Abstract
The trypsin-like protease Der p 3, a major allergen of the house dust mite Dermatophagoides pteronyssinus, is synthesized as a zymogen, termed proDer p 3. No recombinant source of Der p 3 has been described yet, and the zymogen maturation mechanism remains to be elucidated. The Der p 3 zymogen was produced in Pichia pastoris. We demonstrated that the recombinant zymogen is glycosylated at the level of its propeptide. We showed that the activation mechanism of proDer p 3 is intermolecular and is mediated by the house dust mite cysteine protease Der p 1. The primary structure of the proDer p 3 propeptide is associated with a unique zymogen activation mechanism, which is different from those described for the trypsin-like family and relies on the house dust mite papain-like protease Der p 1. This is the first report of a recombinant source of Der p 3, with the same enzymatic activity as the natural enzyme and trypsin. Glycosylation of the propeptide was found to decrease the rate of maturation. Finally, we showed that recombinant Der p 3 is inhibited by the free modified prosequence TP1R.
House dust mite (HDM)6 allergens have been shown to be causative factors of allergic manifestations such as atopic dermatitis, perennial rhinitis, or bronchial asthma. More than 80% of patients suffering from allergic asthma are positive for mite extracts and have large amounts of IgE specific for mite allergens. In Europe, the most prevalent species of house dust mites are Dermatophagoides pteronyssinus and Dermatophagoides farinae, depending on relative humidity and temperature. No less than 23 groups of allergens were identified from extracts of these species (1, 2).
Although the group 1 and 2 allergens were extensively studied, those from group 3 were poorly characterized. In D. pteronyssinus, the allergen of this group has been first identified by Stewart et al. (3) as a trypsin-like protease termed Der p 3. The binding of IgE from sera of allergic patients to Der f 3, a D. farinae protease homologous to Der p 3, appears to depend on the purity of the allergen, the tested patient populations, and the sensitivity of the technique (4). Although the frequency of IgE reactivity measured toward Der p 3 was ∼70–80%, suggesting that Der p 3 is a major allergen (5), a recent study determined a lower allergenic potency similar to mite allergen groups 8 and 10 (6). On the basis of both sequence comparisons and enzymatic studies, Der p 3 has been classified among the trypsin-like proteases from the S1A serine proteases family. Two other serine proteases with chymotryptic and collagenolytic activities and showing 36 and 76% of identity with Der p 3, were also identified in D. pteronyssinus extracts and were termed Der p 6 and Der p 9, respectively (7–9).
The trypsin-like protease Der p 3 displays 47% of identity with salmon trypsin and 45% with bovine trypsin (3, 10, 11). All residues involved in the catalytic activity (i.e. the catalytic triad His-51, Asp-96, and Ser-196), in the substrate specificity, and the six cysteines engaged in disulfide bridges formation are highly conserved (10). In addition to their allergenic properties, the enzymatic activity of group 3 allergens has been shown to enhance the inflammatory process, either by activating lung epithelial cells through cleavage of the protease-activated receptor 2, or by proteolytic processing of proteins C3 and C5 leading to formation of anaphylatoxins C3a and C5a. These peptides are known to act directly on small blood vessels, smooth muscles, mast cells, and peripheral blood leukocytes (12, 13). Analysis of Der p 3 cDNA reveals that the protease is synthesized as a prezymogen (preproDer p 3) formed by a signal peptide of 18 amino acids, an N-terminal propeptide of 11 amino acids, and a domain of 232 amino acids (10). After cleavage of the signal peptide, the zymogen (proDer p 3) matures into a protease of 232 residues (Der p 3) that cleaves peptide bonds after arginine or lysine residues.
Maturation of the zymogen is associated with cleavage and release of the inhibitory propeptide, leading to the des-inhibition of the proteolytic activity. Cleavage of the propeptide implies the recognition of a specific activation site, either by another protease (intermolecular mechanism) or by the catalytic domain of the zymogen itself (intramolecular mechanism) (14). Although the proDer p 3 activation process is still unknown, maturation of the human trypsinogen has been widely described and can occur according to different mechanisms. First of all, human trypsinogen is known to be activated by an intermolecular mechanism, in which the duodenal enterokinase, a transmembrane serine protease, can cleave the zymogen into active trypsin (15). An alternative mechanism, associated with hypercalcemia, involves an intramolecular processing and leads to pancreatitis (16, 17).
In trypsin-like proteases, the length of the propeptide varies between 4 and 24 amino acids. The motif commonly found at the C-terminal extremity of propeptide is a polyaspartyl sequence followed by a lysine residue (-DDDDKP1) at the P1 position (18) according to the Schechter and Berger nomenclature (19) for the description of protease subsites. Surprisingly, this polyaspartyl sequence is not conserved in the Der p 3 propeptide (Fig. 1). Furthermore, it contains a potential N-glycosylation site at position P3 and, most interestingly, a threonine at its C-terminal extremity instead of a residue such as lysine or arginine, reported to be associated with removal of the pro-region, suggests a unique activation mechanism (10, 20).
FIGURE 1.
PreproDer p 3 sequence. The 18-residue signal peptide is in boldface type. The propeptide is underlined, and the protease sequence is in italic. Residue numbering starts at the N-terminal residue (Asn) of the propeptide. The arrows indicate the cleavage sites of the signal peptide (1) and of the propeptide (2). The dotted arrow indicates the site where cleavage yields an inactive protein.
Natural Der p 3, like the other mite proteases Der p 6, Der p 9, and Der p 1, another mite protease, is more abundant in fecally enriched dust mite extracts than in body extracts, suggesting their implication in mite digestion (3, 21, 22). Der p 1 is a D. pteronyssinus cysteine protease, which is also synthesized as an inactive zymogen termed proDer p 1. The Der p 1 zymogen is able to recognize its own C-terminal propeptide extremity (-LNAEP1) and can autoactivate at acidic pH (23).
Unfortunately, the quantities of purified Der p 3 allergen from the fecal pellets are very low (10), probably because of autolysis or degradation by other proteases during the purification process, as indicated by the low molecular mass products of Der p 3 found in dust extracts (3). To study the implication of Der p 3 in allergy, it is therefore really important to develop strategies for producing a pure and active recombinant form of the allergen. To date, only two studies reported the expression of proDer f 3 or mature Blomia tropicalis protease, homologous to Der p 3, as fusion proteins with glutathione S-transferase in Escherichia coli. The authors suggested that the proteins did not exhibit the appropriate conformation indicating a possible implication of the propeptide in the correct folding of the protease (20, 24).
In this study, we report the production of correctly folded, N-glycosylated, and inactive recombinant Der p 3 zymogen in Pichia pastoris. With the use of recombinant Der p 1 (rDer p 1) and house dust mite (HDM) extracts, we highlighted a particular activation mechanism of the zymogen, depending on the house dust mite cysteine protease Der p 1. We have determined the impact of proDer p 3 propeptide glycosylation in the activation kinetics. Finally, we have explored the interaction of the enzyme with its prosequence in terms of inhibition of the enzymatic activity.
EXPERIMENTAL PROCEDURES
Chemicals—N-p-Tosyl-Gly-Pro-Arg-7-amido-4-methylcoumarin (MCA) and N-t-Boc-Phe-Ser-Arg-MCA were purchased from Sigma; Boc-Ile-Glu-Gly-Arg-MCA acetate, Boc-Gln-Ala-Arg-MCA acetate salts, Boc-Gln-Ala-Arg-p-nitroanilide (pNA), N-succinyl-Ala-Ala-Pro-Phe-pNA, and N-succinyl-Ala-Ala-Pro-Leu-pNA were purchased from Bachem (Buttendorf, Switzerland). The cysteine protease inhibitor, l-trans-epoxysuccinyl-leucylamido(4-guanidino) butane (E-64), the trypsin inhibitor, 4-amidinophenylmethanesulfonyl fluoride hydrochloride (p-APMSF), and general serine proteases inhibitor, soybean trypsin inhibitor (SBTI), were obtained from Sigma. Unglycosylated recombinant mature Der p 1 (rDer p 1) was obtained from P. pastoris proDer p 1, as described (23). D. pteronyssinus culture extracts were obtained from 2 g of whole mite culture resuspended in 20 ml of PBS, pH 7.5, during 12 h at 4 °C, ultracentrifuged at 45,000 rpm during 40 min at 4 °C. The supernatant containing proteins was collected and frozen at -20 °C.
Expression of Recombinant ProDer p3 in E. coli—The expression plasmid pET 15b containing proDer p 3 sequence7 transformed into E. coli Origami™ 2(DE3) cells (Novagen, Notthingam, UK). The transformants were selected on Luria-Bertani agar plates containing ampicillin (100 μg/ml) at 37 °C. For expression, transformants were grown in an LB solution containing ampicillin (100 μg/ml), tetracycline (12.5 μg/ml), and streptomycin (50 μg/ml) at 20, 28, and 37 °C until an A600 value of 0.5–0.8 was reached. The cultures were induced with 0.5 mm isopropyl β-d-thiogalactopyranoside, and samples were collected after 2, 4, and 16 h. The samples were centrifuged at 4000 × g during 20 min, and cells were resuspended in PBS, pH 7.4, and lysed by a cell disrupter (Constant Systems, Daventry, UK). The lysates were centrifuged at 12,000 × g during 5 min. The supernatants (soluble fraction) and the pellets (insoluble fraction) were analyzed by SDS-PAGE.
