Abstract
FAD synthetases (EC 2.7.7.2) catalyze biosynthesis of FAD from FMN and ATP. Monofunctional FAD synthetases are known to exist in mammals and yeast; bifunctional enzymes also catalyzing phosphorylation of riboflavin to FMN are known to exist in bacteria. Previously known eukaryotic enzymes with FAD synthetase activity have no sequence similarity to prokaryotic enzymes with riboflavin kinase and FAD synthetase activities. Proteins homologous to bacterial bifunctional FAD synthetases, yet shorter and lacking amino acid motifs at the C terminus, were found by bioinformatic analyses in vascular plant genomes, suggesting that plants contain a type of FAD synthetase previously known to exist only in prokaryotes. The Arabidopsis thaliana genome encodes two of such proteins. Both proteins, which we named AtRibF1 and AtRibF2, carry N-terminal extensions with characteristics of organellar targeting peptides. AtRibF1 and AtRibF2 cDNAs were cloned by reverse transcription-PCR. Only FAD synthetase activity was detected in the recombinant enzymes produced in Escherichia coli. FMN and ATP inhibited both enzymes. Kinetic parameters of AtRibF1 and AtRibF2 for the two substrates were similar. Confocal microscopy of protoplasts transformed with enhanced green fluorescence protein-fused proteins showed that AtRibF1 and AtRibF2 are targeted to plastids. In agreement with subcellular localization to plastids, Percoll-isolated chloroplasts from pea (Pisum sativum) synthesized FAD from imported riboflavin. Riboflavin kinase, FMN hydrolase, and FAD pyrophosphatase activities were detected in Percoll-isolated chloroplasts and mitochondria from pea. We propose from these new findings a model for subcellular distribution of enzymes that synthesize and hydrolyze flavin nucleotides in plants.
FMN and FAD are essential cofactors for a variety of enzymes that participate in many metabolic processes in all organisms. In plants, these cofactors are required for photosynthesis, mitochondrial electron transport, fatty acid oxidation, photoreception, DNA repair, metabolism of other cofactors, and biosynthesis of many secondary metabolites. Metabolism of FMN and FAD includes reactions that synthesize and hydrolyze these flavin nucleotides. Enzymes catalyzing flavin nucleotide biosynthesis and hydrolysis, and their subcellular localization, are incompletely understood in plants and in other organisms.
Phosphorylation of riboflavin to FMN and adenylylation of FMN to FAD are, respectively, catalyzed by the enzymes riboflavin kinase (EC 2.7.1.26) and FAD synthetase (EC 2.7.7.2) in the presence of ATP and Mg2+. These enzymes have been found in prokaryotes and eukaryotes.
In prokaryotes, bifunctional enzymes with riboflavin kinase and FAD synthetase activities (1–5), and monofunctional enzymes with only riboflavin kinase activity (6, 7) have been described. No monofunctional FAD synthetases have yet been found in prokaryotes. Bioinformatic evidence suggests that the bifunctional enzymes are prevalent among the prokaryotic species sequenced to date (8).
In mammals and yeast, only monofunctional enzymes with riboflavin kinase or FAD synthetase activity have been described (9–20). Riboflavin kinases, but not FAD synthetases, from mammals and yeast share sequence similarity to the bifunctional enzymes from bacteria.
Subcellular localization of riboflavin kinases and FAD synthetases has been investigated in mammals and yeast. Enzymatic synthesis of FAD was first shown to occur in the cytosol of rat liver (21). Synthesis of FAD from externally added riboflavin or FMN has subsequently provided evidence for riboflavin kinase and FAD synthetase activities in mitochondria in Saccharomyces cerevisiae (22, 23) and rat liver (24, 25). A subcellular localization study provides support for a riboflavin kinase residing in microsomes and the inner mitochondrial membrane in S. cerevisiae (14).
In plants, earlier studies have demonstrated the presence of monofunctional riboflavin kinases (26–28) and FAD synthetases (29). However, none of those enzymes have been fully characterized nor have the corresponding genes been cloned. Thus, it is unknown whether those plant enzymes are sequence homologs of riboflavin kinases and FAD synthetases from mammals and yeast, or from bacteria.
Recently, we have reported bioinformatic evidence showing that plants have sequence homologs of eukaryotic monofunctional enzymes with riboflavin kinase or FAD synthetase activity, and sequence homologs of prokaryotic bifunctional enzymes with both activities (8). We have also reported the first cDNA cloning, recombinant expression, and biochemical characterization of a riboflavin kinase from a plant species (8). This enzyme shares sequence similarity to the monofunctional riboflavin kinases from mammals and yeast.
The only study investigating subcellular localization of flavin nucleotide biosynthesis in plants reported riboflavin kinase activity in the cytosol and in an organellar fraction containing chloroplasts and mitochondria in spinach (30). The riboflavin kinase we recently described probably resides in the cytosol because it lacks an organellar targeting peptide. To our knowledge, subcellular localization of FAD synthetase activity in plants remains to be explored. Bioinformatic evidence suggests that plant cells have FAD synthetase activity in organelles and the cytosol (8), but this remains to be established experimentally.
Hydrolysis of FMN to riboflavin and inorganic phosphate is catalyzed by acid phosphatases (EC 3.1.3.2) in plants (30–32) and in other organisms (33–38). Acid phosphatases are most active in an acidic environment, quickly loosing activity at pH > 5.5–6.0 (30, 31, 34). They also hydrolyze substrates other than FMN (30, 32). We have recently described a novel type of FMN hydrolase that belongs to the haloacid dehalogenase enzyme superfamily (8). This Mg2+-dependent enzyme is fused to the N terminus of the riboflavin kinase described above, is highly specific for FMN,2 and, unlike acid phosphatases, catalyzes FMN hydrolysis effectively at pH 6.0–8.0.
Mammalian cells contain FMN-hydrolyzing enzymes in the cytosol (33, 34) and in the intermembrane space of mitochondria (38). Subcellular localization of FMN-hydrolyzing acid phosphatases in plants remains to be explored. As discussed earlier, the Mg2+-dependent FMN hydrolase we reported is probably cytosolic.
Hydrolysis of FAD to FMN and AMP is catalyzed by the enzyme FAD pyrophosphatase (EC 3.6.1.18) in plants (39–43) and in other organisms (38, 44–46). None of the genes for FAD pyrophosphatases have yet been cloned. All known FAD pyrophosphatases also hydrolyze other metabolites such as NAD+, NADP+, ATP, ADP, CoA, and nucleotide sugars (39, 43, 44, 46). Subcellular localization of these enzymes remains unexplored. The only exception is a study providing evidence for an FAD pyrophosphatase in the outer mitochondrial membrane in rat liver (38).
We report here on cDNA cloning, recombinant expression, purification, and biochemical characterization of two novel FAD synthetases (AtRibF1 and AtRibF2). We provide evidence using confocal microscopy of protoplasts transformed with EGFP3-fused proteins that AtRibF1 and AtRibF2 reside in plastids. In agreement with the confocal microscopy data, we show that Percoll-isolated chloroplasts from pea (Pisum sativum) synthesize FAD from externally added riboflavin. Also, we show that protein extracts of Percoll-isolated chloroplasts and mitochondria from pea have riboflavin kinase, FMN hydrolase, and FAD pyrophosphatase activities. We propose from these new findings a revised model for subcellular localization of enzymes that synthesize and hydrolyze flavin nucleotides in plants.
EXPERIMENTAL PROCEDURES
Materials and Plant Growth Conditions—Riboflavin, [G-3H]riboflavin, FMN, FAD, and ATP were obtained from Sigma; BugBuster reagent, Benzonase nuclease, and recombinant enterokinase were from Novagen (Madison, WI); Percoll, Biodegradable Counting Scintillant, and the Gel Filtration Calibration kit were from GE Healthcare; silicone oil AR 200 was from Fluka; oligonucleotides were from MWG (High Point, NC).
