Abstract
The most severe form of bone autosomal recessive osteopetrosis both in humans and in the gray-lethal (gl/gl) mouse is caused by mutations in the Ostm1 gene. Although osteopetrosis is usually associated with a defect in the hematopoietic-derived osteoclast cells, this study determined that Ostm1 is expressed in many hematopoietic cells of the myeloid and lymphoid B- and T-lineages. Hematopoiesis in gl/gl mice is characterized by a marked expansion of the osteoclast lineage but also by deregulation of the lymphoid lineages with a decrease in B-lymphoid cell populations and altered distribution in T-lymphoid double and single CD4 CD8-positive cells. In committed gl/gl osteoclasts, specific Ostm1 transgene targeting showed a requirement of additional factors and/or cells for normal osteoclast function, and importantly, defined the gl osteopetrotic defect as non-cell autonomous. By contrast, gl/gl osteoclast, B- and T-lymphoid lineage phenotypes were rescued when Ostm1 is expressed under PU.1 regulation from a bacterial artificial chromosome transgene, which established an essential role for Ostm1 in hematopoietic cells in addition to osteoclasts. Together these experiments are the first to demonstrate the existence of hematopoietic crosstalk for the production of functional and active osteoclasts.
A strict balance between bone formation and resorption in vertebrates is required throughout adult life to maintain a constant bone mass (1–3). The cells responsible for bone resorption are the osteoclasts that are formed through the fusion of hematopoietic myeloid cells. Failure of appropriate hematopoietic progenitors to differentiate or to mature into functional osteoclasts results in abnormal accumulation of mineralized osteoid and leads to osteopetrosis (4).
Severe human osteopetrosis is characterized by dense sclerotic bone, accumulation of mineralized osteoid and cartilage (5–8), and causes drastic reduction of bone marrow (1, 9). Associated with the limited bone marrow compartment, patients display defective hematopoiesis with anemia, thrombocytopenia, high susceptibility to infections, and die at a young age. The only therapy for severe cases of osteopetrosis is bone marrow transplantation (10, 11). Allogeneic bone marrow transplantation is possible for these patients, but this approach has major limitations. Alternatively, autologous hematopoietic stem cell transplantation upon gene therapy correction could circumvent these drawbacks. However, development of gene therapy protocols requires proper knowledge of hematopoiesis in osteopetrotic disorders.
At present only mutation in OSTM1 and in three other genes, TCIRG1, ClCN7, and RANKL have been directly associated with severe autosomal recessive osteopetrosis in human (12–16). The human OSTM1 gene has been characterized, and OSTM1 patients display the most severe recessive osteopetrotic phenotype and die at early ages (17–19).
Osteopetrotic mouse mutations have been characterized and associated with osteoclast differentiation or activation defects, including numerous transcription factors, such as PU.1, that play a crucial role in osteoclast lineage differentiation (20), as well as proteins important in osteoclast activation, the murine Ostm1 (21). This murine Ostm1 gene was first identified in the gray-lethal (gl)2 mouse mutant (17) and encodes a unique transcript. Based on protein structural analysis, the Ostm1 protein likely corresponds to a type I transmembrane protein (17). The gl mutation consists of a deletion that results in a null phenotype with absence of transcript and protein expression. gl mice produce osteoclasts, defined by multinucleated cells positive for the classic TRAP marker, but which are functionally inactive (21). Interestingly, gl mice demonstrated a significant increase in the mature bone-resident osteoclast population (21). Although the osteoclast defects in gl mice have been well analyzed histologically and ex vivo, it is important for the design of appropriate therapeutic approaches, for example autologous gene therapy, to understand the fundamental hematopoietic cellular defects and the role of Ostm1 in hematopoietic lineage differentiation and maturation.
This study first characterized, in gl/gl hematopoietic phenotypes, an expansion of the osteoclast lineage as a compensatory mechanism and major lymphopoiesis anomalies with reduced B-lymphoid cell population and altered T-lymphoid cell differentiation pattern. Second, our results on Ostm1 hematopoietic-targeted transgenic mice support that Ostm1 is required not only in osteoclast lineage but also in other lineages to fully rescue gl/gl osteopetrosis and defective hematopoiesis.
MATERIALS AND METHODS
Mice
The mouse strain GL/Le dlJ +/+gl was obtained from The Jackson Laboratory (Bar Harbor, ME) and maintained by heterozygous brother × sister mating for ∼150 generations. Homozygous gl/gl mice used for analysis had a healthy appearance. Experiments with animals complied and were approved by the institutional animal care committee and Canadian Committee for Animal Protection.
Production and Analysis of Transgenic Mice
TRAP-Ostm1—The TRAP-Ostm1 construction was produced with the Ostm1 cDNA (1.055 kb) linked to the TRAP promoter (1.8 kb, XhoI/XbaI) upstream (22), and to the human hGH poly(A) signal (2.1 kb) downstream. Linearized transgene (XhoI/NotI) was injected into fertilized oocytes from F2 (C3H × C57BL/6) (23). Transgenic mice were identified by PCR with TRAP forward (5′-GTCCTCACCAGAGACTCTGAAC-3′) and Ostm1 reverse (5′-CAAGTCCTGCACCTCCAACAG-3′) primers. PCR amplification conditions were 94 °C, 5 min, followed by 30 cycles of 94 °C for 0.5 min, 65 °C for 0.5 min, and 72 °C for 0.5 min. Transgene integrity and copy number were determined by Southern analysis. Each transgenic line was successively crossed with heterozygous gl/+ mice to generate gl/gl TRAP-Ostm1 transgenic progenies.
PU.1-Ostm1—Three PU.1 BAC clones were first isolated from a 129Sv mouse pBelo11 BAC genomic library (Research Genetics). These BAC clones contained, based on two PCR assays, >19 kb of upstream regulatory sequences of PU.1 with primers at ∼–19 kb (forward, 5′-TCTTGAATGCCTGCTTGTG-3′; reverse, 5′-TTGGAGCTGGAGAGATGGTTC-3′) and the PU.1 second exon with another primer set (forward, 5′-CGGATGACTTGGTTACTTACG-3′; reverse, 5′-GGGCTGACATTGTGTGGATAC-3′). From pulse field gel electrophoresis, Southern blot, and end-junction sequencing analysis, one of these BAC clones (∼100 kb) was selected for further experiments and contained >30 kb of PU.1 regulatory sequence, the full-length PU.1 gene as well as the Tbp1 gene.
Two targeting constructs for homologous recombination were produced in pLD53.SC1 BAC recombination vector to modify the original PU.1 BAC clone. The first construct was designed to delete the Tbp1 gene from the original BAC. Two homologous regions on each side of the Tbp1 gene, 31 kb apart, were PCR-amplified from PU.1 BAC DNA and cloned in pLD53.SC1 vector (24). The second construct was designed to replace the PU.1 genomic sequence by the Ostm1 open reading frame and an in-frame iEGFP-PGK poly(A) sequences (1.7 kb). Homologous PU.1 5′ and 3′ regions were amplified from the original PU.1 BAC DNA and inserted upstream and downstream of the Ostm1-pA cloned in the pLD53.SC1 vector (supplemental Table S1).