Construction of the ProDer p 3 (N9Q) Expression Vector—The N9Q proDer p 3 mutant was constructed by PCR from the proDer p 3 sequence using primers introducing the restriction sites for EcoRI and XbaI, respectively, 5′-ATC-GAA-TTC-AAT-CCG-ATC-CTG-CCG-GCA-TCC-CCG-CAA-GCG-ACC-ATC-GTT-GGC-GGC-GAA-AAA-GCA-CTG-3′ and 5′-CGA-TTG-GAT-TGA-ATC-TAA-ACG-TAG-CCA-GTG-ATC-TAG-AAT-A-3′. The amplified fragment was cloned into pGEM-T easy vector (Promega, Madison, WI). The presence of the N9Q mutation was verified by DNA sequencing. The proDer p 3 (NP3Q) sequence was isolated by digestion of the pGEM-T easy vector with EcoRI-XbaI and was subsequently cloned into the pPICZα vector (Invitrogen) previously restricted with the same enzymes.
Expression of Recombinant ProDer p3 in P. pastoris—Expression plasmid pPIC9K contained the proDer p 3 gene7 cloned downstream from the Saccharomyces cerevisiae α factor signal peptide. P. pastoris SMD1168 strain was transformed with expression vector pPIC9K-proDer p 3 by electroporation. Transformants carrying the HIS4 gene were grown on histidine-deficient medium (RDB). Clones with multiple integrated copies were further selected for resistance to increasing geneticin (G418, Invitrogen) concentrations (0.25–3 mg/ml). For production of proDer p 3, the most resistant clone was grown in 400 ml of Buffered Glycerol-complex Medium (BMGY) at 30 °C up to an A600 value of 2–6. The culture was then transferred into 3 liters of BMGY in a 5-liter Sartorius fermentor and grown during 24 h at 30 °C. During the next 12 h, glycerol feed rate was regulated by following the dissolved oxygen level. At an A600 value of ∼100, culture was induced with methanol to a final concentration of 0.5%. During 24 h, the methanol feed rate was regulated to maintain a dissolved oxygen level of minimum 30%. Culture was then centrifuged at 13,000 × g during 20 min, and the supernatant was stored at -20 °C.
After electroporation of P. pastoris SMD1168 strain with the pPICZα expression vector containing the proDer p 3 (N9Q) sequence, transformants were selected on Yeast extract Peptone Dextrose (YPD) medium containing Zeocin (50 μg/ml) (Invitrogen). In the pPICZα, the sequence of interest was also cloned downstream from the S. cerevisiae α factor signal peptide. The expression of the mutant was then tested in 100 ml of BMGY at 28 °C up to an A600 value of ∼1. The culture was centrifuged during 10 min at 5000 × g. The pellet was resuspended into 100 ml of Buffered Methanol-complex Medium (BMMY, 0.5% methanol) for the expression at 28 °C during 3 days. The culture was centrifuged at 10,000 × g during 10 min and the supernatant stored at -20 °C.
Purification of Recombinant WT ProDer p 3 and Der p 3—1 liter of the culture supernatant containing proDer p 3 was filtered through a 0.45-μm filter (Millipore, Billerica, MA) and dialyzed overnight at 4 °C against a 20 mm ethanolamine buffer, pH 9.5 (buffer A). The solution was then stirred with 200 ml of Qstreamline exchanger (GE Healthcare) equilibrated with buffer A at 4 °C. The exchanger was washed with buffer A and then packed into a 250-ml column (GE Healthcare). Bound proteins were eluted stepwise with buffer A added with 1 m NaCl. After SDS-PAGE analysis, fractions containing proDer p 3 were pooled and dialyzed at 4 °C against a 20 mm ethanolamine buffer, pH 9 (buffer B). Solution was then loaded onto a Q-HP-Sepharose column (60 ml) (2.6 × 10 cm; GE Healthcare) equilibrated with buffer B. The flow-through containing proDer p 3 was dialyzed at 4 °C against 20 mm sodium citrate, pH 6.5 (buffer C), before purification on an S-HP-Sepharose column (25 ml) (1.6 × 10 cm; GE Healthcare) equilibrated with buffer C. The proDer p 3 in the flow-through was concentrated by ultrafiltration (cutoff, 10 kDa) and stored at -20 °C. The concentration of proDer p 3 was estimated by the BCA assay (Pierce).
After activation of proDer p 3 by the rDer p 1 protease, mature rDer p 3 was isolated by a fourth purification step with a 1-ml MonoQ column (0.5 × 5 cm; GE Healthcare) equilibrated with 20 mm Tris-HCl, pH 8.5, buffer (buffer D). Elution was performed with a linear gradient of buffer D added with 1 m NaCl over 10 column volumes. Fractions containing Der p 3 activity were pooled and dialyzed against 20 mm sodium acetate, pH 4, before storage at -20 °C.
Determination of Secondary Structure Contents by Attenuated Total Reflection (ATR) Fourier Transform Infrared—Purified protein samples (natural Der p 3, P. pastoris proDer p 3 and E. coli proDer p 3 solubilized in 50 mm Tris-HCl containing 500 mm NaCl and 6 m urea) (1–3 μg) were micro-dialyzed against 1 mm Hepes, pH 7.2, and spread on a micro-ATR crystal for measurement in attenuated total reflection mode. Spectra (64 acquisitions averaged, 2 cm-1 resolution) were recorded on a Brucker IFS-55 Fourier transform infrared spectrophotometer equipped with a liquid nitrogen-cooled mercury cadmium telluride detector. Acquired spectra were corrected for water vapor and amino acid side-chain contribution in the amide I and II bands and smoothed with a 4-cm-1 width gaussian band. For secondary structure comparisons, deuterium-saturated nitrogen was continuously flushed in the chamber containing the ATR crystal, and spectra were recorded after reaching a plateau phase in the amide II exchange kinetics (60 min).
Western Blot Analysis—Mutant N9Q and purified proDer p 3, deglycosylated proDer p 3, rDer p 3, and rDer p 1 were denatured at 100 °C in the presence of denaturing buffer and separated by SDS-PAGE (15%). The proteins were electroblotted onto a polyvinylidene difluoride membrane. Immunoblot analyses using polyclonal anti-Der p 3 and anti-Der p 1 antibodies at dilutions of 1:2000 and 1:2500, respectively, were carried out, and mouse antibodies were detected with 5-bromo-4-chloro-3-indolyl phosphate and nitro blue tetrazolium by using rabbit alkaline phosphatase-conjugated anti-mouse antibodies.
N-terminal Sequencing—The proDer p 3 zymogen, mature rDer p 3, and bands resulting from activation of proDer p 3 by the house dust mite extracts, rDer p 1 and rDer p 3, were sequenced on an Applied Biosystems 476A protein sequencer (Applied Biosystems), based on Edman degradation. Samples were analyzed by SDS-PAGE followed by electrophoretic transfer onto a polyvinylidene difluoride membrane (Millipore).
Autoactivation of Recombinant Glycosylated and Unglycosylated ProDer p 3 by pH and Ca2+—ProDer p 3 (65 μm final concentration in 300 μl final volume) was incubated at 37 °C for 16 h in 50 mm polybuffer (mixture of 50 mm Tris, citrate, CAPS, and potassium chloride) adjusted to desired pH ranging from 2 to 12 with 1 m NaOH or 1 m HCl. Assays were analyzed by SDS-PAGE, and rDer p 3 enzymatic activity was measured as described below.
To analyze the putative autoactivation of proDer p 3 in the presence of Ca2+ ions, the zymogen (65 μm final concentration) was incubated at 37 °C in 50 mm polybuffer, pH 8, in the presence of increasing CaCl2 concentrations (0–20 mm). After periods of time ranging from 15 min to 12 h, aliquots were withdrawn, and the rDer p 3 activity was measured. The different samples were analyzed by SDS-PAGE.
HDM Extracts Activity Measurements—The different enzymatic activities of the extracts (125 μg/ml) were measured by following the hydrolysis of Boc-Gln-Ala-Arg-pNA, N-succinyl-Ala-Ala-Pro-Phe-pNA, and N-succinyl-Ala-Ala-Pro-Leu-pNA 1 mm in 20 mm Tris-HCl, pH 7.8, or in PBS, pH 7.4, containing 1 mm DTT and 1 mm EDTA during 1 h at 25°C in a Power-WaveX spectrophotometer (λ = 410 nm) (Bio-Tek Instruments, Inc., Winooski, VT). The hydrolysis of Boc-Gln-Ala-Arg-pNA by rDer p 3 (20 nm) and rDer p 1 (100 nm) in 20 mm Tris-HCl, pH 7.8, and in PBS, pH 7.4, containing 1 mm DTT and 1 mm EDTA, respectively, were used as controls.
Activation of Recombinant Glycosylated ProDer p 3 by HDM Extracts, rDer p 1, and rDer p 3—ProDer p 3 (150 μm) was incubated at 37 °C for several periods of time with rDer p 1 (1 μm) in 20 mm sodium citrate buffer, pH 6.5, containing 1 mm DTT and 1 mm EDTA, with rDer p 3 (0.05–0.6 μm) in 20 mm Tris-HCl, pH 7.8, or with HDM extracts (2 mg/ml) in PBS, pH 7.4, containing 1 mm DTT and 1 mm EDTA. Samples were analyzed by SDS-PAGE and N-terminal sequencing. For activation of proDer p 3 by rDer p 1 and rDer p 3, the rDer p 3 activity was measured in samples diluted 5000-fold as described below. To obtain mature rDer p 3, after the incubation of the glycosylated zymogen (150 μm) with rDer p 1 during 90 min at 37 °C, the reaction was stopped with 1 mm E-64, and rDer p 3 was purified as described above.
