not appear to be related to the extent to which TCTP expression had been depleted.
If TCTP acts as a positive regulator of mTORC1, then knocking it down would be expected to impair S6 phosphorylation. However, reducing TCTP expression did not consistently decrease the phosphorylation of S6 (as shown in Fig. 5C). In some experiments we actually saw an increase in S6 phosphorylation (not shown). The data are not consistent with the idea that TCTP acts as a limiting upstream regulator of mTORC1.
Because the mTORC1-dependent phosphorylation of S6 requires both insulin and amino acids (44) (Fig. 5C), the interpretation of these experiments would be compromised if knocking down TCTP affected the control of Akt, the link between insulin and mTORC1 (e.g. by affecting mTORC2, which phosphorylates Ser-473 in Akt (37)). Phosphorylation of Akt at Ser-473 (a substrate for mTORC2 (37)) was also not altered in TCTP knockdown cells (Fig. 5D), indicating that TCTP is not limiting for mTORC2 function either.
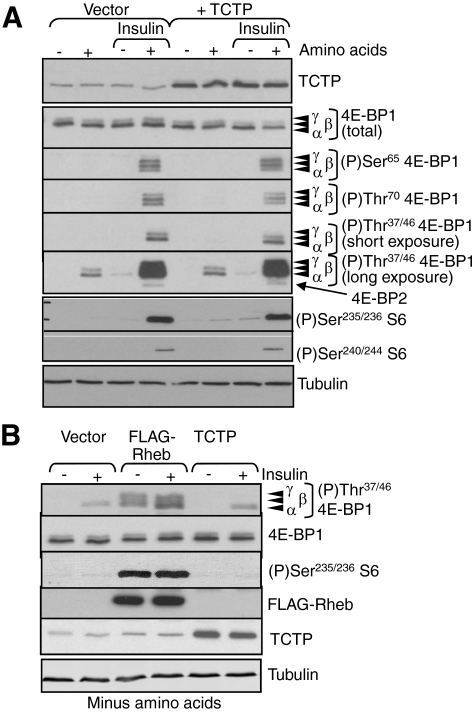
Overexpression of Rheb, but Not TCTP, Enhances mTORC1 Signaling—The regulation of mTORC1 by amino acids does not require the Rheb-GAP TSC2 (12, 13). However, it is possible that amino acids regulate GDP-GTP exchange, rather than GTP hydrolysis, on Rheb, perhaps by promoting TCTP (Rheb-GEF) activity (Fig. 1A, arrow 1). If this were the case, then overexpressing TCTP should activate mTORC1 signaling in amino acid-starved cells where the Rheb-GEF activity of TCTP might be limiting.
In these experiments, we expressed untagged TCTP, as we were concerned that the addition of a tag might interfere with the function of this small protein. In multiple experiments using amino acid-starved cells, no increase in 4E-BP1 phosphorylation was observed even upon substantial overexpression of TCTP (Fig. 6A). In contrast, and consistent with earlier work (6, 45), overexpressing Rheb did markedly increase the phosphorylation of S6 and 4E-BP1 (Fig. 6B).
FIGURE 6.
Overexpression of TCTP does not affect mTORC1 signaling. A, HEK293 cells were transfected with a vector encoding TCTP, Rheb or, as control, the empty vector. Cells were starved overnight of serum and, where indicated, also for amino acids. In some cases, cells were treated with insulin (100 nm, 30 min). Cells were lysed and samples analyzed by SDS-PAGE/Western blot using the indicated antibodies (with long and short exposures for 4E-BP1 Thr(P)-37/46). B, HEK293 cells were transfected with a vector encoding FLAG-Rheb or TCTP, or the empty vector as indicated. Cells were starved of serum and for amino acids. In some cases, cells were treated with insulin (100 nm, 30 min). Samples of cell lysates were analyzed by Western blot using the indicated antibodies. Tubulin was used as a loading control.
Thus, endogenous TCTP activity is apparently not limiting for mTORC1 signaling, even when the opposing GAP (TSC1/2) is active (i.e. without insulin or serum) or in amino acid-starved cells. The observation that overexpression of Rheb markedly increased the phosphorylation of 4E-BP1 at Thr-37/46 (Fig. 6B) confirms that Rheb activates mTORC1 signaling. However, these data show that overexpressing the proposed Rheb-GEF, TCTP, does not do so, whether in the presence or absence of amino acids and/or insulin.
