Abstract
The mapping of the photosynthetic membrane of Rhodobacter sphaeroides by atomic force microscopy (AFM) revealed a unique organization of arrays of dimeric reaction center-light harvesting I-PufX (RC-LH1-PufX) core complexes surrounded and interconnected by light-harvesting LH2 complexes (Bahatyrova, S., Frese, R. N., Siebert, C. A., Olsen, J. D., van der Werf, K. O., van Grondelle, R., Niederman, R. A., Bullough, P. A., Otto, C., and Hunter, C. N. (2004) Nature 430, 1058–1062). However, membrane regions consisting solely of LH2 complexes were under-represented in these images because these small, highly curved areas of membrane rendered them difficult to image even using gentle tapping mode AFM and impossible with contact mode AFM. We report AFM imaging of membranes prepared from a mutant of R. sphaeroides, DPF2G, that synthesizes only the LH2 complexes, which assembles spherical intracytoplasmic membrane vesicles of ∼53 nm diameter in vivo. By opening these vesicles and adsorbing them onto mica to form small, ≤120 nm, largely flat sheets we have been able to visualize the organization of these LH2-only membranes for the first time. The transition from highly curved vesicle to the planar sheet is accompanied by a change in the packing of the LH2 complexes such that approximately half of the complexes are raised off the mica surface by ∼1 nm relative to the rest. This vertical displacement produces a very regular corrugated appearance of the planar membrane sheets. Analysis of the topographs was used to measure the distances and angles between the complexes. These data are used to model the organization of LH2 complexes in the original, curved membrane. The implications of this architecture for the light harvesting function and diffusion of quinones in native membranes of R. sphaeroides are discussed.
The biological membrane at its simplest is a lipid bilayer that divides cellular contents from the exterior medium. Yet the bilayer performs more than a simple barrier function as it plays host to a wide variety of integral membrane proteins that perform transport, sensing, motility, biosynthesis, energy generation (in the form of ATP), as well as energy harvesting in the case of phototrophic organisms. We have structural information about the membrane proteins involved in photosynthesis, e.g. the bacterial reaction center (1), the peripheral light-harvesting complex (2), and the photosystem I (3) and photosystem II (4) supercomplexes and in transport (5), sensing (6), and electron transport (7). However, there is a need now to gain information upon the organization of these proteins within the membrane in vivo. The use of AFM2 to directly visualize membrane proteins in situ allows us to make precise measurements of the disposition of any protein within the membrane (8) under nearly native conditions. We have demonstrated that the comparatively disordered intracytoplasmic membrane from the model photosynthetic bacterium Rhodobacter sphaeroides can be successfully imaged by AFM (9) to reveal the hidden architecture that nature has developed to harvest, transfer, and finally utilize light energy with great efficiency. Scheuring and co-workers have also produced high resolution images of the photosynthetic membranes of Blastochloris viridis (10), Rhodospirillum photometricum (11), Rhodobacter blasticus (12), and Phaeospirillum molischianum (13), allowing a comparison of the differing strategies that purple bacteria have evolved for the harvesting and utilization of light energy (14).
It has been observed by Scheuring and Sturgis (15) that the presence of RC-LH1 core complexes affects the packing of the peripheral, LH2, complexes in native membranes of R. photometricum. The availability of a wide array of genetic tools for R. sphaeroides (16) can be exploited for AFM studies of membrane organization, because it allows control to be exerted over the composition of the proteins present in the membrane. Mutants of R. sphaeroides, which form highly ordered tubular membranes containing only RC-LH1-PufX dimers, have been used to study the organization of complexes by electron microscopy (17, 18), and similar techniques have been applied to photosynthetic proteins reconstituted into two-dimensional crystals (19–21). However, because the purification process removes the native lipids, and, in the case of R. sphaeroides, reconstitutes the complexes in an alternating, and therefore nonbiological, “up-down-up” topology, such work is unsuitable for examining the native organization of the complexes.
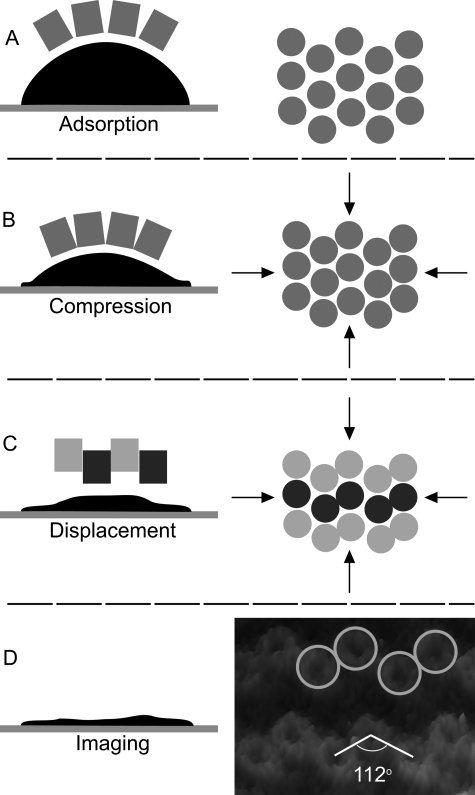
In this paper we present data on membranes of a R. sphaeroides deletion strain that contain only the peripheral light-harvesting complex, LH2, thus eliminating the core complex and its effect upon long range membrane protein packing in the membrane. This simple system can be used as a model for studying the packing of integral membrane proteins in a curved membrane. The R. sphaeroides WT photosynthetic membrane is curved into spherical intracytoplasmic vesicles by the LH2 and RC-LH1-PufX complexes embedded within it (22, 23). It has not been possible to image the LH complexes in such highly curved, ∼60-nm diameter (24), flexible membranes, and limited exposure to a mild detergent is utilized to generate opened vesicles that flatten when they are adsorbed onto mica for AFM imaging (9). The lateral dimensions of these patches vary from 50 to 120 nm and present a challenge to image at high resolution; early attempts to use contact mode AFM resulted in displacement of the patches from the mica. The use of tapping mode AFM at low amplitudes has allowed us to image large numbers of patches to locate examples that are well adsorbed and sufficiently flat for collection of high resolution data. The adsorption process leads to the regular vertical displacement of the LH2 molecules within the membrane patches such that they form a series of “zigzag” corrugations. Quantification of these displacements by AFM reveals that the corrugations in the newly flattened membrane arise from the vertical displacement of ∼1 nm of the LH2 complexes in the upper versus the lower lines. An intriguing observation is that within these zigzag corrugations pairs of LH2 complexes are apparent, suggesting a close association possibly involving maximal protein-to-protein contacts at the mutual interface. The average center-to-center distance between LH2 complexes in the membrane patches is 8.5 nm, and the average angle subtended by three complexes is 112°, consistent with a hexagonal organization.
