Abstract
Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) has diverse biological functions including its nuclear translocation in response to oxidative stress. We show that GAPDH physically associates with APE1, an essential enzyme involved in the repair of abasic sites in damaged DNA, as well as in the redox regulation of several transcription factors. This interaction allows GAPDH to convert the oxidized species of APE1 to the reduced form, thereby reactivating its endonuclease activity to cleave abasic sites. The GAPDH variants C152G and C156G retain the ability to interact with but are unable to reactivate APE1, implicating these cysteines in catalyzing the reduction of APE1. Interestingly, GAPDH-small interfering RNA knockdown sensitized the cells to methyl methane sulfonate and bleomycin, which generate lesions that are repaired by APE1, but showed normal sensitivity to 254-nm UV. Moreover, the GAPDH knockdown cells exhibited an increased level of spontaneous abasic sites in the genomic DNA as a result of diminished APE1 endonuclease activity. Thus, the nuclear translocation of GAPDH during oxidative stress constitutes a protective mechanism to safeguard the genome by preventing structural inactivation of APE1.
The evolutionary conserved enzyme glyceraldehyde-3-phosphate dehydrogenase (GAPDH)3 exists as a tetramer that catalyzes a critical reaction in the second stage of the glycolytic pathway (1). It uses the oxidized form of nicotinamide adenine dinucleotide (NAD+) and converts glyceraldehyde-3-phosphate to 1,3-bisphosphoglycerate with the concomitant release of NADH in an oxido-reduction reaction (1). GAPDH is also a key redox-sensitive protein that possesses an active site cysteine sulfhydryl that is susceptible to oxidation (2). Under oxidative stress, GAPDH rapidly undergoes disulfide bond formation leading to reduction in its enzymatic activity (2, 3). GAPDH has the propensity to interact with several proteins that are vulnerable to aggregation and are associated with neurodegenerative disorders such as in the case of the pro-oxidant amyloid β peptide involved in Alzheimer disease (4). Recent studies have documented that GAPDH is also involved in several other nuclear processes that include histone H2B gene expression, nuclear RNA export, apoptosis, and cellular response to DNA damage (5–8).
Several lines of evidence support a role for GAPDH in DNA damage and repair (5, 9). For example, GAPDH can translocate from the cytoplasm to the nucleus when cells are challenged with the potent chemical oxidant and DNA-damaging agent H2O2, although it is not clear what is the function executed by GAPDH under this stress condition (10). However, a more recent study documented that nitric oxide can also induce nuclear localization of GAPDH where it is acetylated by the acetyltransferase p300/CBP via direct protein interaction, which in turn causes stimulation of the catalytic activity of p300/CBP, resulting in the activation of downstream targets such as p53 (11). Other studies have shown that GAPDH is associated with DNA-containing genotoxic lesions such as thioguanylated DNA generated by the chemotherapeutic agent mercaptopurine used for treating acute lymphoblastic leukemia (12, 13). Thus, it seems that GAPDH response to oxidative stress might be linked to a role in maintaining the integrity of the genome.
In this study, we report that human GAPDH directly interacts with APE1, an essential enzyme that functions in the base excision DNA repair pathway to process spontaneous and drug-induced abasic or apurinic/apyrimidinic (AP) sites, as well as to regulate the redox state of a number of transcriptional factors such as p53, AP-1, c-Jun, c-Fos, and NF-κB (14–18). Using recombinant proteins, we show that the active site cysteine 152 of GAPDH is not required for the interaction with APE1 but indispensable for converting the oxidized forms of APE1 to its reduced form. The catalytic action of GAPDH on oxidized APE1 re-establishes a key biological function of the protein, that is, the ability to cleave AP sites, indicating that the redox-sensitive cysteine of GAPDH can function to reduce APE1. Concomitantly, this reduction process greatly enhanced the detection of APE1 by anti-APE1 antibodies consistent with the oxidized form of the protein undergoing a structural change. We further demonstrate that siRNA knockdown of GAPDH in HCT116 cells caused sensitivity to agents known to create DNA lesions that are processed by APE1, including the alkylating DNA-damaging agent methyl methane sulfonate (MMS), which produces AP sites, but not to 254 nm of UVC (19, 20). Importantly, the GAPDH knockdown cells accumulated a higher level of spontaneous AP sites in the genomic DNA. We propose that GAPDH serves a critical role in maintaining genomic stability by protecting APE1 from aberrant structural changes caused by oxidative stress.
EXPERIMENTAL PROCEDURES
Bacteria Strains—The Escherichia coli strains used in this work were BW528 [Δ(xth-pnc), nfo1::kan] (kindly provided by B. Weiss, Emory University, Atlanta, GA) and BL21(DE3)pLysS [dcm ompT hsdS (rB-mB-) galλ(DE3) (pLysS Camr)] (Stratagene).
Cell Culture—Primary human diploid lung fibroblast LF1 (low passage) and human colon carcinoma HCT116 cell lines were kindly provided by Dr. Drobetsky (University of Montreal, Montreal, Canada), and the human colon carcinoma DLD1 cell lines stably transfected with either pcDNA3.1/GFP-(Gly5)-GAPDH or pcDNA3.1/EYFP were generously provided by Dr. W. E. Evans (University of Tennessee, Memphis, TN). The HDLF1, HCT116, and DLD1 cells were maintained in Ham's F10 nutrient medium (Sigma-Aldrich), McCoy5A medium (Invitrogen), and RPMI 1640 medium (BioWhittaker Inc., Walkersville, MD), respectively, supplemented with 10% fetal bovine serum, 0.1 mg/ml penicillin, and 0.1 mg/ml streptomycin. For DLD1 cells, 1 mg/ml of G418 (USB Corporation) was added in the RPMI 1640 medium (21). The cells were incubated at 37 °C with 5% CO2.
AP Endonuclease Assay—This assay was performed as previously described using a 42-mer substrate U21·G containing a single AP site (22).
Purification of a Protein with Associated AP Endonuclease Activity—Approximately 5 × 106 human lung fibroblast cells were broken in a Dounce homogenizer with 10 strokes (glass and Teflon) in 1 ml of buffer A (50 mm Tris-HCl, pH 7.0, 20 mm NaCl, 10% glycerol, and protease inhibitor mixture (EDTA-free; Roche Applied Science). The cell extracts were centrifuged at 14000 rpm at 4 °C for 10 min, and the supernatants were incubated with 1.6 ml of pre-equilibrated DEAE-Sepharose resin (Amersham Biosciences) in buffer A by rotating at 4 °C for 15 min. The unbound proteins were then loaded onto a 500-μl single-stranded DNA agarose column (Amersham Biosciences) pre-equilibrated in buffer A. The proteins were eluted by a step gradient of 100, 200, 300, and 500 mm NaCl in buffer A in fraction sizes of 250 μl. Fractions (200–500 mm) containing AP endonuclease activity were pooled and concentrated (Amicon Ultra Centricon, Millipore Corp.) while exchanging buffer A to B (50 mm Tris-HCl, pH 7.5, 20 mm NaCl, 0.5 mm dithiothreitol, and 0.5 mm EDTA) with a 10-fold dilution repeated three times. The concentrated fraction was diluted 10-fold in buffer B and loaded onto a 200 μl of MonoQ column (Bio-Rad) pre-equilibrated in buffer B. The flow-through fraction was concentrated and exchanged with buffer C (50 mm Tris-HCl, pH 8.0, 20 mm NaCl, 0.5 mm dithiothreitol, and 0.5 mm EDTA), as above. The concentrated fraction was diluted 10-fold in buffer C and reloaded onto a 200-μl MonoQ column pre-equilibrated in buffer C. The flow-through fraction was then loaded onto a 200-μl CM-Sepharose column (Amersham Biosciences) pre-equilibrated in buffer C. The column was washed three times with buffer C, and the protein was eluted with 100, 200, 400, and 600 mm NaCl. The eluted proteins from 200, 400, and 600 mm NaCl elution showed AP endonuclease activity and were pooled and concentrated to 25 μl and desalted by centrifugation (Amicon Ultra Centricon (Millipore Corp.) in buffer C. The concentrated proteins were loaded onto a 150-μl MonoS column (Bio-Rad) pre-equilibrated with buffer C. The column was washed three times with 300 μl of buffer C and eluted with 100, 200, 400, and 600 mm NaCl in buffer C. The major AP endonuclease activity was present in the 400 mm NaCl fraction.