Each of the two homologous constructs was used in a two-step RecA strategy for BAC modifications (24). The Tbp1 gene was excised from the original PU.1 BAC creating the new BAC PU.1ΔTbp1. The PU.1 genomic coding sequences were then replaced by those of Ostm1 in the BAC PU.1ΔTbp1, generating the PU.1-Ostm1 BAC. At each step, the new BACs were fully characterized by pulse field gel electrophoresis and Southern blot. Modified Tbp1 and PU.1 regions were sequenced to verify proper gene deletion/substitution and exclude other alterations. The resulting PU.1-Ostm1 BAC (∼70 kb) was microinjected (17), and transgenic founders were identified by PCR using PU.1 forward (5′-GCCTTTCTCCCTCCCAGCC-3′) and Ostm1 reverse (5′-CAAGTCCTGCACCTCCAACAG-3′) primers. PCR amplification conditions were 94 °C, 5 min, followed by 30 cycles of 94 °C for 0.5 min, 65 °C for 0.5 min, and 72 °C for 0.5 min, and transgene integrity and copy number were determined by Southern blot. Each founder was successively crossed with heterozygous gl/+ mice to generate gl/gl PU.1-Ostm1 transgenic progenies.
Cellular Analysis
Hematologic Parameters—Blood from gl/gl and control mice was obtained by cardiac puncture and collected into EDTA-coated tubes (BD Microtainer). Hematological analysis was performed using a Bayer Advia 120 automated cell analyzer with the mouse archetype of multispecies software version 2.206 (CBTR, Montreal, Canada). The spleen and thymus were monitored as percentage of organ to total body weight.
FACS Analysis—Flow cytometric analyses were carried out on spleen, thymus, and bone marrow single cell suspension in phosphate-buffered saline with 1% heat-inactivated fetal bovine serum. Nucleated cells (1.5 × 106) were stained with phycoerythrin-conjugated CD11b, B220, IgD, or CD4 (Pharmingen) and with fluorescein isothiocyanate-conjugated IgM, CD5, Ly6-G, CD8, and CD11c (Cedarlane). Samples were analyzed on a BD FACSCalibur four-color flow cytometer with System II software.
Hematopoietic Progenitor Clonogenic Assays—The number of splenic nucleated cells was quantified from single cell suspension in Iscove's modified Dulbecco's medium containing 1% methylcellulose, 10% fetal calf serum, erythropoietin (1.0 unit/ml), murine IL-3 (0.8% v/v of WEHI-3 supernatant), 1% bovine serum albumin, transferrin (190 mg/ml), and α-monothioglycerol (5 × 10–2 m) (25). Each assay was set up in triplicates, and colony-forming units were defined by morphology, size (cell number), and hemoglobinization. Erythroid colony-forming units (CFU-E) were counted at day 2, whereas burst-forming units (BFU-E), granulocyte-macrophage colony-forming units (CFU-GM), and macrophage colony-forming units (CFU-M) were counted at day 7 and granulocyte, erythroid, macrophage, and megakaryocyte colony-forming units (CFU-GEMM) at day 11.
Ex Vivo Osteoclast Differentiation—Bone marrow-derived macrophage cells were generated ex vivo from bone marrow cells cultured for 5 days in α-minimal essential medium supplemented with 10% fetal bovine serum and macrophage-colony-stimulating factor. Osteoclast-like cells (OCLs) were generated from bone marrow-derived macrophage cell differentiation in vitro for 6 days in α-minimal essential medium supplemented with 10% fetal bovine serum, 10 ng/ml macrophage-colony-stimulating factor, and 50 ng/ml receptor activator of NF-κB ligand. Alternatively, OCLs were generated ex vivo by co-culture of primary calvaria osteoblasts with bone marrow/spleen cells as previously described (21).
Expression Analysis
Total RNA from OCLs, enriched hematopoietic cells, and tissues, including bone marrow, brain, kidney, thymus, liver, and spleen, were isolated with TRIzol (Invitrogen). TRAP-Osmt1 transgene expression was determined by semi-quantitative PCR on 0.5 μg of total RNA. The primers used were: Ostm1 endogenous forward 5′-CCTGCTTTGAGCATAACCTGA-3′ (Ostm1 exon 3) and reverse 5′-CTGCAGTCCCAACATTTCGTGAG-3′ (Ostm1 3′-untranslated region); TRAP-Ostm1 transgene, forward 5′-GTGGTTGCTGTGTCTGTGTTCA-3′ (Ostm1 exon 5), reverse 5′-TTGGGATATAGGCTTCTTCAAACTC-3′ (hGH); Trap gene forward 5′-TGTTCTCTGACCGTGCCCTTC-3′ (Trap exon 3) and reverse 5′-CCCACTCAGCACATAGCCCCA-3′ (Trap exon 5). PCR conditions were 94 °C for 5 min, followed by 30 cycles of 94 °C for 0.5 min, 65 °C for 0.5 min, and 72 °C for 0.5 min.
PU.1-Ostm1 transgene expression was determined by real-time quantitative PCR on 0.1 μg of DNase (Invitrogen)-treated total RNA. The primers used were for Ostm1 endogenous forward 5′-GCTTCCTTCACTCAGAGCAA-3′ (Ostm1 exon 5) and reverse 5′-GTGAGAATGCAACTTGTCCGA-3′ (Ostm1 exon 6); PU.1-Ostm1 transgene, forward 5′-GCTTCCTTCACTCAGAGCAA-3′ (Ostm1 exon 5), reverse 5′-AGTGAATAGGAACTTCGGAA-3′(EGFP); Trap forward 5′-GGTATGTGCTGGCTGGA-3′ (Trap exon 3) and reverse 5′-GACTGGCAAAGTCATCTGA-3′ (Trap exon 5); PU.1 endogenous forward 5′-ATGGAGCTGGAACAGATGCA-3′ (PU.1 exon 3) and reverse 5′-CCAAGCCATCAGCTTCTCCA-3′ (PU.1 exon 4); S16 forward 5′-GCTACCAGGGCCTTTGAGATG-3′ and reverse 5′-AGGACGGATTTGCTGGTGTGG-3′. All reactions were performed in triplicate in a Syber Green Master Mix (Qiagen). PCR conditions were 94 °C, 15 min, followed by 50 cycles of 94 °C for 0.5 min, 55 °C for 0.5 min, and 72 °C for 0.5 min in an MX4000 Mutiplex quantitative PCR analyzer.
Histological and X-ray Analysis
Bone samples from 3-week-old mice were fixed in 10% phosphate-buffered formalin, decalcified in 0.5 m EDTA, and embedded in paraffin. Sections were stained with hematoxylin and eosin (H&E), mounted, and analyzed. X-ray scan was carried out using Faxitron MX20 (18 Kv, 10 s).
Statistical Analysis
Values are expressed as mean ± S.D. Unpaired two-sample Student's t test was used for statistical analysis with p < 0.05 considered significant.