Arabidopsis thaliana plants, ecotype Columbia, were grown in potting soil at 12-h light intervals (100–120 μEm-2 s-1) for 3 weeks at 22 °C. Peas, cv. Bohatyr, were grown in coarse vermiculite at 16-h light intervals (300 μEm-2 s-1) for 10 days at 22 °C during the day or at 18 °C during the night.
cDNA Cloning, Constructs, Sequence Analysis, and Expression in Escherichia coli—AtRibF1 (At5g23330) and AtRibF2 (At5g08340) cDNAs were cloned by reverse transcription PCR. Total A. thaliana RNA was isolated using the RNeasy Plant Mini Kit (Qiagen, Valencia, CA), and reverse-transcribed using Superscript II reverse transcriptase (Invitrogen) and an oligo(dT) primer. The AtRibF1 and AtRibF2 open reading frames were then amplified using Taq2000 DNA polymerase (Stratagene, La Jolla, CA) and using, respectively, the primer pairs 5′-GACGACGACAAGATGTTATGCGGAGGCTC-3′ (AtRibF1 forward) and 5′-GAGGAGAAGCCCGGTTTAACCGAACTCAATCCCTA-3′ (AtRibF1 reverse) or 5′-GACGACGACAAGATGTTGTGCGGAGGCTC-3′ (AtRibF2 forward) and 5′-GAGGAGAAGCCCGGTTCAACCGAACTCAACACTCA-3′ (AtRibF2 reverse). Note that the vector-specific sequences (underlined), needed for cloning into pET Ek/LIC expression vectors (Novagen), flank the gene-specific sequences of the primers. Amplified open reading frames, purified using Wizard PCR columns (Promega, Madison, WI), were cloned into the pGEM-T Easy vector (Promega) to generate the pGEM-AtRibF1 and pGEM-AtRibF2 constructs.
The AtRibF1 and AtRibF2 open reading frames, excluding the regions coding for the putative organellar targeting peptides, were amplified, respectively, from pGEM-AtRibF1 or pGEM-AtRibF2 using Pfu DNA polymerase (Stratagene) and the primer pairs 5′-GACGACGACAAGATGTCGTCGTTTAGGTCTCA-3′ (AtRibF1n forward) and AtRibF1 reverse or 5′-GACGACGACAAGATGATTTCTTCATTTGGGTCTC-3′ (AtRibF2n forward) and AtRibF2 reverse. To generate the expression vectors, resulting PCR fragments were purified using Wizard PCR columns, treated with T4 DNA polymerase, and then ligated into pET-30 Ek/LIC (AtRibF1) or pET-44 Ek/LIC (AtRibF2). All procedures were done in accordance with the manufacturer's protocols. Each construct was verified by DNA sequencing. The expression vectors were then introduced into the Rosetta strain of E. coli (Novagen) to produce the recombinant proteins. Bacteria carrying the expression vectors were cultured at 37 °C in LB medium containing 34 μg/ml chloramphenicol and 100 μg/ml kanamycin (for pET-30–AtRibF1) or 100 μg/ml ampicillin (for pET-44–AtRibF2). Once A600 reached 0.6–1, isopropyl β-d-thiogalactopyranoside was added to a final concentration of 200 μm, and incubation was continued overnight at 15 °C.
Recombinant Protein Isolation and Molecular Weight Determination—Extracts of E. coli cells expressing the recombinant AtRibF1 were prepared for purification as follows. Induced E. coli cells from 200-ml cultures were harvested by centrifugation (5,000 × g, 15 min, 4 °C). Bacterial pellets were resuspended in 5 ml of lysis buffer (50 mm Tris-HCl, pH 8.0, 300 mm NaCl, 10 mm imidazole, 0.5 mm THP, 1× BugBuster reagent, 0.5 μl of Benzonase nuclease, and 10% glycerol). After a 15-min incubation at room temperature, cell lysates were cleared by centrifugation (20,000 × g, 20 min, 4 °C); supernatants were filtered through a 45-μm polyvinylidene fluoride membrane and then purified by Ni-NTA His-Bind (Novagen) affinity chromatography following the manufacturer's protocol. Cleared cell lysates were incubated with 1 ml of Ni-NTA resin (Novagen) for 1 h at 8 °C. Wash and elution buffers contained 50 mm Tris-HCl, pH 8.0, 300 mm NaCl, 20 (wash) or 150 mm (elution) imidazole, 0.5 mm THP, and 10% glycerol. Eluate fractions were immediately desalted on PD-10 columns (GE Healthcare) equilibrated with buffer A (50 mm Tris-HCl, pH 8.5, 1 mm MgCl2, 1 mm THP, 10% glycerol, and 0.1% CHAPS), and then digested with recombinant enterokinase to release the tag. The digested enzyme was purified by ion exchange chromatography using an ÄKTA FPLC system equipped with a Mono Q 5/50 GL column (GE Healthcare). The column was equilibrated with buffer A, and bound proteins were eluted with a linear gradient of 0–500 mm NaCl over 20 column volumes. AtRibF1 eluted at about 110 mm NaCl. AtRibF1-containing fractions were pooled and further purified by gel filtration chromatography using a Superose 12 10/300 GL column (GE Healthcare) equilibrated with buffer A.
To purify the recombinant AtRibF2, cleared lysates of E. coli cells expressing the recombinant protein were prepared as described for AtRibF1. The recombinant enzyme was purified by ion exchange chromatography using a Mono Q 5/50 GL column. The column was equilibrated with buffer A, and bound proteins were eluted with a linear gradient of 0–500 mm NaCl over 20 column volumes. AtRibF2 carrying a Nus tag eluted at about 150 mm NaCl. AtRibF2-containing fractions were pooled, desalted on a PD-10 column equilibrated with buffer A, and then digested with recombinant enterokinase to release the tag. The digested enzyme was further purified by ion exchange chromatography as described for the Nus-tagged protein. Untagged AtRibF2 eluted at about 100 mm NaCl. Purified AtRibF1 or AtRibF2 was frozen in liquid N2 and stored at -80 °C until use. Freezing did not affect the activity of AtRibF1 or AtRibF2.
Molecular weights of native AtRibF1 and AtRibF2 were estimated by gel filtration chromatography using a Superose 12 10/300 GL column equilibrated with buffer A. Reference proteins were aprotinin (6,500), ribonuclease A (13,700), carbonic anhydrase (29,000), and ovalbumin (43,000). Protein concentrations were determined by the Bradford method (47) using bovine serum albumin as standard.
Isolation of Chloroplasts and Mitochondria from Pea—Chloroplasts and root mitochondria were isolated by centrifuging on Percoll density gradients using published procedures (48, 49) as described before (50). Isolated organelles were resuspended in 100 mm Tris-HCl buffer, pH 7.5, containing 1 mm MgCl2 and 1 mm THP; and were broken by four cycles of freezing and thawing. Organelle extracts were cleared by centrifugation (20,000 × g, 20 min, 4 °C), and supernatants were desalted on PD-10 columns equilibrated with 50 mm Tris-HCl buffer, pH 7.5, containing 1 mm MgCl2, 1 mm THP, and 10% glycerol. The mitochondrial marker fumarase and the plastid marker glyceraldehyde-3-phosphate dehydrogenase were assayed as described before (50). Fumarase and FAD synthetase were assayed immediately after desalting. All other enzymes were assayed for activity with extracts that were frozen in liquid N2 and stored at -80 °C.
Purification of FMN and FAD—Aqueous FMN or FAD solution (100 mm) was subjected to reversed-phase preparative chromatography on a SunFire Prep C18 OBD column (19 × 50 mm, 5 μm) using a 626 LC system equipped with a 717plus autosampler (Waters, Milford, MA). The mobile phase contained 100 mm ammonium formate, 100 mm formic acid, and 25% methanol (2). FMN- or FAD-containing fractions were evaporated to dryness; flavin nucleotides were dissolved in water, re-purified as above, and then desalted using Chromabond C18 Hydra columns (Macherey-Nagel, Düren, Germany) with 80% methanol as eluent. FMN- or FAD-containing fractions were evaporated to dryness; purified flavin nucleotides were dissolved in water, and stored at -20 °C until use.