Recently there has been a substantive effort to construct detailed structural models of the intracytoplasmic membrane vesicles of R. sphaeroides as prototypes of membrane protein organization, both for the wild type membrane (25) and for an LH2-minus mutant that assembles tubular membranes formed from helical arrays of RC-LH1-PufX core dimers (23). The AFM data in the present work show that a typical flattened membrane patch is likely to be the product of a single intracytoplasmic vesicle of between 47 and 60 nm in diameter, containing between 100 and 160 LH2 complexes. From these data we have constructed a model of the organization of the LH2 complexes within the native curved membrane that is consistent with the known excitation energy transfer times and that could have implications for quinone diffusion through the native membrane.
EXPERIMENTAL PROCEDURES
The R. sphaeroides deletion strain DPF2G (16) was grown and the membranes were prepared according to the methods in Olsen et al. (26) and further treated with the detergent β-DDM (Glycon Biochemistry, GmbH Biotechnology, Germany), at 0.005 and 0.01% (w/v) concentration, following the protocol of Bahatyrova et al. (9). The membrane patches were adsorbed onto freshly cleaved mica (Agar Scientific) in 20 mm HEPES, pH 7.5, 150 mm KCl, 25 mm MgCl2, 0.5 mm NiCl2 for an hour at room temperature. The sample was then washed twice with 20 mm HEPES, pH 7.5, 100 mm KCl recording buffer. Standard Olympus TR800PSA SiN cantilevers (Atomic Force GmbH, Mannheim, Germany), spring constant 0.15 n/m were used in a standard tapping mode liquid cell at operating frequencies of ∼8.5 KHz, using a Nanoscope IV AFM and E scanner (VEECO). The images were recorded at scan frequencies of 0.5–1 Hz.
The images were processed using the software supplied with the microscope (VEECO) and SPIP (Image Metrology). All of the distances, angles, and areas were measured using ImageJ (open source software). The model vesicle was prepared using Chimera (27) (open source software).
RESULTS
General Vesicle Morphology—Previous work to obtain high resolution AFM images of native membranes of R. sphaeroides used the mild detergent β-DDM; however, for the membranes of the deletion strain DPF2G, we attempted to record data on nondetergent-treated membranes as well as those prepared by existing methodologies. We observed that areas of empty lipid, designated on the basis of their height, were present in both the nondetergent-prepared membrane patches and those at 0.005 and 0.01% β-DDM. In the nondetergent preparations the empty lipid patches can be seen occurring randomly in the images and occasionally appear to be associated with vesicles. Qualitatively there did not seem to be much difference in the occurrence of regions of empty lipid in the 0.005 and 0.01% samples (see supplemental data).
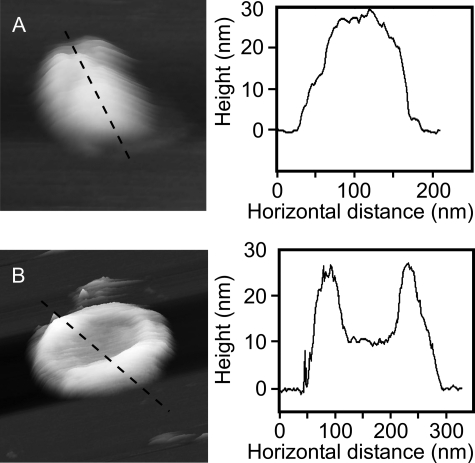
The intact spherical vesicles generated by disruption in a French pressure cell, but not treated with β-DDM, do not adhere strongly to the mica surface because of the limited surface-to-surface contact zone. However, using low tapping amplitudes and slow scan rates, we have been able to gain an estimate of the size range of membrane vesicles and record images showing a limited amount of surface detail (Fig. 1). The DPF2G vesicles collapse onto the mica surface and flatten to a greater extent than the WT vesicles (32.1 ± 8.4 nm and 37.5 ± 4.1 nm high, respectively). An extreme example of this tendency to deform is shown in Fig. 1B, which illustrates a collapsed vesicle with a deep central depression. At its lowest point this vesicle is only ∼10 nm thick, which would equate to the height of two closely appressed lipid bilayers containing LH2 complexes. This level of deformation, which was not observed for WT vesicles, indicates that the LH2-only membranes are more flexible than WT membranes comprising both the RC-LH1-PufX and LH2 complexes.