Der p 3 Enzymatic Activity Measurements and Determination of Kinetic Parameters—Hydrolysis of 10 μm substrate (N-p-tosyl-Gly-Pro-Arg-MCA, N-t-Boc-Phe-Ser-Arg-MCA, Boc-Ile-Glu-Gly-Arg-MCA, Boc-Gln-Ala-Arg-MCA, Asn-Ala-Thr-ACC, or Asn-Ala-Arg-ACC, see below) by rDer p 3 (325 pm) in 50 mm polybuffer, pH 8.5, at 37 °C was followed during 200 s in a fluorimeter LS 50 B (PerkinElmer Life Sciences) with excitation and emission wavelengths of 380 and 460 nm, respectively. Kinetics of hydrolysis were reported as the time course of MCA (μm) released, using an MCA (Sigma) standard curve with concentrations ranging from 0 to 1.8 μm. For determination of the rDer p 3 kinetic parameters, the rate of hydrolysis of increasing substrate concentrations (0 to 300 μm) was measured, and the data analyzed according to the Henri-Michaelis-Menten equation.
Glycosylated and Unglycosylated ProDer p 3 Activation Kinetics—The enzymatic test was adapted from the experimental procedure described for processing of procathepsins L, B, and S (25, 26). Glycosylated and unglycosylated proDer p 3 (130 nm) were activated at 37 °C in the presence of increasing concentrations of rDer p 1 (0–17 nm) in 50 mm phosphate buffer, pH 7.4, containing 150 mm NaCl, 1 mm DTT, and 1 mm EDTA. rDer p 3 enzymatic activity was followed continuously, by measuring hydrolysis of Boc-Ile-Glu-Gly-Arg-MCA 10 μm during 30 min. Data obtained from activation of glycosylated and unglycosylated proDer p 3 were then fitted to Equation 1 (27–31) using the Grafit software version 5.0.10 (Erithacus Software Ltd.), and the first pseudo-order rate constants (kobs) were calculated for each rDer p 1 concentration.
 |
(Eq. 1) |
where P, v0, and vss correspond, respectively, to the amount of MCA produced (μm), the initial rate for product release (μm/s), and the steady-state rate for product release (μm/s).
Solid Phase Synthesis of Asn-Ala-Thr-ACC, Asn-Ala-Arg-ACC, NPILPASPNAT (WT), and NPILPASPNAR (TP1R) Peptides—The peptides were prepared by the solid-phase peptide synthesis strategy on a PS3 automated peptide synthesizer (Protein Technologies, Inc., Tucson, AZ) using N-α-fluorenylmethoxycarbonyl (Fmoc)-based chemistry on 4-methylbenzhydryl resin or Fmoc-l-Arg(Pbf)/Fmoc-l-Thr(tBu)-ACC rink amide resin (supplemental data, Fmoc-l-Arg(Pbf)/Fmoc-l-Thr(tBu)-ACC resin synthesis). The 4-methylbenzhydryl resins (Fmoc-l-Arg(Pbf)-4-methylbenzhydryl resin and Fmoc-l-Thr(tBu)-4-methylbenzhydryl resin) were purchased from CBL, Baltimore, MD. N-α-Fmoc amino acids (0.4 mmol) were purchased from Iris Biotech GmbH, Marktredwitz, Germany. The side chains of Ser and Thr were protected with the t-butyl derivative (tBu), and Arg was protected with the 2Pbf group, and Asn was protected with the trityl group. 2-(1H-benzotriazol-1-yl)-1,1,3,3-Tetramethyluronium hexafluorophosphate (Iris Biotech GmbH) (0.4 mmol) and N-methylmorpholine 0.4 m in N,N-dimethylformamide (DMF) (3 ml, 0.4 mmol) were used as coupling and activating reagents, respectively. Fmoc deprotection at each step was carried out using 20% piperidine/DMF (12 ml). The peptides attached to the resin beads were washed subsequently with DMF, EtOH, and dichloromethane. The side-chain protecting groups were removed, and the peptides were cleaved from the resin using trifluoroacetic acid/anisole/water (6 ml; 10:1:1) during 3 h at room temperature. The uncharged resin was separated from the solution by filtration. The crude peptides were precipitated with Et2O and were then purified by reverse-phase HPLC using a semi-preparative C18 column (10 × 250 mm, 10 μm XTerra Prep RP18, Waters, Milford, MA) to obtain ∼50 mg of each peptide. The solvents consisted of an aqueous 0.1% (v/v) trifluoroacetic acid solution and acetonitrile. The elution was carried out in 20 min at a flow rate of 4 ml/min by using a linear gradient from 0 to 40% acetonitrile. Each peptide was characterized after purification by mass spectrometry (TSQ 7000 Thermoquest Finnigan, positive ionization, 4.5 kV) and the purity was assessed by analytical HPLC. The HPLC chain consisted of a pump (Waters 600) and a UV detector (PDA Waters 996), the absorbance at wavelengths between 198 and 400 nm was constantly recorded. The analytical HPLCs were performed on an XTerra RP18 column (150 × 4.6 mm, 3.5 μm, Waters).
Recombinant Der p 3 Inhibition by Its Propeptide—rDer p 3 activity (600 pm) was measured at 37 °C in 50 mm polybuffer, pH 8.5, with increasing concentrations (10–75 μm) of the IEGR-MCA substrate after the addition of its wild-type (NPIL-PASPNAT) or modified (NPILPASPNAR, TP1R) propeptides (0, 1.25, 2.5, and 5 mm) synthesized as described above. The model of inhibition by TP1R peptide was determined with the help of the Hanes plot, and the inhibition constants, KI and K′I were obtained using Equation 1, Bio-Tek Instruments, Inc.,
 |
(Eq. 2) |
where vi, V, (S), (I), Km, K′I, and KI represent, respectively, the initial rate for product release (μm/s), the maximal rate for product release (μm/s), the concentration of substrate (μm), the concentration of inhibitor (mm), the Km of the enzyme for its substrate (μm), the dissociation constant for the ESI complex (mm), and the dissociation constant for the EI complex (mm).
Modeling—The model of the Der p 3 protease was established by using the program CPHmodels 2.0 Server (32) and was based on the available crystallographic structures of trypsin.
RESULTS
Expression of Recombinant ProDer p 3—Expression of Der p 3 zymogen in E. coli yielded inclusion bodies at all tested temperatures (20, 28, and 37 °C), whereas a soluble form could be obtained in P. pastoris culture supernatants. To determine whether solubilized E. coli-expressed proDer p 3 was correctly folded, the recombinant allergen was analyzed by infrared spectroscopy, and its secondary structure content was compared with that of natural Der p 3 in the amide I and II regions that are highly sensitive to the secondary structure content of proteins. The secondary structure of proDer p 3 was shown to be different from that of natural Der p 3 in the amide I and II regions (supplemental Fig. S1). In particular, the recombinant protein showed an increased content of antiparallel β-pleated sheet. This phenomenon, commonly observed for aggregated and thermally denatured proteins, confirmed that recombinant proDer p3 did not adopt the Der p 3 appropriate folding, most probably because of imperfect denaturation/refolding steps.
In contrast to the zymogen produced in bacteria, the spectra of natural Der p 3 and P. pastoris expressed proDer p 3 were highly similar in shape and position of their maximum absorbance (1642 cm-1), suggesting that secreted proDer p 3 adopted a fold similar to that of the mature Der p 3 (supplemental Fig. S1).
This secreted zymogen could be purified to homogeneity, yielding ∼120 mg of proDer p 3 per liter of culture supernatant. N-terminal sequencing (YVNPILP5-) revealed that the expected sequence was preceded by two additional residues (Tyr and Val), resulting from the SnaBI cloning site in the pPIC-9K plasmid. SDS-PAGE (Fig. 2A, lane 1) indicated that proDer p 3 migrates as a broad band of ∼40 kDa. It contrasts with the molecular mass of 26,327 Da calculated on the basis of the zymogen sequence and suggests the occurrence of large post-translational modifications on the recombinant allergen, likely N-glycosylations because of the presence of one putative N-glycosylation site in the prosequence (-N9AT-). Staining of the electrophoresis gel with the Gel Code® glycoprotein staining kit (Fig. 2B) showed that the 40-kDa band corresponded to the proDer p 3 carried glycosylations. Moreover, incubation of the zymogen for 6 h with N-glycosylase F yielded a sharp band at 29 kDa (Fig. 2A, lane 2). Furthermore, analysis by mass spectrometry indicated that deglycosylated proDer p 3 appears as a single peak with the expected molecular mass of 26,320 Da.
FIGURE 2.
Deglycosylation and staining of glycosylated proDer p 3. A, glycosylated proDer p 3 (200 μm) before (lane 1) and after (lane 2) a 6-h incubation with N-glycosylase F (1 unit per 5 nmol of proDer p 3), in 20 mm citrate buffer, pH 6.5, at 37 °C. B, staining of glycosylated and deglycosylated proDer p 3 (200 μm) with the Gel Code® glycoprotein staining kit (Pierce) according to the manufacturer. C, immunodetection after Western blotting of proDer p 3 (1 μg, lane 1), deglycosylated proDer p 3 (1 μg, lane 2), and mutant proDer p 3 N9Q (lane 3, 25 μl culture supernatant) using anti-proDer p 3 polyclonal antibody. Stained (St), unstained, or prestained protein molecular weight marker (Fermentas GmbH, St. Leon-Rot, Germany).
