Abstract
Protein-tyrosine phosphatase receptor type Z (Ptprz) is preferentially expressed in the brain as a major chondroitin sulfate proteoglycan. Three splicing variants, two receptor isoforms and one secretory isoform, are known. Here, we show that the extracellular region of the receptor isoforms of Ptprz are cleaved by metalloproteinases, and subsequently the membrane-tethered fragment is cleaved by presenilin/γ-secretase, releasing its intracellular region into the cytoplasm; of note, the intracellular fragment of Ptprz shows nuclear localization. Administration of GM6001, an inhibitor of metalloproteinases, to mice demonstrated the metalloproteinase-mediated cleavage of Ptprz under physiological conditions. Furthermore, we identified the cleavage sites in the extracellular juxtamembrane region of Ptprz by tumor necrosis factor-α converting enzyme and matrix metalloproteinase 9. This is the first evidence of the metalloproteinase-mediated processing of a receptor-like protein-tyrosine phosphatase in the central nervous system.
Receptor-like protein-tyrosine phosphatases (RPTPs)2 are a structurally and functionally diverse family of enzymes comprised of eight subfamilies (1). Protein-tyrosine phosphatase receptor type Z (Ptprz, also called PTPζ or RPTPβ) is a RPTP classified in the R5 subfamily and expressed in neuronal and glial cells in the central nervous system (2–4). The physiological importance of this molecule has been demonstrated through studies of Ptprz-deficient mice (4, 5), which display impairments in hippocampal function in a maturation-dependent manner (6, 7). An independently generated knock-out mouse line suggests a fragility of myelin in the central nervous system (8).
It is known that three isoforms of Ptprz are generated by alternative splicing from a single Ptprz gene (on mouse chromosome 6; human chromosome 7), the two transmembrane isoforms Ptprz-A and Ptprz-B and the secretory isoform Ptprz-S (also known as 6B4 proteoglycan or phosphacan) (2, 9–12), all of which are expressed as chondroitin sulfate proteoglycans in the brain (3). However, some inexplicable issues about the molecular profiles of Ptprz have remained in previous studies. For instance, although there exists substantial expression of the respective transcripts for all isoforms (11, 13), full-length Ptprz-A has been scarcely observed at the protein level in the adult brain (3, 4). In addition, several lower molecular species have been detected with a specific antibody against Ptprz in wild-type mice (4). The technical difficulty in removal of the chondroitin sulfate chains to separate their core proteins by SDS-PAGE may induce variability in the signal patterns of this molecule in Western blotting among researchers.
In this study we examined the molecular profile of Ptprz in the adult mouse brain at both protein and mRNA levels in detail and revealed that the proteolytic fragments are abundantly accumulated. The two receptor isoforms were found to undergo ectodomain cleavage by metalloproteinases, releasing their extracellular fragments. The membrane-tethered fragment of Ptprz was further cleaved by presenilin/γ-secretase to release the intracellular fragment, which was consequently detected in the cytoplasm and nucleus. These findings suggest a novel signaling mechanism of Ptprz by the regulated proteolytic processing in the central nervous system.
EXPERIMENTAL PROCEDURES
Pharmacological Reagents—Phorbol 12-myristate 13-acetate (PMA) were purchased from Sigma. GM6001, compound E, lactacystin, and recombinant tumor necrosis factor-α (TNF-α)-converting enzyme (TACE) were from Calbiochem.
Animal Experiments—Adult wild-type C57BL/6 mice and Ptprz-deficient mice (4) backcrossed with the inbred C57BL/6 strain for more than 10 generations were used. Mice were administered GM6001, suspended in saline containing 1.5% carboxyl methyl cellulose, intraperitoneally (100 mg per kg body weight). For intraventricular infusion of GM6001, anesthetized mice were placed in a stereotaxic apparatus, and brain infusion cannulas (brain infusion kit 3, Alza Corp.) were inserted in the cerebral ventricle. The cannula was secured to the skull with an anchoring screw and dental cement. The stereotaxic coordinates were 0.5 mm posterior and 1.0 mm lateral to the bregma and 2.4 mm below the skull surface. An osmotic mini-pump (model 1007D, flow rate = 0.5 μl/h, Alza Corp.) filled with 2.5 mm GM6001 in 50% DMSO was implanted between the scapulae and connected to the infusion cannulas. The brains were separated as described (14). All animal experiments were performed according to the guidelines of Animal Care with approval by the Committee for Animal Research, National Institutes of Natural Sciences.
Expression Constructs for Ptprz Isoforms—Full-length rat Ptprz-A and Ptprz-S (10) were subcloned into the expression vector pZeoSV2 (Invitrogen) to yield pZeo-PTPζ-A and pZeo-PTPζ-S, respectively. The expression plasmid for rat Ptprz-B (pZeo-PTPζ) was described previously (15). This construct was used as a template to generate pZeo-PTPζ-G1631I (a mutant in which glycine at 1631 is substituted with isoleucine) by using a QuikChange multi-site-directed mutagenesis kit (Stratagene). The expression plasmid (pZeoSV-PtprzICR) for the entire intracellular region of rat Ptprz-A/-B (amino acid residues 1665–2316; GenBank™ accession number U09357) was prepared by PCR from pZeo-PTPζ with an initiative methionine encoding primer and cloning the fragment into the NotI site of pZeoSV2.
Cell Culture and DNA Transfection—HEK293T cells (human embryonic kidney epithelial cells) were grown and maintained on dishes coated with rat tail collagen in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum in a humidified incubator at 37 °C with 5% CO2. HEK293 cell lines stably expressing either human wild-type presenilin 1 (PS1 WT) or a dominant-negative PS1 variant (substitution of Asp at 385 for Ala, PS1 D385A) (16) were kindly provided by Takeshi Ikeuchi (Niigata University, Niigata, Japan). These cells were transfected with Ptprz expression plasmids by calcium-phosphate precipitation as described (17).
CHO-M2 (TACE-deficient CHO cell line), CHO-(M2+TACE) (CHO-M2 cells rescued by expression of TACE), and parental CHO-WT cells (18) were kindly provided by Joaquín Arribas (University Hospital Vall d'Hebron, Barcelona, Spain). CHO-M2 and CHO-WT cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and 500 μg/ml of G418. CHO-(M2+TACE) cells were maintained with G418 and hygromycin (500 μg/ml of each). These CHO cells were transfected by using Lipofectamine Plus reagent (Invitrogen; 1 μg of plasmid per 3.5-cm dish). Transfected cells were replated once on 3.5-cm dishes, cultured for 24 h, and then used for the experiments.
Protein Extraction and Chondroitinase ABC (chABC) Treatment—Mouse tissues quickly separated on ice were homogenized with more than 10 volumes of a lysis buffer: 20 mm Tris-HCl, pH 8.0, 1% Nonidet P-40, 137 mm NaCl, 10 mm NaF, 1 mm sodium orthovanadate, and a EDTA-free protease inhibitor mixture (complete EDTA-free, Roche Applied Science). Supernatants were then collected by centrifugation at 15,000 × g for 15 min. Cultured cells were extracted with the same lysis buffer (300 μl per 3.5-cm dish) as above. The samples were stored at -85 °C before use.
For chABC digestion, the protein concentration of each sample was adjusted with the lysis buffer (<4 mg/ml). Aliquots (10 μl) were then incubated with an equal volume of 0.2 m Tris-HCl, 4 mm sodium acetate, pH 7.5, with or without chABC (Seikagaku Co., Tokyo, Japan; the enzyme was added at 60 microunits/μg of protein) for 1 h at 37°C. Protein concentrations were determined with a Micro BCA protein assay kit (Pierce).