FIGURE 1.
Examples of chromatophores adsorbed to mica. A, three-dimensional representation of a WT vesicle on mica with the section across it marked by the dashed line. The vesicle is not perfectly smooth; local depressions may be the result of areas containing only LH2 complexes. B, three-dimensional representation of a DPF2G, LH2-only vesicle on mica with the section across it marked by the dashed line. This vesicle shows the extreme central depression that some vesicles exhibited; the center has collapsed so that the two bilayers are likely to be in contact with each other as evidenced by the ∼10-nm height above the mica of this part of the section.
The most gentle detergent treatment, 0.005% β-DDM, did not result in any patches of membrane that could be imaged to high resolution; moreover, the presence of large intact vesicles suggests that there was insufficient detergent present to open them out. In consequence only images of the 0.01% β-DDM-treated samples are shown in this work. It should be noted that although the vesicles were treated with 0.01% β-DDM, the final concentration of detergent was 0.001% or lower because the samples were always diluted with nondetergent buffer by a minimum of 1:10 prior to adsorption onto the mica surface.
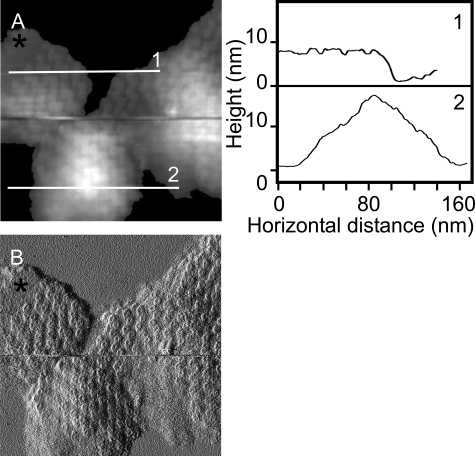
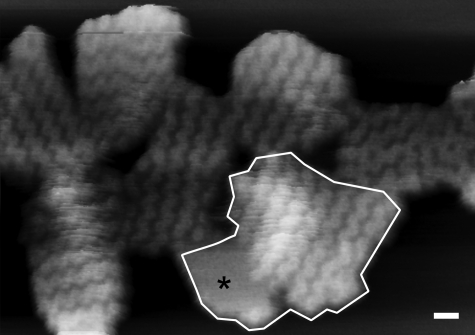
Composition and Morphology of Membrane Patches Containing Only LH2 Complexes—The first noteworthy feature of the AFM images of the membrane patches is that not all of them lie completely flat upon the mica surface, and some display significant curvature, similar to the native nearly spherical condition (Fig. 2). Sections 1 and 2 in Fig. 2 illustrate the varying degrees of membrane patch curvature; section 1 is taken across a flat piece of membrane, whereas section 2 is across a highly curved patch that causes the AFM difficulty in tracking the surface topology. The flat patch in the image was chosen as a possible flattened vesicle. The program ImageJ (open source software) was used to measure the total area within the selected patch; this was found to be 9045 nm2, which equates to a vesicle diameter of 54 nm, in good agreement with the dimensions of WT intracytoplasmic membrane vesicles (also known as chromatophores) of 64 ± 16 nm, measured by dynamic light scattering (28). In other images where the membrane patches have also effectively flattened out onto the mica, it is possible to estimate the number of vertically translated LH2 complexes and thus assess the number in the original vesicle. A total of nine patches was examined where a reasonable estimate of the number of LH2 complexes could be made. In Fig. 3, each patch is distinguished from the others by the orientation of the zigzag lines of LH2 complexes; one example is outlined. For each patch the total area was measured, and the area that could be accounted for solely by LH2 complexes was estimated, based upon a diameter of 8 nm for the LH2 complex from Rhodopseudomonas acidophila (2). The data, summarized in Table 1, show that the average vesicle surface area was 9065 nm2, corresponding to an average vesicle diameter of 53 nm, of which only 6729 nm2 could be accounted for by the 134 LH2 complexes present. The difference between these two figures is ∼26% of the total surface area of the vesicle and is likely to represent the lipid surface area, not including the lipid sequestered inside the LH2 rings.
FIGURE 2.
Height and amplitude AFM topographs of flattened membrane patches. A, a height image of three membrane pieces. Section 1 shows that the upper left patch is essentially flat and well adsorbed to the mica with the contours of LH2 complexes defined in the trace, whereas section 2 across the lower patch has significantly more vertical topology yet still demonstrates the zigzag lines of complexes, which are clear in the amplitude image (B). A region of empty lipid is denoted by the black asterisk. Full gray scale, 20 nm. B, the amplitude image of the same region showing that even the highly curved membrane patch still has the regular zigzag lines of LH2 complexes traversing it.
FIGURE 3.
A group of flattened membrane patches showing regular zigzag lines of LH2 complexes. Three-dimensional representation of a group of membrane pieces all showing the differing orientations of the highly regular zigzag lines that were used to distinguish between patches when demarking possible flattened vesicles. One putative flattened vesicle is outlined. This patch also includes an area of lipid largely free of any complexes, marked by the black asterisk; the origin of these large zones of empty lipid is not known. Scale bar, 20 nm; full gray scale, 15 nm.
TABLE 1.