To confirm that the sugar chain is carried by the propeptide -N9AT-, the asparagine residue at position P3 was replaced by a glutamine by site-directed mutagenesis. The N9Q mutant was also expressed in P. pastoris as a secreted soluble form. As shown by Western blotting (Fig. 2C), the N9Q mutant migrated as a single band of ∼29 kDa, undistinguishable from that corresponding to the N-glycosylase F-treated zymogen.
Absence of Autoactivation of Recombinant ProDer p 3—In analogy with the autocatalytic mechanism of trypsinogen maturation (16, 17), both glycosylated and deglycosylated proDer p 3 were incubated at different pH values (ranging from 2 to 12) and in the presence of increasing Ca2+ concentrations (0–20 mm), but no effect was observed. Indeed, no decrease of the molecular mass and no rDer p 3 activity could be detected suggesting that maturation of proDer p 3 did not follow an autocatalytic process. This observation suggests an intermolecular activation mechanism of proDer p 3 in which the activity of a mature protease could be involved.
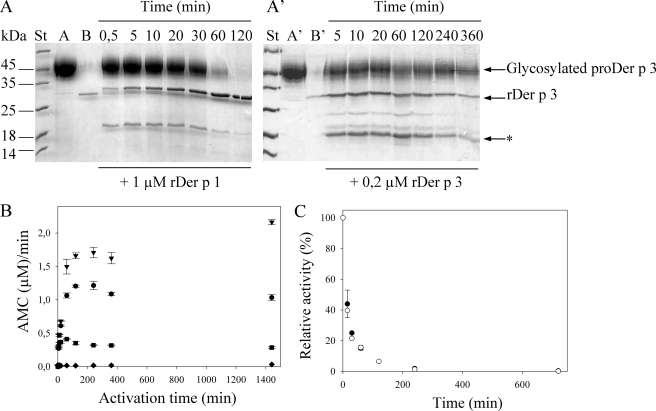
Intermolecular Activation Mechanism of Recombinant ProDer p 3—Upon incubation at pH 6.5 in the presence of the mite cysteine protease rDer p 1 (25 kDa), both glycosylated and deglycosylated proDer p 3 were activated, yielding a single band of ∼29 kDa on SDS-PAGE, with the expected N-terminal sequence of Der p 3 protease (I12VGGEKALAG). With glycosylated zymogen, incubation in the presence of rDer p 1 led to the expected molecular mass decrease of ∼11 kDa, corresponding to the glycosylated propeptide removal (Fig. 3A). Aliquots taken at regular time intervals and analyzed by SDS-PAGE (Fig. 3A) indicate that the propeptide is completely removed within 2 h. This proteolytic cleavage of the propeptide is accompanied by an increase in the catalytic activity of the protease (Fig. 3B), as monitored by hydrolysis of the fluorescent tetrapeptic substrate, IEGR-MCA, specific of trypsin-like proteases (not cleaved by rDer p 1 in these conditions) and described previously as a specific substrate for the natural Der p 3 (Fig. 3B). Propeptide removal, as followed by SDS-PAGE (Fig. 3A), and zymogen activation, as measured by substrate hydrolysis (Fig. 3B), seem to occur over the same time scale, suggesting that intermolecular activation of proDer p 3 occurs in one single step, corresponding to the loss of the entire propeptide.
FIGURE 3.
Inter-molecular activation of glycosylated proDer p 3 by rDer p 1 (A) or rDer p 3(A′) and stability of rDer p 3 are shown. A and A′, SDS-PAGE (15%) analysis; 150 μm of glycosylated proDer p 3 (lanes A and A′) was incubated at 37 °C, in 20 mm sodium citrate buffer, pH 6.5, with 1 μm rDer p 1 (lane B) or in 20 mm Tris-HCl, pH 7.8, with 0.2 μm rDer p 3 (lane B′). Stained (St) and unstained protein molecular weight marker were from Fermentas; *, rDer p 3 degradation band (115AVGLP). B, for each activation time (0–1440 min) of proDer p 3 by rDer p 1 (•), rDer p 3 (♦), rDer p 1 with addition of 100 μm E-64 after 10 min (▪), and rDer p 1 with addition of 10 mm benzamidine (▾), aliquots were diluted 5000-fold, and the rDer p 3 activity was measured using IEGR-MCA as substrate (10 μm). C, after an incubation of rDer p 3 (2.3 μm) in a 50 mm polybuffer, pH 8.5, at 37 °C during increasing times (0–12 h) in presence of rDer p 1 (0.6 μm) or not, respectively, in black and white, the relative activity (%) of rDer p 3 was measured in 1000-fold diluted aliquots using the IEGR-MCA substrate (10 μm). Standard deviations were <10%.
Utilization of rDer p 1 previously incubated with 100 μm E-64, an inhibitor of cysteine proteases, did not result in activation of proDer p 3 (supplemental Fig. S2A). Indeed, even after 6 h of contact, neither propeptide nor Der p 3 activity could be detected (data not shown). This demonstrates the essential role of the rDer p 1 cysteine protease activity in the maturation process of proDer p 3.
In the course of the proDer p 3 activation process, a minor band of ∼19 kDa was also detected (Fig. 3A). It is characterized by the 115AVGLP N-terminal sequence and corresponds to the hydrolysis of -NAK114↓115VGLP- in the mature recombinant Der p 3 (rDer p 3). Interestingly, a corresponding degradation site was previously reported for human trypsin (-NAR117↓118VSTIL-) (33). To determine the sensitivity of rDer p 3 to hydrolysis, pure rDer p 3 was incubated in 50 mm polybuffer, pH 8.5, at 37 °C in the presence or absence of rDer p 1, and its enzymatic activity was monitored after different times ranging from 0 to 240 h. Fig. 3C shows that total inactivation of rDer p 3 occurs within 240 min in these conditions. rDer p 1 appears to have a minor role in this phenomenon, as seen by the inactivation of rDer p 3 in the absence of the cysteine protease. The predominant role of Der p 3 in its own degradation was first confirmed by the addition of 100 μm E-64 10 min after starting proDer p 3 activation in the presence of rDer p 1. SDS-PAGE analysis (supplemental Fig. S2B) shows that upon addition of E-64, rDer p 1 is totally inhibited (i.e. the maturation process of proDer p 3 is stopped), whereas both proDer p 3 and Der p 3 bands diminish continuously in intensity, leading to low molecular mass degradation fragments. Furthermore, the increase in rDer p 3 activity measured for the first 10 min of the experiment was stopped, and a progressive decrease followed, because of Der p 3 autolysis (Fig. 3B). Second, after the activation of proDer p 3 by rDer p 1 in the presence of 10 mm benzamidine, a reversible inhibitor of trypsin-like proteases, a larger Der p 3 activity was recorded after 5000-fold dilution of the samples (Fig. 3B), and proteolysis of the active Der p 3 was decreased (supplemental Fig. S2C).
The possibility of inter-molecular activation of proDer p 3 (150 μm) by rDer p 3 (after its purification, see below) was also tested. Upon SDS-PAGE, rDer p 3 appeared to process proDer p 3 into two major bands of 29 and 19 kDa but more slowly than rDer p 1 (Fig. 3A′). Although the 19-kDa degradation band corresponds to the previously identified fragment of Der p 3 (115AVGLP-), the 29-kDa band has the 18ALAG-N-terminal sequence that follows the -GEK17-residues in the Der p 3 sequence. As expected, rDer p 3 was not able to recognize the Thr at the C-terminal extremity of the proDer p 3 propeptide but cleaved six residues downstream of lysine 17. No Der p 3 activity could be measured, however, suggesting that the truncated form of Der p 3 (18ALAG-) obtained directly from the maturation of the zymogen under these conditions is not active (Fig. 3B).
ProDer p 3 Activation by HDM Extracts—Several proteases with different specificities were identified in the HDM extracts, including the cysteine protease Der p 1 of the papain-like family, the trypsin-like Der p 3, the chymotrypsin-like Der p 6, and the elastase/collagenase-like Der p 9 (3, 7–9, 11, 22). Both cysteine and serine protease activities were detected in the HDM extracts (Table 1) with specific substrates showing a predominant chymotrypsin-like activity in the extracts. The QAR-pNA substrate was used to measure cysteine (Der p 1 (34)) and trypsin-like (Der p 3 (35)) activities, AAPF-pNA, the chymotrypsinlike activity of Der p 6 (but it is also cleaved by Der p 9 (9, 11)), and AAPL-pNA, the elastase activity of Der p 9 (11). The activities of rDer p 1 and rDer p 3 were used as controls. Table 1 shows that 100 μm E-64 inhibits specifically rDer p 1, leading to a decrease of the QAR-pNA hydrolysis in HDM extracts. Indeed, in these conditions, only the contribution of Der p 3 (trypsin-like protease) is detected with this substrate. The specific inactivator of trypsin-like proteases, APMSF (200 μm), inhibits only the Der p 3 activity, whereas the general inhibitor of serine proteases SBTI (10 μm) does not affect Der p 1. Only chymotrypsin-like and elastase activities could be measured in the presence of a combination of E-64 and APMSF.
TABLE 1.