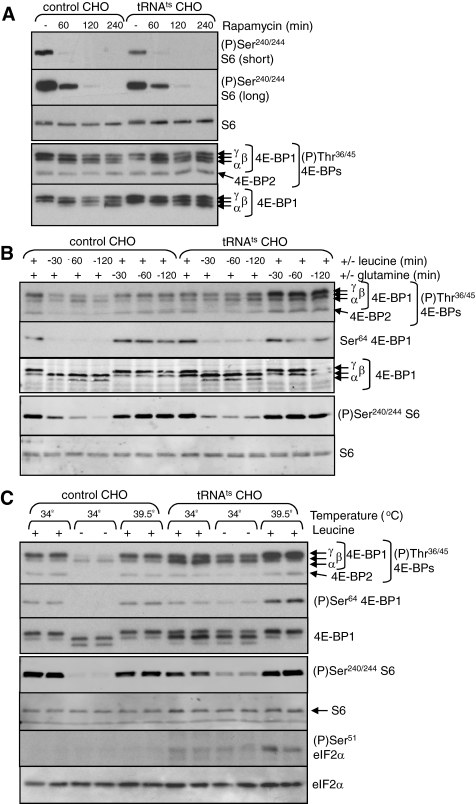
Is mTORC1 Regulated by tRNA Charging Levels?—Starving cells for essential amino acids may lead to the accumulation of uncharged tRNAs. It has been suggested that accumulation of uncharged tRNA in amino acid-starved cells leads to the inhibition of mTORC1 (29), either directly or through (for example) activation of the eIF2α kinase mGcn2. To test this idea, we made use of a CHO cell line (tsH1), which harbors a mutated leucyl-tRNA synthetase (tRNALeu-RS), the activity of which is temperature-sensitive (it is active at 34 °C but not at 39.5 °C (46, 47)). In both the tsH1 cells and the corresponding control (TR-3), CHO cells rapamycin completely blocked the phosphorylation of S6 at Ser-240/244 tested, although it did not impair the phosphorylation of 4E-BP1 at Thr-36/45 (Fig. 7A). This is consistent with earlier data showing that the latter sites, while regulated by mTORC1, are relatively insensitive to rapamycin (6). Rapamycin did cause a shift of 4E-BP1 toward less phosphorylated, faster migrating species (Fig. 7A), suggesting it does elicit the dephosphorylation of certain sites in 4E-BP1, although we have not studied this further.
FIGURE 7.
Regulation of mTORC1 signaling in control and mutant CHO cells. A, control (TR-3) and mutant (tsH1) CHO cells were treated with rapamycin (100 nm) for the indicated times at 34 °C. The cells were lysed, and samples were analyzed by SDS-PAGE and Western blot using the indicated antibodies. The differentially phosphorylated species of 4E-BP1 are indicated (α-γ). B, TR-3 and tsH1 cells were starved for leucine or glutamine for the indicated times at 34 °C. The cells were lysed and samples analyzed as in A. C, TR-3 and tSH1 cells were incubated at 34 or 39.5 °C. The period of leucine starvation or incubation at the higher temperature was 1 h. The cells were lysed and samples analyzed as in A. As a positive control for the effects of uncharged tRNA accumulation, these lysates were also analyzed for eIF2α and eIF2α phosphorylated on Ser-51.
It is particularly appropriate that the mutation in these cells is in the tRNALeu-RS because leucine is generally the amino acid that most strongly regulates mTORC1 signaling in mammalian cells (reviewed in Ref. 48). This is also the case for both the tsH1 cells and the TR-3 control cells, as judged by the phosphorylation of S6 and 4E-BP1 (Fig. 7B). Deprivation of leucine led to the decreased phosphorylation of both proteins in both cell lines (within 30 min; Fig. 7B), whereas starvation for another amino acid, glutamine, had no consistent or substantial effect. The effect of leucine starvation tended to be more modest in the tsH1 cell line. This cell line shows increased activity of the leucine transporter (system L), and this could be the reason for this difference (49).