Average dimensions and LH2 complex content of the patches measured Nine patches of membrane were measured, and the number of LH2 complexes was estimated. The area covered by the LH2 complexes was calculated and compared with the actual area of the patch, the difference being assigned to lipid. Assuming that the original vesicle was a sphere, the diameter was calculated from the area of the patches.
| Estimated number of LH2 complexes per patch | Measured area of patch | Area estimated to be covered by LH2 complexes | Difference (area covered by lipid) | Calculated vesicle diameter |
|---|---|---|---|---|
| nm2 | nm2 | nm2 | Nm | |
| 134 ± 29 | 9065 ± 1988 | 6729 ± 1475 | 2336 ± 1081 | 53 ± 6 |
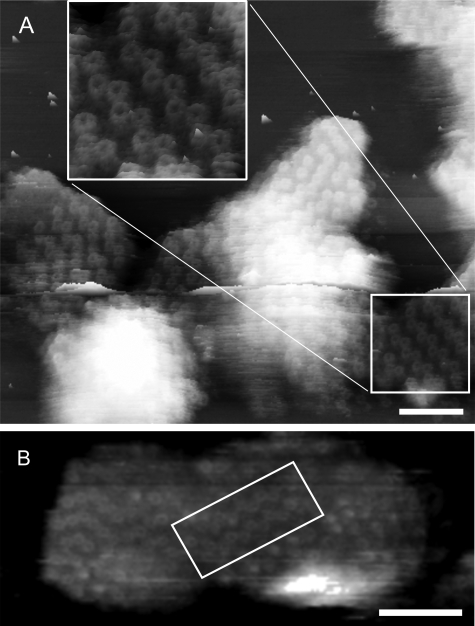
Membrane Reorganization upon Adsorption to Mica—Examination of the membrane patches that show well defined LH2 complexes reveals that the LH2 molecules in the zigzag lines are apparently paired off in “dimers” (Fig. 4A and inset). This visual assignment of dimers was tested by measuring the center-to-center distance of a representative pool of complexes in two different membrane pieces; the results are summarized in Table 2. These data show that LH2 complexes within a pair are closer together than they are to adjacent complexes, having center-to-center distances of 7.7 ± 0.5 nm (n = 22) and 9.2 ± 0.4 nm (n = 23), respectively. Based upon the R. acidophila LH2 structure, the shorter of these two distances can be accommodated if the two complexes pack closely together. It seems likely that in these adsorbed membranes there is protein-to-protein contact between the LH2 complexes in a dimer but that there is lipid between the adjacent LH2 complexes between dimers. The average angle subtended by the dimers in the zigzags is 112.1 ± 7.0° (n = 20) (Fig. 5D), which is consistent with a hexagonal packing, rather than a cubic packing arrangement, which has been observed in reconstituted two-dimensional crystals of LH2 (19, 21, 29, 30). An additional feature of the paired LH2 complexes is that they are tilted with respect to each other in a variety of orientations, which is likely to reflect steric imperatives in the interfacial packing zone.
FIGURE 4.
High resolution AFM topographs showing highly ordered zigzag lines of LH2 complexes. A, three-dimensional representation showing the region of well ordered and defined LH2 complexes used for the measurement of center-to-center distances and the angle subtended by the dimeric complexes. The inset is an enlargement of the area highlighted by the box. The image also shows an area of more disordered LH2 complexes that do not form the regular zigzag lines seen in the majority of the flattened membrane patches. Scale bar, 50 nm; full gray scale, 20 nm. B, three-dimensional representation of a piece of membrane from WT R. sphaeroides, containing RC-LH1-PufX core complexes and LH2 complexes, isolated, and imaged after purely mechanical disruption of the cell in the absence of detergent. The area outlined by the box clearly shows the same zigzag lines of LH2 complexes seen in the DPF2G, LH2-only membrane patches. This is clear evidence that it is the adsorption process that generates the zigzag lines of LH2 complexes. Scale bar, 50 nm; full gray scale, 22 nm.
TABLE 2.
Comparison of the topological data for WT and LH2-only patches High resolution images of flattened patches yielded spatial data on the disposition of the LH2 complexes in the flattened membranes from which it was clear that the complexes formed zigzag rows at two different heights and that in the rows, LH2 complexes paired off in a regular manner. NA, not applicable.
| Intradimer distance center to center | Interdimer distance center to center | Average distance center to center | Vertical displacement | Angle between LH2 dimers | |
|---|---|---|---|---|---|
| nm | nm | nm | nm | ||
| DPF2G with 0.01% β-DDM | 7.7 ± 0.5 (n = 22) | 9.2 ± 0.4 (n = 23) | 8.4 ± 0.9 (n = 45) | 1.2 ± 0.4 (n = 14) | 112.1 ± 7.0° |
| WT with 0.01% β-DDM | 8.0 ± 0.3 (n = 8) | 9.3 ± 0.5 (n = 12) | 8.8 ± 0.8 (n = 20) | 1.2 ± 0.3 (n = 15) | 128.3 ± 6.8° |
| WT with no β-DDM | NA | NA | 7.8 ± 0.5 (n = 11) | NA | 119.4 ± 8.7° |
FIGURE 5.