Enzymatic activities of rDer p 1, rDer p 3, and HDM extracts in presence of PBS, pH 7.4, containing 1 mm EDTA and 1 mm DTT or in 20 mm Tris-HCl, pH 7.8 The activities can be mainly attributed to the various proteins as follows: trypsin-like, Der p 3; cysteine protease, Der p 1; chymotrypsin-like, Der p 6; and elastase-like, Der p 9. Also see the text.
|
Inhibitors
|
rDer p1, substrate QAR-pNA
|
rDer p3, substrate QAR-pNA
|
House dust mites extracts activities (μmpNA/min)
|
||
|---|---|---|---|---|---|
| Trypsin/Cys protease, substrate QAR-pNA | Chymotrypsin, substrate AAPF-pNA | Elastase, substrate AAPL-pNA | |||
| None | 4.5 ± 0.4 | 3.4 ± 0.1 | 3.27 ± 0.02 | 6.2 ± 0.4 | 1.1 ± 0.1 |
| E-64 | 0.06 ± 0.06 | 3.4 ± 0.1 | 1.7 ± 0.1 | 5.8 ± 0.3 | 1.04 ± 0.04 |
| APMSF | 4.4 ± 0.7 | 0.28 ± 0.01 | 1.64 ± 0.04 | 5.3 ± 0.2 | 1.01 ± 0.01 |
| SBTI | 4.9 ± 0.1 | 0.00 ± 0.00 | 1.44 ± 0.01 | 0.01 ± 0.00 | 0.00 ± 0.00 |
| APMSF + SBTI | 5.1 ± 0.4 | 0.00 ± 0.00 | 1.6 ± 0.3 | 0.014 ± 0.002 | 0.004 ± 0.006 |
| APMSF + E-64 | 0.01 ± 0.00 | 0.22 ± 0.07 | 0.07 ± 0.00 | 5.3 ± 0.3 | 0.98 ± 0.00 |
| APMSF + SBTI + E-64 | 0.01 ± 0.00 | 0.003 ± 0.004 | 0.02 ± 0.01 | 0.02 ± 0.00 | 0.1 ± 0.1 |
The contribution of the enzymatic activities of the HDM extracts of the proDer p 3 maturation was explored. To determine specifically the role of the cysteine protease, trypsin-like and chymotrypsin-like/elastase activities, glycosylated proDer p 3 was incubated (5–60 min) with HDM extracts previously inhibited with E-64, APMSF/SBTI, or APMSF/E-64. The incubation of proDer p 3 with uninhibited HDM extracts leads to its complete processing after 15 min (Fig. 4A) into a band of 29 kDa with the Der p 3 N-terminal sequence (12IVGGE-) and in several fragments. In the presence of E-64, the proDer p 3 cleavage was very limited and led to the formation of low proportions of truncated and inactive Der p 3 (18ALAG-) (Fig. 4A). These observations confirm the essential role of the cysteine protease activity in the activation of proDer p 3 into a mature and active protein. The addition of APMSF and SBTI to the HDM extracts did not impair the maturation of proDer p 3 into Der p 3 (12IVGGE-) and further proteolysis was very limited (Fig. 4B) as shown for the maturation of proDer p 3 by rDer p 1 in the presence of benzamidine (see above). These observations indicate that the serine proteases are more involved in Der p 3 inactivation than in its formation. Moreover, upon incubation of proDer p 3 with HDM extracts previously treated with APMSF and E-64, only degradation bands were formed thus excluding the involvement of chymotrypsin-like or elastase activities in the formation of active Der p 3 (Fig. 4B). The N-terminal sequences of Der p 3 or degradation bands obtained in the different proDer p 3 activations experiments are summarized in the Table 2.
FIGURE 4.
Inter-molecular activation of glycosylated proDer p 3 by HDM extracts. A and B, SDS-PAGE (15%) analysis. 150 μm glycosylated proDer p 3 (lanes A and A′) was incubated at 37 °C, in 20 mm PBS, pH 7.4, with 2 mg/ml of HDM extracts (lanes B and B′) in the presence or absence of several inhibitors (100 μm E-64, 200 μm APMSF, 10 μm SBTI). Stained (St) and unstained protein molecular weight marker were from Fermentas.
TABLE 2.
N-terminal sequence analysis of the activation tests of pro-Der p 3 incubated with rDer p 1, rDer p 3, or HDM extracts
| Proteases | N-terminal sequences of Der p 3 | Trypsin-like protease activity of Der p 3 |
|---|---|---|
| rDer p 1 | 12IVGGEKAL | Yes |
| 115AVGLP | No | |
| rDer p 3 | 18ALAGECP | No |
| 115AVGLP | No | |
| Other degradation bands | No | |
| HDM extracts | 12IVGGEKAL | Yes |
| Degradation bands | No | |
| HDM extracts + E-64 | 18ALAGECP | No |
| Degradation bands | No | |
| HDM extracts + APMSF + SBTI | 12IVGGEKAL | Yes |
| Degradation bands | No | |
| HDM extracts + APMSF + E-64 | Degradation bands | No |
Effect of Glycosylation on ProDer p 3 Maturation Rate—Increase of the catalytic activity of rDer p 3 during its intermolecular activation by rDer p 1 could be monitored in real time (see “Experimental Procedures”) (Fig. 5A). Under the experimental conditions described in Fig. 5, proDer p 3 displayed a very low activity, which remained constant over the time scale of the experiment (≤30 min). A similar phenomenon was reported with trypsinogen (36). In contrast, the rate of product formation (MCA) increased during the maturation of proDer p 3, and the rate of the activation process increased with the concentration of rDer p 1 (Fig. 5, A and B). For each proDer p 3 maturation curve, the constant velocities of rDer p 3 reached were different indicating that the real steady state, corresponding to complete activation of the zymogen, was not reached. This phenomenon could be correlated with the degradation of rDer p 3 after its activation until a steady state between the two processes was reached. For these reasons, the results were only interpreted in a qualitative manner. The kinetics were fitted to Equation 1 to give pseudo first-order rate constants (kobs) values. These values were then plotted as a function of the rDer p 1 concentration (Fig. 5C). The observation that kobs values increase linearly with the rDer p 1 concentration suggests an intermolecular activation process. Moreover, in the absence of rDer p 1, extrapolation of the kobs values confirm that no intramolecular process occurs (i.e. kobs = 0). Data in Fig. 5C indicate that the nonglycosylated proDer p 3 is activated faster than its glycosylated counterpart. This can be explained by the situation of the N-glycosylation site in the propeptide of proDer p 3 zymogen, only three residues upstream the activation cleavage site, which could thus well represent a steric hindrance to rDer p 1.
FIGURE 5.
Continuous assay for the glycosylated and deglycosylated proDer p 3 maturation by rDer p 1. Substrate hydrolysis (10 μm IEGR-MCA) versus time curves related to the activation of glycosylated (A) and deglycosylated (B) proDer p 3 (130 nm) by increasing concentrations of rDer p 1 (0, 3, 4, 6.4, 8.5 and 17 nm) in 50 mm phosphate buffer, pH 7.4, containing 150 mm NaCl, 1 mm DTT, and 1 mm EDTA at 37 °C is shown. Curves fitted to Equation 1 are shown in gray. C, variation of the first pseudo order rate constants (kobs) measured in A (open circles, standard deviations were between 0 and 20%) or B (black circles, standard deviations <2%) with rDer p 1 concentration (nm).
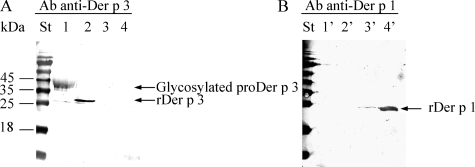
Recombinant Der p 3, a Trypsin-like Protease—Following activation in the presence of rDer p 1, recombinant Der p 3 was purified to homogeneity, using a high resolution Mono Q column. Western blot analysis (Fig. 6) using anti-Der p 1 and anti-Der p 3 antibodies confirmed that the rDer p 1 protease could be completely removed from the preparation (lanes 2 and 2′). Furthermore, N-terminal sequencing resulted in the expected Der p 3 sequence (12IVGGEKALAG-), with no significant amount of contaminant.
FIGURE 6.
Analysis of rDer p 3 purity. Immunodetection of purified proDer p 3 as control (1 μg, lanes 1 and 1′), rDer p 3 after its purification (1 μg, lanes 2 and 2′), and purified rDer p 1 as control (0.5 and 3 μg for lanes 3, 3′, and 4, 4′, respectively, molecular mass; 25 kDa) by Western blotting is shown using anti-Der p 3 (A) or anti-rDer p 1 (B) polyclonal antibodies. Stained (St) and prestained protein molecular weight marker was from Fermentas.
The pH activity profile of rDer p 3 was determined between pH 2 and 12, in a 50 mm polybuffer, using IEGR-MCA (10 μm) as substrate. Fig. 7 shows that Der p 3 retains activity between pH 5 and 11, with a maximum at pH ≈8.5; a similar behavior has been reported for trypsin and natural Der p 3 (11, 35). In addition, the percentage of activity recovered after readjusting the pH value to 8 (using 100 mm polybuffer, pH 8) was measured for all samples incubated in the pH 2–12 range (Fig. 7). Complete return of activity could be achieved for rDer p 3 samples incubated for 1 h at pH values ranging from 4 to 11.
FIGURE 7.
pH activity profile of rDer p 3. Enzymatic activity was measured by using 10 μm IEGR-MCA as substrate. The relative activity (%) of rDer p 3 after a 1-h incubation in 50 mm polybuffer, pH ranging from 2 to 12, at 37 °C, is shown in black. The activity recovered after adjusting the pH back to 8 for 1 h at 37 °C is represented in white.