Shifting the tRNALeu-RS mutant cells to 39.5 °C markedly increased the phosphorylation of eIF2α (Fig. 7C), providing evidence that uncharged tRNA accumulated to the extent that mGcn2 was activated, and consistent with earlier findings (36, 46, 47). As expected, no change in eIF2α phosphorylation was seen in the control cells as a result of the temperature shift. Importantly, although incubation at 39.5 °C did elicit phosphorylation of eIF2α in the tSH1 cells, it did not impair mTORC1 signaling, as assessed by the phosphorylation of S6 or 4E-BP1 (Fig. 7C). Indeed, if anything, an increase in the phosphorylation of 4E-BP1 and S6 was observed. These findings are not consistent with roles for either uncharged tRNA or activated mGcn2 in the control of mTORC1 by amino acid starvation.
DISCUSSION
In this study, we have tested the roles of two recently proposed regulators of mTORC1 signaling, FKBP38 (24) and TCTP (28). We have also evaluated an earlier proposal that amino acid starvation inhibits mTORC1 signaling because of the accumulation of uncharged tRNA (29). Our data lead us to conclude that none of these mechanisms acts to regulate mTORC1 signaling, at least in the cells studied here.
We have been able to confirm that FKBP38 can interact with Rheb and with mTOR, as described in Ref. 24 based on the initial finding that they interact in the yeast two-hybrid system. Interestingly, although the inactive Rheb(C181S) mutant (which shows a very low level of GTP binding (40)) fails to bind FKBP38, the Rheb(S20N) mutant (which likewise does not bind guanine nucleotides) actually bound very well to FKBP38. These data (i) suggest that the reason that Rheb(C181S) cannot bind Rheb is not simply because it cannot bind GTP and (ii) cast doubt on the earlier conclusion that Rheb·GTP preferentially binds FKBP38 and thereby releases the inhibitory effect of FKBP38 on mTORC1 (24). The fact the expression of the Rheb(S20N) mutant is poor suggests it may not adopt a native conformation; this feature might explain its enhanced ability to bind FKBP38, a member of the immunophilin family of peptidylprolyl isomerases.
In this study, we were unable to observe any effect of either amino acid starvation or insulin treatment on the association of Rheb or mTOR with FKBP38. Such data are not consistent with other recent findings (24). The lack of effect on FKBP38/mTOR association could perhaps be a consequence of expressing FKBP38 at higher levels, such that there is insufficient endogenous Rheb to remove FKBP38 from mTOR (which was the function proposed earlier for Rheb (24)). However, if FKBP38 remains bound to mTOR in the presence of amino acids and/or insulin, then that model predicts that mTORC1 signaling should be constitutively inhibited, an effect that we have not observed in any of our extensive range of experiments. Our data do not support the idea that the interaction of mTOR with FKBP38 controls mTORC1 signaling in response to insulin or amino acids.
In the case of TCTP, knocking down this proposed Rheb-GEF did not consistently affect mTORC1 signaling. It did tend to decrease the phosphorylation for 4E-BP1, but this effect was variable and may reflect one of the numerous other roles that have been ascribed previously to TCTP. Indeed, no reproducible effect was observed on the phosphorylation of S6, a protein whose mTORC1-dependent phosphorylation is well characterized and is very sensitive to rapamycin. Our data therefore do not support the idea that TCTP is an upstream activator of mammalian TOR signaling, in contrast to the proposed role of dTCTP in Drosophila (28). Our findings are consistent with the observation that serum-stimulated S6 phosphorylation was not impaired in TCTP knock-out mice (27).
It is possible that the absence of a consistent effect of TCTP knockdown on S6 and 4E-BP1 phosphorylation may reflect factors such as residual levels of TCTP, a rather abundant protein (43), or the substantial spontaneous nucleotide exchange rate on Rheb, at least as studied in vitro (28).4 TCTP is structurally related to the Mss4/Dss4 family of proteins that bind nucleotide-free forms of small G-proteins but display only weak GEF activity (43). Interestingly, TCTP was reported to interact with a GTP-binding protein (eEF1A) and its GEF (eEF1Bβ) but actually inhibited guanine nucleotide release (50) (which would actually impair nucleotide exchange).