Western Blot Analysis—Samples were mixed with an equal volume of 2× SDS-PAGE sample buffer (containing 4% mercaptoethanol), boiled for 5 min, and then separated on a 5–20% gradient polyacrylamide gel (E-R520L, Atto Corp., Tokyo, Japan). Proteins were transferred to a polyvinylidene difluoride membrane (Millipore Corp.) for 1 h using a conventional semidry electrotransfer (1.3 mA per cm2). The membrane was incubated for 1 h in a blocking solution (4% nonfat dry milk and 0.1% Triton X-100 in 10 mm Tris-HCl, pH 7.4, 150 mm NaCl) and incubated overnight with anti-Ptprz-S rabbit serum (1:10,000) (3) in the blocking solution supplemented with 0.04% SDS to prevent nonspecific binding. Mouse monoclonal anti-RPTPβ (the epitope region is amino acid residues 2098–2307 of human Ptprz-A, 250 ng/ml, BD Biosciences) was incubated with the blots in the blocking solution. The binding of these antibodies was detected with an ECL Western blotting system (GE Healthcare).
Subcellular Localization Analysis—Cells were washed 3 times with 10 mm phosphate buffer, pH 7.3, containing 150 mm NaCl, fixed with 10% neutral formalin, and then blocked with the blocking buffer as above and followed by overnight incubation with anti-RPTPβ (1 μg/ml) in the blocking buffer. Bound antibodies were visualized with Alexa488-conjugated anti-mouse antibody (Molecular Probes, Eugene, OR). For the nucleus labeling, the cells were incubated with TO-PRO-3 (Molecular Probes) and then analyzed with a Zeiss LSM-510 confocal scanning laser microscope (Carl Zeiss, Jena, Germany) using a Zeiss water-immersion objective (C-Apochromat 40×/1.20 W Korr).
Northern Blot Analysis—Total RNA was isolated from mouse tissues using TRIzol (Invitrogen), and then poly(A)+ RNA was purified using the Dynabeads mRNA purification kit (Dynal) according to the manufacturer's instructions. Northern blotting was performed as described (19) with slight modifications in the electrophoresis. The poly(A)+ RNA was denatured in 1× MOPS buffer (20 mm MOPS, 2 mm sodium acetate, and 1 mm EDTA, pH 7.0) containing 6.8% formaldehyde, 50% formamide, and 50 μg/ml of ethidium bromide at 67 °C for 10 min, chilled on ice for 5 min, and then 2.5 μl of a loading buffer (50% glycerol, 0.25% bromphenol blue, and 0.25% xylene cyanol) was added. Electrophoresis was performed on a formaldehyde-denatured agarose gel (6.8% formaldehyde and 1% agarose in 1× MOPS buffer) at 5 V/cm for 4 h with the circulation of an electrophoresis buffer (6.8% formaldehyde in 1× MOPS buffer).
Templates of complementary DNA probes were as follows: CAH-FNIII probe (nucleotide residues 93–1215 for rat Ptprz-A; GenBank™ accession number U09357), PTP-D1 probe (nucleotide residues 5047–6081 for rat Ptprz-A), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) probe (nucleotide residues 22–553 for mouse GAPDH; GenBank™ accession number BC096440). Signals of bands on the blot were detected using a BAS-MS 2025 imaging plate (Fuji Photo Film, Tokyo, Japan) and visualized using a Typhoon 9400 scanner (GE Healthcare).
In Vitro Digestion Analyses—Peptides were synthesized on an Applied Biosystems ABI 433A peptide synthesizer by using the standard fluorenylmethoxycarbonyl protocol and purified by high pressure liquid chromatography on a C-18 reverse phase column. These peptides (2 pmol each) were incubated at 37 °C for 30 h in 25 mm Tris-HCl, pH 9.0, 2.5 μm ZnCl2, and 0.005% Brij-35 with or without recombinant TACE (250 ng) in a final volume of 10 μl. After purification using pipette tips packed with a C18 resin (ZipTip, Millipore), the samples were analyzed by matrix-assisted laser desorption ionization-time of flight mass spectrometry (Reflex III, Bruker Daltonics) using α-cyano-4-hydroxycinnamic acid matrix (Sigma).
RESULTS
Expression Profile of Ptprz in the Adult Mouse Brain—It is well known that three splicing variants of Ptprz are expressed in the brain from a single gene (see Fig. 1A). All three isoforms expressed in the brain are highly glycosylated with chondroitin sulfate (3). Therefore, the removal of the chondroitin sulfate chains beforehand is necessary to resolve their core proteins by SDS-PAGE (20).
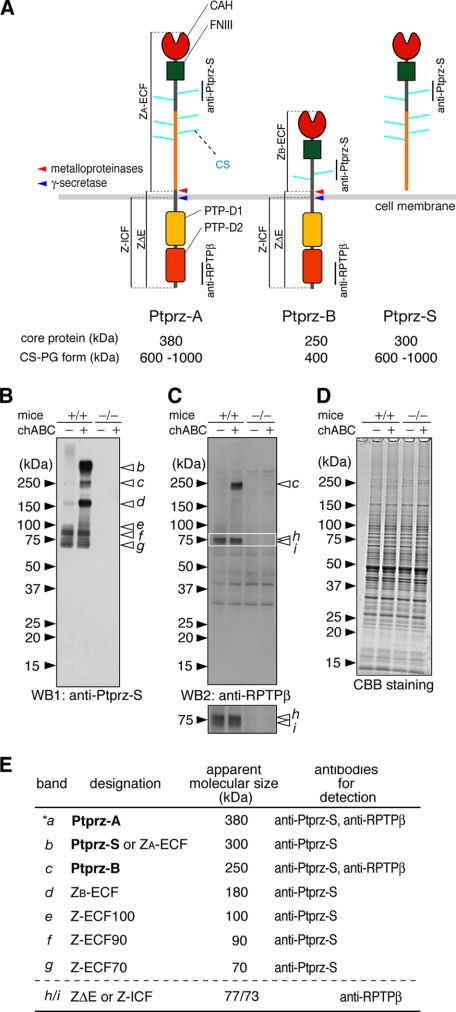
FIGURE 1.
Novel protein species of Ptprz in the adult mouse brain. A, schematic representation of Ptprz isoforms. Molecular sizes of the chondroitin sulfate proteoglycan forms (CS-PG) and their core proteins after treatment with chABC are shown (3). Regions corresponding to the epitopes of antibodies used in this study are indicated by vertical lines. We designated the proteolytic fragments as follows: ZA-ECF or ZB-ECF, the extracellular fragment of Ptprz-A or Ptprz-B; ZΔE, the membrane-tethered fragment of Ptprz-A and Ptprz-B; Z-ICF, the intracellular fragment cleaved from ZΔE. The cleavage sites are indicated by arrows (red, metalloproteinases including TACE and MMP-9; blue, presenilin/γ-secretase). Domains are highlighted in different colors: CAH, carbonic anhydrase-like domain; FNIII, fibronectin type III domain; PTP-D1 and PTP-D2, tyrosine phosphatase domains. B–D, brain extract (5 μg of protein) of wild-type mice (+/+) and Ptprz-deficient mice (-/-) was treated with (+) or without (-) chABC. The samples were separated on a 5–20% gradient gel followed by Western blotting (WB) using anti-Ptprz-S (B). The same blot was stripped and reprobed with anti-RPTPβ (C). The lower image is a vertical enlargement of the area enclosed by a rectangle in the upper image. Staining with Coomassie Brilliant Blue R-250 to check the amounts of protein applied (D). The figures are representative of five separate experiments. E, summary of the immunoreactive bands (b–i). Their designation, apparent molecular size, and specific antibodies for detection are shown. The core protein of Ptprz-A (band *a) was scarcely detected in the brain extract by Western blotting, but it was evident in immunoprecipitation assays (data not shown).