RESULTS
Ostm1 Expression in Several Mature Hematopoietic Cell Populations
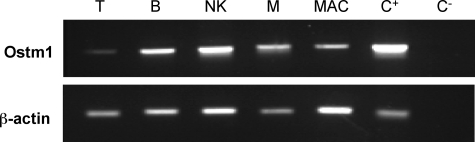
Ostm1 expression profile was determined in wild-type mature hematopoietic cells derived from myeloid and lymphoid populations. Enriched ex vivo hematopoietic cell populations were obtained from cultured C57BL/6J spleen, bone marrow, and peritoneal cavity cells (26, 27). Semi-quantitative expression analysis of endogenous Ostm1 showed high levels in B, NK, and mast cells and lower levels in T cells and activated macrophages relative to actin control (Fig. 1), suggesting that Ostm1 could play a role in several hematopoietic cell types.
FIGURE 1.
Ostm1 expression in hematopoietic cells. Semi-quantitative Ostm1 expression from enriched hematopoietic cells detected high levels in B-lymphocyte (B), natural killer (NK), mast (M) cells and lower expression in T-lymphocyte (T) and activated macrophage (MAC) in comparison to positive brain tissue control (C+), negative control (C–), and normalized to β-actin.
Altered Hematology and Hematopoiesis in gl/gl Mice
Anemia and Leukopenia in gl/gl Mice—To determine the impact of the gl mutation on hematopoietic cell populations, we evaluated hematological parameters of gl/gl and wild-type control littermates. The gl/gl mice showed a significant reduction in the number of red blood cells compared with wild-type mice, as well as a significant reduction in hematocrit and hemoglobin indicating a mild anemia (Table 1). In parallel, the number of white blood cells and lymphocytes was significantly decreased, indicating leukopenia and lymphopenia in gl/gl mice (Table 1). Homozygous gl/gl mice displayed a spleen to body weight ratio comparable to wild-type mice despite anemia (Table 2). Consistently, histological examination of the spleen did not show any profound architectural alteration of the white or red pulps where B cell maturation and myelopoiesis occur respectively (data not shown).
TABLE 1.
Hematological parameters of grey-lethal (gl/gl) mice
Data are expressed as the means ± S.D.
| Mice | n | RBC | Hematocrit | Hemoglobin | WBC | Lymphocytes | Monocytes | PLT |
|---|---|---|---|---|---|---|---|---|
| 109 cells/ml | % | g/dl | 106 cells/ml | 106 cells/ml | 104 cells/ml | 106 cells/ml | ||
| +/+ | 10 | 6.3 ± 1.7 | 34.1 ± 8.9 | 10.0 ± 2.7 | 4.1 ± 0.9 | 3.2 ± 0.6 | 11.1 ± 6.5 | 1000 ± 460 |
| gl/gl | 9 | 4.2 ± 2.1a | 22.0 ± 10.7b | 6.2 ± 2.8b | 2.8 ± 1.7a | 1.6 ± 1.3c | 11.4 ± 6.5 | 707 ± 586 |
p < 0.05.
p < 0.02.
p < 0.001.
TABLE 2.
Altered hematopoietic progenitors in spleen of grey-lethal (gl/gl) mice
Data are expressed as the means ± S.D.
| Mice | n | Body weight | Spleen weight | Spleen/body WT | Nucleated cell | n | CFU-E | BFU-E | CFU-GM | CFU-M | CFU-GEMM |
|---|---|---|---|---|---|---|---|---|---|---|---|
| g | g | % | (×106/spleen) | ×102 | ×102 | ×102 | ×102 | ×102 | |||
| +/+ | 10 | 13.0 ± 2.4 | 0.096 ± 0.007 | 0.72 | 80.1 ± 3.6 | 3 | 298 ± 50 | 75 ± 10 | 61 ± 9 | 32 ± 7 | 3.9 ± 2 |
| gl/gl | 9 | 6.1 ± 1.3a | 0.051 ± 0.017a | 0.84 | 70.6 ± 9.4 | 3 | 287 ± 20 | 87 ± 11 | 147 ± 10a | 33 ± 6 | 2.9 ± 1.3 |
p < 0.05.
Analysis of Myeloid Lineages: Stimulation of gl Osteoclast Progenitors—Since gl/gl mice exhibit altered hematological parameters, spleen hematopoietic progenitor populations of homozygous gl/gl mice were evaluated using clonogenic assays, because there is a massive depletion of bone marrow cells in this severe osteopetrotic phenotype. The level of multipotent CFU-GEMM cell population in gl/gl and age-matched control littermate mice was not significantly affected (Table 2). Similarly, erythroid differentiation potential quantified by early BFU-E and late CFU-E spleen progenitor cell populations in gl/gl mice showed, despite the anemia, no significant increase relative to age-matched controls (Table 2). In contrast, all gl/gl mice showed a marked 2.5-fold expansion of granulocyte-macrophage progenitors (CFU-GM), from which osteoclasts are derived, relative to wild-type control (Table 2). The size of CFU-GM colonies in gl/gl mice compared with control mice appeared larger, but maintained similar morphology. In addition, the levels of the CFU-M progenitor cells, from which macrophages are derived, were comparable in gl/gl and control mice, indicating that only the CFU-GM are affected.
To further characterize and quantify the osteoclast cell populations, we performed flow-cytometric analysis (FACS) on spleen cell populations using CD11b+ and Ly6-G+ antibodies. As shown in Table 3, the analysis revealed a significant increase of splenic CD11b+ and CD11b+/Ly6-G+ cells that include osteoclast, macrophage, monocyte, and granulocyte populations in gl/gl mice relative to controls. In fact, the CD11b+/Ly6-G+ cell subpopulation was elevated ∼4-fold in homozygous gl/gl mice, consistent with increased CFU-GM progenitors (Table 2). By contrast, analysis of the population of splenic dendritic CD11b+/CD11c+ cells was unchanged in gl/gl mice (data not shown).
TABLE 3.
Altered splenic myeloid and lymphoid cell populations of grey-lethal (gl/gl) mice
Data are expressed as the means % ± S.D.
|
Mice
|
n
|
Myeloid lineage
|
Lymphoid lineages
|
|||||
|---|---|---|---|---|---|---|---|---|
| CD11b+ | CD11b+Ly6-G+ | B220+ IgM+ | IgM+ IgD+ | CD5+ | CD4+ | CD8+ | ||
| +/+ | 6 | 3.7 ± 0.3 | 1.3 ± 0.2 | 23.1 ± 1.9 | 16.1 ± 2.5 | 9.2 ± 1.8 | 6.1 ± 1.4 | 3.1 ± 0.9 |
| gl/gl | 6 | 8.7 ± 2.2a | 5.9 ± 2.5a | 13.4 ± 1.6a | 12.4 ± 1.6a | 7.5 ± 1.2 | 9.4 ± 3.0 | 6.5 ± 2.6 |
p < 0.001.