Enzyme Assays—Riboflavin kinase, FAD synthetase, FMN hydrolase, and FAD pyrophosphatase activities were measured using an Alliance 2695 HPLC system with a 2475 fluorescence detector (Waters). Riboflavin, FMN, and FAD concentrations were determined spectrophotometrically (14). Unless otherwise indicated, the procedures described below were used. Initial reaction rates at steady state were measured. Substrates were saturating, and product formation was proportional to enzyme concentration and time. Less than 5% of the substrates were typically consumed. Final assay volumes were 50 μl. Riboflavin kinase and FAD synthetase activities were assayed in 100 mm Tris-HCl buffer, pH 7.5 or 8.5, containing 15 mm MgCl2, 1 mm THP, 3 mm ATP, and 0.05 mm flavin. FMN hydrolase and FAD pyrophosphatase activities were assayed in 100 mm Tris-HCl buffer, pH 7.5 or 8.5, containing 15 mm MgCl2, 1 mm THP, and 0.05 mm flavin.
After incubation at 30 °C for 20 min, reactions were stopped by adding saturated formic acid (5% of final assay volume), and then centrifuged (20,000 × g, 15 min, 4 °C) to remove the precipitated protein. Reaction products were separated by reversed-phase chromatography using a Waters SunFire C18 column (4.6 × 150 mm, 3.5 μm) and were measured by fluorescence detection using an excitation wavelength of 470 nm and an emission wavelength of 530 nm. The mobile phase was as described for flavin nucleotide purification. Product formation was determined from fluorescence after subtraction of a blank, wherein the enzyme was added after incubation.
The kinetic parameters (Km, Ki, and Vmax) were calculated from experiments at pH 8.5 in which both substrates (FMN and ATP) were varied as follows. Apparent Vmax values at fixed concentrations of the first substrate were calculated by varying the second substrate and then fitting the initial reaction rates to a model of uncompetitive substrate inhibition using nonlinear regression (Enzyme Kinetics Module 1.2, SigmaPlot 9.0). Resulting apparent Vmax values, adjusted for the effect of inhibition by the second substrate, were then plotted against corresponding concentrations of the first substrate to create secondary plots. Km, Ki, and Vmax for the first substrate were calculated by fitting the secondary plot data to a model of uncompetitive substrate inhibition using nonlinear regression. The process was repeated for the second substrate. The standard error for catalytic efficiency, kcat/Km, was calculated by error propagation (51).
Transient Expression of EGFP-fused Proteins in A. thaliana Protoplasts—Constructs carrying C-terminal EGFP fusions to the putative signal peptides of AtRibF1 and AtRibF2 in the p2GWF7 vector (52) were generated using the Gateway cloning system (Invitrogen) as instructed by the manufacturer. Putative signal peptides at the N termini of AtRibF1 (MLCGGSRASVHLWDHRHPPRLGAKVLRKSSFMLRPCSAISQQRIKSSFRSHCKTPRKIPAPLDCFSQGDDHPELSAEGLSPVAGGIVALGKFDALHIGHRE) or AtRibF2 (MLCGGSRVLQHLSDHNHHNSIGLGLGFCGAKIVQLSSFFLRPSQAMAKSHHFSRKLRQRMISSFGSHCRTSGEVPILHNCFSQREDDPELPVEGLSPVSGGIVALGKFDALHIGHRE) were amplified using Pfu DNA polymerase and using, respectively, the primer pairs 5′-AAAAAGCAGGCTATGTTATGCGGAGGCTC-3′ and 5′-AGAAAGCTGGGTGCTCTCGATGACCGATATG-3′ or 5′-AAAAAGCAGGCTATGTTGTGCGGAGGCTC-3′ and 5′-AGAAAGCTGGGTGCTCTCGATGGCCGATATG-3′. Resulting PCR fragments were re-amplified using attB adapter primers (Invitrogen), purified using Wizard PCR columns, and subcloned into the pDONR 221 vector (Invitrogen) using BP recombination. The putative signal peptides of AtRibF1 and AtRibF2 were then introduced into the p2GWF7 vector from pDONR 221 using LR recombination.
A. thaliana protoplasts were isolated from leaves of 3-week-old plants, and transformed using published methods (53). Fluorescence was monitored using an LSM 510 confocal laser scanning microscope (Carl Zeiss MicroImaging, Thornwood, NY). EGFP fluorescence was excited at 488 nm, and measured at 505–530 nm. Chlorophyll fluorescence was excited at 488 nm, and measured at >650 nm.
In Vivo Analysis of FAD Biosynthesis in Chloroplasts—Percoll-isolated chloroplasts (0.9 mg of chlorophyll/ml) were resuspended in import buffer (50 mm Hepes-KOH, pH 8.0, and 330 mm d-sorbitol). [3H]Riboflavin (25 Ci/mmol; 50% ethanol solution) was evaporated to dryness, and resuspended in import buffer. Chloroplast suspensions (100 μl) were incubated with 5.8 μm [3H]riboflavin (3.45 Ci/mmol, adjusted with unlabeled riboflavin) over selected time intervals at 22 °C, with mixing by pipetting every 20 min. After incubation, chloroplasts were purified by silicone oil centrifugation (54), with the following modifications. Chloroplast suspensions were overlaid on 100-μl silicone oil layers in 300-μl microtubes, and centrifuged for 10 min using a microcentrifuge for PCR strips (Fisher). Live chloroplasts appeared as a pellet at the bottom of the tube, below the layer of silicone oil; broken chloroplasts and the incubation medium remained above the layer of silicone oil. Sample microtubes were frozen in liquid nitrogen, and stored in a vertical position at -80 °C until analysis.
We determined incorporation of the radiolabel into flavin nucleotides as follows. Sample microtubes were re-frozen in liquid nitrogen to solidify silicone oil. Chloroplast pellets were cut off near the bottom of the microtubes, and then resuspended in 100 μl of mobile phase described under “Enzyme Assays”; suspensions were heated for 10 min at 80 °C, and cleared by centrifugation (20,000 × g, 15 min, 4 °C). Supernatants were separated by reversed-phase chromatography as described under “Enzyme Assays.” To quantify [3H]FMN and [3H]FAD formed during the incubation of chloroplasts with [3H]riboflavin, four post-column fractions (100 μl each) containing these flavin nucleotides were collected; 80 μl of each fraction was mixed with 5 ml of scintillant, and the radioactivity was counted in a Tri-Carb 2100TR liquid scintillation analyzer (PerkinElmer). Peak identity was confirmed by co-elution with authentic standards. The radiolabel incorporated into FMN or FAD in each sample was calculated by adding the disintegrations per minute of all four fractions, and then subtracting the blank values calculated as described below.
To determine blank values, chloroplasts were incubated with [3H]riboflavin, purified using silicone oil, resuspended in 5 ml of scintillant, and the radiolabel was counted as before. Next, an equal amount of [3H]riboflavin was added to silicone oil-isolated chloroplasts incubated without [3H]riboflavin. Then the chloroplast pellet was extracted, the supernatant was separated by reversed-phase chromatography, and the radioactivity in the collected fractions was counted as before. Chlorophyll content in chloroplast pellets was determined spectrophotometrically (55).
Phylogenetic Analysis—We selected protein sequences from plants and bacteria for use in the phylogeny as follows.
For plant protein sequences, we conducted similarity searches of the GenBank™ protein, genome, and EST databases as well as the DOE Joint Genome Institute Eukaryotic Genomics data base (www.genome.jgi-psf.org) using the BLASTP and TBLASTN prediction programs with AtRibF1 as a query sequence. Then we selected RibF protein sequences from Oryza sativa, monocot (OsRibF, Os03g0801700); Picea glauca, gymnosperm (PgRibF, ESTs Ex437360.1 and Ex318592.1); Physcomitrella patens, moss (PpRibF1, fgenesh1_pg.scaffold_150000010; PpRibF2, estExt_Genewise1.C_2290051); Chlamydomonas reinhardtii, unicellular green alga (Chlorophyceae; CrRibF, EDO98955.1); and Ostreococcus tauri, unicellular green alga (Prasinophyceae; OtRibF, CAL54923.1). The P. glauca sequence was generated by translating the overlapping ESTs listed above. The P. patens sequences were obtained from the DOE Joint Genome Institute Eukaryotic Genomics data base. The N terminus of the C. reinhardtii protein sequence appeared to be incorrectly predicted. Hence, it was manually corrected based on alignments between RibF protein sequences from other plants, and that obtained by transcribing C. reinhardtii genomic DNA.