Furthermore, the overexpression of TCTP did not overcome the inhibitory effect of amino acid starvation on 4E-BP1 phosphorylation, implying that amino acid insufficiency does not render TCTP activity more limiting for mTORC1 signaling. Thus, although TCTP positively affects mTORC1 signaling, amino acids do not impair mTORC1 signaling by a mechanism involving the impairment of TCTP function. (This possible mechanism is indicated by arrow 1 in Fig. 1A.) This is consistent with the finding that amino acid starvation barely affects Rheb·GTP levels (12, 51, 52). Amino acid starvation may act “downstream” of Rheb·GTP to block mTORC1 signaling, as suggested earlier (51), either by modulating the ability of Rheb to activate mTORC1 or by controlling the function of mTORC1 itself (Fig. 1A, arrows 2 and 3).
Another mechanism previously proposed for the regulation of mTOR is the accumulation of uncharged tRNA under conditions of amino acid starvation (29), which was suggested to inhibit mTORC1. However, in the CHO tsH1 cells used here, which have a temperature-sensitive mutant of tRNALeu-RS (46, 47), elevated temperature did not inhibit mTORC1 signaling, although it did enhance the phosphorylation of eIF2α. The latter observation is consistent with earlier data for these cells, and most likely indicates that uncharged tRNAs (tRNALeu) have accumulated and activated the eIF2α kinase mGcn2. The fact that mTORC1 signaling was not affected under these conditions suggests that neither uncharged tRNALeu nor activated mGcn2 is responsible for the impairment of mTORC1 signaling that occurs when CHO cells are starved of leucine. The present findings are consistent with our own recent data from mouse embryonic fibroblasts that lack mGcn2 (53); in such cells, amino acid starvation does inhibit mTORC1 signaling (more strongly than in GCN2+/+ cells), illustrating that mGcn2 is not required for this response. In fact, such inhibition was actually greater in the mGCN2 knock-out cells.
Our data do not support the previous suggestion, also partly based on the use of a temperature-sensitive aminoacyl-tRNA synthetase mutant, that uncharged tRNA inhibits p70 S6k activity through regulation of mTOR (29). There are a number of possible explanations for this discrepancy, including the use of different cell lines, a different aminoacyl-tRNA synthetase mutant, and the measurement of different targets of mTOR activity. However, our conclusions are in agreement with those of Pham et al. (54) who showed that, in adipocytes, the phosphorylation of 4E-BP1 is not impaired by amino alcohols (agents that inhibit tRNA charging).
It is probable that free amino acids (or perhaps amino acid metabolites) regulate mTORC1. This would be consistent with our earlier conclusions (55). Such a mechanism may be physiologically more appropriate for cells than relying on a build up of uncharged tRNA, which could result in detrimental consequences to the cell, as the lack of a given set of aminoacylated tRNAs could lead to missense incorporation or premature termination events during polypeptide chain elongation. It is also worth noting that although shifting cells to the nonpermissive temperature does increase eIF2α phosphorylation, likely because of activation of mGcn2 by uncharged tRNA, starving the same cells of leucine does not. This lack of effect of amino acid starvation on eIF2α phosphorylation is consistent with our other recent data (53).
Because mammalian cells can respond to amino acid starvation by both increasing the phosphorylation of eIF2α and inhibiting the activity of mTOR, the issue arises as to which of these mechanisms is the more important for the inhibition of overall protein synthesis under such conditions. This may depend on the cell type. In this study we have shown that accumulation of uncharged tRNA in tsH1 cells (which is well known to inhibit protein synthesis) stimulates eIF2α phosphorylation but does not inhibit mTORC1 signaling. In fact, mTORC1 signaling actually tended to be activated at the nonpermissive temperature. The reason for this is unclear; perhaps the inhibition of protein synthesis caused by eIF2α phosphorylation leads to a larger pool of cytoplasmic amino acids, thus maintaining mTORC1 signaling. (Our earlier data from CHO cells indicate that mTORC1 signaling is regulated by intracellular amino acid levels (55).)