Impaired gl Lymphopoiesis—Because homozygous gl/gl mice have reduced white blood cell count and human osteopetrotic patients have higher susceptibility to infections possibly due to altered immune response, the lymphoid lineages were investigated in gl/gl spleen and thymus. Quantification of the “late” B-lymphoid cell population by FACS revealed a significant decrease of ∼40% of the B220+/IgM+ cell population in the gl/gl mice compared with control. Similarly, the more mature IgM+/IgD+ cell population was also significantly reduced in gl/gl mice (Table 3). Because the B-lymphoid compartment is constituted of two distinct B1 and B2 cell populations (28), we have quantified the B1 cell population separately using the specific CD5 (Ly-1) marker to monitor B-cell differentiation potential. In gl/gl mice, the B1 self-renewing cells (CD5+) that differentiate independently from bone marrow stroma were slightly reduced (∼18%) relative to control mice (Table 3).
The gl/gl thymus was consistently smaller in comparison to age-matched littermates and the ratio of thymus to body weight was significantly reduced (Table 4). Quantification of the major thymic T-cell populations in gl/gl and wild-type littermate animals showed altered thymic cellular subpopulations. Although the gl/gl double negative CD4–CD8– cell subpopulation was unchanged, an important depletion in the double positive CD4+CD8+ subpopulation was determined relative to wild type (Table 4). Concurrently, the gl/gl mice displayed a major 4- to 5-fold increase in single CD4- and CD8-positive cells (Table 4), suggesting enhanced thymic cell maturation.
TABLE 4.
Thymus analysis and altered distribution in cell populations of grey-lethal (gl/gl) mice
Data are expressed as the means % ± S.D.
| Mice | n | Body weight | Thymus weight | Thymus/body WT | Nucleated cells | CD4+CD8+ | CD4+ | CD8+ |
|---|---|---|---|---|---|---|---|---|
| g | g | % | ×106/thymus | |||||
| +/+ | 4 | 13.5 ± 2.1 | 0.058 ± 0.001 | 0.45 | 86.8 ± 13 | 80.5 ± 6.4 | 11.7 ± 5.1 | 5.1 ± 1.2 |
| gl/gl | 4 | 5.7 ± 1.0a | 0.020 ± 0.003a | 0.35b | 10.5 ± 2.2a | 9.5 ± 4.1a | 63.7 ± 5.0a | 21.7 ± 6.3a |
p < 0.001.
p < 0.05.
Non-cell Autonomous Ostm1 Role in Committed Osteoclast
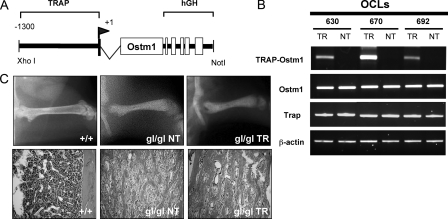
In gl/gl mice, the increased progenitor and mature osteoclast population, concomitant with inactive osteoclasts (21), indicated that Ostm1 expression was required in late committed precursors of the osteoclast lineage. To investigate this hypothesis, transgenic mice expressing Ostm1 under the control of the committed osteoclasts TRAP gene promoter were generated (Fig. 2A). Three TRAP-Ostm1 transgenic lines (2–10 transgene copies) were assessed for osteoclast-specific expression, precursor frequency, and differentiation potential. Expression of the transgene was slightly below endogenous expression in two lines (630 and 692), and above in one line (670) (Fig. 2B). Transgenic OCLs from all lines showed similar frequency and differentiation potential as non-transgenic OCLs (data not shown). Importantly, the TRAP-Ostm1 transgenic mice did not exhibit any bone defects.
FIGURE 2.
Production, expression, and analysis of TRAP-Ostm1 transgenic mice. A, schematic representation of the transgene used to generate TRAP-Ostm1 mice. The TRAP regulatory sequences (1.3 kb), transcription initiation site (+1), first exon (500 bp), and intron 1 were cloned upstream of Ostm1 (1.055 kb) open reading frame followed by the 3′ and poly(A) of human growth hormone gene (hGH; 2.1kb). B, transgene expression in OCLs (TR) from each TRAP-Ostm1 transgenic lines was compared with non-transgenic OCL controls (NT). Endogenous Trap, Ostm1, and β-actin expression were used as controls. C, persistence of osteopetrosis was observed in homozygous gl/gl TRAP-Ostm1 transgenic mice (gl/gl TR) similar to gl/gl non-transgenic littermate (gl/gl NT) in contrast to wild-type control (+/+) as detected by x-ray analysis (top panels) and on histological bone sections (bottom panels, magnification, ×50).
The TRAP-Ostm1 transgenic mice from the three lines then served to test for osteopetrosis complementation by successive mating to heterozygous gl/+ mice. Transgenic gl/gl TRAP-Ostm1 mice generated from these matings were genotyped for the Ostm1 mutation (17) and the transgene. All gl/gl TRAP-Ostm1 transgenic progenies displayed an osteopetrotic phenotype similar to gl/gl non-transgenic mice with reduced bone marrow space and lack of tooth eruption (Fig. 2C). This indicated that Ostm1 expression in committed osteoclast precursors to mature cells is not sufficient to rescue the gl/gl osteoclast defect.
gl Hematopoiesis/Osteopetrosis Correction by Ostm1 Expression in Multihematopoietic Cells
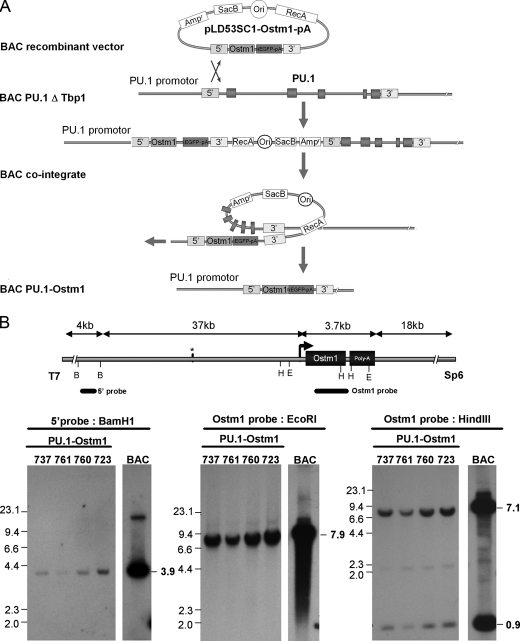
Lack of complementation in gl/gl TRAP-Ostm1 transgenic mice and the multihematopoietic lineage defects in gl/gl mice suggested that Ostm1 expression is required in other hematopoietic cell or lineage at earlier stage. Because the PU.1 transcriptional factor is essential in the early osteoclast lineage as well as in several early hematopoietic progenitors, including erythropoietic and lymphoid lineages (20, 29), regulatory elements of the PU.1 (Sfpi1) gene were selected to target Ostm1 expression in transgenic mice. A 100-kb PU.1 BAC clone was isolated that contains regulatory sequences with the CAAT/enhancer-binding protein-α enhancer binding site (30), localized 14 kb upstream of PU.1 essential for expression in transgenic mice (31–33). This PU.1 BAC was modified by homologous recombination. The first modification specifically deleted the adjacent Tbp1 (Pmsc3) gene (∼30 kb) to have exclusively PU.1 gene in the BAC. This PU.1-ΔTbp1 BAC was then subjected to a second round of recombination, to replace the PU.1 coding sequence by that of Ostm1 (Fig. 3A). The resulting ∼70 kb PU.1-Ostm1 BAC clone contained ∼37 kb of 5′ PU.1 regulatory sequence and 18 kb of 3′ sequence as determined by restriction pattern analysis and sequencing.