The catalytic parameters of rDer p 3 were measured with four different synthetic substrates (Table 3). In agreement with the behavior reported for trypsin (35), rDer p 3 shows little specificity for residues in P2 and P3 positions. Interestingly, its catalytic efficiency is ∼50-fold higher than that of the rDer p 1 protease measured with the substrate QAR-MCA. We synthesized the NAT-ACC substrate corresponding to the C-terminal extremity of the proDer p 3 propeptide and NAR-ACC as control to highlight the potential recognition of its own propeptide by Der p 3. Although the catalytic efficiencies of rDer p 3, measured with the QAR-MCA and NAR-ACC are similar, the NAT-ACC substrate was not cleaved by the protease. This observation is in agreement with a specific trypsin-like activity (i.e. specific cleavage after Arg or Lys (35)) and corroborates the absence of the release of the 12IVGG-Der p 3 form after cleavage of proDer p 3 by rDer p 3.
TABLE 3.
Kinetic parameters for rDer p 3 in the presence of 50 mm polybuffer, pH 8.5
| Substrate | Km | kcat | kcat/Km |
|---|---|---|---|
| μm | s–1 | 105m–1 s–1 | |
| Boc-IEGR-AMC | 27 ± 1 | 17.3 ± 0.2 | 6.4 ± 0.3 |
| Boc-GPR-AMC | 14 ± 1 | 12.4 ± 0.2 | 8.9 ± 0.7 |
| Boc-FSR-AMC | 11 ± 1 | 9.9 ± 0.3 | 9 ± 0.9 |
| Boc-QAR-AMC | 33 ± 3 | 8.6 ± 0.2 | 2.6 ± 0.2 |
| NAR-ACC | 190 ± 30 | 70 ± 3 | 3.7 ± 0.6 |
| NAT-ACC | NCa | NC | NC |
NC means not cleaved
Specific Inhibition of rDer p 3 by the Wild-type (WT) and Modified (TP1R) Propeptides—Initial rate measurements in the presence of increasing concentrations (up to 5 mm) of the wild-type propeptide indicated only very poor inhibition of the enzyme activity (inhibition constant (KI) >> 5 mm). The single substitution of Thr by Arg at the C-terminal extremity of the propeptide (TP1R mutant) was sufficient, however, to dramatically enhance the inhibitory properties of the peptide. The Hanes plot suggests that the TP1R behaves as a noncompetitive inhibitor (Fig. 8), with dissociation constants KI (for the rDer p 3-TP1R complex, EI) and K′I (for the rDer p 3-IEGR-MCA-TP1R complex, ESI) of 1.2 ± 0.2 and 1.4 ± 0.2 mm, respectively. The difference between these values is probably nonsignificant, and this indicates that the ternary rDer p 3-substrate-TP1R complex (ESI) formed should be as stable as rDer p 3-TP1R (EI). The linearity of the Dixon plots (1/v versus I) showed that this ternary complex was not catalytically competent (data not shown), but the binding mode of the propeptide analogue to the active enzyme remains undetermined.
FIGURE 8.
Hanes plot of the inhibition of rDer p 3 (0. 6 nm) by the TP1R peptide. Substrate concentration (IEGR-MCA)/initial hydrolysis rate versus substrate concentrations (10, 30, 50, 75 μm) in the presence of increasing concentrations of TP1R peptide are shown as follows: 0 (•), 1.25 (○), 2.5 (▾), and 5 mm (▿) in 50 m m polybuffer, pH 8.5, at 37 °C.
DISCUSSION
To date, the trypsin-like allergen Der p 3 was poorly characterized, most probably because the Der p 3 content of whole mite cultures is very low (50 μg/g exhausted mite cultures). In addition, the purified protease isolated from house dust mite extract is highly unstable because of its degradation (3, 10). Consequently, production of recombinant Der p 3 is highly desirable.
This study reports the successful production of recombinant proDer p 3 in P. pastoris. By site-directed mutagenesis and glycosidase F treatment, we clearly demonstrated that proDer p 3, produced in yeast, is glycosylated at the level of the potential N-glycosylation site present in the propeptide.
Infrared spectroscopy clearly detected important differences at the level of the overall secondary structures between renatured E. coli proDer p 3 and natural Der p 3, as judged by the increase of the β-sheet content in proDer p 3. It must be pointed out that transformation of α-helical structure into intermolecular antiparallel β-sheet is usually observed in aggregated proteins (37, 38).
We provided evidence that the Der p 3 precursor form produced in P. pastoris adopted the global fold of trypsin-like proteases. As expected, the zymogen was inactive indicating that its propeptide could act as an inhibitor of the protease domain. Indeed, many proteases are synthesized as inactive zymogens. This allows a spatially and temporally controlled activation of the proteases and avoids uncontrolled digestions (14). During the course of evolution of its sequence, the trypsin ArgP1 or LysP1 residues have been selected in the propeptide probably to permit an autoactivation mechanism of trypsinogen. In vertebrates, this intramolecular activation phenomenon has decreased with the appearance of a repetition of two to four Asp residues preceding the P1 amino acid. This motif is associated with the recognition of the propeptide by the enterokinase, a duodenal serine protease that can activate trypsinogen (18). Moreover, these acidic residues permit the control of the trypsinogen autoactivation in defined conditions such as in presence of Ca2+, which can neutralize the carboxylate negative charges (18). This mechanism occurs in humans affected by pancreatitis because of hypercalcemia. In this pathology, a high Ca2+ concentration (>1 mm) leads to a premature autoactivation of the trypsinogen associated with an uncontrolled digestion of the pancreatic cells (16, 17, 39). Our results indicate that proDer p 3 cannot be activated in the presence of Ca2+ and/or by incubation at different pH values. The lack of a polyaspartyl motif in its prosequence and the replacement of the ArgP1 or LysP1 residue by a threonine residue at the C-terminal extremity of the propeptide (Fig. 1) can explain our observations. Indeed, the trypsin-like proteases are known to specifically cleave peptide bonds after arginine or lysine residues (18, 35). In addition, the expression of the recombinant homologous proDer f 3 in fusion with glutathione S-transferase was previously described by Nishiyama et al. (20). In this study, proDer f 3 activation was unsuccessful suggesting that another protease present in dust mites could be responsible for the zymogen activation (20).
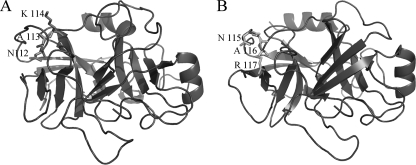
The mite Der p 1 cysteine protease belonging to the papainlike protease family, abundant in mite feces, could be co-localized with the proDer p 3 zymogen during the digestion of the mite. We evaluated the putative maturation of proDer p 3 under the action of rDer p 1 in vitro. Under our experimental conditions, the incubation of recombinant proDer p 3 with rDer p 1 led to mature rDer p 3 (Fig. 3A). This activation mechanism occurred in one step. It corresponded to the recognition and cleavage of the propeptide of proDer p 3 as seen by N-terminal sequencing, which revealed only the sequence corresponding to the mature Der p 3 form (12IVGGEKALAG-). The enzymatic activity of rDer p 3 released, after increasing times of incubation with rDer p 1 (Fig. 3B), was correlated to the molecular mass changes (Fig. 3A). This proDer p 3 maturation was totally abolished in the presence of E-64, suggesting an essential role of the cysteine protease activity in the intermolecular activation of the zymogen. Moreover, we demonstrated that the Der p 1 activity in the HDM extracts was necessary to process proDer p 3 into the mature and active form (12IVGGE-) (Fig. 4A). Another band of ∼19 kDa corresponding to a degradation of rDer p 3 appeared during the activation of proDer p 3. It corresponds to a cleavage of the peptide bond between the Lys-114 and Ala-115 residues in the Der p 3 sequence -NAK114↓115AVGLP-. This proteolysis of Der p 3 was not influenced by the presence of E-64, although it was significantly decreased by the addition of benzamidine (Fig. 3B and supplemental Fig. S2, B and C), demonstrating the role of the protease Der p 3 in its own degradation. A similar site of degradation is observed with human trypsin (-NAR117↓118VSTIL-) and could control the half-life of the active enzyme (33, 40). Indeed, a natural R117H mutant of human trypsin seems to be involved in hereditary pancreatitis (40). The structure model of Der p 3 (Fig. 9) indicates that the -NAR117- and -NAK114-degradation sites, in the trypsin and Der p 3, respectively, are situated on the surface of the proteins and are thus accessible to degradation by proteases. The K114H mutation in Der p 3 could likely abolish this degradation.
FIGURE 9.
Der p 3 model structure and human cationic trypsin x-ray structure. A, model of Der p 3. The -NAK115-degradation site is indicated by the lateral chain and their corresponding numbers. B, x-ray structure of human cationic trypsin (Protein Data Bank code 2R9P). The -NAR117-autolysis site is indicated above.
The incubation of proDer p 3 with rDer p 3 clearly indicated that the protease did not recognize the Thr at the C-terminal extremity of its own propeptide but processed the zymogen into two major forms (Fig. 3A′) as follows: a nearly complete form, 18ALAGE- and the degradation fragment 115AVGLP-. This observation correlates with the specificity of the trypsinlike proteases (see below). The Der p 3 form devoid of the six N-terminal residues, previously identified with the natural enzyme (3), was shown to be inactive (Fig. 3B). X-ray structures of trypsin and trypsinogen indicate that the proteins display ∼85% of structural identity (41–43). These studies show that upon activation of trypsinogen by removal of its propeptide, the new N-terminal Ile-16 folds into a pocket and establishes a salt bridge with Asp-194 liberating the S1-binding site and the oxyanion hole. More recent investigations demonstrated the essential role of the Ile-16 in the stabilization of the catalytic domain for the activity of trypsin (44, 45). The lack of activity of the deleted form of Der p 3 obtained after zymogen cleavage by the protease itself could thus be explained by the fact that an N-terminal Ala-18 is unable to replace the N-terminal Ile-12 to induce the ordering of the catalytic domain of the protease.