Although considerable effort has been made over the last decades to unravel the details of the mTOR signaling network, several aspects of the control of mTORC1 activity still remain unsolved. Novel players in mTORC1 signaling have been proposed recently that would be potential targets for regulation by amino acids and other physiological stimuli. However, our data are not compatible with the notions that TCTP, FKBP38, or deacylated tRNA are involved in the control of mTORC1. Further work is required to identify conclusively the mechanisms by which stimuli such as insulin and amino acids (especially leucine) activate signaling to this important protein kinase complex.
Acknowledgments
We thank Dr. Yanni Wang for help in cloning FKBP38.
This work was supported by grants from the Canadian Institutes of Health Research, the Ajinomoto Amino Acid Research Program, and the University of British Columbia (to C. G. P.), from the Wellcome Trust (to U. A. B., A. E., and M. J. C.), and from the Ralph Bates Cancer Research Fund (to A. E. and M. J. C.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: mTOR, mammalian target of rapamycin; 4E-BP, eukaryotic initiation factor 4E-binding protein; eIF, eukaryotic initiation factor; GAP, GTPase-activator protein; Gcn, general control of amino acid biosynthesis, nonderepressing; GEF, guanine nucleotide-exchange factor; mTORC1/2, mTOR complex 1 (or 2); Rheb, Ras homolog enriched in brain; RS, [tRNA] synthetase; p70 S6K, p70 S6 kinase; TCTP, translationally controlled tumor protein; TSC, tuberous sclerosis complex; HA, hemagglutinin; siRNA, short interfering RNA; CHO, Chinese hamster ovary.
X. Wang and C. G. Proud, unpublished observations.
References
- 1.Wullschleger, S., Loewith, R., and Hall, M. N. (2006) Cell 124 471–484 [DOI] [PubMed] [Google Scholar]
- 2.Yang, Q., and Guan, K. L. (2007) Cell Res. 17 666–681 [DOI] [PubMed] [Google Scholar]
- 3.Easton, J. B., and Houghton, P. J. (2006) Oncogene 25 6436–6446 [DOI] [PubMed] [Google Scholar]
- 4.Lee, C. H., Inoki, K., and Guan, K. L. (2007) Annu. Rev. Pharmacol. Toxicol. 47 443–467 [DOI] [PubMed] [Google Scholar]
- 5.Wang, X., and Proud, C. G. (2006) Physiol. (Bethesda) 21 362–369 [DOI] [PubMed] [Google Scholar]
- 6.Wang, X., Beugnet, A., Murakami, M., Yamanaka, S., and Proud, C. G. (2005) Mol. Cell. Biol. 25 2558–2572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fonseca, B. D., Smith, E. M., Lee, V. H., MacKintosh, C., and Proud, C. G. (2007) J. Biol. Chem. 282 24514–24524 [DOI] [PubMed] [Google Scholar]
- 8.Avruch, J., Hara, K., Lin, Y., Liu, M., Long, X., Ortiz-Vega, S., and Yonezawa, K. (2006) Oncogene 25 6361–6372 [DOI] [PubMed] [Google Scholar]
- 9.Long, X., Lin, Y., Ortiz-Vega, S., Yonezawa, K., and Avruch, J. (2005) Curr. Biol. 15 702–713 [DOI] [PubMed] [Google Scholar]
- 10.Land, S. C., and Tee, A. R. (2007) J. Biol. Chem. 282 20534–20543 [DOI] [PubMed] [Google Scholar]
- 11.Manning, B. D., and Cantley, L. C. (2003) Trends Biochem. Sci. 28 573–576 [DOI] [PubMed] [Google Scholar]
- 12.Smith, E. M., Finn, S. G., Tee, A. R., Browne, G. J., and Proud, C. G. (2005) J. Biol. Chem. 280 18717–18727 [DOI] [PubMed] [Google Scholar]