A schematic representation of the reorganization of a curved piece of membrane as it adsorbs to mica. The LH2 complex is depicted as a smooth cylinder for the sake of clarity, but it is widely accepted that the R. sphaeroides LH2 causes membrane curvature because of its shape. This may be due to an effectively conical profile of the complex or the complex plus bound lipids having a conical shape. A, initial adsorption can only take place at the rim of the membrane; this binding effectively sets the maximum horizontal surface area that the membrane can occupy once flattened. B, more of the membrane adsorbs onto the surface, thus increasing the pressure within the remainder, which squeezes the lipids from between the LH2 complexes allowing limited contact of the complexes. In a hexagonally packed organization the 9-fold symmetric LH2 complexes will preferentially “contact” two of their six neighbors; ramification of this selective contact across the membrane patch causes the formation of the zigzag lines. Further compression of the membrane pushes the rows up and down to pack them more closely together. C, as the hydrophobic core becomes partially exposed by the vertical displacement the LH2 complexes form dimers in close protein-to-protein contact, tilting to pack the interface as close as possible. The lipids will flow around the complexes to fill any voids opened by the displacements of the complexes. Light gray circles are the raised complexes, and the dark gray circles are the complexes that are lower and most likely in contact with the mica surface. D, the flattening process is completed, but the patch is rarely totally flat when it is imaged, which is likely to reflect local disorder in the membrane when irregularly packed LH2 complexes are forced into a roughly hexagonal packing order. The final packing order in C is seen in the three-dimensional representation of AFM data, which illustrates the close packed dimers and the angle subtended by the complexes in the zigzag lines.
The organized zigzag lines that appear upon adsorption onto mica are also seen in the topograph in Fig. 4B that was acquired from a patch of WT photosynthetic membrane prepared without any detergent. The LH2-rich area (boxed in Fig. 4B) displays the same zigzag architecture as occurs in the LH2-only membrane patches. These patterns were analyzed for their center-to-center distance (7.8 ± 0.5 nm; n = 11) and angle subtended by three complexes (119 ± 8.7°; n = 12). The images were not sufficiently well resolved to assign distances to intra- or interdimer; thus the 7.8 nm represents the average of these two distances.
DISCUSSION
General Vesicle Morphology—The presence of empty areas of lipid in the AFM images, whether as isolated patches or intimately associated with LH2-containing membrane patches, appears to be a normal phenomenon because we have observed them in preparations of membranes that were never exposed to detergent as well as those treated with β-DDM (see supplemental data). Moreover Scheuring et al. (31) have reported large areas of empty lipid in photosynthetically grown membranes of Rhodopseudomonas palustris that were prepared without the use of detergents.
The AFM data indicate that intact LH2-only vesicles are more easily deformed upon adsorption to the mica surface than adsorbed WT vesicles, as shown by the lower, wider profile. This is a reasonable result because the WT vesicles contain rows of core complex dimers that are likely to “stiffen” the WT membrane, both along the long axis of the RC-LH1-PufX core dimer (23) and also perpendicular to it, because dimer-dimer associations are favored, as shown by AFM topographs of WT membranes (9). The occurrence of deeply indented vesicles (see for example Fig. 1B) was only observed in the LH2-only strain, a further indication that these membranes are more flexible. In Fig. 1B the indentation is so deep that the lowest point of the depression is only ∼10 nm above the surface of the mica, consistent with the thickness expected of two bilayers containing membrane proteins; two empty lipid bilayers stacked upon each other would not have a thickness exceeding 8 nm. The flexibility of these membranes is probably the reason why, even with very low tapping amplitudes and slow scan rates, no details of the LH2 complexes could be recorded.
Formation of the Zigzag Lines of LH2 Complexes in Flattened Membranes—The very regular zigzag lines that appear in nearly all of the membrane pieces adsorbed onto mica are a striking feature of the AFM topographs, but this pattern of complexes does not arise from an effect of the detergent. We have observed this phenomenon in WT membrane patches even though there was no detergent treatment at any stage of membrane preparation (see the boxed area in topograph in Fig. 4B).
The most straightforward explanation for the appearance of highly regular zigzag lines of complexes is that the adsorption process leads to the adjustment of complexes sitting within a fluid, curved membrane as it is flattened on mica. Lateral pressure generated in the membrane during adsorption causes a rearrangement where ∼50% of the LH2 complexes have been vertically translated downwards as the extrinsic regions of LH2 are electrostatically attracted to the mica. The other 50% of the complexes are vertically translated upwards in the membrane. The lateral forces are distributed equally across the whole membrane, so that the height displacement between the upper and lower zigzag lines is consistent for all of the LH2 complexes in a particular patch and between all patches measured to date.
In Fig. 5 (A–C) we suggest how the process of adsorption leads to this remarkable change in the disposition of the embedded complexes. The pieces of membrane are initially highly curved and can only contact the mica around the periphery (Fig. 5A); in such membranes the LH2 complexes are likely to be equally distributed. The edges of the patch are “anchored” by the adsorption process, which effectively restricts the ability of the patch to spread out as it flattens, and thus lateral pressure is generated in the membrane. Reordering of the complexes is probably initiated at or soon after this point, because curved membranes can be observed in our AFM images in Fig. 2 that already show evidence of zigzag lines. It should be noted that imaging to molecular resolution of small, curved areas of membrane by AFM poses many challenges and has only been achieved using low amplitude tapping mode AFM, because the lateral forces generated by contact mode AFM either dislodge or disrupt the membranes (“nanodissection”). As membrane adsorption progresses, more of the edge of the patch becomes flattened onto the mica (Fig. 5B), and the complexes begin to be tightly compressed, leading to the maximum vertical disposition and tight interlocking of the dimers, until ultimately a nearly flat topology is achieved (Fig. 5C). It is these flat patches that are imaged to molecular resolution (Fig. 5D). We propose that the driving force for the reordering is the increase in the lateral pressure in the membrane during the change from a highly curved to a largely planar final topology. There are likely to be additional factors in determining the final disposition, i.e. vertical translation and tilt of each LH2 complex within the flattened membrane. The LH2 complex is not a smooth cylinder; it has a very irregular surface topology in the hydrophobic transmembrane region that will allow significant interlocking of adjacent complexes in a dimer as they are squeezed together. The ∼1-nm height difference between the two layers of LH2 complexes may represent the maximum vertical shift of the complexes that occurs before the belt of hydrophobic residues of the transmembrane helices of the complex becomes exposed to the buffer. This mechanism would operate upon all the LH2 complexes irrespective of whether they are closely packed together with their neighbors, as seen in the dimer pairs, or whether lipid molecules separate them from adjacent LH2 complexes, as would seem likely for complexes in adjacent dimers. Indeed the formation of LH2 dimers through protein-to-protein contacts may also be driven by the requirement to minimize the exposure of the hydrophobic central belt of the complexes to the buffer solution.