Protein Sequencing—The enriched polypeptide band (37-kDa) from the 400 mm NaCl MonoS fraction was excised from preparative 12% SDS-PAGE, washed with 50% acetonitrile in water, and sequenced by liquid chromatography-mass spectrometry (Harvard Microchemistry).
Plasmids and Site-directed Mutagenesis—PCR was used to amplify the entire GAPDH cDNA from the plasmid template (ATCC no. 817954R, accession number M33197, NCBI), digested with BamHI and EcoRI, and cloned into the E. coli expression vector pGEX-4T-1 (Amersham Biosciences) to produce the plasmid pGST-GAPDH. Site-directed mutagenesis (QuikChange kit, Stratagene, La Jolla, CA) was used to mutate the three cysteines to glycine in GAPDH using pGST-GAPDH as the template and the following primers C152G-F1 (5′-CATCAGCAATGCCTCCGGCACCACCAACTGCTTAGC-3′), GAPDH C156G-F2 (5′-GCCTCCTGCACCACCAACGGCTTAGCACCCCTGGC-3′), and GAPDH C247G-F3 (5′-GTGGTGGACCTGACCGGCCGTCTAGAAAAACCTGCC-3′). The resulting plasmids were pGST-GAPDH C152G, pGST-GAPDH C156G, and pGST-GAPDH C247G, and the mutations were verified by DNA sequence analysis.
Purification of GST and His-tagged Proteins—GST-GAPDH fusion proteins were overexpressed in E. coli BW528 strain and purified by glutathione-Sepharose 4B mini columns as previously described (23), and the His6-APE1 was purified using Talon affinity beads according to the manufacturer's protocol (Clontech).
APE1 Pulldown Assay—One ml of matrix slurry (Talon beads) was placed in a plastic disposable 10-ml gravity-flow column (Bio-Rad). The beads were equilibrated with 10 ml of buffer B (50 mm sodium phosphate, pH 7.0, and 300 mm NaCl). At least 10 μg of purified His-APE1 protein was then incubated with the beads for 30 min at room temperature with gentle shaking. The beads were washed with 30 ml of buffer B, and 100-μl aliquots with the equivalent of ∼400 ng of bound His-APE1 were mixed with either 2 μg of purified GST-PNKP, GST-GAPDH, GST-GAPDH C152G, GST-GAPDH C156G, or GST-GAPDH C247G and incubated for 1 h at room temperature. The beads were washed three times with 3 ml of buffer B, and the aliquots were analyzed directly for bound proteins by Western blot.
GST Pulldown Assays—Glutathione-Sepharose 4B beads (100 μl) alone or bound to 10 μg of the indicated GST-tagged proteins were mixed with purified N-terminal His-tagged APE1 (1 μg) in 0.5 ml of buffer A and incubated for 30 min at room temperature with gentle rotation. The beads were washed three times with buffer A, and aliquots were analyzed for bound proteins by Western blots probed with either anti-His or anti-GST monoclonal antibody.
Immunoprecipitation—Immunoprecipitation was performed on whole cell lysates prepared in radioimmune precipitation assay buffer (24). Following preclearing with 30 μl of Aminolink beads plus coupling gel (Pierce), 1.5 mg of total protein was incubated with anti-APE1 antibodies overnight at 4 °C and then incubated with 50 μl of Aminolink beads for another 1 h at 4 °C. The beads were washed four times with radioimmune precipitation assay buffer and then boiled in 2× SDS sample buffer. The immunoprecipitates were then detected by immunoblotting with the indicated antibodies.
Extract Preparation and Immunodetection—Total cells extracts were prepared and analyzed by Western blots as previously described (24). The antibodies used in this study were monoclonal anti-His (Santa Cruz), anti-GAPDH (Chemicon), anti-APE1 (R & D Systems), polyclonal anti-GST (Sigma), and anti-APE1 (kindly provided by Dr. Bruce Demple, Harvard University and subsequently purchased from Santa Cruz).
Oxidation of APE1 Protein with H2O2—The His-APE1 protein bound to the TALON metal affinity column (1 ml; BD Biosciences) was treated with H2O2 (5 μm for 5 min), washed three times with buffer B (50 mm sodium phosphate, pH 7.0, and 300 mm NaCl), and then eluted using buffer B containing 150 mm imidazole. The eluted proteins were dialyzed in a Spectra/Por 1 dialysis tube (molecular weight cut-off of 6,000–8,000; Spectrum) by repeated concentration and dilution with 20 mm Tris-HCl (pH 7.5) and 50 mm NaCl for five times. The concentrations of the proteins were estimated on a Coomassie Blue-stained 12% SDS-PAGE gel.
siRNA-mediated GAPDH Silencing—siRNA (Ambion) was used to knock down the expression of GAPDH in DLD1 and HCT116 cells. The cells were seeded on Petri plates and transfected with 75 nm of GAPDH siRNA using Lipofectamine 2000 (Invitrogen) in Opti-MEM (Invitrogen) according to the manufacturer's protocol. Negative control (75 nm) that has no significant homology to any known gene sequences from mouse or human was also included. The plates were incubated at 37 °C in 5% CO2 for 4 h, after which the complete growth medium was added, and the cells were processed as indicated.
Drug Exposure and Clonogenic Assay—The siRNA transfected cells (1 × 106) were plated in 60-mm × 15-mm Petri plates and incubated for 24 h before exposure to different drugs. The adherent cells were incubated with the indicated concentrations of either bleomycin for 1 h or methyl methane sulfonate for 30 min in fetal bovine serum-free incomplete medium. The colony-forming unit assay was performed as described previously (24).
UV Irradiation—The cells were irradiated with monochromatic 254-nm UV (UVC from G25T8 germicidal lamp (Sankyo Denki) at a dose ranging from 0–20 J/m2 in 1× phosphate-buffered saline. UVC 254-nm UV fluorescence was measured with a Spectroline DRC 100× digital radiometer (Spectronics, Westbury, NY) equipped with 254 sensors (25).
DNA Extraction and DNA Damage Quantification—Briefly, ∼5.0 × 106 of the indicated cells transfected with either the control or GAPDH-siRNA were subjected to genomic DNA extraction as previously described (25). Formation of AP sites was measured by using the DNA damage quantification kit according to the manufacturer's protocol (Biovison Inc.).