When the chABC-treated extract of the wild-type mouse brain (+/+) was analyzed with anti-Ptprz-S, which recognizes the extracellular region of all three isoforms (see Fig. 1A), six bands (bands b–g) in the range from 300 to 70 kDa were clearly detected (Fig. 1B). Because these bands are not present in Ptprz-deficient mice (-/-), all these molecular species are considered to be derived from Ptprz gene products. Among them, the 300-kDa (band b) and 250-kDa (band c) species represent the core proteins of Ptprz-S and Ptprz-B, respectively (3); however, the other four species (bands d–g) have not been characterized; of note is that the band of Ptprz-A at 380 kDa is scarcely detected (the band a with an asterisk in Fig. 1E; see also Ref. 4). The signal intensity of the three lower molecular species (bands e–g) was not changed by chABC treatment (Fig. 1B), indicating that they are not modified with chondroitin sulfate. In contrast, the larger bands b–d were almost missing without chABC-treatment because they could not enter the gel.
To identify the receptor isoforms and their derivatives, the same blot was reprobed with anti-RPTPβ, which recognizes the intracellular region (see Fig. 1A). As shown in Fig. 1C, the 250-kDa species (band c, Ptprz-B) was detected by anti-RPTPβ as expected along with additional bands at around 75 kDa. The enlarged view of the 75-kDa band (lower panel), demonstrated that the signal consists of two adjacent bands of 77 kDa (band h) and 73 kDa (band i). On the other hand, anti-RPTPβ did not recognize the other species (bands b, d, e–g) detected by anti-Ptprz-S.
Although the uncharacterized species of Ptprz (bands d–i) appears to be processing products of the mature three isoforms of Ptprz, we addressed the possibility with the best of care that unknown novel Ptprz transcripts might be detected by Northern blotting. Probe 1 for the CAH-FNIII region, which should detect all Ptprz transcripts, demonstrated that the three transcripts of 8.5 kb (Ptprz-A), 7.5 kb (Ptprz-S), and 5.8 kb (Ptprz-B) are expressed only in the wild-type, not in the knock-out mice (Fig. 2). Probe 2 for the PTP-D1 region, which detects the transcripts for the receptor isoforms, showed the 8.5-kb (Ptprz-A) and 5.8-kb (Ptprz-B) transcripts as expected. Although full-length Ptprz-A protein (380 kDa) was hardly detected in the adult brain lysate, its mRNA was, thus, expressed in a significant amount. Importantly, other transcripts corresponding to the smaller Ptprz proteins such as the 180- or 75-kDa species were not detected.
FIGURE 2.
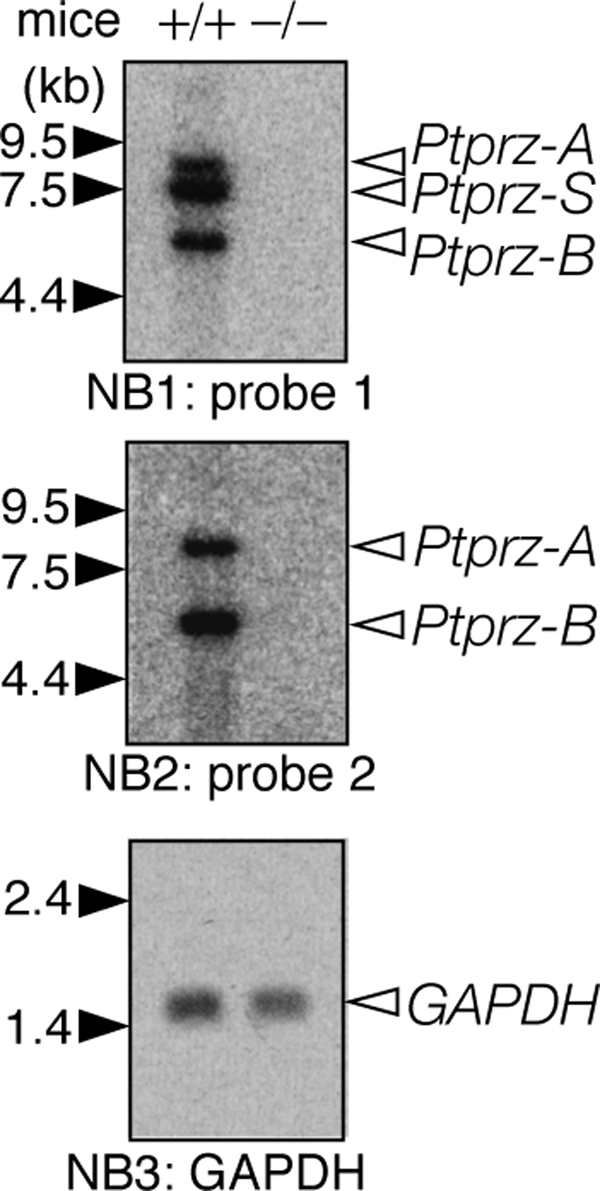
Northern blot analyses of Ptprz transcripts in the adult mouse brain. Poly(A)+ RNA (2 μg) from the adult mouse brain was hybridized with a 32P-labeled cDNA probe for the CAH-FNIII region (probe 1). The same blot was stripped and then reprobed for the PTP-D1 region (probe 2). The amount of RNA loaded was confirmed with a probe for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) in the same blot. The figures are representative of three separate experiments.
Ectodomain Shedding of Ptprz by Metalloproteinases—In our studies to exogenously express Ptprz-B in mammalian cells, we noticed that an immunoreactive species of 180 kDa is secreted into the culture medium. Because this size corresponded to that of the whole extracellular region of Ptprz-B (band d observed in the brain in Fig. 1B), we assumed that this is generated by the ectodomain shedding, a specialized type of limited proteolysis releasing the extracellular domain of a variety of cell surface receptors (21, 22); it occurs in the vicinity of the cell surface, generally dependent upon the actions of matrix metalloproteinases (MMPs) or adamalysins (ADAMs, a disintegrin and metalloproteinases).
We tested this possibility by using a protein kinase C activator, PMA, which is known to trigger ectodomain shedding in various cells. The treatment of HEK293T cells expressing Ptprz-B with PMA resulted in an increase in the 180-kDa species in the conditioned medium, which was inversely correlated with the decrease in Ptprz-B in cell extracts (Fig. 3A). This event occurred dependent on the concentration of PMA (Fig. 3B), strongly suggesting that the band of 180 kDa represents the ectodomain (ZB-ECF) of Ptprz-B. Intriguingly, in the presence of a broad-spectrum metalloproteinase inhibitor, GM6001, the generation of the 180-kDa species was clearly inhibited under both unstimulated and stimulated conditions with PMA (Fig. 3A, right panel).
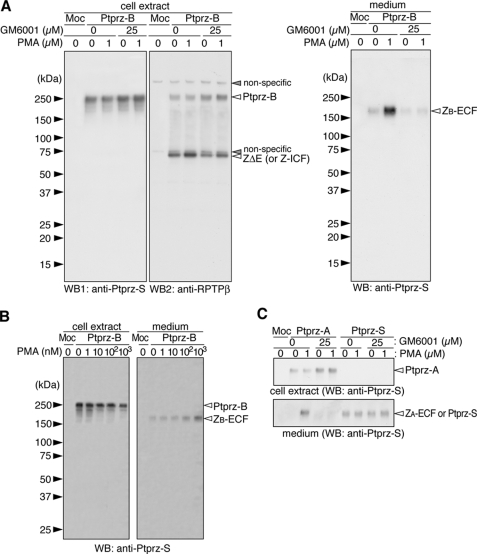
FIGURE 3.