Membrane Organization of Flattened Vesicles in Comparison with Reconstituted Two-dimensional Crystals—The patterns observed in our AFM topographs have not been seen in any AFM images of other purple bacterial photosynthetic membranes. These other data originate from bacteria that elaborate a lamellar or stacked morphology in vivo (10–13, 31), whereas R. sphaeroides produces a system of spherical intracytoplasmic membrane vesicles (32). We suggest that the distinctive zigzag patterns observed in the present study would not be expected to form when lamellar sheets encounter the mica surface, because there is not likely to be lateral compression of the membrane during adsorption. However, zigzag lines of LH2 complexes, as well as other regular alternating patterns, have been observed in two-dimensional crystals formed from LH2 complexes that have been purified in their detergent-solubilized, monomeric state, then reconstituted into ordered crystalline sheets following removal of the detergent (19, 21, 29, 30). In such cases, the alternating arrangements of LH2 complexes are not caused by association with a planar substrate because the crystals are formed in liquid suspension and are seen even when suspended in glucose for cryo-electron microscopy (19). Detailed AFM studies of two-dimensional crystals of LH2 complexes from R. sphaeroides have quantified the height difference of the protruding faces of the complexes with respect to each other and the lipid bilayer and the angles subtended between the complexes (21, 29, 30). In two-dimensional reconstituted crystals, the height difference is 0.5 nm compared with our measured height difference of 1.2 nm, and the angles subtended in zigzag pattern two-dimensional crystals were 94° (29) and 90° (30) compared with 112° in this work. These values differ significantly as they arise from the alternating topology of complexes in reconstituted two-dimensional crystals and the unidirectional topology of complexes in native LH2-only membranes.
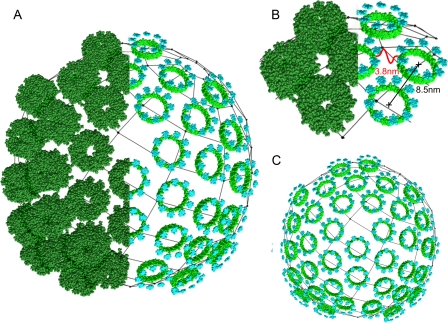
A Model LH2-only Vesicle—A previous AFM study on WT R. sphaeroides membranes (9) revealed the architecture around the RC-LH1-PufX core complexes but only poorly resolved the associated regions of LH2, because they were highly curved and not adequately adsorbed onto the mica. Now that we have obtained information on LH2-only regions, we can use this architecture to inform us about the original packing of the LH2 complexes within the native curved membrane. In compressed, flattened membranes, the average center-to-center distances between complexes within a dimer is 7.7 nm, whereas the average between complexes in adjacent dimers is 9.2 nm. It is reasonable to assume that if the flattened membrane was allowed to relax back to a curved topology, then the separation of all the complexes would become approximately equal in the absence of any isotropic lateral pressure. For R. sphaeroides DPF2G membranes, this distance is probably close to the average of the two distances measured in the flattened membrane, i.e. 8.5 nm. If this is the case, then there is sufficient space between the complexes to accommodate a continuous ring of lipid around them. Such a “greasy” layer would facilitate the rearrangement of the complexes during the flattening process, indeed if no such lubricating layer existed it could be argued that protein-to-protein contacts would seriously impede any vertical displacement of the order we have observed. The angle formed by the dimers in the zigzags averages 112.1 ± 7.0° (Fig. 5D); this is consistent with a hexagonal packing arrangement in the original curved membrane, subsequently distorted during flattening. Using Chimera (27), we have modeled a representative piece of membrane in its native curved state using an 8.5-nm LH2 center-to-center distance and a vesicle diameter of 50 nm (Fig. 6). The model, Fig. 6A, only portrays a portion of the chromatophore surface consisting of 37 LH2 complexes in a largely hexagonal packing arrangement. It should be noted that a complete sphere cannot be formed using hexagonal packing and that some pentagonal packing is essential, as shown. In this view we have reproduced half the complexes as only their BChl macrocycles; Fig. 6B shows a close-up of one area of hexagonally packed complexes to illustrate the closest approach of the B850 BChls that the model predicts to be 3.8 nm. The model also predicts a minimum separation between the complexes of ∼0.5 nm, a space that we propose is occupied by lipids in the membranes. In Fig. 6C, we have substituted just the B800 and B850 BChl macrocycles for all of the complexes to more clearly show the predominantly hexagonal packing.
FIGURE 6.