- 13.Nobukuni, T., Joaquin, M., Roccio, M., Dann, S. G., Kim, S. Y., Gulati, P., Byfield, M. P., Backer, J. M., Natt, F., Bos, J. L., Zwartkruis, F. J., and Thomas, G. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 14238–14243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Byfield, M. P., Murray, J. T., and Backer, J. M. (2005) J. Biol. Chem. 280 33076–33082 [DOI] [PubMed] [Google Scholar]
- 15.Findlay, G. M., Yan, L., Procter, J., Mieulet, V., and Lamb, R. F. (2007) Biochem. J. 403 13–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walker, V. E., Atanasiu, R., Lam, H., and Shrier, A. (2007) J. Biol. Chem. 282 23509–23516 [DOI] [PubMed] [Google Scholar]
- 17.Edlich, F., Maestre-Martinez, M., Jarczowski, F., Weiwad, M., Moutty, M. C., Malesevic, M., Jahreis, G., Fischer, G., and Lucke, C. (2007) J. Biol. Chem. 282 36496–36504 [DOI] [PubMed] [Google Scholar]
- 18.Edlich, F., Weiwad, M., Erdmann, F., Fanghanel, J., Jarczowski, F., Rahfeld, J. U., and Fischer, G. (2005) EMBO J. 24 2688–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edlich, F., Erdmann, F., Jarczowski, F., Moutty, M. C., Weiwad, M., and Fischer, G. (2007) J. Biol. Chem. 282 15341–15348 [DOI] [PubMed] [Google Scholar]
- 20.Erdmann, F., Jarczowski, F., Weiwad, M., Fischer, G., and Edlich, F. (2007) FEBS Lett. 581 5709–5714 [DOI] [PubMed] [Google Scholar]
- 21.Shirane, M., and Nakayama, K. I. (2003) Nat. Cell Biol. 5 28–37 [DOI] [PubMed] [Google Scholar]
- 22.Wang, H. Q., Nakaya, Y., Du, Z., Yamane, T., Shirane, M., Kudo, T., Takeda, M., Takebayashi, K., Noda, Y., Nakayama, K. I., and Nishimura, M. (2005) Hum. Mol. Genet. 14 1889–1902 [DOI] [PubMed] [Google Scholar]
- 23.Rosner, M., Hofer, K., Kubista, M., and Hengstschlager, M. (2003) Oncogene 22 4786–4798 [DOI] [PubMed] [Google Scholar]
- 24.Bai, X., Ma, D., Liu, A., Shen, X., Wang, Q. J., Liu, Y., and Jiang, Y. (2007) Science 318 977–980 [DOI] [PubMed] [Google Scholar]
- 25.Bommer, U. A., and Thiele, B. J. (2004) Int. J. Biochem. Cell Biol. 36 379–385 [DOI] [PubMed] [Google Scholar]
- 26.Gachet, Y., Tournier, S., Lee, M., Lazaris-Karatzas, A., Poulton, T., and Bommer, U. A. (1999) J. Cell Sci. 112 1257–1271 [DOI] [PubMed] [Google Scholar]
- 27.Chen, S. H., Wu, P. S., Chou, C. H., Yan, Y. T., Liu, H., Weng, S. Y., and Yang-Yen, H. F. (2007) Mol. Biol. Cell 18 2525–2532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hsu, Y. C., Chern, J. J., Cai, Y., Liu, M., and Choi, K. W. (2007) Nature 445 785–788 [DOI] [PubMed] [Google Scholar]
- 29.Iiboshi, Y., Papst, P. J., Kawasome, H., Hosoi, H., Abraham, R. T., Houghton, P. J., and Terada, N. (1999) J. Biol. Chem. 274 1092–1099 [DOI] [PubMed] [Google Scholar]
- 30.Proud, C. G. (2005) Semin. Cell Dev. Biol. 16 3–12 [DOI] [PubMed] [Google Scholar]
- 31.Berlanga, J. J., Santoyo, J., and de Haro, C. (1999) Eur. J. Biochem. 265 754–762 [DOI] [PubMed] [Google Scholar]