For bacterial protein sequences, we conducted similarity searches of the GenBank protein data base using BLASTP with AtRibF1 as a query sequence. Then we selected protein sequences from one species per phylum for nine phyla of bacteria. This selection comprises E. coli K12, Proteobacteria (NP_414566.1); Synechococcus elongatus PCC 6301, Cyanobacteria (YP_171738.1); Bacillus subtilis, Firmicutes (BsRibC, P54575); Propionibacterium acnes KPA171202, Actinobacteria (Pa, YP_056179.1); Dehalococcoides ethenogenes 195, Chloroflexi (YP_181344.1); Deinococcus radiodurans R1, Deinococcus thermus (NP_294732.1); Thermotoga maritima MSB8, Thermotogae (AAD35939.1); Chlorobium tepidum TLS, Chlorobi (NP_661148.1); and Rhodopirellula baltica SH 1, Planctomycetes (NP_868283.1). The selection process resulted in 17 protein sequences, which were further processed to eliminate the N- and C-terminal protrusions and were aligned using ClustalW (www.ebi.ac.uk/Tools/clustalw/index.html).
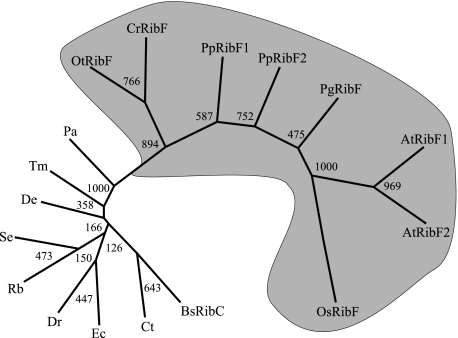
The phylogenetic trees were assembled using the PHYLIP (3.6.3) program package for MacOS X (56) as described before (8). The RibC protein from B. subtilis was used as outgroup for the phylogenetic analysis because it was the only protein of the group that had been biochemically characterized.
In Silico Expression Analysis—Gene expression was analyzed in silico using the publicly available A. thaliana microarray data from Affymetrix and the Meta Analyzer tool of the GENEVESTIGATOR software package (www.genevestigator.ethz.ch) (57).
RESULTS
Bioinformatic Sequence Analyses—We previously reported bioinformatic evidence that plant genomes encode sequence homologs of bifunctional enzymes catalyzing adenylylation of FMN to FAD, as well as phosphorylation of riboflavin to FMN (8). Such enzymes were previously known to exist only in prokaryotes. Current evidence suggests that mammals and yeast have only monofunctional enzymes catalyzing adenylylation of FMN to FAD, and that these eukaryotic enzymes have no sequence similarity to the bifunctional enzymes from bacteria.
BLAST searches of the A. thaliana data base using the protein sequence of the bifunctional riboflavin kinase–FAD synthetase from B. subtilis revealed two homologs encoded by genes At5g23330 and At5g08340 (8). Riboflavin kinase–FAD synthetase from B. subtilis is designated RibC, and the same enzyme from E. coli is designated RibF. Because plant riboflavin synthases (58) were previously named following the E. coli nomenclature, the proteins encoded by genes At5g23330 and At5g08340 were, respectively, named AtRibF1 and AtRibF2, also following the E. coli nomenclature.
Searches of genome and EST databases using TBLASTN and the RibC protein sequence revealed protein homologs present in angiosperms, gymnosperms, mosses, and green algae; and absent in other eukaryotes. A multiple sequence alignment of representative bacterial riboflavin kinases–FAD synthetases and their plant homologs is shown in Fig. 1. Presence of one or more genes encoding RibF proteins in angiosperms, gymnosperms, mosses, and green algae suggests that the RibF proteins originated early in plant evolution.
FIGURE 1.
Multiple alignment of selected FAD synthetase protein sequences. The protein sequences used for the multiple alignment are listed under “Phylogenetic Analysis” under “Experimental Procedures.” Multiple alignment was done using Multalin (61), and shading using Boxshade 3.21 (www.ch.embnet.org/software/BOX_form.html). Identical residues are shaded in black, similar residues in gray. Dashes are gaps introduced to maximize alignment. The arrowhead marks the second amino acid of the AtRibF1 expression construct.
RibF protein sequences from A. thaliana (AtRibF1 and AtRibF2), O. sativa (OsRibF), P. glauca (PgRibF), and P. patens (PpRibF1 and PpRibF2) were found to have N-terminal extensions relative to their sequence homologs from bacteria. Previous predictions using full-length RibF proteins from flowering plants (8) led us to hypothesize that these proteins reside in organelles in plants. Subsequent predictions using WoLF PSORT (www.wolfpsort.org) and TargetP 1.1 (www.cbs.dtu.dk/services) assigned high scores for organellar targeting to PpRibF1 and PpRibF2 from the moss P. patens (not shown).
Subcellular localization of algal RibF proteins could not be predicted because of insufficient sequence data. Full-length EST sequences were not available, and poor sequence similarity hindered the prediction of putative targeting peptides using the translated genomic sequences for the RibF proteins from C. reinhardtii and O. tauri.
A predicted RibF protein from the unicellular green alga O. tauri was fused to a putative protein of unknown function at the N terminus. Also, a gene encoding a sequence homolog of this unknown protein is present upstream of the predicted RibF gene in C. reinhardtii. However, the O. tauri RibF gene is transcribed in the opposite direction to the C. reinhardtii RibF gene. Thus, the prediction that the O. tauri RibF protein is fused to another protein is likely incorrect. More accurate predictions of algal RibF protein sequences will become possible when full-length EST sequences for these proteins become available.
Most bacteria contain bifunctional enzymes with riboflavin kinase and FAD synthetase activities. Shared sequence similarity to monofunctional riboflavin kinases from eukaryotes suggests that determinants for riboflavin kinase activity are located in the C-terminal regions of the bifunctional enzymes from bacteria (8). Low sequence conservation between plant and bacterial proteins in that region (Fig. 1) suggested that plant RibF proteins lack riboflavin kinase activity. Indeed, we show below that AtRibF1 and AtRibF2 lack this activity.
The phylogenetic analysis showed that sequences of plant RibF proteins cluster together, and apart from sequences of bacterial riboflavin kinases–FAD synthetases (Fig. 2). Three lines of evidence, the sequence alignment (Fig. 1), the phylogenetic analysis (Fig. 2), and the experimental results below, collectively suggest that plant RibF proteins have only FAD synthetase activity, and that they lost their ability to phosphorylate riboflavin to FMN before the speciation of vascular plants.
FIGURE 2.
Molecular phylogenetic tree of selected FAD synthetase protein sequences. The sequences used for the molecular phylogeny are listed under “Phylogenetic Analysis” under “Experimental Procedures.” Bootstrap numbers are given at the branch nodes. Plant sequences are shaded.
Cloning and Recombinant Expression of AtRibF1 and AtRibF2—Full-length cDNAs for AtRibF1 and AtRibF2 were cloned by reverse transcription-PCR using mRNA isolated from A. thaliana stems as template. Resulting cDNA fragments encoding putative mature proteins were amplified by PCR, subcloned into the pET-30 Ek/LIC (AtRibF1) or pET-44 Ek/LIC (AtRibF2) vector, and then functionally expressed in E. coli.
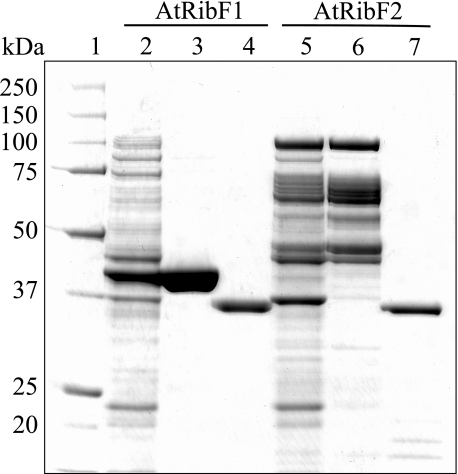
Purification and Biochemical Characterization of AtRibF1 and AtRibF2—The recombinant AtRibF1 carrying S-protein and hexahistidine tags at the N terminus was purified by Ni-NTA His-Bind affinity chromatography, and the recombinant AtRibF2 carrying Nus and hexahistidine tags at the N terminus by ion exchange chromatography. Both proteins were then digested with recombinant enterokinase to cleave the tags. Uncleaved AtRibF1 or AtRibF2, and the tag, were removed by ion exchange chromatography (Fig. 3). Untagged recombinant enzymes were used in all subsequent work.