A model showing the idealized packing of LH2 complexes in a spherical vesicle. A model vesicle based upon the data in this work generated using Chimera (27) is shown. Spacefill representations of the LH2 complex (1NKZ) of R. acidophila have been placed on the surface of a 50-nm-diameter sphere at center-to-center separations of 8.5 nm in a predominantly hexagonal organization. Locally pentagonal packing has been used to achieve ordered packing of the hexagonally arranged complexes; the resultant pattern is as in C60 “Buckminsterfullerene” and icosahedral viral capsids. A, a composite vesicle in spacefill representation of LH2 complexes and of only the B800 and B850 BChl macrocycles showing how the pigments are arranged. B, a small section has been enlarged and the predicted distance (3.8 nm) between the closest B850 BChls illustrated. C, the same model vesicle as in A, but only showing the BChls, in which the predominantly hexagonal packing can clearly be seen.
We can test our model curved LH2-only membrane by examining the distances that it predicts between the B850 BChls in adjacent complexes and relating this to the observed energy transfer lifetimes. Three Pulse Photon Echo Peak Shift data obtained on LH2-only membranes showed a decay component of ∼5 ps, which was assigned to energy transfer between LH2 complexes (33). The equivalent rate for energy transfer from LH2 to LH1 complexes gave a range between 3 and 5 ps (34, 35), which was consistent with an average distance between B850 (LH2) and B875 (LH1) BChl rings of 3 nm. The closest B850 BChl to B850 BChl distance in our model of the membrane is 3.8 nm (Fig. 6B), which is in reasonable agreement with the estimated distance based upon energy transfer lifetimes and with the distances used in the modeling of the bacterial photosynthetic unit by Sener et al. (25). This value is also within the range calculated by Noy et al. (36) for efficient excitation energy transfer, where a 4-nm separation between pigment molecules and a Förster radius of 8 nm would give rise to an energy transfer quantum yield of 98.5% or better.
From the membrane model in Fig. 6, it is evident that the complexes are not tightly packed together, thus allowing lipid to surround each complex. When nine independent patches were measured, and the average figures were calculated (excluding any empty lipid; see Table 1), it was estimated that ∼26% of the vesicle surface can be accounted for by lipid. Thus, assuming that this arrangement of complexes holds true for LH2-only regions of WT membranes (Fig. 4B), there is more than adequate space between LH2 complexes to allow diffusion of quinone and quinol molecules into and through the peripheral antenna without compromising energy transfer migration between complexes. Our LH2-only model has important implications for cyclic electron transport in the cell because it would allow electron carriers to shuttle between RC-LH1-PufX core complexes and physically separated cytochrome bc1 complexes, a process that in the native membrane is necessary to foster efficient cyclic electron flow. It should be noted that no AFM evidence has been published that establishes the location of the cytochrome bc1 complex in relation to the core complex, which is suggestive of a “remote” position away from the core complex. The AFM study of P. molischianum by Goncalves et al. (13) concluded that the LH2 complexes are in protein-to-protein contact and thus prevent any movement of electron carriers through the peripheral antenna. However, the two photosystems of R. sphaeroides and P. molischianum are sufficiently different in their architectures that they are likely to utilize alternative strategies for maximizing photosynthetic growth.
We have proposed a reorganization rationale for the highly regular zigzag lines formed when membrane patches of R. sphaeroides are adsorbed onto mica, and using the data accumulated from flattened patches of membrane we have derived an average center-to-center distance of the LH2 complexes of 8.5 nm. A model of a section of curved membrane constructed using these data indicates that the closest B850 BChls in neighboring complexes are ∼3.8 nm apart; this distance is in agreement with known energy transfer lifetimes but is still a sufficient separation of the complexes (∼0.5 nm) to allow electron carriers to freely diffuse between them. Such an arrangement in the WT membrane would allow quinones and quinols to diffuse between RC-LH1-PufX core complexes and physically remote cytochrome bc1 complexes elsewhere in native chromatophores of R. sphaeroides.
Supplementary Material
This work was supported by funding from the Biotechnology and Biological Sciences Research Council UK (to J. D. O., J. D. T., P. Q., J. A. T., and C. N. H.) and from the Engineering and Physical Sciences Research Council UK (to C. V.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains a supplemental figure.
Footnotes
The abbreviations used are: AFM, atomic force microscopy; RC, reaction center; LH, light harvesting; WT, wild type; BChl, bacteriochlorophyll; β-DDM, n-dodecyl-β-maltoside.