RESULTS
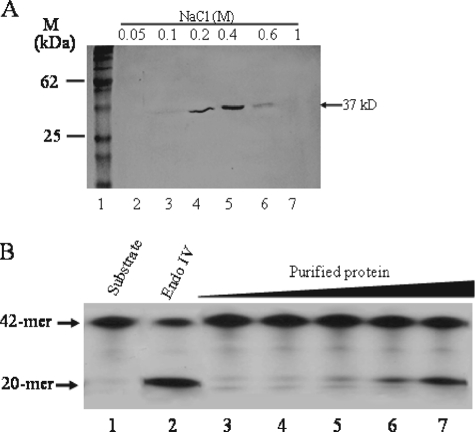
A Minor AP Endonuclease Activity Is Associated with the Glycolytic Enzyme GAPDH—We initially set out to examine whether mammalian cells contained a Mg2+-independent AP endonuclease(s) belonging to the E. coli endonuclease IV family, because such an activity has not been previously identified. Using total cell extracts derived from cultured human lung fibroblast, we detected a weak Mg2+-independent AP endonuclease activity that cleaved a 42-mer double stranded γ-32P-labeled synthetic oligonucleotide substrate containing a single AP site at position 21 to produce a 20-mer product (22). The activity was enriched by subjecting the total extract to a 6-step column chromatography purification scheme, followed by a NaCl discontinuous gradient on a MonoS column (see “Experimental Procedures”). Fractions from the MonoS column yielded a single major polypeptide of ∼37 kDa as visualized by staining with Coomassie Blue dye (Fig. 1A). The fraction (0.4 m NaCl) with the most protein exhibited the highest level of Mg2+-independent AP endonuclease activity (data not shown). Moreover, increasing the amount of this fraction resulted in an increase production of the 20-mer product that was identical to that generated by Endo IV (Fig. 1B, lanes 3–7 versus lane 2) but not the more slowly migrating 3′-α,β-unsaturated aldehyde 20-mer product created by endonuclease III (data not shown), suggesting that the purified protein may contain AP endonuclease activity. The protein cannot cleave the 42-mer double-stranded oligonucleotide containing the U·G mismatch, unless the uracil is removed, consistent with the notion that the purified protein fraction has associated AP endonuclease and no hint of uracil-DNA glycosylase activity nor trace amounts of other enzymes that belong to the base-excision DNA repair pathway such as AP lyases, DNA polymerase, and DNA ligase (data not shown).
FIGURE 1.
Copurification of a Mg2+-independent AP endonuclease activity with a 37-kDa protein. A, the Mg2+-independent AP endonuclease activity detected in total cell extracts derived from lung fibroblast cells was subjected to several purification steps followed by a discontinuous NaCl gradient on MonoS. The activity was monitored by nicking of the AP site 5′-32P-labeled 42-mer substrate (80 ng), and the final active fractions were analyzed by SDS-PAGE stained with Coomassie Blue. Lane 1, molecular mass standards; lanes 2–7, MonoS fractions from NaCl step gradient. B, increasing concentrations of the 0.4 m NaCl MonoS fraction was incubated with the AP site substrate and monitored for the formation of the 20-mer product. Lane 1, substrate alone; lane 2, purified Endo IV (10 ng); lanes 3–7, increasing concentrations (25–300 ng) of the purified 0.4 m NaCl MonoS fraction. The arrows indicate the positions of the 42-mer substrate and the 20-mer product.
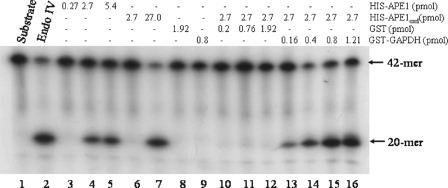
We next determined the identity of the 37-kDa purified protein by excising the Coomassie-stained polypeptide from the SDS-PAGE and subjecting it to sequencing using microcapillary reverse-phase high pressure liquid chromatography coupled to a nano-electrospray ionization source of an ion trap mass spectrometer. Unexpectedly, of 59 individual sequences analyzed, all showed 95–100% identity to the human GAPDH enzyme. This finding suggests that the Mg2+-independent AP endonuclease activity is likely the result of a minor protein that copurified with GAPDH. To ascertain that the purified protein preparation indeed contained GAPDH, we tested for the enzyme activity by monitoring the reduction of NAD+. As anticipated, the purified protein displayed a significant level of GAPDH activity when compared with a commercial preparation or a recombinant form (GST-GAPDH) of the human GAPDH expressed and purified from E. coli (supplemental Figs. S1 and S2). Because APE1 is the major AP endonuclease present in mammalian cells and the recombinant source of human GAPDH (GST-GAPDH) lacked AP endonuclease activity (see Fig. 5), we considered that a minor fraction of APE1 with a different mobility might be copurifying with GAPDH. This copurification might indicate a link between the two proteins.
FIGURE 5.
GAPDH reactivates the AP endonuclease activity of oxidized APE1. A fixed amount (2.7 pmol) of oxidized His-APE1oxd was incubated with increasing concentrations of either purified GST (lanes 10–12) or purified GST-GAPDH (lanes 13–16) for 5 min at room temperature and then assayed for AP endonuclease activity. Shown are the controls, substrate (80 ng) alone (lane 1) and the following purified proteins Endo IV (0.3 pmol, lane 2), His-APE1 (lanes 3–5), oxidized His-APE1oxd (lanes 6 and 7), GST (lane 8), and GST-GAPDH (lane 9).
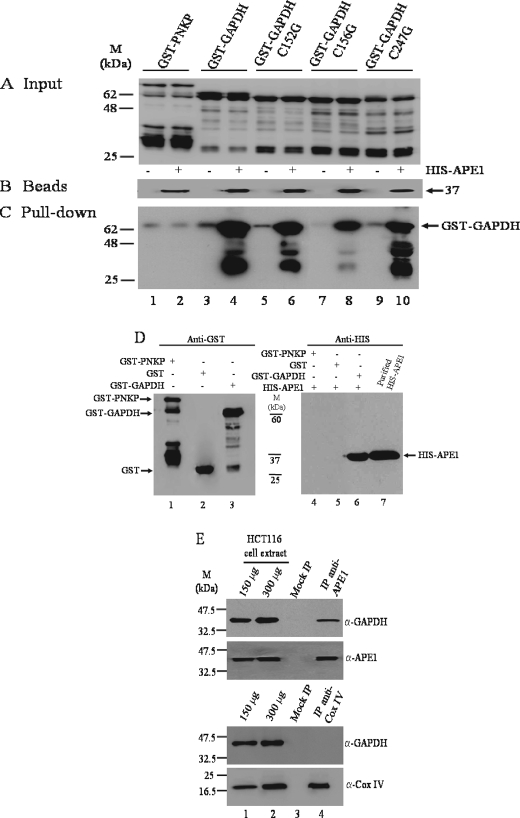
GAPDH and Its Variants Interact with APE1—To directly test whether GAPDH is associated with APE1, we used Talon affinity beads consisting of functionally active APE1 tagged at the N-terminal end with the His tag and examined for retention of the purified GST-GAPDH (see “Experimental Procedures”). When purified GST-GAPDH (Fig. 2A, lane 4) was mixed with the His-APE1 affinity beads (Fig. 2B, lane 4), followed by extensive washing, a significant amount of the protein was pulled down by the beads as detected by Western blot probed with anti-GST monoclonal antibody (Fig. 2C, lane 4). Likewise, three cysteine variants of GST-GAPDH: the presumed active site cysteine 152 (C152G), the adjacent cysteine 156 (C156G), and cysteine 247 (C247G) with no documented role in the redox function of GAPDH (Fig. 2A), were also retained by the His-APE1 affinity beads (Fig. 2C). In controls, the purified DNA repair protein GST-PNKP (Fig. 2A, lane 2) was not retained by the His-APE1 affinity beads (Fig. 2C, lane 2). In these experiments, a very minute amount of either GST-GAPDH or the variants (Fig. 2A) was pulled down by the empty beads (Fig. 2B), suggesting a possible weak and nonspecific interaction of the fusion proteins with the beads. Of note, the fragmented forms of GST-GAPDH (∼35–40 kDa), and not those of GST-PNKP, were enriched by the His-APE1 affinity beads, suggesting that the lower molecular mass forms of GST-GAPDH may contain a possible interacting region with His-APE1. Several additional control experiments revealed that GST-GAPDH was not pulled down by beads carrying a different protein His-protein phosphatase 2A activator, a peptidyl prolyl cis/trans isomerase (data not shown). Moreover, the glycolytic enzymes pyruvate dehydro-genase and 6-phosphofructokinase obtained from a commercial source were not retained by the His-APE1 affinity beads (data not shown). Thus, these combined data support the notion that GAPDH may interact with APE1 and that none of the three cysteine residues of GAPDH is essential for the association, contrasting the interaction of GAPDH with inositol 1,4,5-triphosphate receptors requiring the cysteine residues of both proteins (26).