FIGURE 3.
Purification of the recombinant ARibF1 and AtRibF2 under native conditions. The enzymes were purified as described under “Experimental Procedures.” Samples were separated by SDS-PAGE on a 10% NuPAGE BisTris gel and stained with Coomassie Blue. Lane 1, 5 μg of molecular mass standards; lane 2, 10 μg of crude E. coli extract expressing AtRibF1; lane 3, 10 μg of partially purified AtRibF1 after Ni-NTA affinity chromatography; lane 4, 2.5 μg of the final AtRibF1 enzyme preparation; lane 5, 10 μg of crude E. coli extract expressing AtRibF2; lane 6, 10 μg of partially purified AtRibF2 after Mono Q anion-exchange chromatography; lane 7, 2.5 μg of the final AtRibF2 enzyme preparation. Molecular masses of the standards (kDa) are indicated at the left.
Mobility of purified AtRibF1 and AtRibF2 on the SDS-PAGE gel agreed, respectively, with the theoretical molecular weight values of 34,200 and 33,900, which were calculated from the amino acid sequences. Purified AtRibF1 and AtRibF2 had FAD synthetase activity, but not riboflavin kinase activity (not shown).
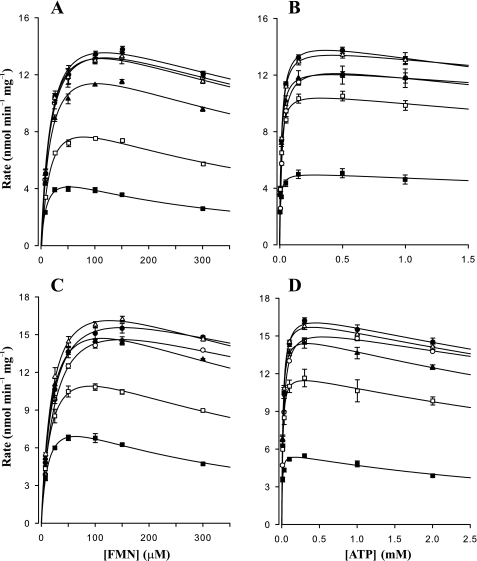
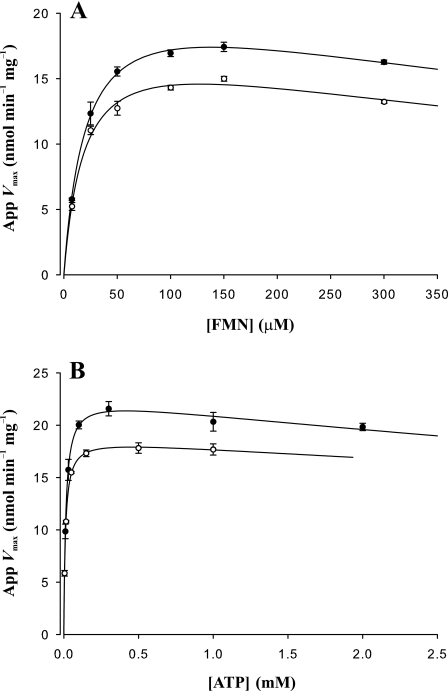
Primary and secondary plots of steady-state kinetic data for AtRibF1 and AtRibF2 are shown in Figs. 4 and 5. These plots showed that FMN and ATP inhibit AtRibF1 and AtRibF2. Primary plots of initial reaction rates against corresponding FMN (Fig. 4, A and C) or ATP (Fig. 4, B and D) concentrations were used to calculate the apparent Vmax values. These apparent Vmax values were then plotted against the second substrate (Fig. 5, A and B) to calculate the final Vmax, Km, and Ki values.
FIGURE 4.
Primary plots of steady-state kinetic data for AtRibF1 and AtRibF2. A–D, primary plots of initial reaction rates against FMN and ATP concentrations. A and B, AtRibF1; C and D, AtRibF2. Solid to open squares, triangles, and circles designate increasing order of substrate concentrations. [FMN] = 7.5, 25, 50, 100, 150, and 300 μm; [ATP] = 0.005, 0.015, 0.05, 0.15, 0.5, and 1.0 mm for AtRibF1; [ATP] = 0.01, 0.03, 0.1, 0.3, 1.0, and 2.0 mm for AtRibF2. Data points are mean ± S.E. of three triplicate determinations. Curves are nonlinear best fits to a model of uncompetitive substrate inhibition (SigmaPlot 9.0).
FIGURE 5.
Secondary plots of steady-state kinetic data for AtRibF1 and AtRibF2. A and B, secondary plots of apparent Vmax values against FMN and ATP concentrations. AtRibF1, open circles; AtRibF2, solid circles. Apparent Vmax values were obtained from data presented in Fig. 4. Substrate concentrations are as described in the legend to Fig. 4. Data points are mean ± S.E. of three triplicate determinations. Curves are nonlinear best fits to a model of uncompetitive substrate inhibition (SigmaPlot 9.0).
Substrate inhibition by FMN and ATP was reported for the enzyme from Corynebacterium ammoniagenes (1), but not for the enzymes from B. subtilis (5), rat (15, 17), and human (19, 20). Directly comparing catalytic properties of AtRibF1 and AtRibF2 (Table 1) to those of other reported FAD synthetases (Table 2) is unattainable because of differences in assay buffer composition and incubation temperature.
TABLE 1.
Kinetic parameters of AtRibF1 and AtRibF2 for FMN and ATP FAD synthetase activities were measured at pH 8.5 using recombinant enzymes purified under native conditions. Data are best parameter fits to a model of uncompetitive substrate inhibition as described under “Experimental Procedures.” Data are mean ± S.E. of three triplicate determinations.
| Enzyme (substrate) | Km | Ki × 103 | kcat × 10–2 | kcat/Km |
|---|---|---|---|---|
| μm | μm | s–1 | s–1m–1 | |
| AtRibF1 (FMN) | 18.9 ± 2.4 | 0.85 ± 0.20 | 1.08 ± 0.05 | 570 ± 77 |
| AtRibF1 (ATP) | 11.1 ± 0.8 | 18.68 ± 10.89 | 1.07 ± 0.02 | 967 ± 73 |
| AtRibF2 (FMN) | 20.8 ± 2.6 | 0.92 ± 0.23 | 1.28 ± 0.06 | 616 ± 81 |
| AtRibF2 (ATP) | 13.1 ± 1.7 | 13.02 ± 4.85 | 1.28 ± 0.04 | 979 ± 129 |
TABLE 2.
Summary of published kinetic parameters for FAD synthetases The top of the table shows kinetic parameters for prokaryotic FAD synthetases, which share sequence similarity to AtRibF1 and AtRibF2. The bottom of the table shows kinetic parameters for mammalian FAD synthetases, which share no sequence similarity to AtRibF1, AtRibF2, or prokaryotic FAD synthetases.
| Organism (substrate) | Km | Ki × 103 | kcat × 10–2 | Ref. |
|---|---|---|---|---|
| μm | μm | s–1 | ||
| C. ammoniagenes (FMN) | 1 | 30 | Efimov et al. (1) | |
| C. ammoniagenes (ATP) | 37 | 0.0216 | 30 | Efimov et al. (1) |
| 160 | 45 | Manstein and Pai (3) | ||
| B. subtilis (FMN) | 0.38 | Kearney et al. (5) | ||
| B. subtilis (ATP) | 300 | Kearney et al. (5) | ||
| Rat (FMN) | 9.1 | Yamada et al. (15) | ||
| 9.6 | Oka and McCormick (17) | |||
| Rat (ATP) | 71 | Yamada et al. (15) | ||
| 53 | Oka and McCormick (17) | |||
| Human 1 (FMN) | 1.5 | Brizio et al. (19) | ||
| Human 2 (FMN) | 0.4 | 0.36 | Galluccio et al. (20) |
Molecular weights of native AtRibF1 and AtRibF2 were estimated by gel filtration chromatography. The activities of AtRibF1 and AtRibF2 migrated as symmetrical peaks with apparent molecular weight values of 28,800 ± 250 and 29,200 ± 200 (mean ± S.E., n = 3), respectively, in close agreement with the theoretical values of 34,200 and 33,900. Thus, our data indicate that AtRibF1 and AtRibF2 are active as monomers. Bifunctional enzymes with riboflavin kinase and FAD synthetase activities from B. subtilis (2) and C. ammoniagenes (3) are also active as monomers.