Metalloproteinase-mediated ectodomain cleavage of the receptor isoforms of Ptprz. A, HEK293T cells were transiently transfected with the expression construct of Ptprz-B or control vector (Moc). Twenty-four hours after transfection, cells were washed and incubated with or without PMA in fresh serum-free medium for 1 h. GM6001 was added 20 min before the stimulation by PMA or vehicle. The cell extracts (left panels) were analyzed by Western blotting (WB) using anti-Ptprz-S. The same membrane was then reprobed with anti-RPTPβ. Conditioned media (right panel) were analyzed with anti-Ptprz-S. B, cells were incubated with the indicated amount of PMA in fresh serum-free medium for 1 h and analyzed by Western blotting as above. C, HEK293T cells were transiently transfected with the Ptprz-A or Ptprz-S expression construct and treated as described in A. Before SDS-PAGE, the samples were treated with chABC. The figures are representative of three separate experiments, and the results of the densitometric analyses are shown in supplemental Fig. S1. The designations of the detected bands are shown in Fig. 1, A and E.
Similar results were observed with the Ptprz-A isoform. Unlike Ptprz-B, when Ptprz-A was expressed in HEK293T cells, the mature receptor protein was highly modified with chondroitin sulfate (data not shown). Therefore, the samples were treated with chABC before SDS-PAGE. As with Ptprz-B, the release of the entire extracellular fragment of Ptprz-A, ZA-ECF (300 kDa), into the culture medium was clearly enhanced by PMA and suppressed in the presence of GM6001 (Fig. 3C). On the other hand, when the secretory isoform (Ptprz-S) was expressed, the full-length Ptprz-S was detected exclusively in the medium, and the amount was not affected by the treatment of cells either with PMA or with GM6001 (Fig. 3C). It is recognizable here that ZA-ECF is indistinguishable from Ptprz-S in size and antigenicity.
TACE-mediated Shedding of Ptprz in Cultured Cells—Because GM6001 is a broad-spectrum metalloproteinase inhibitor, additional experiments were required to define the specific proteinase(s) involved in the ectodomain shedding of Ptprz-A/-B. TACE (also known as ADAM-17) is a GM6001-sensitive, membrane-anchored, zinc-dependent metalloproteinase. TACE functions as a membrane sheddase to release the ectodomain portions of many transmembrane proteins including TNF-α and Notch (23). To determine whether TACE is involved in the ectodomain cleavage of Ptprz, we took advantage of CHO-M2 cells which are defective in TACE-mediated shedding (18). Parental wild-type CHO cells (CHO-WT) and CHO-M2 cells which stably express a functional TACE, CHO-(M2+TACE), were also used for comparison.
Transfection of the expression construct of Ptprz-B yielded similar expression levels of Ptprz-B in these cells (Fig. 4, left panels), and similar amounts of ZB-ECF were accumulated in the conditioned media during 1 h of incubation (Fig. 4, right panels); this basal level of accumulation of ZB-ECF was also observed in CHO-M2 and similarly suppressed by GM6001, indicating that the basal amount of ectodomain shedding of Ptprz-B is independent of TACE. PMA-stimulated ectodomain cleavage was reproduced in CHO-WT and CHO-(M2+TACE) cells and was inhibited by GM6001. However, the PMA-stimulated shedding was not observed in CHO-M2 cells. Similar results were observed with Ptprz-A (data not shown). TACE is, thus, highly responsible for the PMA-inducible cleavage of Ptprz for the generation of ZB-ECF and ZA-ECF but not for the constitutive cleavage in these cell lines.
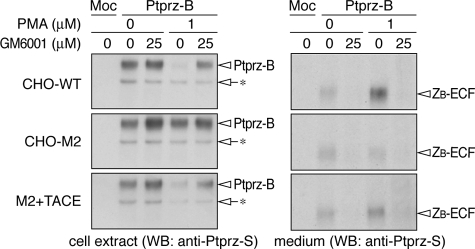
FIGURE 4.
Involvement of TACE in PMA-stimulated ectodomain cleavage of Ptprz. CHO-WT (wild-type), CHO-M2 (TACE defective), and CHO-(M2+TACE) (M2 cells expressing functional TACE) cells were transiently transfected with the Ptprz-B expression construct. Twenty-four hours after transfection cells were washed and incubated in fresh serum-free medium with or without PMA for 1 h. GM6001 was added 20 min before PMA or vehicle. The cell extracts (left panels) and conditioned media (right panels) were analyzed by Western blotting (WB) with anti-Ptprz-S. The arrows with asterisks indicate immature forms of Ptprz-B accumulated in cells. The designations are shown in Fig. 1, A and E. The figures are representative of three separate experiments, and the results of the densitometric analyses are shown in supplemental Fig. S2.
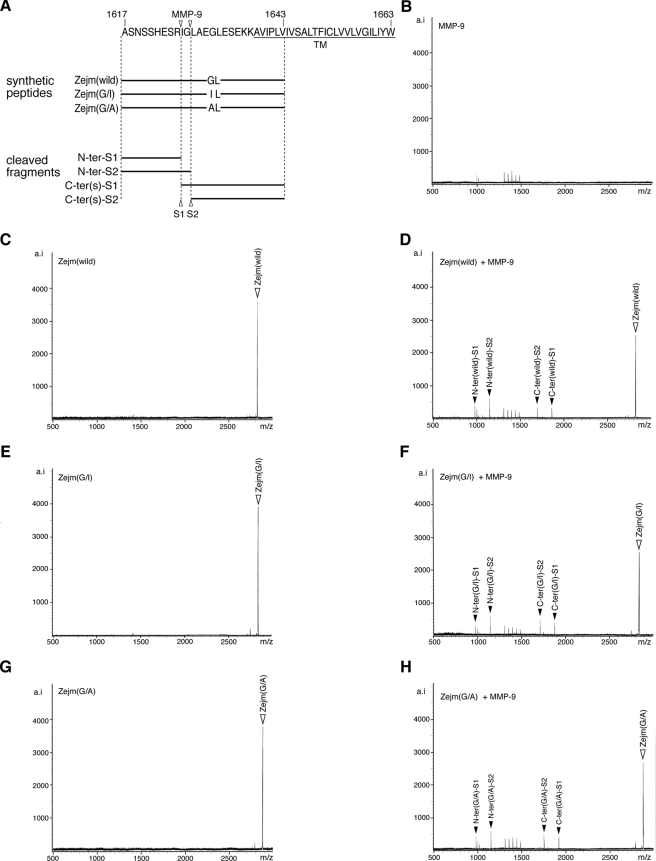
Because the Ptprz-A and Ptprz-B isoforms have a common short sequence in the extracellular membrane-proximal region, the cleavage site was expected within this region. When a synthetic peptide corresponding to the juxtamembrane sequence was incubated with recombinant TACE in vitro, TACE indeed induced the cleavage of the substrate peptide into the two fragments. The molecular mass of them indicated that the enzymatic cleavage occurs between Gly at 1631 (P1 site) and Leu at 1632 (P1′ site) (see Fig. 5A). Previous studies with peptide substrates of TNF-α indicated that TACE has a strong preference for cleavage at Ala-Val sequences and cannot cleave a TNF-α-based peptide with the substitution of Ala with Ile at the P1 position (24). Consistently, a mutant peptide, Zejm(G/I), in which Gly at P1 is replaced with Ile, was hardly cleaved by TACE (Fig. 5C). In contrast, the cleavage was highly enhanced by substitution with Ala at P1, Zejm (G/A), the same as TNF-α (Fig. 5D).