FIGURE 2.
GAPDH interacts with APE1 in vitro and in vivo. A, the indicated GST-tagged input proteins were isolated from E. coli expression system, purified on a GST affinity column, and analyzed by Western blot probed with anti-GST polyclonal antibodies. Lanes 1–10 each contained 200 ng of the purified proteins. B, Talon beads (30 μl each lane) empty (lanes 1, 3, 5, 7, and 9) and with bound His-APE1 (lanes 2, 4, 6, 8, and 10) used for pulling down the GST-tagged proteins in A were probed with anti-His monoclonal antibody. C, the empty Talon beads (lanes 1, 3, 5, 7, and 9) or Talon beads with His-APE1 (400 ng) (lanes 2, 4, 6, 8, and 10) were mixed with the indicated purified GST-tagged proteins (2000 ng each) and washed, and the aliquots (30 μl) were directly assessed for the bound GST-tagged proteins by Western blot probed with anti-GST polyclonal antibodies. D, GST affinity beads containing either purified GST-PNKP (lane 1), GST (lane 2), or GST-GAPDH (lane 3) were mixed with purified His-APE1 (400 ng), washed, and directly assessed for bound His-APE1 (lanes 4–6) by Western blot probed with anti-His monoclonal antibody. Lane 7, purified His-APE1 (150 ng). Molecular mass standards (kDa) are shown in the middle. E, total cell extract (1.5 mg) derived from HCT116 cells was incubated without and with either anti-APE1 or anti-COX IV antibodies and subjected to immunoprecipitation followed by Western blot probed with the indicated antibodies. Lanes 1 and 2, 150 and 300 μg of total cell extracts; lane 3, mocked immunoprecipitation (no antibodies); lane 4, coimmunoprecipitation with either anti-APE1 or anti-COX IV antibodies.
To further confirm the GAPDH-APE1 interaction, we conducted the reciprocal experiment by preparing GST-GAPDH affinity beads (Fig. 2D, lane 3) and examined for the retention of His-APE1. As shown in Fig. 2D, purified His-APE1 was pulled down by the GST-GAPDH affinity beads as detected by Western blot probed with anti-His monoclonal antibodies (lane 6). In contrast, neither GST-PNKP nor GST beads (Fig. 2D, lanes 1 and 2) pulled down the His-APE1 protein (Fig. 2D, lanes 4 and 5). Thus, the serendipitous isolation of GAPDH with AP endonuclease activity is due to the interaction of GAPDH with APE1. It is therefore not surprising that commercial preparations of GAPDH, but not pyruvate dehydrogenase and 6-phosphofructokinase obtained from mammalian sources, contained AP endonuclease activity (data not shown).
To assess whether GAPDH is associated with APE1 in vivo, an immunoprecipitation reaction was performed with total cell extracts derived from HCT116 cells using anti-APE1 polyclonal antibodies. After eliminating the nonspecific interaction by washing, the immunoprecipitated complexes were analyzed by Western blot using anti-GAPDH monoclonal antibody. As shown in Fig. 2E, the endogenous GAPDH was coimmunoprecipitated with anti-APE1 antibodies (lane 4), but not by the mock reaction lacking anti-APE1 antibodies (lane 3). We note that anti-GAPDH antibody did not cross-react with APE1, nor did anti-APE1 antibody cross-reacted with GAPDH (supplemental Fig. S3). Because GAPDH is also localized to the mitochondria and can interact with many proteins, for example, the voltage-dependent anion channel of the mitochondrial membrane and the inositol 1,4,5-triphosphate receptor involved in the release of calcium from endoplasmic reticulum (26, 27), it raises the possibility that GAPDH interaction with APE1 could be the result of a “sticky” nature. As such, we checked whether GAPDH could be coimmunoprecipitated with a subunit COX IV of the membrane-associated cytochrome c oxidase complex of the mitochondria. Anti-COX IV antibodies did not coimmunoprecipitate GAPDH (Fig. 2E, lane 4), dismissing the possibility of a generalized sticky feature of this protein and underscoring a relevant physiological interaction with APE1, as reported recently for the GAPDH-p300/CBP interaction (11). In other controls, neither PNKP nor protein phosphatase 2A activator (with cytoplasmic and nuclear distribution) (24) was coimmunoprecipitated with GAPDH (data not shown). Thus, we conclude that the data from whole cell extracts is consistent with the in vitro observation with purified proteins that GAPDH interacts with APE1.
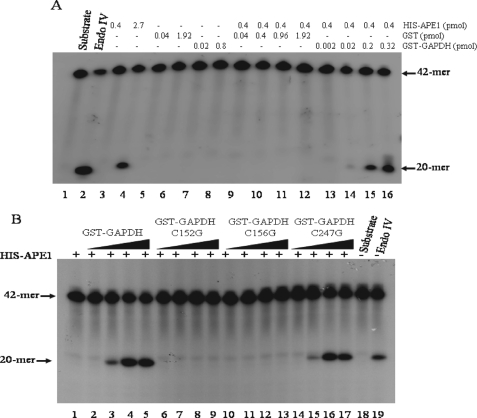
Native GAPDH, and Not the Variant C152G or C156G, Activates the AP Endonuclease Activity of APE1—It has been shown that APE1 can exist in the oxidized and reduced forms and that the reduced form is necessary to activate transcription factors including p53, NF-κB, and c-Jun and c-Fos, subunits of AP-1 (16, 18, 28). Moreover, thioredoxin is required to physically interact with APE1, and this association potentiates AP-1 transcriptional activity (29). We therefore explored whether GAPDH could function to activate the AP endonuclease activity of APE1. In this experiment, we first determined the minimal amount of purified His-APE1 that showed no detectable AP endonuclease activity. Purified His-APE1 in the range of 0–0.4 pmol showed no measurable AP endonuclease activity (Fig. 3A, lane 3), whereas the activity was detected if the protein was in excess of 2.7 pmol (Fig. 3A, lane 4). Interestingly, preincubation of a fixed amount (0.4 pmol) of His-APE1 with increasing amounts of GST-GAPDH resulted in the detection of the AP endonuclease activity (Fig. 3A, lanes 13–16). In control experiments, neither purified GST nor GST-PNKP activated the AP endonuclease activity of His-APE1 (lanes 9–12 and data not shown). Because the purified GST-GAPDH from E. coli is free of contaminating AP endonuclease activity (Fig. 3A, lanes 7 and 8), the above data therefore suggest that GAPDH may play a distinct role by reactivating an inactive form of the purified His-APE1.
FIGURE 3.
GAPDH, but not the variant C152G or C156G, stimulates APE1 AP endonuclease activity. A, increasing concentrations of either purified GST (lanes 9–12) or GST-GAPDH (lanes 13–16) were incubated with purified His-APE1 (lanes 9–16) for 5 min at room temperature before assaying for AP endonuclease activity. Shown are the controls, substrate (80 ng) alone (lane 1), and the following purified proteins Endo IV (0.15 pmol, lane 2), His-APE1 (lanes 3 and 4), GST (lanes 5 and 6), and GST-GAPDH (lanes 7 and 8). The arrows indicate positions of the 42-mer substrate and the 20-mer product. B, increasing amounts (0, 0.16, 0.64, and 0.8 pmol) of either GST-GAPDH (lanes 2–5), or the variants C152G (lanes 6–9), C156G (lanes 10–13), and C247G (lanes 14–17) were incubated with a fixed amount of purified His-APE1 (0.4 pmol each, lanes 1–17) assayed for AP endonuclease activity. Also shown are substrate alone (lane 18) and Endo IV (0.06 pmol, lane 19).