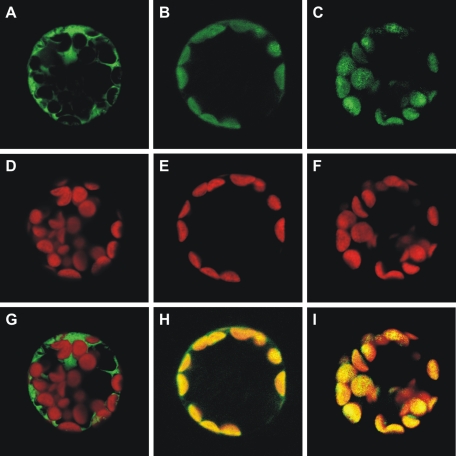
Transient Expression of EGFP-fused Proteins in A. thaliana Protoplasts—Subcellular localization of AtRibF1 and AtRibF2 was studied by fusing their putative N-terminal targeting peptides to EGFP. Green fluorescence produced by expression of the fusion proteins in A. thaliana protoplasts colocalized with the red autofluorescence of chloroplasts (Fig. 6). Also, green fluorescence was visible throughout the cytoplasm in control protoplasts expressing EGFP alone (Fig. 6). These findings thus establish that AtRibF1 and AtRibF2 have functional N-terminal peptides for targeting to plastids.
FIGURE 6.
AtRibF1 and AtRibF2 localize to chloroplasts. N-terminal EGFP fusions to putative organellar targeting peptides of AtRibF1 and AtRibF2 were transiently expressed in A. thaliana protoplasts. Non-fused EGFP in the pUC18-GFP5T-sp plasmid was used as a control for targeting to the cytosol. Subcellular localization was analyzed by fluorescence microscopy. A–C, EGFP fluorescence; D–F, chlorophyll autofluorescence; G–I, merged images.
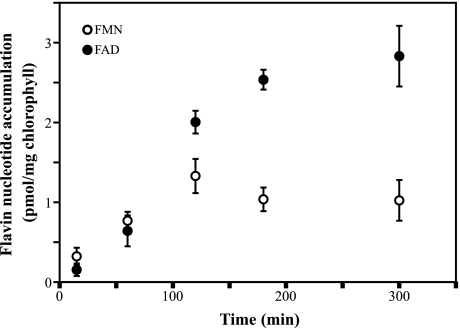
Pea Chloroplasts Synthesize FAD from Externally Added Riboflavin—Our confocal microscopy data indicated the presence of FAD synthetase in plastids. To provide independent evidence for FAD synthetase activity in plastids, we incubated Percoll-isolated chloroplasts from pea with [3H]riboflavin over selected time intervals, and determined accumulation of radiolabeled FMN and FAD (Fig. 7). Our results showed a time-dependent synthesis of radiolabeled FMN and FAD from externally added [3H]riboflavin and endogenous ATP in pea chloroplasts, thus providing evidence that these organelles have not only FAD synthetase, but also riboflavin kinase activity.
FIGURE 7.
Pea chloroplasts synthesize FMN and FAD from externally added riboflavin. Incorporation of [3H]FMN and [3H]FAD was measured in Percoll-isolated pea chloroplasts, incubated with [3H]riboflavin for 15, 60, 120, 180, or 300 min at 22 °C. Data points are mean ± S.E. of four feeding experiments.
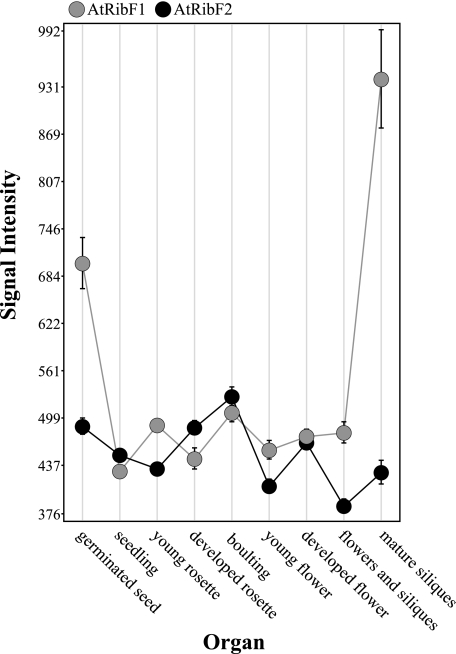
In Silico Expression Analysis— Organ- and development-specific expression of AtRibF1 and AtRibF2 was studied in silico using publicly available microarray data and the GENEVESTIGATOR software package. AtRibF1 and AtRibF2 were expressed in all plant organs and at all developmental stages examined (Fig. 8). Relative expression levels of AtRibF1 and AtRibF2 showed little or no variation among the organs and at the developmental stages examined. Two exceptions were mature siliques and germinated seeds, which expressed 50–100% more AtRibF1 mRNA than AtRibF2 mRNA. These findings suggest that AtRibF1 and AtRibF2 are house-keeping enzymes needed by all plant organs throughout development.
FIGURE 8.
In silico analysis of AtRibF1 and AtRibF2 gene expression in plant organs. Public Affymetrix expression analysis microarray data were analyzed using the GENEVESTIGATOR reference expression data base and meta-analysis system. GENEVESTIGATOR obtains the signal intensity values from raw array data by normalization using the MAS5 algorithm available in the Affymetrix software package. Error bars denote standard errors.
Enzyme Activities Catalyzing Synthesis and Hydrolysis of FMN and FAD in Chloroplasts and Mitochondria—To test for the presence of riboflavin kinase, FAD synthetase, FMN hydrolase, and FAD pyrophosphatase activities, we used chloroplasts and root mitochondria from pea because the recovery of intact organelles from A. thaliana has in our hands been inefficient. We have previously used Percoll-isolated pea organelles, which contain little or no cross-contamination from other subcellular compartments, to study subcellular distribution of enzyme activities (50, 59).
We tested for riboflavin kinase, FAD synthetase, FMN hydrolase, and FAD pyrophosphatase activities in soluble protein fractions from pea chloroplasts and mitochondria (Table 3). All four activities were assayed at pH 7.5 or at pH 8.5; FAD synthetase was further assayed by adding or omitting one of two detergents, 0.1% CHAPS and 0.1% Tween 20. All enzymes except FAD synthetase had detectable activities, suggesting that both organelles are autonomous for biosynthesis of FMN, and for hydrolysis of FMN and FAD.
TABLE 3.
Riboflavin kinase, FMN hydrolase, and FAD pyrophosphatase activities in pea organelles Enzyme activities were measured in protein extracts of pea chloroplasts and mitochondria at two pH values. Substrate concentrations were 50 μm for riboflavin, FMN, and FAD and 3 mm for ATP. Data are mean ± S.E. of three independent organelle preparations, each assayed in triplicate.
| Organelle | Riboflavin kinase | FMN hydrolase | FAD pyrophosphatase |
|---|---|---|---|
| pmol min–1 mg–1 | |||
| Chloroplasts | |||
| pH 7.5 | 0.79 ± 0.03 | 244 ± 35 | 30.0 ± 2.4 |
| pH 8.5 | 2.47 ± 0.08 | 151 ± 4 | 38.4 ± 4.7 |
| Mitochondria | |||
| pH 7.5 | 7.8 ± 0.8 | 1393 ± 58 | 343 ± 44 |
| pH 8.5 | 15.9 ± 1.7 | 446 ± 22 | 482 ± 41 |
DISCUSSION
Flavin nucleotides are required in many metabolic processes in plastids, mitochondria, and the cytosol. Presence of N-terminal extensions with characteristics of chloroplast targeting peptides in plant enzymes of the riboflavin biosynthesis pathway (60–62), and efficient import of lumazine synthase from spinach by pea chloroplasts (62), suggest that riboflavin is synthesized only in plastids. This raises the questions whether flavin nucleotides are synthesized from riboflavin in one or multiple subcellular compartments, and whether riboflavin or flavin nucleotides are transported across organellar membranes in plants. We are addressing these questions using bioinformatics, molecular biology, and biochemistry.