- 32.Sood, R., Porter, A. C., Olsen, D., Cavener, D. R., and Wek, R. C. (2000) Genetics 154 787–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang, X., Campbell, L. E., Miller, C. M., and Proud, C. G. (1998) Biochem. J. 334 261–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thompson, L. H., Harkins, J. L., and Stanners, C. P. (1973) Proc. Natl. Acad. Sci. U. S. A. 70 3094–3098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pollard, J. W., and Stanners, C. P. (1979) J. Cell. Physiol. 98 571–585 [DOI] [PubMed] [Google Scholar]
- 36.Pollard, J. W., Galpine, A. R., and Clemens, M. J. (1989) Eur. J. Biochem. 182 1–9 [DOI] [PubMed] [Google Scholar]
- 37.Sarbassov, D. D., Guertin, D. A., Ali, S. M., and Sabatini, D. M. (2005) Science 307 1098–1101 [DOI] [PubMed] [Google Scholar]
- 38.Browne, G. J., and Proud, C. G. (2004) Mol. Cell. Biol. 24 2986–2997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Castro, A. F., Rebhun, J. F., Clark, G. J., and Quilliam, L. A. (2003) J. Biol. Chem. 278 32493–32496 [DOI] [PubMed] [Google Scholar]
- 40.Li, Y., Inoki, K., and Guan, K. L. (2004) Mol. Cell. Biol. 24 7965–7975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Basso, A. D., Mirza, A., Liu, G., Long, B. J., Bishop, W. R., and Kirschmeier, P. (2005) J. Biol. Chem. 280 31101–31108 [DOI] [PubMed] [Google Scholar]
- 42.Oldfield, S., and Proud, C. G. (1992) Eur. J. Biochem. 208 73–81 [DOI] [PubMed] [Google Scholar]
- 43.Thaw, P., Baxter, N. J., Hounslow, A. M., Price, C., Waltho, J. P., and Craven, C. J. (2001) Nat. Struct. Biol. 8 701–704 [DOI] [PubMed] [Google Scholar]
- 44.Avruch, J., Belham, C., Weng, Q., Hara, K., and Yonezawa, K. (2001) Prog. Mol. Subcell. Biol. 26 115–154 [DOI] [PubMed] [Google Scholar]
- 45.Tee, A. R., Manning, B. D., Roux, P. P., Cantley, L. C., and Blenis, J. (2003) Curr. Biol. 13 1259–1268 [DOI] [PubMed] [Google Scholar]
- 46.Austin, S. A., Pollard, J. W., Jagus, R., and Clemens, M. J. (1986) Eur. J. Biochem. 157 39–47 [DOI] [PubMed] [Google Scholar]
- 47.Clemens, M. J., Galpine, A., Austin, S. A., Panniers, R., Henshaw, E. C., Duncan, R., Hershey, J. W., and Pollard, J. W. (1987) J. Biol. Chem. 262 767–771 [PubMed] [Google Scholar]
- 48.Kimball, S. R., and Jefferson, L. S. (2006) J. Nutr. 136 S227–S231 [Google Scholar]
- 49.Moore, P. A., Jayme, D. W., and Oxender, D. L. (1977) J. Biol. Chem. 252 7427–7430 [PubMed] [Google Scholar]
- 50.Cans, C., Passer, B. J., Shalak, V., Nancy-Portebois, V., Crible, V., Amzallag, N., Allanic, D., Tufino, R., Argentini, M., Moras, D., Fiucci, G., Goud, B., Mirande, M., Amson, R., and Telerman, A. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 13892–13897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Long, X., Ortiz-Vega, S., Lin, Y., and Avruch, J. (2005) J. Biol. Chem. 280 23433–23436 [DOI] [PubMed] [Google Scholar]
- 52.Roccio, M., Bos, J. L., and Zwartkruis, F. J. (2006) Oncogene 25 657–664 [DOI] [PubMed] [Google Scholar]
- 53.Wang, X., and Proud, C. G. (2008) Mol. Cell. Biol. 28 1429–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pham, P. T., Heydrick, S. J., Fix, H. L., Kimball, S. R., Jefferson, L. S., and Lynch, C. J. (2000) J. Cell. Biochem. 79 427–441 [DOI] [PubMed] [Google Scholar]
- 55.Beugnet, A., Tee, A. R., Taylor, P. M., and Proud, C. G. (2002) Biochem. J. 372 555–566 [DOI] [PMC free article] [PubMed] [Google Scholar]