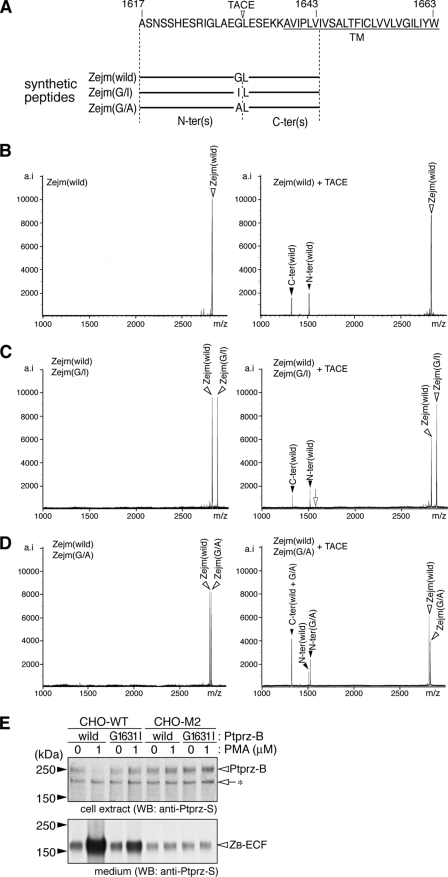
FIGURE 5.
Identification of the TACE-cleavage site in Ptprz. A, amino acid sequence of the common extracellular membrane-proximal region in Ptprz-A and Ptprz-B. Amino acid numbers refer to the sequence of rat Ptprz-A. Three synthetic peptides are shown under the sequence. The arrowhead indicates the TACE cleavage site deduced by in vitro peptide digestion as below. TM, transmembrane segment. B–D, synthetic peptides only (left panels) or synthetic peptides with recombinant TACE (right panels) were incubated for 30 h at 37 °C, and the products were analyzed by mass spectrometry. The combination of peptides is shown at the top left corner of each panel. Peaks are labeled with arrowheads as in A. The expected m/z value for the N-terminal fragment when cleaved at the Ile-Leu bond of Zejm(G/I) is indicated by an open arrow (C, right panel). The figures are representative of two separate experiments. E, CHO-WT and CHO-M2 were transiently transfected with expression constructs for wild-type or G1631I mutant of Ptprz-B. Twenty-four hours after transfection cells were washed and incubated in fresh serum-free medium with or without PMA for 1 h. The cell extracts (upper panel) and conditioned media (lower panel) were analyzed by Western blotting (WB) with anti-Ptprz-S. The figure is representative of three separate experiments, and the results of the densitometric analyses are shown in supplemental Fig. S3. The designations are shown in Fig. 1, A and E.
We confirmed the validity of this in vitro result by generating the mutant construct of Ptprz-B. Changing Gly at 1631 to Ile (G1631I) did not affect the efficiency of the basal constitutive ectodomain cleavage; however, the efficiency of the ectodomain cleavage enhanced by PMA was markedly decreased in the G1631I-mutant of Ptprz-B (Fig. 5E). In TACE-defective CHO-M2 cells, both wild-type Ptprz-B and the G1631I mutant showed a similar accumulation of ZB-ECF at a low level with and without PMA stimulation, indicating that TACE indeed cleaves the Gly-Leu bond in CHO cells (Fig. 5E).
The finding that TACE-independent basal (constitutive) shedding was not affected by the G1631I mutation suggests that there exist multiple cleavage sites within the membrane proximal region by different ADAMs or MMPs other than TACE. In line with this view, we found that recombinant MMP-9 can cleave the peptide substrate Zejm (wild) at two sites, between Arg-1625—Ile-1626 and Gly-1627—Leu-1628, located in the vicinity of the TACE cleavage site (Fig. 6). We confirmed that the peptide cleavage by MMP-9 is not affected by substitution of either Gly to Ile (Fig. 6, E and F) or Gly to Ala (Fig. 6, G and H). The ectodomain shedding, thus, occurs within the common membrane proximal region of the two receptor isoforms in both a constitutive and a PMA-stimulated manner.
FIGURE 6.
In vitro cleavage of Ptprz by MMP-9. A, the amino acid sequence of the extracellular membrane-proximal region common to Ptprz-A and Ptprz-B and synthetic peptides used are shown below. Arrowheads indicate the MMP-9 cleavage site deduced by in vitro peptide digestion with MMP-9 as below. B–H, samples were incubated in 10 μl of 50 mm Tris-HCl, pH 7.6, 150 mm NaCl, 5 mm CaCl2, and 0.01% Brij-35 for 6 h at 37 °C (2 pmol, each peptide; 0.1 μg recombinant MMP-9, Calbiochem) and then analyzed by mass spectrometry. The combination of peptide and MMP-9 is shown at the top left corner of each panel. Peaks are labeled as in A. MMP-9 can cleave the Zejm peptides at two sites, Arg-1625—Ile-1626 and Gly-1627—Leu-1628 of Ptprz-A. The figures are representative of two separate experiments.
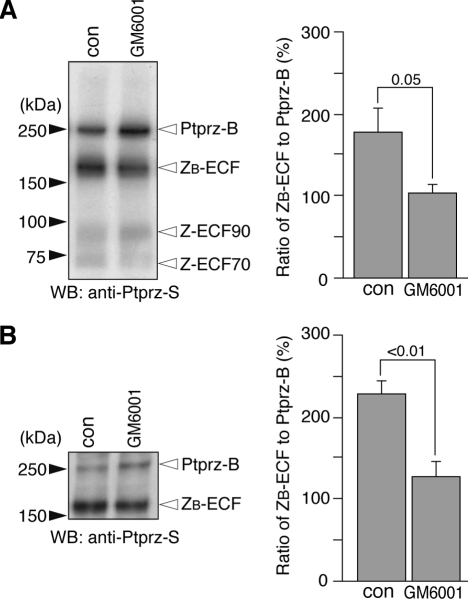
Metalloproteinase-mediated Proteolytic Cleavage of Ptprz in Vivo— To verify the occurrence of the metalloproteinase-mediated cleavage of Ptprz in vivo, mice were administered with GM6001. We tested the effect of GM6001 in the eyeball first, because Ptprz-B mRNA is exclusively expressed in the retina (data not shown). When the extract prepared from the eyeball was analyzed, the signal intensity of the 180-kDa species (ZB-ECF) was found to be markedly decreased as compared with that of Ptprz-B (250 kDa) at 48 h by single administration of GM6001 (Fig. 7A). The decrease in the relative amount of ZB-ECF to Ptprz-B was also observed in the brain by continuous intraventricular infusion of GM6001 for 4 days (Fig. 7B). This indicates that metalloproteinase-mediated processing of Ptprz occurs under physiological conditions.
FIGURE 7.
Metalloproteinase-mediated cleavage of Ptprz in vivo. A, tissue extracts of eye balls were prepared from wild-type mice 2 days after administration of GM6001 or vehicle (con). The extracts (5 μg protein) were treated with chABC and then analyzed by Western blotting (WB) using anti-Ptprz-S. The designations are shown in Fig. 1, A and E. Densitometric analysis indicated a significant decrease in the amount of ZB-ECF relative to that of Ptprz-B in mice treated with GM6001 as compared with those treated with vehicle (Student's t test, n = 4 each). Values are expressed as the mean ± S.E. B, wild-type mice were received a continuous intracerebroventricular infusion of GM6001 or vehicle (con) for 4 days. Tissue extracts of the midbrain region (corresponding to the midbrain, thalamus, and subthalamus) (see Ref. 14) were analyzed as above. There was a significant decrease in the amount of ZB-ECF relative to that of Ptprz-B in mice treated with GM6001 as compared with those treated with vehicle (con) treatments (Student's t test, n = 3 each). Values are expressed as the mean ± S.E.