Other proteases have been identified in the HDM extracts. Der p 6 has been described as a chymotrypsin-like protease that can cleave substrates containing Phe or Tyr in the P1 position (7, 8). Der p 9 cleaves SA2PL-pNA and SA2PF-pNA, like the cathepsin G, but also type III collagen (9, 11). We explored the potential specific roles of Der p 6 and Der p 9 in the activation mechanism of proDer p 3 by incubation in the presence of the HDM extracts previously treated with APMSF/E-64. We showed that these compounds respectively and specifically inhibit the trypsin-like and cysteine protease activities in the HDM extracts leaving intact the chymotrypsin-like and elastase activities (Table 1). The action of these serine proteases on proDer p 3 resulted in the appearance of degradation bands without formation of Der p 3 (Fig. 4B) confirming the essential role of Der p 1 in the activation of proDer p 3 but also excluding the participation of Der p 6 and Der p 9 in this mechanism. However, a recent study described that, like Der p 3, Der p 9 cleaves the protease-activated receptor 2 receptor of lung epithelial cells after Arg-36 (13) suggesting a trypsin-like specificity of Der p 9. In this case, a protease with the same specificity as Der p 3 should cleave proDer p 3 yielding the same major and inactive forms (18ALAGE- and 115AVGLP-). A study on the recombinant Der p 9 should be necessary to further clarify its specificity. In consequence, we can conclude that the cysteine protease Der p 1 is probably the major activator of proDer p 3 in the house dust mite D. pteronyssinus, whereas the serine proteases (Der p 3, Der p 6 and Der p 9) are involved in the degradation phenomenon.
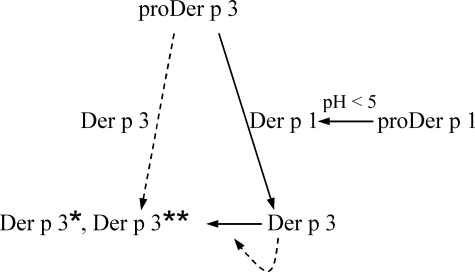
The intermolecular activation mechanism of proDer p 3 by the cysteine protease Der p 1 could represent one important step of a potential protease activation cascade, which could occur in D. pteronyssinus. This proteolytic cascade, depicted in Fig. 10, could imply the initial intramolecular maturation of the Der p 1 zymogen under acidic conditions, followed by proDer p 3 maturation, through Der p 1 action. In turn, Der p 3 could be involved both in the mite digestive function and in its own inactivation. The maturation of the proDer p 3 zymogen by the Der p 1 protease could be explained by the specificity of the protease previously studied by Harris et al. (46). Indeed, they showed that Der p 1 was very specific for a P2 alanine residue, a P4 proline residue, and polar amino acids in P1 and P3 positions. These observations fit the sequence of the C-terminal extremity of the proDer p 3 propeptide (Fig. 1). Moreover, the proDer p 1 and proDer p 3 zymogens belonging to the papainlike and trypsin-like families have conserved residues at the extremity of their prosequences, -LNAEP1 and -PNATP1 respectively. From these results and in vivo observations, the involvement of Der p 1 in the maturation of the proDer p 3 zymogen during digestion in the house dust mite appears to be quite likely. In humans, activation of trypsinogen by a papainlike protease was observed in a type of pancreatitis. In this pathology, cathepsin B, a lysosomal cysteine protease, prematurely activates trypsinogen in trypsin in the pancreas leading to the cell lysis (47–49).
FIGURE 10.
Proposed proteolytic cascade occurring in the D. pteronyssinus house dust mite. The inter-molecular activation of the Der p 3 zymogen by the Der p 1 protease, the autoactivation of proDer p 1 at acidic pH, and the degradation of Der p 3 in Der p 3** (115AVGLP...) are represented by solid arrows. The involvement of Der p 3 in cleavage of its own zymogen in inactive Der p 3* (18ALAGE...) and degradation (Der p 3**) is suggested by dotted arrows.
The enzymatic test used to measure the inter-molecular activation of proDer p 3 by the rDer p 1 protease clearly showed that no intramolecular activation occurred during the proDer p 3 processing and that the propeptide glycosylation decreased the maturation rate. Unfortunately, in the dust mite extracts, the natural Der p 3 enzyme is only present in its mature form making the study of the potential propeptide glycosylation of the natural zymogen impossible. However, we observed that, in vitro, the nonglycosylated proDer p 3 (the N9Q mutant) was more sensitive to degradation. It has also been shown that the dust mite proDer p 1 precursor contains two potential sites of N-glycosylation, one in the propeptide and one in the mature enzyme, and that the mature protease is glycosylated in its natural and recombinant forms (23, 50).
Glycosylation of the propeptide decreases the rate of autoactivation of proDer p 1 (23, 50) as well as the rate of proDer p 3 activation by Der p 1. It is very likely that the two phenomena are correlated because, in both propeptides, the glycosylation site is only three residues away from the cleavage site. This could constitute an additional protection against attacks by other proteases or premature activation in the mite.
The optimum activity pH of rDer p 3 is ∼8.5, which is in agreement with previous studies on trypsin and natural Der p 3 (11, 35). The specificity of rDer p 3 was also explored for the P1, P2, and P3 residues using six different synthetic substrates (IEGR-MCA, GPR-MCA, FSR-MCA, QAR-MCA, NAR-ACC, and NAT-ACC). Our data indicate that the residues in P2 and P3 positions have little effect on the activity as demonstrated for trypsin (35). Indeed, the specificity pocket of trypsin-like proteases rests on the presence of an aspartate residue in the P1-binding site, which in consequence shows a strong preference for the basic side chains of arginine and lysine (35, 42). Indeed, rDer p 3 was not able to cleave the NAT-ACC substrate. This is in agreement with the observed processing of proDer p 3 by rDer p 3, which does not cleave its zymogen after the C-terminal residue of the propeptide (Thr-11) but after the first lysine (Lys-17) in the Der p 3 domain.
We finally investigated the interaction between the mature rDer p 3 and its free prosequence. We observed that, even at a propeptide concentration of 5 mm, no major decrease of protease activity could be detected. This result could be explained by local differences in the secondary structures of the zymogen and the mature protease. Indeed, Gombos et al. (51) showed that the maturation of trypsinogen resulted in a small irreversible conformation change in the protease domain without affecting the content of the secondary structures. A similar event is likely to occur during the maturation process of proDer p 3, and the absence of strong inhibition by the propeptide could be correlated with a modification of the interactions between the protease and its propeptide after hydrolysis of the Thr-11 to Ile-12 bond. A replacement of Thr by Arg at the C-terminal extremity of the propeptide enhanced its affinity for rDer p 3. Interestingly, although no autoactivation mechanism was observed for the proDer p 3 zymogen and for its homologous proDer f 3, Nishiyama et al. (20) showed that the proDer f 3 TP1R mutant was activated during the fermentation process. Therefore, the C-terminal extremity of the proDer p 3 propeptide is really important for the zymogen inhibition and activation mechanisms. Complementary studies with different propeptide mutants would be interesting to determine the amino acids that control this interaction.
Supplementary Material
This work was supported in part by Fonds de la Recherche Fondamentale et Collective Grants 2.4561.07, 1.5.209.07, 2.4511.06 and 2.4550.05 by the Direction Générale des Technologies de la Recherche et de l'Energie, Région Wallonne, and by GlaxoSmithKline Biologicals (Rixensart, Belgium). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental text and additional references and Figs. 1 and 2.
Footnotes
The abbreviations used are: HDM, house dust mite; rDer p 1, recombinant Der p 1; rDer p 3, recombinant Der p 3; E-64, l-trans-epoxysuccinyl-leucylamido(4-guanidino) butane; APMSF, 4-amidinophenylmethanesulfonyl fluoride hydrochloride; SBTI, soybean trypsin inhibitor; WT, wild-type; Boc, t-butyloxycarbonyl; CAPS, 3-(cyclohexylamino)-1-propanesulfonic acid; MCA, methoxycoumarin acetic acid; DTT, dithiothreitol; DMF, dimethylformamide; Fmoc, 9-fluorenylmethyloxycarbonyl; Pbf, 2,2,4,6,7-pentamethyldihydrobenzofuran-5-sulfonyl; tBu, tert-butyl; PBS, phosphate-buffered saline; HPLC, high pressure liquid chromatography; ATR, attenuated total reflection; pNA, p-nitroanilide.