FIGURE 3.
Production and analysis of BAC PU.1-Ostm1 transgene. A, schematic representation illustrates the BAC PU.1 ΔTbp1 to the BAC PU.1-Ostm1 recombination step. The PU.1 genomic sequences in BAC PU.1 ΔTbp1 were replaced by Ostm1-EGFP-poly(A) with pLD53-Ostm1-PA vector containing 5′ and 3′ PU.1 homology cassettes on each side of Ostm1-EGFP-pA. The BAC cointegrant recombination removed the pLD53.SC1 vector and produced the new BAC PU.1-Ostm1. B, the BAC PU.1-Ostm1 was used to generate transgenic lines. Southern blot of genomic DNA analysis with the following restriction digests are shown: left panel, BamHI (B) digest hybridized with a 5′ probe that detect a 3.9-kb band for the transgene; middle and right panels, EcoRI (E) or HindIII (H) hybridized with an Ostm1 probe (exon1–exon6) with one band at 7.9 kb for EcoRI digestion and two bands at 7.1 kb and 0.9 kb for HindIII digests for the transgene. Genomic DNA from the original BAC PU.1-Ostm1 was used as control.
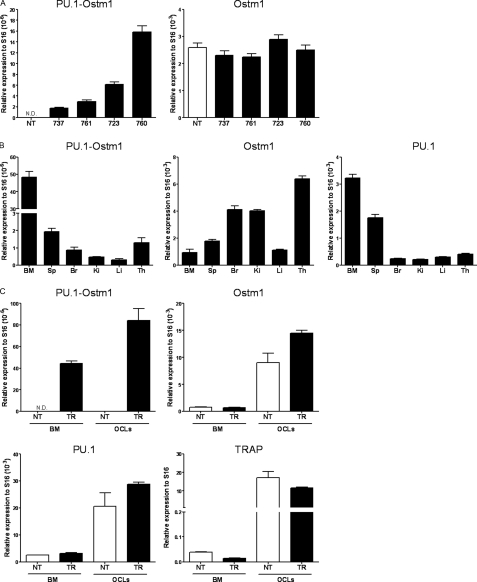
Four transgenic PU.1-Ostm1 founders were generated carrying between 2 and 10 copies of the transgene. Transgene integrity in these lines (723, 737, 761, and 760) was monitored by analysis of vector/insert junctions and hybridization pattern with four specific probes (Fig. 3B and data not shown). Transgene quantitative expression was detected in spleen of all transgenic lines but at a much lower level compared with endogenous Ostm1 in transgenic and in non-transgenic controls (Fig. 4A). In two representative lines, the spleen, bone marrow, and thymus displayed highest expression, whereas liver, kidney, and brain had lower levels (Fig. 4, A and B). In comparison to endogenous Ostm1 and PU.1 expression, transgene expression levels were notably lower but displayed a similar expression pattern as PU.1 (Fig. 4, A and B). Transgene expression in bone marrow differentiated OCLs also showed ex vivo expression as observed for the endogenous Ostm1, PU.1, and TRAP controls (Fig. 4C). Notably, differentiated OCLs from transgenic PU.1-Ostm1 bone marrow and spleen were produced at similar frequency as non-transgenic controls (data not shown). These transgenic mice did not develop any detectable phenotype.
FIGURE 4.
Expression analysis of the PU. 1-Ostm1 transgene. A, real-time quantitative spleen PU.1-Ostm1 transgene and endogenous Ostm1 expression from the four PU.1-Ostm1 transgenic lines (TR) and non-transgenic (NT) was established relative to S16 as internal control. B, real-time quantitative PU.1-Ostm1 transgene and Ostm1, PU.1 endogenous genes expression was carried out on different tissues of PU.1-Ostm1 transgenic (TR) mice line #761 (n = 3) relative to S16 as internal control. High expression was detected in bone marrow (BM), spleen (Sp), and thymus (Th) and lower levels in brain (Br), liver (Li), and kidney (Ki). C, real-time quantitative transgene expression was performed on bone marrow (BM) and osteoclast like cells (OCLs) generated in culture from PU.1-Ostm1 transgenic #737 (TR) and non-transgenic (NT) mice (n = 3) relative to S16 as internal control and compared with endogenous expression of Ostm1, PU.1, and TRAP.
To functionally monitor whether the PU.1-Ostm1 transgene could rescue gl/gl osteopetrosis, three transgenic PU.1-Ostm1 lines (737, 760, and 761) were successively mated to heterozygous gl/+ mice. Transgenic gl/gl PU.1-Ostm1 mice from these matings were genotyped at the Ostm1 locus and for the BAC transgene. Although non-transgenic homozygous gl/gl littermates exhibited the gl osteopetrotic phenotypes, all gl/gl transgenic progenies from the three lines tested displayed full tooth eruption and appropriate bone marrow development, providing evidence for functional rescue of osteoclastogenesis (Fig. 5).
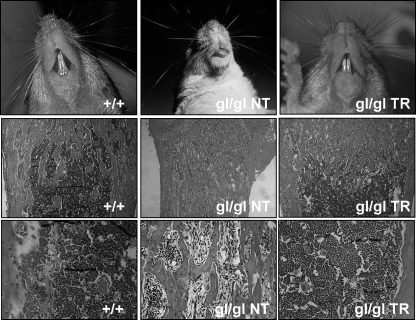
FIGURE 5.
Complementation of osteopetrosis in gl/gl PU. 1-Ostm1 transgenic mice. Rescue of osteopetrosis in gl/gl PU.1-Ostm1 transgenic (gl/gl TR) relative to non-transgenic (gl/gl NT) mice as shown by tooth eruption (top panels) and normal development of medullary space on bone sections stained with H&E (middle and bottom panels).
To determine whether the entire spectrum of gl/gl hematopoiesis defects was corrected in gl/gl PU.1-Ostm1 BAC transgenic mice, the myeloid and lymphoid cell populations of transgenic and age-matched non-transgenic gl/gl mice from two representative transgenic lines (737 and 761) were quantified by FACS analysis. In transgenic gl/gl PU.1-Ostm1 mice, splenic CD11b+ and CD11b+/Ly6-G+ cell populations showed levels similar to wild-type littermates, whereas these populations were significantly increased in homozygous gl/gl littermates (Table 5). Notably, correction of the myeloid population was obtained with both PU.1-Ostm1 transgenic lines tested (Table 5). Analysis of transgenic bone marrow cell populations also showed control wild-type levels for the osteoclast lineage. In addition, the mature B cell populations in transgenic gl/gl PU.1-Ostm1 evaluated in spleen and bone marrow returned to control levels in contrast to gl/gl mice. Interestingly, the thymic structure and the cellular distribution profile of single and double CD4+CD8+-positive T lymphoid cell populations in gl/gl PU.1-Ostm1 transgenic were indistinguishable from controls (Table 5 and supplemental Fig. S1), indicating that the PU.1-Ostm1 had fully rescued gl/gl thymic defect.