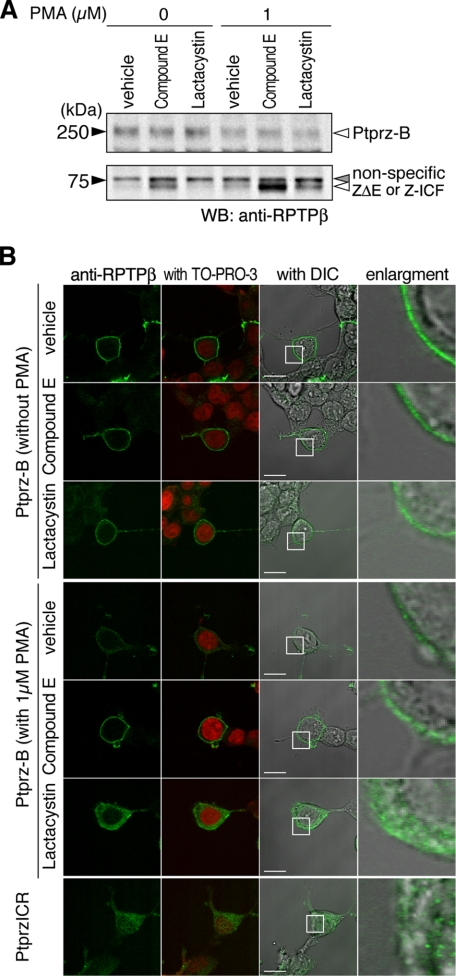
Regulated Intracellular Processing of Ptprz—In most cases membrane-associated fragments that are subsequently generated by metalloproteinase-mediated cleavage of transmembrane proteins are converted to cytosolic fragments by regulated intramembrane proteolysis (RIP) with an active γ-secretase complex (25). However, the intracellular domain released by the two-step proteolytic cleavage has been seldom visualized because of proteasome digestion (26). In the extracts of HEK293T cells expressing Ptprz-B, the 73-kDa band was observed with anti-RPTPβ (Fig. 3A); the same band was observed also in the cell extracts expressing Ptprz-A (data not shown). To determine whether the 73-kDa band represents either the membrane-tethered fragment ZΔE or the intracellular fragment Z-ICF (see Fig. 1A), cells were treated with a γ-secretase inhibitor, compound E, which was expected to induce the accumulation of ZΔE by suppressing the conversion of ZΔE to Z-ICF. In the presence of compound E, the signal intensity of the 73-kDa band was increased in both vehicle-stimulated and PMA-stimulated conditions (Fig. 8A). On the other hand, upon treatment with a proteasome inhibitor, lactacystin, which is expected to inhibit the degradation of Z-ICF, the signal intensity of the 73-kDa band was again increased in PMA-stimulated cells (Fig. 8A). These results indicate that the band of ZΔE overlaps with that of Z-ICF and that Z-ICF produced from ZΔEby γ-secretase is rapidly degraded by the proteasome under normal conditions.
FIGURE 8.
Presenilin/γ-secretase-mediated intramembrane cleavage of Ptprz in HEK293T cells. A, HEK293T cells expressing Ptprz-B were pretreated with vehicle, compound E (1 μm), or lactacystin (5 μm) for 3 h, then cultured with or without PMA stimulation for 1 h. Cell extracts were analyzed by Western blotting (WB) using anti-RPTPβ. The figure is representative of three separate experiments, and the result of the densitometric analysis is shown in supplemental Fig. S4. The designations are shown in Fig. 1, A and E. B, HEK293T cells treated as described in A were immediately fixed with formalin and stained with anti-RPTPβ. In addition, HEK293T cells that were transiently transfected with the expression construct of the entire intracellular region of Ptprz (PtprzICR) were also analyzed as above. The fluorescence images of anti-RPTPβ staining (green) and merged images with TO-PRO-3-stained nuclei (red) or with differential interference contrast images (DIC) are shown. The rightmost images are enlargements of the area enclosed by a square in the adjacent images. Scale bars, 10 μm. The figures are representative of three separate experiments.
For discrimination between ZΔE and Z-ICF, their subcellular localization by immunofluorescent staining using a confocal microscope should be helpful. As shown in Fig. 8B, in the basal condition, anti-RPTPβ immunoreactivity was exclusively localized to the cell surface in Ptprz-B-positive cells, whereas the cell surface signal was considerably decreased by PMA treatment along with a slight increase in the intracellular staining. In the presence of compound E to inhibit γ-secretase cleavage, PMA-induced decrease in the cell surface signal was apparently prevented, indicating that the cell membrane-tethered ZΔE occupies the majority of the 73-kDa species in this condition.
In contrast, when cells were stimulated with PMA in the presence of lactacystin to inhibit proteasomal degradation, the immunoreactivity was mainly localized to the intracellular compartment, and furthermore, several immunoreactive puncta were observed in the nucleus (Fig. 8B). Therefore, in this condition, the intracellular fragment Z-ICF is substantially accumulated in the cell, whereas some amounts of Ptprz-B and ZΔE remain on the cell surface. Of note, a similar distribution was observed when the entire intracellular region of Ptprz (PtprzICR) was expressed using a cDNA construct, where the signal in the nucleus was more evident (Fig. 8B). There was no signal of PtprzICR at the cell membrane, indicating that the intracellular domain of Ptprz cleaved by γ-secretase is not tethered to the membrane anymore.
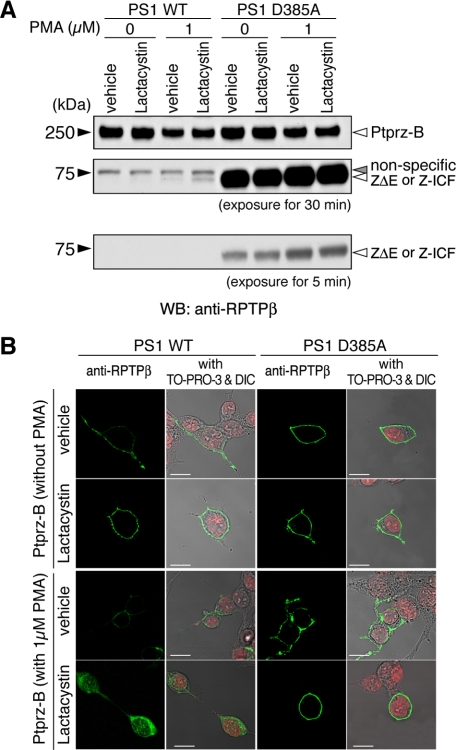
To verify that the generation of Z-ICF is dependent on the activity of PS, the catalytic subunit of the γ-secretase, Ptprz-B, was transiently expressed in HEK293 cells stably expressing wild-type PS1 (PS1 WT) or a dominant negative mutant (PS1 D385A) (16). In PS1 D385A cells, the signal intensity of the 73-kDa band was drastically increased as compared with PS1 WT cells (Fig. 9A), indicating rapid conversion of ZΔE to Z-ICF by RIP and instability of the Z-ICF. Consistently, in the presence of lactacystin, PMA-enhanced intracellular staining was markedly enhanced in cells expressing wild-type PS1 but not in cells expressing PS1 D385A (Fig. 9B). This also reinforces the view that the generation of Z-ICF is dependent on the PS activity.
FIGURE 9.
Intramembrane cleavage of Ptprz in HEK cells stably expressing presenilin or its dominant-negative variant. A, HEK293 cells stably expressing either wild-type presenilin 1 (PS1 WT) or a dominant-negative PS1 variant (PS1 D385A) were transiently transfected with the Ptprz-B expression construct. Twenty-four hours after transfection cells were treated and analyzed by Western blotting (WB) using anti-RPTPβ as described in Fig. 8. The figure is representative of three separate experiments, and the result of the densitometric analysis is shown in supplemental Fig. S5. The designations are shown in Fig. 1, A and E. B, the cells treated as described in A were fixed and stained with anti-RPTPβ (green). Merged images with TO-PRO-3-stained nuclei (red) and differential interference contrast images (DIC) are also shown. Scale bars, 10 μm. The figures are representative of three separate experiments.