We next checked whether the cysteine variants would interfere with the ability of GAPDH to activate APE1. The purified variants C152G and C156G showed nearly 60% reduction in the glycolytic activity, whereas C247G retained full parental GST-GAPDH activity (supplemental Fig. S1). Increasing amounts of either the variant C152G or C156G did not activate the AP endonuclease activity of His-APE1 (Fig. 3B, lanes 6–9 and 10–13), as compared with the native GST-GAPDH (Fig. 3B, lanes 2–5). Moreover, H2O2-oxidized GAPDH did not stimulate the AP endonuclease activity of His-APE1 (supplemental Fig. S4). Unlike C152G and C156G, the variant C247G possessed full ability to reactivate the AP endonuclease activity of His-APE1 (Fig. 3B, lanes 14–17). Because Cys152 is the active site cysteine that corresponds to the rabbit GAPDH Cys149, recently shown to play an essential role in oxidative stress-induced disulfide bridge formation and aggregation of the protein (30), we reasoned that the cysteines Cys152 and Cys156, but not Cys247, of human GAPDH may participate in a redox reaction to reduce the oxidized form of APE1.
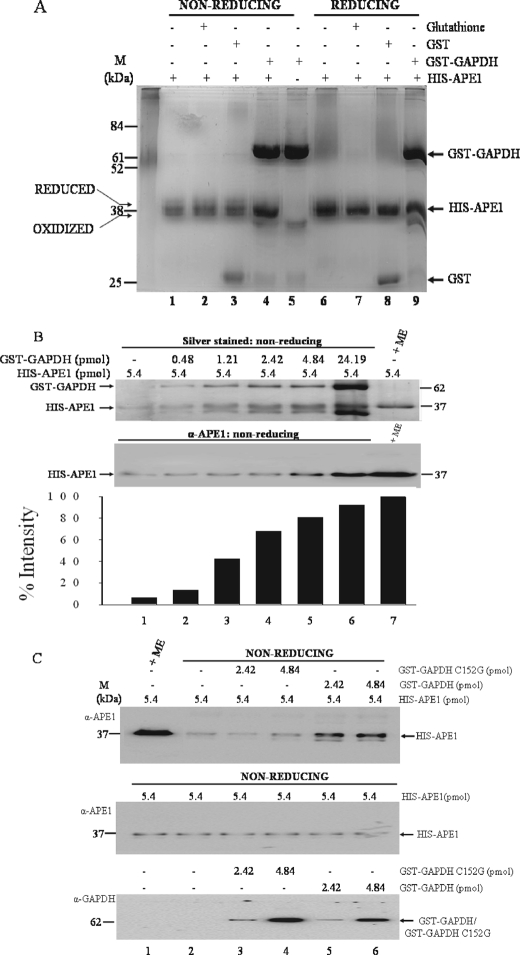
GAPDH, but Not C152G, Functions to Reduce the Oxidized Form of APE1—APE1 composed of oxidized and reduced forms can be visualized on nonreducing SDS-PAGE gels as a set of heterogeneous polypeptide species (29). We tested whether these species of APE1 are affected by GST-GAPDH. In this experiment, the purified His-APE1 was incubated with and without purified GST-GAPDH and examined for mobility changes under nonreducing conditions. When purified His-APE1 (8.11 pmol) was incubated with a 3-fold excess of GST-GAPDH, the heterogeneous species became more intensely stained with silver nitrate (Fig. 4A, lane 4). This phenomenon was not observed when His-APE1 was incubated with either glutathione or purified GST (Fig. 4A, lanes 2 and 3). Under reducing gel migration conditions, i.e. with 2-mercaptoethanol, the same amount of His-APE1 was intensely stained by silver nitrate and independently of GST-GAPDH (Fig. 4A, lane 6 versus lane 9). This observation strongly suggests that GAPDH might act to reduce APE1 leading to its enhanced staining with silver nitrate.
FIGURE 4.
GAPDH, but not the variant C152G, reduces APE1 and increases its detection by both silver staining and anti-APE1 antibodies. A, purified His-APE1 (8.11 pmol each for lanes 1–4 and 6–9) was incubated without any addition (lanes 1 and 6) or with either glutathione (10 mm, lanes 2 and 7), GST (3.84 pmol, lanes 3 and 8), or GST-GAPDH (6.45 pmol, lanes 4 and 9) for 5 min at room temperature. The samples were analyzed on 12% SDS-PAGE under nonreducing conditions (lanes 1–4) and reducing conditions with 2-mercaptoethanol (250 mm) (lanes 6–9) and stained with silver. Lane 5 contained only GST-GAPDH (6.45 pmol). The left arrows indicate the positions of the oxidized and reduced forms of His-APE1. M, molecular mass standards. B, increasing concentrations of purified GST-GAPDH (lanes 1–6) was incubated with fixed amount (5.41 pmol) of purified His-APE1 for 5 min at room temperature and analyzed by SDS-PAGE under nonreducing conditions. Lane 7, purified His-APE1 analyzed under reducing conditions with 2-mercaptoethanol (ME). The proteins were detected either by silver staining (top panel) or by Western blot (middle panel) using anti-APE1 polyclonal antibodies. The histogram (bottom panel, bars 1–6) depicts the quantitative increase silver staining intensity of His-APE1, relative to His-APE1 (bar 7) under reducing condition with 2-mercaptoethanol. C, top panel, fixed amount of purified His-APE1 (lanes 2–6) was incubated for 5 min at room temperature with either the purified variant C152G (lanes 3 and 4) or purified GST-GAPDH (lanes 5 and 6). The samples were analyzed by Western blot under nonreducing conditions for His-APE1 staining intensity with anti-APE1 polyclonal antibodies. Middle panel (loading controls), lanes 1–6 each contained 5.4 pmol of purified His-APE1 alone and analyzed by Western blot under nonreducing conditions. Bottom panel (loading controls), the variant C152G (lanes 3 and 4) and GST-GAPDH (lanes 5 and 6) analyzed by Western blot using anti-GST monoclonal antibody. His-APE1, GST-GAPDH, and GST-GAPDH C152G are indicated with arrows at 37 and 62 kDa.
In separate experiments, quantitative analysis revealed that at least 1 pmol of GAPDH was required to increase the silver staining intensity of 2.23 pmol of His-APE1 by 50% during 5 min of incubation at room temperature (Fig. 4B), indicating that GAPDH is not needed in excess to rapidly reduce His-APE1. Interestingly, the incubation of GST-GAPDH with His-APE1 also significantly enhanced its detection with the anti-APE1 polyclonal antibodies (Fig. 4B, lane 5 versus lane 1). For these experiments, 2-mercaptoethanol treatment was used for generating maximal reduction of APE to be detected by sliver staining and anti-APE1 antibody (Fig. 4B, lane 7, top and middle panels, respectively). Thus, it would appear that the GST-GAPDH-induced reduction of His-APE1 is associated with the protein undergoing structural alterations. Moreover, we note that the observation leading to the enhanced immunodetection of APE1 provided an additional tool to monitor the action of GAPDH on APE1.
We next examined whether the variant C152G could act on His-APE1 to enhance its detection by the anti-APE1 antibodies. As shown in Fig. 4C, the C152G variant lacked the ability to induce detection of a fixed amount of His-APE1 by the anti-APE1 antibodies, as compared with the same amount of the native GST-GAPDH (Fig. 4C, lanes 3 and 4 versus lanes 5 and 6). Similarly, the C156G variant also was defective in enhancing detection of His-APE1 by anti-APE1 antibodies, but not the C247G variant (data not shown). These data are consistent with the notion that the cysteine residues Cys152 and Cys156 of GAPDH might be required to catalyze the reduction of oxidized species of APE1, thereby causing a conformational change in the protein.