We previously reported bioinformatic evidence that plants have sequence homologs of monofunctional enzymes with FAD synthetase or riboflavin kinase activity, previously found only in mammals and yeast, and sequence homologs of bifunctional enzymes with both activities, previously found only in bacteria (8). Presence of N-terminal extensions with characteristics of organellar targeting signals in sequence homologs of the bifunctional enzymes suggests organellar localization; absence of such extensions in sequence homologs of the monofunctional enzymes suggests cytosolic localization of these enzymes in plants.
Previously, we cloned the cDNA and characterized a sequence homolog from A. thaliana of monofunctional riboflavin kinases from yeast. We showed that this enzyme is fused to a novel type of Mg2+-dependent FMN hydrolase that belongs to the haloacid dehalogenase enzyme superfamily. We also showed that this FMN hydrolase is an effective catalyst at neutral pH, in agreement with its putative localization in the cytosol, and in contrast with the previously known FMN-hydrolyzing acid phosphatases that are poor catalysts at pH > 5.5–6.0.
In this study, we report cDNA cloning and characterization of two enzymes from A. thaliana (AtRibF1 and AtRibF2), which are sequence homologs of bifunctional enzymes with riboflavin kinase and FAD synthetase activities from bacteria. A protein sequence comparison to their bacterial homologs revealed that plant RibF proteins are shorter and lack conserved C-terminal motifs (Fig. 1), suggesting that AtRibF1 and AtRibF2 might have only one catalytic activity. Subsequent biochemical characterization of the recombinant enzymes produced in E. coli showed that AtRibF1 and AtRibF2 have only FAD synthetase activity; sequence similarity lead us to hypothesize that all plant RibF enzymes have only this activity. Thus, plants are unique among eukaryotes in that they have sequence homologs of FAD synthetases previously found only in prokaryotes. These FAD synthetases differ from the prokaryotic enzymes in that they lack riboflavin kinase activity.
Transient expression pattern of EGFP-fused proteins in A. thaliana protoplasts is consistent with AtRibF1 and AtRibF2 being localized in plastids. In agreement with this finding is synthesis of FMN and FAD from imported riboflavin in Percoll-isolated pea chloroplasts (Fig. 7). Taken together, these findings provide to our knowledge the first experimental evidence of FAD synthesis in chloroplasts.
Contradictory to the confocal microscopy data and to the evidence for FAD biosynthesis in chloroplasts in vivo is for now our inability to detect FAD synthetase activity in plastid extracts. One explanation to these seemingly conflicting findings is that FAD pyrophosphatase, which is highly active in chloroplasts (Table 3), is masking the endogenous FAD synthetase activity in these organelles. Two other possibilities are that FAD synthetase activity is below the detection limit of the assay, or that the enzyme dies during extraction. To rule out these possibilities, we tested different protein extraction and enzyme assay conditions, and assayed for activity in freshly prepared organelle extracts, with similar results. Conflicting results regarding subcellular localization of FAD synthetase were also reported in S. cerevisiae: FAD synthetase enzyme activity was not detected (13), but FAD synthesis from imported riboflavin was observed (22, 23) in isolated mitochondria.
The evidence that AtRibF1 and AtRibF2 reside in plastids raises two possibilities about the origin of FAD in plant mitochondria: FAD is imported from the cytosol, or synthesized inside mitochondria by an as-yet-undiscovered FAD synthetase. Our attempts to detect FAD synthetase activity in organelle extracts of pea were inconclusive, as mentioned above. Further experiments to investigate the origin of FAD in plant mitochondria are planned for the future, but are beyond the scope of this article.
Absence of riboflavin kinase activity in the recombinant AtRibF1 and AtRibF2 raises the question whether chloroplasts and mitochondria obtain FMN by biosynthesis from riboflavin or by import from the cytosol in plants. Presence of riboflavin kinase activity in Percoll-isolated chloroplasts and mitochondria from pea (Table 3), as well as the ability of chloroplasts to synthesize FMN from imported riboflavin (Fig. 7), suggest the former, in agreement with earlier evidence in spinach (30) showing riboflavin kinase activity in an organellar fraction containing chloroplasts and mitochondria. Our inability to find candidate sequences for organellar riboflavin kinases in plant databases suggests that enzymes with little or no sequence similarity to known riboflavin kinases exist in plants.
Toward our goal to obtain a complete picture of flavin nucleotide metabolism inside plant cells, we also show here that Percoll-isolated chloroplasts and mitochondria from pea have FMN hydrolase and FAD pyrophosphatase activities, and that these activities are detectable at two physiologically relevant pH values, pH 7.5 and 8.5. These data provide to our knowledge the first experimental evidence of flavin nucleotide hydrolysis in plant organelles.
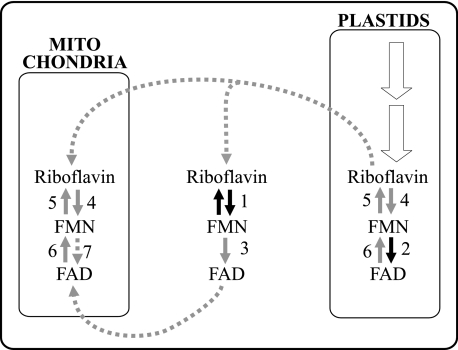
From findings presented here and previously published by other authors, we propose a model for biosynthesis, hydrolysis, and transport of flavin nucleotides inside plant cells (Fig. 9). In this model, riboflavin is synthesized in plastids; FMN is synthesized and hydrolyzed in plastids, mitochondria, and the cytosol; and FAD is synthesized in plastids and the cytosol, and hydrolyzed in plastids and mitochondria. This model is supported by the present biochemical and bioinformatic evidence suggesting that riboflavin is synthesized in plastids (60–62), and that FMN is synthesized in plastids, mitochondria, and the cytosol (Ref. 8 and this article, Fig. 7 and Table 3). We hypothesize from this evidence that plastids export riboflavin to the cytosol, and that mitochondria import riboflavin from the cytosol.
FIGURE 9.
Proposed model for biosynthesis, hydrolysis, and transport of flavin nucleotides inside plant cells. Black arrows indicate those enzymes whose cDNAs have been cloned and recombinant proteins have been biochemically characterized; solid gray arrows indicate enzymes whose existence is supported by biochemical or bioinformatic evidence; dotted gray arrows indicate putative mitochondrial FAD synthetase and the predicted membrane transport steps; white arrows indicate the riboflavin biosynthesis pathway. 1, FMN/FHy; 2, RibF; 3, cytosolic FAD synthetase; 4, organellar riboflavin kinase; 5, organellar FMN hydrolase; 6, organellar FAD pyrophosphatase; 7, mitochondrial FAD synthetase.
Biochemical and confocal microscopy evidence presented here (Figs. 6 and 7) support the existence of plastid FAD synthetases; earlier bioinformatic evidence supports the existence of a cytosolic FAD synthetase (8). Our attempts to find FAD synthetase in mitochondria yielded inconclusive results. Thus, it is unclear for now whether mitochondria obtain FAD by import from the cytosol, or by endogenous biosynthesis from FMN. Biochemical evidence presented here (Table 3) also supports the existence of FMN hydrolases and FAD pyrophosphatases in plastids and mitochondria; earlier biochemical and bioinformatic evidence support the existence of an FMN hydrolase in the cytosol (8).
The results we report contribute new knowledge to the overall understanding of flavin nucleotide metabolism in plants, yet they also raise new questions. Some of those questions are: do mitochondria obtain FAD by import from the cytosol, or by endogenous biosynthesis; what are the physiological roles of the enzymes that hydrolyze flavin nucleotides; and what flavin transporters operate in the organellar membranes of plant cells. Our future work will focus on answering those questions.