DISCUSSION
In the present study we demonstrated by cell-based assays that the two receptor isoforms of Ptprz, Ptprz-A and Ptprz-B, undergo metalloproteinase-mediated ectodomain shedding, which releases the extracellular fragment, ZA/B-ECF, from the cell surface and produces the membrane-tethered counterpart ZΔE. Importantly, administration of GM6001 to mice demonstrated its physiological occurrence. ZΔE is subsequently digested by PS/γ-secretase, and the cytoplasmic fragment (Z-ICF) is released from the plasma membrane to not only the cytoplasm but also the nucleus, suggesting a novel signaling pathway of Ptprz. We summarized their molecular natures (Fig. 1A) and corresponding bands observed in the brain (Fig. 1E). As for the lower molecular species (bands e–g) observed in the brain, we have already revealed that they are produced by the proteolytic cleavage of the extracellular region of Ptprz-A/-B and Ptprz-S by plasmin (27). We have also identified that the upper species (77 kDa, band h) of the doublet bands is produced by proteolytic cleavage of exon 16 (a cytoplasmic exon)-deleted form of Ptprz-A/-B.3
Several groups (11, 13) including us have reported that three transcripts corresponding to Ptprz-A, Ptprz-B, and Ptprz-S are expressed in the adult brain. On the other hand, a cDNA clone for phosphacan short isoform, an isoform of Ptprz that roughly corresponds to the extracellular fragment of Ptprz-B, was reported recently from a neonatal mouse brain cDNA library (28). However, we could not detect the corresponding transcript of 4 kb for phosphacan short isoform in the adult mouse brain (Fig. 2). Moreover, we could not find out the sequence corresponding to the 3′-untranslated region of phosphacan short isoform (GenBank™ accession number AJ428208) in or near the mouse Ptprz gene (the Ensembl data base; release 48, Dec 2007). We suspect that the phosphacan short isoform cDNA clone is a cloning artifact obtained by ligation with an unrelated cDNA fragment. In addition, we did not find any evidence for the presence of PTPRZ2, which was reported as a familial gene on human chromosome 1 at p36 (29), by searching databases of the human and mouse genomes.
The core protein of Ptprz-A (380 kDa) has been hardly detected in the adult brain (Fig. 1B) as described (3) despite the significant expression at the mRNA level (Fig. 2 and Refs. 11 and 13). This strongly suggests that almost all Ptprz-A is constitutively processed to ZA-ECF and ZΔE (or Z-ICF) in the brain. Proteolytically released ZA-ECF has almost the same structure as Ptprz-S. Ptprz-S (phosphacan/6B4 proteoglycan) has been considered as the sole component of phosphate-buffered saline-soluble chondroitin sulfate proteoglycan with a 300-kDa core protein in the brain (10, 11). However, densitometric analyses of the Western (Fig. 1B) and Northern (Fig. 2) blots suggested that ZA-ECF released may account for approximately one-third of the total amount of the phosphacan/6B4 proteoglycan fraction.
The data obtained from TACE-defective CHO-M2 cells clearly showed that the PMA-stimulated shedding of the Ptprz receptor isoforms is mediated in part by TACE. Although release of the extracellular domains by metalloproteinase-mediated processing has been observed for several transmembrane proteins, the mechanism by which MMPs or ADAMs recognize and cleave the substrates is not well defined (21). The peptide digestion experiments in vitro identified the site of Ptprz cleavage by TACE as between Gly-1631 and Leu-1632 (Fig. 5), corresponding to between the 6th and 7th residues outside the transmembrane domain of Ptprz-A/-B. This is consistent with the finding that the cleavage site in a variety of substrates is located within the ectodomain stalk region, at residues 2–20 from the transmembrane region. The mutation of Gly at position 1631 to Ile suppressed the TACE-mediated cleavage of Ptprz both in cultured cells and in vitro, whereas TACE-independent basal (constitutive) shedding was not affected thereby. We presume that there exist multiple cleavage sites within the membrane proximal region, which is common to the two Ptprz receptor isoforms, by metalloproteinases.
One conspicuous defect in adult Ptprz-deficient mice is a significant enhancement of long term potentiation (LTP) in the CA1 region of the hippocampus (6) and learning deficits (6, 7). LTP is a long-lasting augmentation of synaptic strength that has been suggested as a cellular mechanism underlying learning and memory. We recently reported that the tyrosine phosphorylation level of a GTPase-activating protein (GAP) for Rho GTPase, p190 RhoGAP, a substrate molecule of Ptprz, is decreased 1 h after the conditioning in the hippocampus of wild-type mice but not of Ptprz-deficient mice (7), suggesting that the PTP activity of Ptprz may be up-regulated during learning. Dimerization-induced inactivation of RPTPs is a well known mechanism. Ligand binding to the extracellular region induces the dimerization (or oligomerization) of Ptprz and thereby inhibits the PTP activity (30). The removal of the extracellular region by metalloproteinase-mediated processing presumably abolishes this ligand-induced inactivation mechanism. Of note is that the intracellular region of Ptprz (PtprzICR) efficiently dephosphorylates substrates in cultured cells (data not shown).
LTP exhibits two distinct phases (31). The initial early phase (E-LTP), which is rapidly induced, only lasts ∼1–2 h. This form does not require protein synthesis and reflects at least in part posttranslational modifications including phosphorylation and translocation of the synaptic proteins. The second, more slowly emerging late phase (L-LTP), lasts many hours to days or longer. Interestingly, the MMP-9 protein level and accordingly the proteolytic activity are rapidly increased by stimuli that induce L-LTP in the CA1, and the pharmacological blockade of MMP-9 prevents the induction of L-LTP (32). Thus, there is a possibility that Ptprz-B is a target of the MMP(s) associated with L-LTP. Ptprz receptor isoforms interact with the postsynaptic density-95 (PSD95) family including PSD95, SAP97 (synapse-associated protein 97), and SAP102 (synapse-associated protein 102) through the carboxyl-terminal PDZ (PSD95/Disc large/zona occludens1) binding motif (33, 34). In this context TACE is also interesting, as this sheddase harbors a PDZ binding motif at the C terminus and thereby associates with members of the PSD95 family (35). Notably, the C-terminal counterparts of Ptprz receptor isoforms (ZΔE or Z-ICF) are enriched in the PSD fraction of the adult mouse brain.3
It is tempting to speculate that Ptprz is implicated in L-LTP through the regulation of gene expression by regulated shedding in addition to the simple dephosphorylation of the substrate molecules including p190 RhoGAP (7) for E-LTP at central synapses. Our cell-based assays indicated that the intracellular fragment of Ptprz (Z-ICF) is translocated into the nucleus in cultured cells (Figs. 8 and 9). Interestingly, overexpression of Z-ICF by pZeoSV-PtprzICR in glioblastoma cells stimulates the morphological differentiation (our preliminary observations). Elucidating the functional roles of Z-ICF in the nucleus will be highly challenging themes for future studies.
Supplementary Material
Acknowledgments
We thank J. Arribas for providing CHO-M2 cells and T. Ikeuchi for HEK293 (PS1 D385A) cells. We also thank A. Kodama for secretarial assistance. ABI 431 peptide synthesizer, high performance liquid chromatography, and Reflex III were used at the NIBB Center for Analytical Instruments.
This work was supported by grants from the Ministry of Education, Science, Sports, and Culture of Japan. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
Footnotes
The abbreviations used are: RPTP, receptor-like protein-tyrosine phosphatase (PTP); ADAM, a disintegrin and metalloproteinase; CHO, Chinese hamster ovary cells; HEK293 cells, human embryonic kidney cells; LTP, long term potentiation; MMP, matrix metalloproteinase; PMA, phorbol 12-myristate 13-acetate; PS, presenilin; PSD95, postsynaptic density-95; RIP, regulated intramembrane proteolysis; TACE, tumor necrosis factor-α (TNF-α) converting enzyme; WT, wild type; chABC, chondroitinase ABC; MOPS, 4-morpholinepropanesulfonic acid; kb, kilobase(s).