GAPDH Reactivates Chemically Oxidized APE1—We next assessed whether GST-GAPDH could reactivate the AP endonuclease activity of His-APE that was inactivated by pretreatment with the chemical oxidant H2O2. Following H2O2 treatment of His-APE1, the resulting His-APE1oxd (2.7 pmol) showed no detectable AP endonuclease activity when compared with the same amount of untreated His-APE1 (2.7 pmol) that exhibited a measurable level of AP endonuclease activity (Fig. 5, lane 6 versus lane 4). The conditions of H2O2 treatment did not completely inactivate the His-APE1oxd, because a 10-fold higher concentration of the protein (27 pmol) displayed a significant level of AP endonuclease activity (Fig. 5, lane 7 versus lane 6). When a fixed amount of the H2O2-inactivated His-APE1oxd (2.7 pmol) was incubated with increasing concentrations of GST-GAPDH for 5 min, the AP endonuclease activity was rapidly regenerated (Fig. 5, lanes 13–16). In contrast, increasing amounts of either N-acetylcysteine, glutathione, dithiothreitol, or purified GST did not reactivate the AP endonuclease activity of His-APE1oxd (supplemental Fig. S5 and data not shown), indicating that GAPDH performs a specific role in the reactivation of His-APE1oxd. In other control experiments, the addition of various metal ions including MgCl2 to the His-APE1oxd did not reactivate the AP endonuclease activity, excluding the possibility that GAPDH is acting as a donor of metal ions (supplemental Fig. S6). On the basis of these observations, we conclude that GAPDH is endowed with the ability to reduce the oxidized species of APE1, thereby maintaining the enzyme in its active state.
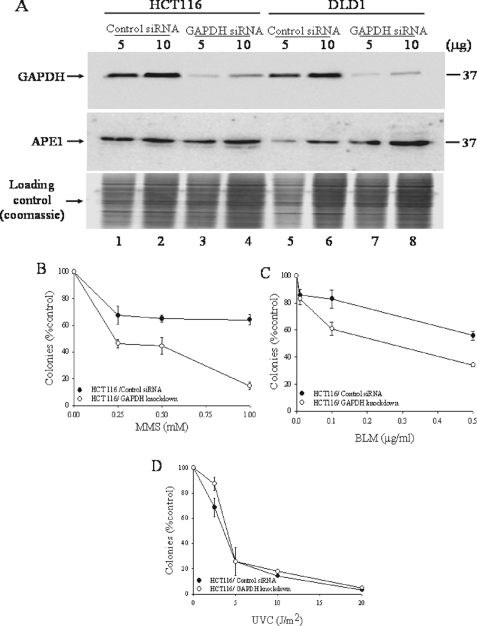
Cellular Sensitivity to a Subset of DNA-damaging Agents Is Dependent upon GAPDH Endogenous Levels—Because APE1 is an essential enzyme that plays a key role in processing several genotoxic DNA lesions such as AP sites, we reasoned that altering the endogenous level of GAPDH would interfere with the DNA repair functions of APE1. To assess this, we examined whether siRNA knockdown of GAPDH would sensitize cells to DNA-damaging agents that produce lesions repairable by APE1. When HCT116 cells were challenged with siRNA against GAPDH for 48 h, the level of the GAPDH protein (monitored by anti-GAPDH monoclonal antibody) present in total cell extract was sharply reduced by nearly 80%, as compared with the same amount of total cell extract derived from cells exposed to the scrambled control siRNA (Fig. 6A, upper panel, lanes 3 versus lane 1). Similar reduction in GAPDH level was also observed in the DLD1 cells challenged with the GAPDH siRNA (Fig. 6A, lanes 7 versus lane 5). Under this treatment condition, neither the control nor the siRNA against GAPDH altered the endogenous level of APE1 as determined by Western blot probed with anti-APE1 antibodies (Fig. 6A, middle panel, lanes 3 versus lane 1), suggesting that GAPDH does not influence the expression level of APE1. However, the GAPDH siRNA decreased the survival of a fraction (∼15–20%) of the cells, as compared with cells transfected with the control siRNA (data not shown). Interestingly, these GAPDH knockdown cells were hypersensitive to the alkylating agent MMS and the oxidant bleomycin (BLM), but not to 254-nm UV or cisplatin (Fig. 6, B–D, and supplemental Fig. S7). The GAPDH knockdown cells exhibited 55 and 40% decreased survival by colony assay when challenged with MMS (0.25 mm for 30 min) and BLM (0.1 μg/ml for 1 h), as compared with cells treated with the control siRNA which showed 30 and 15% decreased survival, respectively (Fig. 6, B and C). Thus, the sensitivity of the GAPDH knockdown cells to MMS and BLM could be related to an inability to process MMS-induced AP sites and BLM-induced DNA strand breaks with blocked 3′-termini. Both of these lesions are substrates for the APE1 DNA repair activities.
FIGURE 6.
GAPDH depletion influences survival of mammalian cells following exposure to genotoxic agents. A, cells were incubated with 75 nm of either control siRNA or GAPDH siRNA and total cell extracts analyzed for GAPDH and APE1 levels by Western blot analysis. B–D, control and siRNA-treated cells were challenged with increasing doses of either MMS (B), bleomycin (C), or UVC (D) and monitored for survival by clonogenic assay. The means and standard deviations of at least three independent experiments are shown.
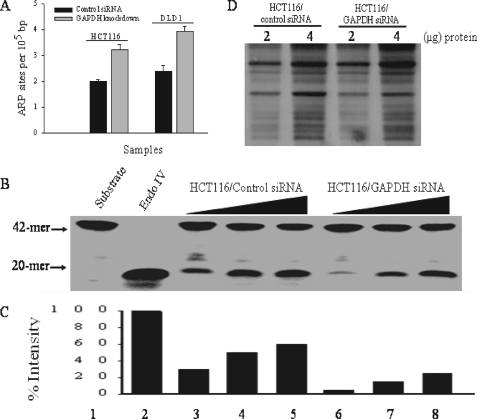
Accumulation of Spontaneous AP Sites in GAPDH Knockdown Cells Is Associated with Decreased AP Endonuclease Activity—If indeed GAPDH governs the DNA repair function of APE1, we anticipate that this would have a direct consequence on the repair of endogenous AP sites. To test this, we measured the spontaneous level of AP sites in the genomic DNA isolated from HCT116 and DLD1 cells treated with either the control siRNA or siRNA against GAPDH. Briefly, AP sites were quantified using an aldehyde-reactive probe that binds to AP sites followed by detection with biotin coupled streptavidin. As shown in Fig. 7A, GAPDH siRNA knockdown caused nearly a 1.5-fold increase in the level of spontaneous AP sites in the genomic DNA derived from either cell lines, as compared with the control siRNA treatment. Consistent with this observation, total extracts prepared from the GAPDH depleted cells showed a decrease in AP endonuclease activity, as compared with the same amount of protein extract derived from the control (Fig. 7, B–D). Because APE1 is the major enzyme responsible for processing AP sites in mammalian cells, we conclude that the accumulated lesions are a direct result of decreased APE1 activity in the GAPDH knockdown cells.
FIGURE 7.
Accumulation of AP sites is associated with decreased AP endonuclease activity in GAPDH-deficient cells. A, genomic DNA was isolated from the control and GAPDH siRNA transfected cells and analyzed for abasic (AP) site lesions using the aldehyde reactive probe assay. The bar graph shows the number of AP sites detected per 100,000 base pairs. The means and standard deviation of three independent experiments are shown. B, total cell extracts (250–750 ng, lanes 3–5 and 6–8) from the indicated cells were monitored for AP enoduclease activity using the 42-mer substrate (80 ng). Shown are the substrate (lane 1) and purified Endo IV (lane 2). C, quantification of the formation of the 20-mer product in B. D, silver-stained gel showing equal amount of total protein in the extracts used for the AP endonuclease assay.