A. Jacquet, unpublished work.
References
- 1.Thomas, W., Smith, W. A., Hales, B., Mills, K., and O'Brien, R. (2002) Int. Arch. Allergy Immunol. 129 1-18 [DOI] [PubMed] [Google Scholar]
- 2.Platt-Mills, T., and Chapman, M. (1987) J. Allergy Clin. Immunol. 80 755-775 [DOI] [PubMed] [Google Scholar]
- 3.Stewart, G., Ward, L., Simpson, R., and Thompson, P. (1992) Immunology 75 29-35 [PMC free article] [PubMed] [Google Scholar]
- 4.Ando, T., Homma, R., Ino, Y., Ito, G., Miyahara, A., Yanagihara, T., Kimura, H., Ikeda, S., Yamakawa, H., Iwaki, M., Okumura, Y., Suko, M., Haida, M., and Okudaira, H. (1993) Clin. Exp. Allergy 23 777-784 [DOI] [PubMed] [Google Scholar]
- 5.Smith, W., and Thomas, W. R. (1996) Int. Arch. Allergy Immunol. 109 133-140 [DOI] [PubMed] [Google Scholar]
- 6.Hales, B. J., Martin, A. C., Pearce, L. J., Laing, I. A., Hayden, C. M., Goldblatt, J., Le Souëf, P. N., and Thomas, W. R. (2006) J. Allergy Clin. Immunol. 118 361-367 [DOI] [PubMed] [Google Scholar]
- 7.Yasueda, H., Mita, H., and Akiyama, K. (1993) Clin. Exp. Allergy 23 384-390 [DOI] [PubMed] [Google Scholar]
- 8.Bennett, B. J., and Thomas, W. R. (1996) Clin. Exp. Allergy 26 1150-1154 [PubMed] [Google Scholar]
- 9.King, C., Simpson, R. J., Moritz, R. L., Reed, G. E., Thompson, P. J., and Stewart, G. A. (1996) J. Allergy Clin. Immunol. 98 739-747 [DOI] [PubMed] [Google Scholar]
- 10.Smith, W., Chuna, K., Kuo, M., Rogers, B., and Thomas, W. (1994) Clin. Exp. Allergy 24 220-228 [DOI] [PubMed] [Google Scholar]
- 11.Stewart, G., Kollinger, M., King, C., and Thompson, P. (1994) Allergy (Cph.) 49 553-560 [DOI] [PubMed] [Google Scholar]
- 12.Maruo, K., Akaike, T., Ono, T., Okamoto, T., and Maeda, H. (1997) J. Allergy Clin. Immunol. 100 253-260 [DOI] [PubMed] [Google Scholar]
- 13.Sun, G., Stacey, M., Schmidt, M., Mori, L., and Mattoli, S. (2001) J. Immunol. 167 1014-1021 [DOI] [PubMed] [Google Scholar]
- 14.Lazure, C. (2002) Curr. Pharm. Des. 8 125-133 [DOI] [PubMed] [Google Scholar]
- 15.Vernon, H. (1914) J. Biol. Chem. 8 494-529 [Google Scholar]
- 16.Brodrick, J., Largman, C., Johnson, J., and Geokas, M. (1978) J. Biol. Chem. 253 2732-2736 [PubMed] [Google Scholar]
- 17.Frick, T., Fernandez-del-Castillo, C., Bimmler, D., and Warshaw, A. (1997) Gut 41 339-343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen, J.-M., Kukor, Z., Le Maréchal, C., Toth, M., Tsakiris, L., Raguenes, O., Ferec, C., and Sahin-Toth, M. (2003) Mol. Biol. Evol. 20 1767-1777 [DOI] [PubMed] [Google Scholar]
- 19.Schechter, I., and Berger, A. (1967) Biochem. Biophys. Res. Commun. 27 157-162 [DOI] [PubMed] [Google Scholar]
- 20.Nishiyama, C., Yasuhara, T., Yuuki, T., and Okumura, Y. (1995) FEBS Lett. 377 62-66 [DOI] [PubMed] [Google Scholar]
- 21.Jacquet, A., Magi, M., Petry, H., and Bollen, A. (2002) Clin. Exp. Allergy 32 1048-1053 [DOI] [PubMed] [Google Scholar]
- 22.Stewart, G. A., Thompson, P. J., and Simpson, R. J. (1989) Lancet ii 154-155; Correction (1989) Lancet ii, 462 [Google Scholar]
- 23.Chevigné, A., Barumandzadeh, R., Groslambert, S., Cloes, B., Dehareng, D., Filée, P., Marx, J.-C., Frère, J.-M., Matagne, A., Jacquet, A., and Galleni, M. (2007) J. Mol. Biol. 374 170-185 [DOI] [PubMed] [Google Scholar]
- 24.Cheong, N., Yang, L., Lee, B. W., and Chua, K. Y. (2003) Allergy (Cph.) 58 352-356 [DOI] [PubMed] [Google Scholar]
- 25.Ménard, R., Carmenon, E., Takebe, S., Dufour, E., Plouffe, C., Mason, P., and Mort, J. (1998) J. Biol. Chem. 273 4478-4484 [DOI] [PubMed] [Google Scholar]
- 26.Quraishi, O., and Storer, A. C. (2001) J. Biol. Chem. 276 8118-8124 [DOI] [PubMed] [Google Scholar]
- 27.Cha, S. (1975) Biochem. Pharmacol. 24 2177-2185 [DOI] [PubMed] [Google Scholar]
- 28.Morrison, J. F. (1969) Biochim. Biophys. Acta 185 269-286 [DOI] [PubMed] [Google Scholar]
- 29.Morrison, J. F. (1982) Trends Biochem. Sci. 7 102-105 [Google Scholar]
- 30.Morrison, J. F., and Stone, S. R. (1985) Comments Mol. Cell. Biophys. 2 347-368 [Google Scholar]
- 31.Morrison, J. F., and Walsh, C. T. (1988) Adv. Enzymol. Relat. Areas Mol. Biol. 61 201-301 [DOI] [PubMed] [Google Scholar]
- 32.Lund, O., Nielsen, M., Lundegaard, C., and Worning, P. (2002) CASP5 Conference, Asimolar (CA), Protein Structure Prediction Center, A102-A104, University of California, Davis
- 33.Kukor, Z., Toth, M., Pal, G., and Sahin-Toth, M. (2002) J. Biol. Chem. 277 6111-6117 [DOI] [PubMed] [Google Scholar]
- 34.Schulz, O., Sewell, H. F., and Shakib, F. (1998) J. Clin. Pathol. (Lond.) 51 222-231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Halfon, S., Baird, T., and Craik, C. (2004) in Handbook of Proteolytic Enzymes (Barrett, A. J., Rawlings, N. D., and Woessner, J. D.) 2nd Ed., pp. 1483-1488, Elsevier Academic Press, New York
- 36.Kay, J., and Kassell, B. (1971) J. Biol. Chem. 246 6661-6665 [PubMed] [Google Scholar]
- 37.van Stokkum, I., Linsdell, H., Hadden, J. M., Haris, P. I., Chapman, D., and Bloemendal, M. (1995) Biochemistry 34 10508-10518 [DOI] [PubMed] [Google Scholar]
- 38.Arrondo, J., and Goni, F. M. (1999) Prog. Biophys. Mol. Biol. 72 367-405 [DOI] [PubMed] [Google Scholar]
- 39.Kukor, Z., Toth, M., and Sahin-Toth, M. (2003) Eur. J. Biochem. 270 2047-2058 [DOI] [PubMed] [Google Scholar]
- 40.Whitcomb, D., Gorry, M. C., Preston, R. A., Furey, W., Sossenheimer, M. J., Ulrich, C. D., Martin, S. P., Gates, L. K., Jr., Amann, S. T., Toskes, P. P., Liddle, R., McGrath, K., Uomo, G., Post, J. C., and Ehrlich, G. D. (1996) Nat. Genet. 14 141-145 [DOI] [PubMed] [Google Scholar]
- 41.Huber, R., and Bode, W. (1978) Acc. Chem. Res. 11 114-122 [Google Scholar]
- 42.Fehlhammer, H., Bode, W., and Huber, R. (1977) J. Mol. Biol. 111 415-438 [DOI] [PubMed] [Google Scholar]
- 43.Brünger, A. T., Huber, R., and Karplus, M. (1987) Biochemistry 26 5153-5162 [DOI] [PubMed] [Google Scholar]
- 44.Hedstrom, L., Lin, T.-Y., and Fast, W. (1996) Biochemistry 35 4515-4523 [DOI] [PubMed] [Google Scholar]
- 45.Pasternak, A., Liu, X., Lin, T.-Y., and Hedstrom, L. (1998) Biochemistry 37 16201-16210 [DOI] [PubMed] [Google Scholar]
- 46.Harris, J., Mason, D., Li, J., Burdick, K., Backes, B., Chen, T., Shipway, A., Van Heeke, G., Cough, L., Ghaemmaghami, A., Shakib, F., Debaene, F., and Winssinger, N. (2004) Chem. Biol. 11 1361-1372 [DOI] [PubMed] [Google Scholar]
- 47.Greenbaum, L., Hirshkowitz, A., and Shoichet, I. (1959) J. Biol. Chem. 234 2885-2890 [PubMed] [Google Scholar]
- 48.Halangk, W., Lerch, M., Brandt-Nedelev, B., Roth, W., Ruthenbuerger, M., Reinheckel, T., Domschke, W., Lippert, H., Peters, C., and Deussing, J. (2000) J. Clin. Investig. 106 773-781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Szilagyi, L., Kenesi, E., Katona, G., Kaslik, G., Juhasz, G., and Graf, L. (2001) J. Biol. Chem. 276 24574-24580 [DOI] [PubMed] [Google Scholar]
- 50.Takai, T., Mizuuchi, E., Kikuchi, Y., Nagamune, T., Okumura, K., and Ogawa, H. (2006) Int. Arch. Allergy Immunol. 139 181-187 [DOI] [PubMed] [Google Scholar]
- 51.Gombos, L., Kardos, J., Patthy, A., Medveczky, P., Szilagyi, L., Malnasi-Csizmadia, A., and Graf, L. (2008) Biochemistry 47 1675-1684 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.