J. P. H. Chow, A. Fujikawa, H. Shimizu, R. Suzuki, and M. Noda, unpublished data.
References
- 1.Tonks, N. K. (2006) Nat. Rev. Mol. Cell Biol. 7 833-846 [DOI] [PubMed] [Google Scholar]
- 2.Levy, J. B., Canoll, P. D., Silvennoinen, O., Barnea, G., Morse, B., Honegger, A. M., Huang, J. T., Cannizzaro, L. A., Park, S. H., Druck, T., Huebner, K., Sap, J., Ehrlich, M., Musacchio, J. M., and Schlessinger, J. (1993) J. Biol. Chem. 268 10573-10581 [PubMed] [Google Scholar]
- 3.Nishiwaki, T., Maeda, N., and Noda, M. (1998) J. Biochem. (Tokyo) 123 458-467 [DOI] [PubMed] [Google Scholar]
- 4.Shintani, T., Watanabe, E., Maeda, N., and Noda, M. (1998) Neurosci. Lett. 247 135-138 [DOI] [PubMed] [Google Scholar]
- 5.Fujikawa, A., Shirasaka, D., Yamamoto, S., Ota, H., Yahiro, K., Fukada, M., Shintani, T., Wada, A., Aoyama, N., Hirayama, T., Fukamachi, H., and Noda, M. (2003) Nat. Genet. 33 375-381 [DOI] [PubMed] [Google Scholar]
- 6.Niisato, K., Fujikawa, A., Komai, S., Shintani, T., Watanabe, E., Sakaguchi, G., Katsuura, G., Manabe, T., and Noda, M. (2005) J. Neurosci. 25 1081-1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tamura, H., Fukada, M., Fujikawa, A., and Noda, M. (2006) Neurosci. Lett. 399 33-38 [DOI] [PubMed] [Google Scholar]
- 8.Harroch, S., Palmeri, M., Rosenbluth, J., Custer, A., Okigaki, M., Shrager, P., Blum, M., Buxbaum, J. D., and Schlessinger, J. (2000) Mol. Cell. Biol. 20 7706-7715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krueger, N. X., and Saito, H. (1992) Proc. Natl. Acad. Sci. U. S. A. 89 7417-7421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maeda, N., Hamanaka, H., Shintani, T., Nishiwaki, T., and Noda, M. (1994) FEBS Lett. 354 67-70 [DOI] [PubMed] [Google Scholar]
- 11.Maurel, P., Rauch, U., Flad, M., Margolis, R. K., and Margolis, R. U. (1994) Proc. Natl. Acad. Sci. U. S. A. 91 2512-2516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sakurai, T., Friedlander, D. R., and Grumet, M. (1996) J. Neurosci. Res. 43 694-706 [DOI] [PubMed] [Google Scholar]
- 13.Canoll, P. D., Petanceska, S., Schlessinger, J., and Musacchio, J. M. (1996) J. Neurosci. Res. 44 199-215 [DOI] [PubMed] [Google Scholar]
- 14.Glowinski, J., and Iversen, L. L. (1966) J. Neurochem. 13 655-669 [DOI] [PubMed] [Google Scholar]
- 15.Kawachi, H., Fujikawa, A., Maeda, N., and Noda, M. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 6593-6598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kasuga, K., Kaneko, H., Nishizawa, M., Onodera, O., and Ikeuchi, T. (2007) Biochem. Biophys. Res. Commun. 360 90-96 [DOI] [PubMed] [Google Scholar]
- 17.Fujikawa, A., Chow, J. P. H., Shimizu, H., Fukada, M., Suzuki, R., and Noda, M. (2007) J. Biochem. (Tokyo) 142 343-350 [DOI] [PubMed] [Google Scholar]
- 18.Borroto, A., Ruíz-Paz, S., de la Torre, T. V., Borrell-Pagès, M., Merlos-Suárez, A., Pandiella, A., Blobel, C. P., Baselga, J., and Arribas, J. (2003) J. Biol. Chem. 278 25933-25939 [DOI] [PubMed] [Google Scholar]
- 19.Suzuki, R., Shintani, T., Sakuta, H., Kato, A., Ohkawara, T., Osumi, N., and Noda, M. (2000) Mech. Dev. 98 37-50 [DOI] [PubMed] [Google Scholar]
- 20.Maeda, N., Hamanaka, H., Oohira, A., and Noda, M. (1995) Neuroscience 67 23-35 [DOI] [PubMed] [Google Scholar]
- 21.Arribas, J., and Borroto, A. (2002) Chem. Rev. 102 4627-4637 [DOI] [PubMed] [Google Scholar]
- 22.Anders, L., Mertins, P., Lammich, S., Murgia, M., Hartmann, D., Saftig, P., Haass, C., and Ullrich, A. (2006) Mol. Cell. Biol. 26 3917-3934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blobel, C. P. (2005) Nat. Rev. Mol. Cell Biol. 6 32-43 [DOI] [PubMed] [Google Scholar]
- 24.Jin, G., Huang, X., Black, R., Wolfson, M., Rauch, C., McGregor, H., Ellestad, G., and Cowling, R. (2002) Anal. Biochem. 302 269-275 [DOI] [PubMed] [Google Scholar]
- 25.Selkoe, D., and Kopan, R. (2003) Annu. Rev. Neurosci. 26 565-597 [DOI] [PubMed] [Google Scholar]
- 26.Gupta-Rossi, N., Le Bail, O., Gonen, H., Brou, C., Logeat, F., Six, E., Ciechanover, A., and Israël, A. (2001) J. Biol. Chem. 276 34371-34378 [DOI] [PubMed] [Google Scholar]
- 27.Chow, J. P. H., Fujikawa, A., Shimizu, H., and Noda, M. (2008) Neurosci. Lett. 442 208-212 [DOI] [PubMed] [Google Scholar]
- 28.Garwood, J., Heck, N., Reichardt, F., and Faissner, A. (2003) J. Biol. Chem. 278 24164-24173 [DOI] [PubMed] [Google Scholar]
- 29.Onyango, P., Lubyova, B., Gardellin, P., Kurzbauer, R., and Weith, A. (1998) Genomics 50 187-198 [DOI] [PubMed] [Google Scholar]
- 30.Fukada, M., Fujikawa, A., Chow, J. P. H., Ikematsu, S., Sakuma, S., and Noda, M. (2006) FEBS Lett. 580 4051-4056 [DOI] [PubMed] [Google Scholar]
- 31.Reymann, K. G., and Frey, J. U. (2007) Neuropharmacology 52 24-40 [DOI] [PubMed] [Google Scholar]
- 32.Nagy, V., Bozdagi, O., Matynia, A., Balcerzyk, M., Okulski, P., Dzwonek, J., Costa, R. M., Silva, A. J., Kaczmarek, L., and Huntley, G. W. (2006) J. Neurosci. 26 1923-1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kawachi, H., Tamura, H., Watakabe, I., Shintani, T., Maeda, N., and Noda, M. (1999) Mol. Brain Res. 72 47-54 [DOI] [PubMed] [Google Scholar]
- 34.Fukada, M., Kawachi, H., Fujikawa, A., and Noda, M. (2005) Methods 35 54-63 [DOI] [PubMed] [Google Scholar]
- 35.Peiretti, F., Deprez-Beauclair, P., Bonardo, B., Aubert, H., Juhan-Vague, I., and Nalbone, G. (2003) J. Cell Sci. 116 1949-1957 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.