DISCUSSION
In this study, we show for the first time that GAPDH interacts with a key DNA repair enzyme, APE1, which functions in the base excision repair pathway. We believe that a possible physiological relevance of the GAPDH-APE1 interaction is to promote the reactivation of oxidized species of APE1 that occur during normal aerobic metabolism. This role seems logical from several standpoints: GAPDH (i) possesses a redox active cysteine in its catalytic center, (ii) translocates from the cytoplasm to the nucleus in response to stress causing oxidative damage to the DNA, and (iii) binds to damaged DNA. Moreover, siRNA knockdown of GAPDH (i) sensitizes cells to agents that produce genotoxic lesions that are processed by APE1 and, more importantly, (ii) engenders relatively higher levels of spontaneous AP site formation (5, 10). This latter finding correlates with the GAPDH knockdown cells displaying an inability to proliferate, as well as exhibiting a profound defect in cell cycle progression, largely arresting in the G1 phase,4 where these AP site lesions would need to be repaired (31). Because AP sites are mutagenic, we postulate that GAPDH could serve an essential role in protecting cells against genomic instability that may arise as a result of oxidative stress by maintaining APE1 in its reduced state. Alternatively, GAPDH may function to prevent cell death by ensuring the efficient repair of spontaneous AP sites, which if left unprocessed are also known to trigger cell death (14). Furthermore, the observation that GAPDH knockdown cells exhibit an ∼15–20% reduction in viability in the absence of exogenous treatment may be accounted for by the concomitant 2-fold increase in spontaneous AP site formation. In this respect it remains to be verified whether the diminished survival of GAPDH knockdown cells can be avoided by ectopic expression of a redox-insensitive AP endonuclease such as the yeast Apn1 (14).
The mechanism by which GAPDH executes its redox function could involve the active site cysteine of GAPDH, because C152G, but not C247G, was incapable of reactivating the AP endonuclease activity of oxidized APE1. In a similar manner, the GAPDH variant C156G lacks the ability to reactivate the AP endonuclease activity of oxidized APE1, suggesting that Cys156 also might be involved in the mechanism by which GAPDH reduces APE1. The specific role performed by Cys152 and Cys156 in reducing oxidized APE1 warrants further study, because the functions of these residues cannot be substituted by the tripeptide glutathione. In our studies, we did not monitor the fate of GAPDH following reduction of oxidized APE1, although it is likely that the redox state of the former protein in turn is altered. Consistent with this notion, a very recent study documented that the redox active site cysteine (Cys149) of rabbit GAPDH (corresponding to Cys152 of human) can be oxidized when cells are challenged with nitric oxide or hydroxyl radical donors to trigger Cys149–Cys149 intermolecular disulfide bond formation leading to aggregation of the molecule (30, 32). The GAPDH aggregates display amyloid fibril-like structures, which upon accumulation serve to trigger oxidative stress-induced cell death (30, 32). These aggregates are barely detectable in the rabbit GAPDH variant C149S following exposure to oxidative stress, clearly implying the essential role of this residue in the aggregation process. It is noteworthy that the nearby Cys153 (corresponding to Cys156 of human) does not appear to perform a similar role in forming GAPDH aggregates (30, 32). It is possible that GAPDH could form aggregates following incubation with oxidized APE1, although this reaction could be limited by several factors.
To date, GAPDH is known to interact with a myriad of proteins with nuclear function. For example, GAPDH is one of seven polypeptides that constitute the OCA-S complex, a coactivator of the DNA binding transcription factor OCT1 (8). GAPDH interacts directly with the POU transactivation domain of OCT1 to play an essential role during the S phase-dependent transcription of histone H2B. Moreover, the interaction between GAPDH and OCT1 is stimulated by NAD+ but inhibited by NADH, suggesting that GAPDH serves a redox sensing role in the H2B gene transcription (8). A more recent study documented that in response to nitric oxide, rat GAPDH becomes S-nitrosylated at the catalytic cysteine (Cys150, corresponding to Cys152 in human) causing the modified GAPDH to bind and stabilize the ubiquitin-protein isopeptide ligase SIAH1 (33). The GAPDH-SIAH1 complex translocates to the nucleus and degrades SIAH1 substrates. It is not clear how GAPDH stabilizes SIAH1, but it is noteworthy that SIAH1 is a cysteine-rich protein. Whether GAPDH uses Cys156 to reduce SIAH1 and protect it from structural deformation caused by oxidation remains to be investigated.
APE1 has an active site cysteine residue (Cys65) that is required for the regulation of c-Jun protein, which possesses a redox sensitive cysteine (Cys252) that can be readily oxidized leading to the inhibition of its DNA binding ability (16, 34). It has been proposed that oxidized APE1 contains a disulfide bridge between Cys65 and Cys93, thereby preventing Cys65 from participating in the redox reactivation of the DNA binding activity of oxidized Jun. Because the APE1 variant C65A displayed the equivalent level of AP endonuclease activity as the native enzyme, it is unlikely that GAPDH exerts a redox function on Cys65 (28). In fact, oxidation of Cys310 has been shown to interfere with APE1 repair activities because it is located adjacent to His309, which is required for catalytic function of the enzyme (28). Because APE1 contains five additional cysteine residues (Cys93, Cys99, Cys138, Cys208, and Cys296), we reason that Cys310 may form various intramolecular disulfide bridges with any of these including Cys65 to generate a population of heterogeneous oxidized molecules compromised for AP endonuclease activity. Thus, creating cysteine to alanine variants of APE1 is likely to reveal the key cysteine(s) that participate in the disulfide bridge formation leading to aberrant molecules with altered AP endonuclease activity. Such APE1 variants are expected to resist oxidation and remain fully active as an AP endonuclease independently of GAPDH (28). Nonetheless, it is clear that oxidized species of APE1 that are reactivated by GAPDH possess distinct physical properties from the native enzyme with respect to mobility on SDS-PAGE and lack of requirement for the metal ion Mg2+. Whether oxidized forms of APE1 bind more tightly to Mg2+ or lose the metal ion requirement warrants further studies (35). This is especially important because it is well established that the substrate specificity and protein conformation of APE1 can be modulated by the concentration of Mg2+ (36–38).
In addition to GAPDH, thioredoxin (TRX), a pleiotropic cellular factor possessing thiol-mediated redox activity, can also interact with APE1 (29). Like GAPDH, TRX can translocate to the nucleus upon phorbol 12-myristate 13 acetate treatment and associates with APE1 to promote activation of the AP-1 transcriptional factor (29). This activation process requires the cysteine residues in the catalytic center of TRX. However, it is not known whether TRX specifically reduces the redox active cysteine (Cys65) of APE1 that is required to maintain AP-1 in the reduced state. Herein, we did not examine whether GAPDH association with APE1 might potentiate the DNA binding activity of c-Jun, although it remains possible that TRX and GAPDH may perform distinct redox functions on oxidized APE1.
In short, we postulate that GAPDH may serve to maintain the redox state of numerous proteins that are susceptible to oxidative stress-induced structural changes. Perhaps this might explain the diverse activities of GAPDH, unrelated to glycolysis, including its implication in some oxidative stress related neurodegenerative diseases such as Alzheimer and Parkinson diseases (39). Our key observation that GAPDH is responsible for preserving the activity of APE1 may suggest that one of the essential roles of the former protein is to maintain genetic stability.
Supplementary Material
Acknowledgments
We thank Drs. E. Drobetsky, J. Filep, and E. B. Affar for helpful comments.
This work was supported partially by Natural Sciences and Engineering Research Council of Canada Grant 202432-01 and Canadian Institute of Health Research Grant MOP-13152 (to D. R.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S7.