TABLE 5.
Normalization of myeloid, B- and T-lymphoid cell populations in gl/gl PU.1-Ostm1 transgenic mice
Data are expressed as the means % ± S.D. compared to wild type non-transgenic (+/+ NT).
|
Mice
|
n
|
Spleen
|
Bone marrow
|
n
|
Thymus
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Myeloid lineage
|
B-lymphoid lineage
|
Myeloid lineage
|
B-lymphoid lineage
|
|||||||||
| CD11b+ | CD11b+Ly6-G+ | B220+ | CD11b+ | CD11b+Ly6-G+ | B220+ | Nucleated cells × 106/thymus | CD4+CD8+ | CD4+ | CD8+ | |||
| +/+ NT | 6 | 11.5 ± 1.6 | 5.2 ± 2.0 | 61.9 ± 17.2 | 11.8 ± 3.1 | 21.1 ± 6.4 | 50.1 ± 9.3 | 6 | 70.6 ± 9.3 | 88.6 ± 3.8 | 7.0 ± 0.8 | 2.7 ± 1.4 |
| +/+ TR | 4 | 9.4 ± 4.9 | 4.3 ± 1.6 | 55.1 ± 14.2 | 9.7 ± 3.1 | 21.5 ± 7.3 | 51.4 ± 5.7 | 4 | 86.6 ± 17.4 | 89.0 ± 3.5 | 6.5 ± 1.6 | 2.8 ± 1.4 |
| gl/gl NT | 5 | 15.6 ± 2.0a | 10.2 ± 4.7a | 39.9 ± 8.9b | NAc | NA | NA | 4 | 24.4 ± 7.0d | 36.2 ± 13.1a | 45.3 ± 20.4b | 16.0 ± 7.2b |
| gl/gl TR 737 | 5 | 8.7 ± 1.2 | 4.0 ± 1.2 | 59.2 ± 6.3 | 10.0 ± 1.7 | 26.7 ± 4.5 | 47.2 ± 2.7 | 5 | 66.0 ± 9.3 | 87.6 ± 1.1 | 7.5 ± 1.3 | 3.4 ± 0.3 |
| gl/gl TR 761 | 3 | 11.3 ± 1.5 | 4.2 ± 1.4 | 55.6 ± 7.8 | 14.9 ± 2.9 | 28.2 ± 1.8 | 41.0 ± 6.9 | 3 | 71.9 ± 8.7 | 89.4 ± 0.5 | 7.0 ± 0.2 | 2.3 ± 0.7 |
p < 0.005.
p < 0.05.
NA, not applicable.
p < 0.001.
DISCUSSION
Ostm1 mutations lead to the most severe form of bone autosomal recessive osteopetrotic disorder in both humans and gl/gl mice and are associated with a defect in the hematopoietic osteoclast cell. Herein, Ostm1 analysis also uncovered expression in several myeloid and lymphoid lineages and when absent in gl mice resulted in altered hematopoietic differentiation and/or maturation with anemia, leukopenia, and lymphopenia. Characterization of Ostm1 in vivo function by sole targeting to the gl/gl committed osteoclasts revealed the existence of additional factors or cells essential for osteoclast functional activation. Significantly, these findings established a non-cell autonomous mechanism for the gl defect. Moreover, complementation by early multilineage hematopoietic Ostm1 expression in gl/gl mice determined that Ostm1 is required in other hematopoietic cells, including osteoclast precursors, for full osteoclast function. Our studies demonstrated that Ostm1 plays a major role in myelopoiesis and lymphopoiesis and provided evidence of a crosstalk mechanism between hematopoietic cells for osteoclast activation.
Characterization of severe leukopenia and mild anemia in gl/gl mice pointed to defective hematopoietic differentiation. Such hematopoietic anomalies were likely downstream of the early hematopoietic multipotent CFU-GEMM progenitor stage, because this clonogenic population in gl/gl mice was unaffected. The unaltered erythroid lineage differentiation of the early BFU-E and late CFU-E progenitors argued that gl anemia results from hampered late erythropoiesis due to the severely reduced medullary space. By contrast, the gl/gl spleen CFU-GM, from which the osteoclasts are derived, were specifically increased while the more committed CFU-M progenitors, that give rise to macrophages but not to osteoclasts (34, 35), were unchanged. Further, a marked expansion of splenic early CD11b+/Ly6-G+ and late CD11b+ osteoclast precursor stages in homozygous gl/gl indicated appropriate osteoclast lineage differentiation potential. This increase in osteoclast progenitors and precursors correlated with the presence of abundant mature gl/gl TRAP+ osteoclasts (21). Such stimulation of cell populations throughout the gl/gl osteoclast lineage to the mature cells supports the existence of a compensatory mechanism in response to the inactive osteoclasts.
In addition to the osteoclast lineage expansion, the gl/gl lymphoid cell differentiation is altered. The reduced white blood cell counts in peripheral blood were consistent with decrease immature and mature B-lymphoid cell populations in gl/gl mice. Similar reduction in immature B-cells was observed previously in osteopetrosis, at least transiently at ∼4 weeks of age, in the other osteopetrotic mice including op,c-fos, oc, and c-src mutants (36–38). Although these mutated genes have different functions and properties, it appears that reduced B-cell population is a hallmark of osteopetrosis and points to the absence of bone marrow microenvironment/compartment as a cause for this phenotype. However, the results of reduced B1 self-renewing cell population (CD5 marker) in gl/gl independently of the marrow and of the “conventional” splenic B cell population support the notion that the B-cell differentiation potential per se might be affected. Consequently, the B-cell phenotype in gl/gl mice may result from combined direct and indirect roles of Ostm1 in B-cell precursors and on bone microenvironment. Because B-cells normally produce osteoprotegerin, an inhibitor of osteoclastogenesis (39), such a decrease in B-cell population is likely to impinge on osteoprotegerin release and thereby be responsible for the increased osteoclast cell population of gl/gl mice. Similar to the B-cell population, the total gl/gl thymic T cell population was reduced and also displayed an altered distribution pattern. Indeed, the unchanged proportion of the immature gl/gl double negative CD4–CD8– cell population compared with the depleted double positive CD4+CD8+ population implied that Ostm1 is very important during these differentiation stages. The subsequent significant increase in gl/gl mature CD4 and CD8 single positive cell populations contrasted with the decrease in double positive cells, as detected in two osteopetrotic mice c-fos and oc (36, 37). This cellular differentiation pattern could be an indirect physiological response to maintain sufficient T-cell production. Alternatively, Ostm1 under normal conditions could slow down the positive selection mechanism of T-cell differentiation or, less likely, promote lymphocyte egression from the thymus (40). Together this hematopoietic characterization of gl/gl mice supports Ostm1 playing critical roles in multiple lineage differentiation at immature and/or mature cellular stages.