References
- 1.Michel, H., Epp, O., and Deisenhofer, J. (1986) EMBO J. 5 2445-2451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McDermott, G., Prince, S. M., Freer, A. A., Hawthornthwaite-Lawless, A. M., Papiz, M. Z., Cogdell, R. J., and Isaacs, N. W. (1995) Nature 374 517-521 [DOI] [PubMed] [Google Scholar]
- 3.Jordan, P., Fromme, P., Witt, H. T., Klukas, O., Saenger, W., and Krauss, N. (2001) Nature 411 909-917 [DOI] [PubMed] [Google Scholar]
- 4.Zouni, A., Witt, H. T., Kern, J., Fromme, P., Krauss, N., Saenger, W., and Orth, P. (2001) Nature 409 739-743 [DOI] [PubMed] [Google Scholar]
- 5.Abramson, J., Smirnova, I., Kasho, V., Verner, G., Kaback, H. R., and Iwata, S. (2003) Science 301 610-615 [DOI] [PubMed] [Google Scholar]
- 6.Bass, R. B., Strop, P., Barclay, M., and Rees, D. C. (2002) Science 298 1582-1587 [DOI] [PubMed] [Google Scholar]
- 7.Xia, D., Yu, C. A., and Kim, H. (1997) Science 277 60-66 [DOI] [PubMed] [Google Scholar]
- 8.Muller, D. J., Engel, A., Matthey, U., Meier, T., Dimroth, P., and Suda, K. (2003) J. Mol. Biol. 327 925-930 [DOI] [PubMed] [Google Scholar]
- 9.Bahatyrova, S., Frese, R. N., Siebert, C. A., Olsen, J. D., van der Werf, K. O., van Grondelle, R., Niederman, R. A., Bullough, P. A., Otto, C., and Hunter, C. N. (2004) Nature 430 1058-1062 [DOI] [PubMed] [Google Scholar]
- 10.Scheuring, S., Seguin, J., Marco, S., Levy, D., Robert, B., and Rigaud, J. L. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 1690-1693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scheuring, S., Rigaud, J. L., and Sturgis, J. (2004) EMBO J. 23 4127-4133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scheuring, S., Busselez, J., and Levy, D. (2005) J. Biol. Chem. 280 1426-1431 [DOI] [PubMed] [Google Scholar]
- 13.Goncalves, R. P., Bernadac, A., Sturgis, J. N., and Scheuring, S. (2005) J. Struct. Biol. 152 221-228 [DOI] [PubMed] [Google Scholar]
- 14.Sturgis, J. N., and Niederman, R. A. (2008) Photosyn. Res. 95 269-278 [DOI] [PubMed] [Google Scholar]
- 15.Scheuring, S., and Sturgis, J. N. (2006) Biophys. J. 91 3707-3717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones, M. R., Fowler, G. J. S., Gibson, L. C. D., Grief, G. G., Olsen, J. D., Crielaard, W., and Hunter, C. N. (1992) Mol. Microbiol. 6 1173-1184 [DOI] [PubMed] [Google Scholar]
- 17.Jungas, C., Ranck, J. L., Rigaud, J. L., Joliot, P., and Verméglio, A. (1999) EMBO J. 18 534-542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Siebert, C. A., Qian, P., Fotiadis, D., Engel, A., Hunter, C. N., and Bullough, P. A. (2004) EMBO J. 23 690-700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walz, T., Jamieson, S. J., Bowers, C. M., Bullough, P. A., and Hunter, C. N. (1998) J. Mol. Biol. 282 833-845 [DOI] [PubMed] [Google Scholar]
- 20.Scheuring, S., Reiss-Husson, F., Engel, A., Rigaud, J. L., and Ranck, J. L. (2001) EMBO J. 20 3029-3035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scheuring, S., Seguin, J., Marco, S., Levy, D., Breyton, C., Robert, B., and Rigaud, J. L. (2003) J. Mol. Biol. 325 569-580 [DOI] [PubMed] [Google Scholar]
- 22.Hunter, C. N., Pennoyer, J. D., Sturgis, J. N., Farrelly, D., and Niederman, R. A. (1988) Biochemistry 27 3459-3467 [Google Scholar]
- 23.Qian, P., Bullough, P., and Hunter, C. N. (2008) J. Biol. Chem. 283 14002-14011 [DOI] [PubMed] [Google Scholar]
- 24.Lommen, M. A., and Takemoto, J. (1978) J. Bacteriol. 136 730-741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sener, M. K., Olsen, J. D., Hunter, C. N., and Schulten, K. (2008) Proc. Natl. Acad. Sci. U. S. A. 104 15273-15278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olsen, J. D., Sockalingum, G. D., Robert, B., and Hunter, C. N. (1994) Proc. Natl. Acad. Sci. U. S. A. 91 7124-7128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C., and Ferrin, T. E. (2004) J. Comput. Chem. 25 1605-1612 [DOI] [PubMed] [Google Scholar]
- 28.Timpmann, K., Woodbury, N. W., and Freiberg, A. (2000) J. Phys. Chem. B 104 9769-9771 [Google Scholar]
- 29.Bahatyrova, S., Frese, R. N., van der Werf, K. O., Otto, C., Hunter, C. N., and Olsen, J. D. (2004) J. Biol. Chem. 279 21327-21333 [DOI] [PubMed] [Google Scholar]
- 30.Liu, L.-N., Aartsma, T. J., and Frese, R. N. (2008) FEBS J. 275 3157-3166 [DOI] [PubMed] [Google Scholar]
- 31.Scheuring, S., Goncalves, R. P., Prima, V., and Sturgis, J. N. (2006) J. Mol. Biol. 358 83-96 [DOI] [PubMed] [Google Scholar]
- 32.Brunisholz, R. A., and Zuber, H. (1992) J. Photochem. Photobiol. B Biol. 15 113-140 [DOI] [PubMed] [Google Scholar]
- 33.Fleming, G. R., Agarwal, R., and Rizvi, A. H. (2002) J. Phys. Chem. A 106 7573-7578 [Google Scholar]
- 34.Hess, S., Chachisvilis, M., Timpmann, K., Jones, M. R., Fowler, G. J. S., Hunter, C. N., and Sundström, V. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 12333-12337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nagarajan, V., and Parson, W. W. (1997) Biochemistry 36 2300-2306 [DOI] [PubMed] [Google Scholar]
- 36.Noy, D., Moser, C. C., and Dutton, P. L. (2006) Biochim. Biophys. Acta 1757 90-105 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.