Footnotes
The abbreviations used are: GAPDH, glyceraldehyde-3-phosphate dehydrogenase; siRNA, small interfering RNA; AP, apurinic/apyrimidinic; MMS, methyl methane sulfonate; GST, glutathione S-transferase; BLM, bleomycin; TRX, thioredoxin; PNKP, polynucleotide kinase 3′-phosphatase.
N. Jouvet and D. Ramotar, manuscript in preparation.
References
- 1.Tatton, W. G., Chalmers-Redman, R. M., Elstner, M., Leesch, W., Jagodzinski, F. B., Stupak, D. P., Sugrue, M. M., and Tatton, N. A. (2000) J. Neural Transm. Suppl. 77-100 [DOI] [PubMed]
- 2.Brodie, A. E., and Reed, D. J. (1990) Arch. Biochem. Biophys. 276 212-218 [DOI] [PubMed] [Google Scholar]
- 3.Cumming, R. C., Andon, N. L., Haynes, P. A., Park, M., Fischer, W. H., and Schubert, D. (2004) J. Biol. Chem. 279 21749-21758 [DOI] [PubMed] [Google Scholar]
- 4.Schulze, H., Schuler, A., Stuber, D., Dobeli, H., Langen, H., and Huber, G. (1993) J. Neurochem. 60 1915-1922 [DOI] [PubMed] [Google Scholar]
- 5.Sirover, M. A. (1999) Biochim. Biophys. Acta 1432 159-184 [DOI] [PubMed] [Google Scholar]
- 6.Singh, R., and Green, M. R. (1993) Science 259 365-368 [DOI] [PubMed] [Google Scholar]
- 7.Saunders, P. A., Chen, R. W., and Chuang, D. M. (1999) J. Neurochem. 72 925-932 [DOI] [PubMed] [Google Scholar]
- 8.Zheng, L., Roeder, R. G., and Luo, Y. (2003) Cell 114 255-266 [DOI] [PubMed] [Google Scholar]
- 9.Meyer-Siegler, K., Mauro, D. J., Seal, G., Wurzer, J., deRiel, J. K., and Sirover, M. A. (1991) Proc. Natl. Acad. Sci. U. S. A. 88 8460-8464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dastoor, Z., and Dreyer, J. L. (2001) J. Cell Sci. 114 1643-1653 [DOI] [PubMed] [Google Scholar]
- 11.Sen, N., Hara, M. R., Kornberg, M. D., Cascio, M. B., Bae, B. I., Shahani, N., Thomas, B., Dawson, T. M., Dawson, V. L., Snyder, S. H., and Sawa, A. (2008) Nat. Cell Biol. 10 866-873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krynetski, E. Y., Krynetskaia, N. F., Gallo, A. E., Murti, K. G., and Evans, W. E. (2001) Mol. Pharmacol. 59 367-374 [DOI] [PubMed] [Google Scholar]
- 13.Xing, C., LaPorte, J. R., Barbay, J. K., and Myers, A. G. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 5862-5866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fung, H., and Demple, B. (2005) Mol. Cell 17 463-470 [DOI] [PubMed] [Google Scholar]
- 15.Xanthoudakis, S., and Curran, T. (1992) EMBO J. 11 653-665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walker, L. J., Robson, C. N., Black, E., Gillespie, D., and Hickson, I. D. (1993) Mol. Cell Biol. 13 5370-5376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jayaraman, L., Murthy, K. G., Zhu, C., Curran, T., Xanthoudakis, S., and Prives, C. (1997) Genes Dev. 11 558-570 [DOI] [PubMed] [Google Scholar]
- 18.Xanthoudakis, S., Miao, G., Wang, F., Pan, Y. C., and Curran, T. (1992) EMBO J. 11 3323-3335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Demple, B., Herman, T., and Chen, D. S. (1991) Proc. Natl. Acad. Sci. U. S. A. 88 11450-11454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walker, L. J., Craig, R. B., Harris, A. L., and Hickson, I. D. (1994) Nucleic Acids Res. 22 4884-4889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brown, V. M., Krynetski, E. Y., Krynetskaia, N. F., Grieger, D., Mukatira, S. T., Murti, K. G., Slaughter, C. A., Park, H. W., and Evans, W. E. (2004) J. Biol. Chem. 279 5984-5992 [DOI] [PubMed] [Google Scholar]
- 22.Shatilla, A., and Ramotar, D. (2002) Biochem. J. 365 547-553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jilani, A., Vongsamphanh, R., Leduc, A., Gros, L., Saparbaev, M., and Ramotar, D. (2003) Biochemistry 42 6436-6445 [DOI] [PubMed] [Google Scholar]
- 24.Azam, S., Drobetsky, E., and Ramotar, D. (2007) Apoptosis 12 1243-1255 [DOI] [PubMed] [Google Scholar]
- 25.Mathonnet, G., Leger, C., Desnoyers, J., Drouin, R., Therrien, J. P., and Drobetsky, E. A. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 7219-7224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patterson, R. L., van Rossum, D. B., Kaplin, A. I., Barrow, R. K., and Snyder, S. H. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 1357-1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tarze, A., Deniaud, A., Le Bras, M., Maillier, E., Molle, D., Larochette, N., Zamzami, N., Jan, G., Kroemer, G., and Brenner, C. (2007) Oncogene 26 2606-2620 [DOI] [PubMed] [Google Scholar]
- 28.Kelley, M. R., and Parsons, S. H. (2001) Antioxid. Redox. Signal. 3 671-683 [DOI] [PubMed] [Google Scholar]
- 29.Hirota, K., Matsui, M., Iwata, S., Nishiyama, A., Mori, K., and Yodoi, J. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 3633-3638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakajima, H., Amano, W., Fujita, A., Fukuhara, A., Azuma, Y. T., Hata, F., Inui, T., and Takeuchi, T. (2007) J. Biol. Chem. 282 26562-26574 [DOI] [PubMed] [Google Scholar]
- 31.Branzei, D., and Foiani, M. (2008) Nat. Rev. Mol. Cell Biol. 9 297-308 [DOI] [PubMed] [Google Scholar]
- 32.Jenkins, J. L., and Tanner, J. J. (2006) Acta Crystallogr. Sect. D Biol. Crystallogr. 62 290-301 [DOI] [PubMed] [Google Scholar]
- 33.Hara, M. R., Agrawal, N., Kim, S. F., Cascio, M. B., Fujimuro, M., Ozeki, Y., Takahashi, M., Cheah, J. H., Tankou, S. K., Hester, L. D., Ferris, C. D., Hayward, S. D., Snyder, S. H., and Sawa, A. (2005) Nat. Cell Biol. 7 665-674 [DOI] [PubMed] [Google Scholar]
- 34.Georgiadis, M. M., Luo, M., Gaur, R. K., Delaplane, S., Li, X., and Kelley, M. R. (2008) Mutat. Res., in press [DOI] [PMC free article] [PubMed]
- 35.Hess, D. T., Matsumoto, A., Kim, S. O., Marshall, H. E., and Stamler, J. S. (2005) Nat. Rev. Mol. Cell Biol. 6 150-166 [DOI] [PubMed] [Google Scholar]
- 36.Strauss, P. R., Beard, W. A., Patterson, T. A., and Wilson, S. H. (1997) J. Biol. Chem. 272 1302-1307 [DOI] [PubMed] [Google Scholar]
- 37.Erzberger, J. P., and Wilson, D. M., 3rd. (1999) J. Mol. Biol. 290 447-457 [DOI] [PubMed] [Google Scholar]
- 38.Gros, L., Ishchenko, A. A., Ide, H., Elder, R. H., and Saparbaev, M. K. (2004) Nucleic Acids Res. 32 73-81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cumming, R. C., and Schubert, D. (2005) FASEB J. 19 2060-2062 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.