An important outcome from the study of transgenic Ostm1 expression in mice targeted to gl/gl committed and mature inactive osteoclasts was the absence of osteoclast correction. Despite substantial TRAP-Ostm1 transgene expression in gl/gl mature osteoclasts, all gl/gl TRAP-Ostm1 transgenic progenies displayed severe osteopetrosis similar to the gl/gl phenotype. This finding infers that the gl osteoclast defect is non-cell autonomous. Such data contrasts with another osteopetrotic phenotype caused by the ablation of the chloride channel ClC-7, a potential partner of Ostm1 in mature osteoclasts (41). In fact, re-expression of Clcn7 with the same TRAP regulatory sequences in transgenic mice, as those used herein for Ostm1 on a Clcn7–/– background, was sufficient to produce active osteoclasts and rescue osteopetrosis (42). Our analysis indicates that the function of Ostm1 is not only in late osteoclastogenesis like CLC-7, but also in a wider hematopoietic cellular repertoire and implies a broader partner spectrum.
The complete hematopoietic rescue by the multilineage-targeted Ostm1 expression in PU.1 BAC transgenic of the gl/gl mice is compatible with a non-cell autonomous defect for the osteoclast and uncovered an essential role for Ostm1 in more than one hematopoietic cell type. Of significance, all gl/gl PU.1-Ostm1 BAC transgenic mice demonstrated rescue of gl/gl osteopetrosis and hematopoietic defects even with a low level of transgene expression. This phenotypic correction provided additional and definitive evidence that Ostm1 is the gene responsible for the gl defect. Most importantly, these results along with those of TRAP-Ostm1 showed that Ostm1 expression in a cell(s) from the myeloid and/or lymphoid lineages contributes to correction of the osteoclast activation defect. In fact, involvement of more than one hematopoietic cell type in gl osteopetrosis is consistent with a previous rescue of gl phenotype by total bone marrow transplantation (43). To date no cell(s) other than osteoclasts, nor any underlying cellular process essential for osteoclast activation, have been described. Our experiments restricts the cell(s) essential for osteoclast resorptive function to those concomitantly expressing Ostm1- and PU.1-targeted cells that are early precursors, multipotent granulocyte-macrophage, pro-T, and/or pro B-cells (29, 44). Based on Ostm1 type I transmembrane structure, the cellular process likely implicates cell-cell interactions or secreted factors for osteoclast activation. Although several cytokines and chemokines secreted from hematopoietic cells (45, 46) and osteoblasts are critical for osteoclastogenesis, none have been shown physiologically or in cell culture to be necessary for osteoclast activation. Nevertheless, we propose that Ostm1 participates in a cell trafficking pathway with a partner repertoire of hematopoietic receptors and factors that will subsequently trigger a crosstalk mechanism for osteoclast activation. Based on this finding, autologous gene therapy for osteopetrosis will require adapted strategy design targeted to several hematopoietic lineages for correction of the disease.
In summary, loss-of-function of Ostm1 resulted in deregulation of multiple hematopoietic lineages in addition to osteoclast lineage. The osteopetrotic phenotype with Ostm1 expression specifically targeted to committed/mature gl/gl osteoclasts highlighted gl defect non-cell autonomous. Complementation of gl/gl osteopetrosis by additionally targeting Ostm1 to early hematopoietic lineages provided definitive evidence that Ostm1 is essential in early precursors to produce functional mature osteoclasts. Most importantly, these studies have identified a novel intercellular hematopoietic crosstalk mechanism.
Supplementary Material
Acknowledgments
We thank Drs. M. E. Hatten, N. Heintz, A. Veillette, and D. Roodman for materials.
This work was supported by Canadian Institutes of Health Research and National Sciences and Engineering Council of Canada grants (to J. V.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Fig. S1.
Footnotes
The abbreviations used are: gl, gray-lethal mouse; BAC, bacterial artificial chromosome; TRAP, tartrate-resistant acid phosphatase; OCLs, osteoclast-like cells; FACS, fluorescence-activated cell sorting; hGH, human growth hormone; CFU-E, erythroid colony-forming units; CFU-GM, granulocyte-macrophage CFUs; CFU-M, macrophage CFUs; CFU-GEMM, granulocyte, erythroid, macrophage, and megakaryocyte CFUs.
References
- 1.Teitelbaum, S. L., and Ross, P. (2003) Nat. Rev. Genet. 4 638–649 [DOI] [PubMed] [Google Scholar]
- 2.Boyle, W. J., Simonet, W. S., and Lacey, D. L. (2003) Nature 423 337–342 [DOI] [PubMed] [Google Scholar]
- 3.Zaidi, M. (2007) Nat. Med. 13 791–801 [DOI] [PubMed] [Google Scholar]
- 4.Janssens, K., and Van Hul, W. (2002) Hum. Mol. Genet. 11 2385–2393 [DOI] [PubMed] [Google Scholar]
- 5.Lazner, F., Gowen, M., Pavasovic, D., and Kola, I. (1999) Hum. Mol. Genet. 8 1839–1846 [DOI] [PubMed] [Google Scholar]
- 6.Wilson, C. J., and Vellodi, A. (2000) Arch. Dis. Child 83 449–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tolar, J., Teitelbaum, S. L., and Orchard, P. J. (2004) N. Engl. J. Med. 351 2839–2849 [DOI] [PubMed] [Google Scholar]
- 8.Balemans, W., Van Wesenbeeck, L., and Van Hul, W. (2005) Calcif. Tissue Int. 77 263–274 [DOI] [PubMed] [Google Scholar]
- 9.Helfrich, M. H. (2003) Microsc. Res. Tech. 61 514–532 [DOI] [PubMed] [Google Scholar]
- 10.Gerristen, E. J. A., Vossen, J., Fasth, A., Friedrich, W., Morgan, G., Padmos, A., Vellodi, A., Porras, O., O'Meara, A., Porta, F., Bordigoni, P., Cant, A., Hermans, J., Griscelli, C., and Fisher, A. (1994) J. Pediatr. 125 896–902 [DOI] [PubMed] [Google Scholar]
- 11.Eapen, M., Davies, S. M., Ramsay, N. K. C., and Orchard, P. J. (1998) Bone Marrow Transplant. 22 941–946 [DOI] [PubMed] [Google Scholar]
- 12.Sobacchi, C., Frattini, A., Guerrini, M. M., Abinun, M., Pangrazio, A., Susani, L., Bredius, R., Mancini, G., Cant, A., Bishop, N., Grabowski, P., Del Fattore, A., Messina, C., Errigo, G., Coxon, F. P., Scott, D. I., Teti, A., Rogers, M. J., Vezzoni, P., Villa, A., and Helfrich, M. H. (2007) Nat. Genet. 39 960–962 [DOI] [PubMed] [Google Scholar]
- 13.Sobacchi, C., Frattini, A., Orchard, P., Porras, O., Tezcan, I., Andolina, M., Babul-Hirji, R., Baric, I., Canham, N., Chitayat, D., Dupuis-Girod, S., Ellis, I., Etzioni, A., Fasth, A., Fisher, A., Gerristen, B., Gulino, V., Horwitz, E., Klamroth, V., Lanino, E., Mirolo, M., Musio, A., Matthijs, G., Nonomaya, S., Notarangelo, L. D., Ochs, H. D., Furga, A. S., Valiaho, J., van Hove, J. L. K., Vihinen, M., Vujic, D., Vezzoni, P., and Villa, A. (2001) Hum. Mol. Genet. 10 1767–1773 [DOI] [PubMed] [Google Scholar]
- 14.Michigami, T., Kageyama, T., Satomura, K., Yamaoka, K., Nakayama, M., and Ozono, K. (2002) Bone 30 436–439 [DOI] [PubMed] [Google Scholar]
- 15.Scimeca, J.-C., Quincey, D., Parinello, H., Romatet, D., Grogeorges, J., Gaudray, P., Philip, N., Fisher, A., and Carle, G. F. (2003) Hum. Mut. 21 151–157 [DOI] [PubMed] [Google Scholar]
- 16.Kornak, U., Kasper, D., Bösl, M. R., Kaiser, E., Schweizer, M., Schulz, A., Friedrich, W., Delling, G., and Jentsch, T. J. (2001) Cell 104 205–215 [DOI] [PubMed] [Google Scholar]
- 17.Chalhoub, N., Benachenhou, N., Rajapurohitam, V., Pata, M., Ferron, M., Frattini, A., Villa, A., and Vacher, J. (2003) Nat. Med. 9 399–406 [DOI] [PubMed] [Google Scholar]
- 18.Quarello, P., Forni, M., Barbereis, L., Defilippi, C., Campagnoli, M. F., Frattini, A., Chalhoub, N., Vacher, J., and Ramenghi, U. (2004) J. Bone Miner. Res. 19 1194–1199 [DOI] [PubMed] [Google Scholar]
- 19.Maranda, B., Chabot, G., Décarie, J.-C., Pata, M., Azeddine, B., Moreau, A., and Vacher, J. (2008) J. Bone Miner. Res. 23 296–300 [DOI] [PubMed] [Google Scholar]
- 20.Tondravi, M. M., McKercher, S. R., Anderson, K., Erdmann, J. M., Quiroz, M., Maki, R., and Teitelbaum, S. L. (1997) Nature 386 81–84 [DOI] [PubMed] [Google Scholar]
- 21.Rajapurohitam, V., Chalhoub, N., Benachenhou, N., Neff, L., Baron, R., and Vacher, J. (2001) Bone 28 513–523 [DOI] [PubMed] [Google Scholar]
- 22.Reddy, S. V., Scarcez, T., Windle, J. J., Leach, R. J., Hundley, J. E., Chirgwin, J. M., Chou, J. Y., and Roodman, G. D. (1993) J. Bone Min. Res. 8 1263–1270 [DOI] [PubMed] [Google Scholar]
- 23.Ferron, M., and Vacher, J. (2005) Genesis 41 138–145 [DOI] [PubMed] [Google Scholar]
- 24.Gong, S., Yang, X. W., Li, C., and Heintz, N. (2002) Genome Res. 12 1992–1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iscove, N. N., Sieber, F., and Whintherhalter, K. H. (1974) J. Cell. Physiol. 83 309–320 [DOI] [PubMed] [Google Scholar]
- 26.Davidson, D., and Veillette, A. (2001) EMBO J. 20 3414–3426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cao, M. Y., Davidson, D., Yu, J., Latour, S., and Veillette, A. (1999) J. Exp. Med. 190 1527–1534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kantor, A. B., and Herzenberg, L. A. (1993) Annu. Rev. Immunol. 11 501–538 [DOI] [PubMed] [Google Scholar]
- 29.Anderson, M. K., Hernandez-Hoyos, G., Diamond, R. A., and Rothenberg, E. V. (1999) Development 126 3133–3148 [DOI] [PubMed] [Google Scholar]
- 30.Yeamans, C., Wang, D.-Z., Paz-Priel, I., Torbett, B. E., Tenen, D. G., and Friedman, A. D. (2007) Blood 110 3136–3142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li, Y., Okuno, Y., Zhang, P., Radomska, H. S., Chen, H.-M., Iwasaki, H., Akashi, K., Klemsk, M. J., McKercher, S. R., Maki, R. A., and Tenen, D. G. (2001) Blood 98 2958–2965 [DOI] [PubMed] [Google Scholar]
- 32.Okuno, Y., Huang, G., Rosenbauer, F., Evans, E. K., Radomska, H. S., Iwasaki, H., Akashi, K., Moreau-Gachelin, F., Li, Y., Zhang, P., Göttgens, B., and Tenen, D. G. (2005) Mol. Cell. Biol. 25 2832–2845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosenbauer, F., Owens, B. M., Yu, L., Tumang, J. R., Steidl, U., Kutok, J. L., Clayton, L. K., Wagner, K., Scheller, M., Iwasaki, H., Liu, C., Hackanson, B., Akashi, K., Leutz, A., Rothstein, T. L., Plass, C., and Tenen, D. G. (2006) Nat. Genet. 38 27–37 [DOI] [PubMed] [Google Scholar]
- 34.Hayase, Y., Muguruma, Y., and Lee, M. Y. (1997) Exp. Hematol. 25 19–25 [PubMed] [Google Scholar]
- 35.Menaa, C., Kurihara, N., and Roodman, G. D. (2000) Biochem. Biophys. Res. Commun. 267 943–946 [DOI] [PubMed] [Google Scholar]
- 36.Tagaya, H., Kunisada, T., Yamazaki, H., Yamane, O., Tokuhisa, T., Wagner, E. F., Sudo, T., Shultz, L. D., and Hayashi, S.-I. (2000) Blood 95 3363–3370 [PubMed] [Google Scholar]
- 37.Blin-Wakkach, C., Wacchach, A., Sexton, P. M., Rochet, N., and Carle, G. F. (2004) Leukemia 18 1505–1511 [DOI] [PubMed] [Google Scholar]
- 38.Nilsson, S. K., and Bertoncello, I. (1994) Dev. Biol. 164 456–462 [DOI] [PubMed] [Google Scholar]
- 39.Li, Y., Toraldo, G., Li, A. G., Yang, X., Zhang, H., Qian, W.-P., and Weitzmann, M. N. (2007) Blood 109 3839–3848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Allende, M. L., Dreier, J. L., Mandala, S., and Proia, R. L. (2004) J. Biol. Chem. 279 15396–15401 [DOI] [PubMed] [Google Scholar]
- 41.Lange, P., Wartosch, L., Jentsch, T., and Fuhrmann, J. (2006) Nature 440 220–223 [DOI] [PubMed] [Google Scholar]
- 42.Kasper, D., Planells-Cases, R., Fuhrmann, J. C., Scheel, O., Zeitz, O., Ruether, K., Schmitt, A., Poët, M., Steinfeld, R., Schweizer, M., Kornak, U., and Jentsch, T. J. (2005) EMBO J. 24 1079–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walker, D. G. (1975) Science 190 784–785 [DOI] [PubMed] [Google Scholar]
- 44.Nutt, S. L., and Kee, B. L. (2007) Immunity 26 715–725 [DOI] [PubMed] [Google Scholar]
- 45.Lorenzo, J. (2000) J. Clin. Invest. 106 749–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weitzmann, M. N., and Pacifici, R. (2005) Immunol. Rev. 208 154–168 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.