Acknowledgments
We thank Dr. Matthew Willmann from the University of Pennsylvania for advice with the transformation of A. thaliana protoplasts. We also thank Dr. Mechthild Tegeder for providing the pUC18-GFP5T-sp plasmid; Craig Whitney and Julianna Gothard-Szamosfalvi for growing pea and A. thaliana plants; and Dr. Michael Knoblauch, Dr. Christine Davitt, and Dr. Valerie Lynch-Holm from the Franceschi Microscopy and Imaging Center at Washington State University for help with the confocal laser scanning microscope.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBank™/EBI Data Bank with accession number(s) EU687457 (AtRibF1) and EU687458 (AtRibF2).
This work was supported by United States Department of Agriculture National Research Initiative Grant 2007-03559 (to S. R.) and the Murdoch Foundation. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
F. J. Sandoval and S. Roje, unpublished data.
The abbreviations used are: EGFP, enhanced green fluorescence protein; THP, tris(hydroxypropyl)phosphine; EST, expressed sequence tag; CHAPS, 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate; Ni-NTA, nickel-nitrilotriacetic acid; BisTris, 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)-1,3-propanediol.
References
- 1.Efimov, I., Kuusk, V., Zhang, X., and McIntire, W. S. (1998) Biochemistry 37 9716-9723 [DOI] [PubMed] [Google Scholar]
- 2.Mack, M., van Loon, A. P., and Hohmann, H. P. (1998) J. Bacteriol. 180 950-955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Manstein, D. J., and Pai, E. F. (1986) J. Biol. Chem. 261 16169-16173 [PubMed] [Google Scholar]
- 4.Mayhew, S. G., and Wassink, J. H. (1980) Methods Enzymol. 66 323-327 [DOI] [PubMed] [Google Scholar]
- 5.Kearney, E. B., Goldenberg, J., Lipsick, J., and Perl, M. (1979) J. Biol. Chem. 254 9551-9557 [PubMed] [Google Scholar]
- 6.Clarebout, G., Villers, C., and Leclercq, R. (2001) Antimicrob. Agents Chemother. 45 2280-2286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Solovieva, I. M., Kreneva, R. A., Leak, D. J., and Perumov, D. A. (1999) Microbiology 145 67-73 [DOI] [PubMed] [Google Scholar]
- 8.Sandoval, F. J., and Roje, S. (2005) J. Biol. Chem. 280 38337-38345 [DOI] [PubMed] [Google Scholar]
- 9.Nakano, H., and McCormick, D. B. (1991) J. Biol. Chem. 266 22125-22128 [PubMed] [Google Scholar]
- 10.Bauer, S., Kemter, K., Bacher, A., Huber, R., Fischer, M., and Steinbacher, S. (2003) J. Mol. Biol. 326 1463-1473 [DOI] [PubMed] [Google Scholar]
- 11.Karthikeyan, S., Zhou, Q., Mseeh, F., Grishin, N. V., Osterman, A. L., and Zhang, H. (2003) Structure (Camb.) 11 265-273 [DOI] [PubMed] [Google Scholar]
- 12.Karthikeyan, S., Zhou, Q., Osterman, A. L., and Zhang, H. (2003) Biochemistry 42 12532-12538 [DOI] [PubMed] [Google Scholar]
- 13.Wu, M., Repetto, B., Glerum, D. M., and Tzagoloff, A. (1995) Mol. Cell. Biol. 15 264-271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Santos, M. A., Jiménez, A., and Revuelta, J. L. (2000) J. Biol. Chem. 275 28618-28624 [DOI] [PubMed] [Google Scholar]
- 15.Yamada, Y., Merrill, A. H., Jr., and McCormick, D. B. (1990) Arch. Biochem. Biophys. 278 125-130 [DOI] [PubMed] [Google Scholar]
- 16.Merrill, A. H., Jr., and McCormick, D. B. (1980) J. Biol. Chem. 255 1335-1338 [PubMed] [Google Scholar]
- 17.Oka, M., and McCormick, D. B. (1987) J. Biol. Chem. 262 7418-7422 [PubMed] [Google Scholar]
- 18.Bowers-Komro, D. M., Yamada, Y., and McCormick, D. B. (1989) Biochemistry 28 8439-8446 [DOI] [PubMed] [Google Scholar]
- 19.Brizio, C., Galluccio, M., Wait, R., Torchetti, E. M., Bafunno, V., Accardi, R., Gianazza, E., Indiveri, C., and Barile, M. (2006) Biochem. Biophys. Res. Commun. 344 1008-1016 [DOI] [PubMed] [Google Scholar]
- 20.Galluccio, M., Brizio, C., Torchetti, E. M., Ferranti, P., Gianazza, E., Indiveri, C., and Barile, M. (2007) Protein Expression Purif. 52 175-181 [DOI] [PubMed] [Google Scholar]
- 21.Deluca, C., and Kaplan, N. O. (1958) Biochim. Biophys. Acta 30 6-11 [DOI] [PubMed] [Google Scholar]
- 22.Pallotta, M. L., Brizio, C., Fratianni, A., De Virgilio, C., Barile, M., and Passarella, S. (1998) FEBS Lett. 428 245-249 [DOI] [PubMed] [Google Scholar]
- 23.Bafunno, V., Giancaspero, T. A., Brizio, C., Bufano, D., Passarella, S., Boles, E., and Barile, M. (2004) J. Biol. Chem. 279 95-102 [DOI] [PubMed] [Google Scholar]
- 24.Barile, M., Passarella, S., Bertoldi, A., and Quagliariello, E. (1993) Arch. Biochem. Biophys. 305 442-447 [DOI] [PubMed] [Google Scholar]
- 25.Barile, M., Brizio, C., Valenti, D., De Virgilio, C., and Passarella, S. (2000) Eur. J. Biochem. 267 4888-4900 [DOI] [PubMed] [Google Scholar]
- 26.Sobhanaditya, J., and Rao, N. A. (1981) Biochem. J. 197 227-232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sadasivam, S., and Shanmugasundaram, E. R. (1966) Enzymologia 31 203-208 [PubMed] [Google Scholar]
- 28.Giri, K. V., Rao, N. A., Cama, H. R., and Kumar, S. A. (1960) Biochem. J. 75 381-386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giri, K. V., Krishnaswamy, P. R., and Rao, N. A. (1957) Nature 179 1134-1135 [DOI] [PubMed] [Google Scholar]
- 30.Mitsuda, H., Tsuge, H., Tomozawa, Y., and Kawai, F. (1970) J. Vitaminol. (Kyoto) 16 52-57 [DOI] [PubMed] [Google Scholar]
- 31.Mitsuda, H., Tomozawa, Y., Tsuboi, T., and Kawai, F. (1965) J. Vitaminol. (Kyoto) 11 20-29 [DOI] [PubMed] [Google Scholar]
- 32.Tejera García, N. A., Olivera, M., Iribarne, C., and Lluch, C. (2004) Plant Physiol. Biochem. 42 585-591 [DOI] [PubMed] [Google Scholar]
- 33.Fuchs, K. R., Shekels, L. L., and Bernlohr, D. A. (1992) Biochem. Biophys. Res. Commun. 189 1598-1605 [DOI] [PubMed] [Google Scholar]
- 34.Granjeiro, J. M., Ferreira, C. V., Juca, M. B., Taga, E. M., and Aoyama, H. (1997) Biochem. Mol. Biol. Int. 41 1201-1208 [DOI] [PubMed] [Google Scholar]
- 35.Sensabaugh, G. F., and Golden, V. L. (1978) Am. J. Hum. Genet. 30 553-560 [PMC free article] [PubMed] [Google Scholar]
- 36.Taga, E. M., and Van Etten, R. L. (1982) Arch. Biochem. Biophys. 214 505-515 [DOI] [PubMed] [Google Scholar]
- 37.Zhang, Z. Y., and Van Etten, R. L. (1990) Arch. Biochem. Biophys. 282 39-49 [DOI] [PubMed] [Google Scholar]
- 38.Barile, M., Brizio, C., De Virgilio, C., Delfine, S., Quagliariello, E., and Passarella, S. (1997) Eur. J. Biochem. 249 777-785 [DOI] [PubMed] [Google Scholar]
- 39.Kumar, S. A., Rao, N. A., and Vaidyanathan, C. S. (1965) Arch. Biochem. Biophys. 111 646-652 [DOI] [PubMed] [Google Scholar]
- 40.Ravindranath, S. D., and Rao, N. A. (1969) Arch. Biochem. Biophys. 133 54-59 [DOI] [PubMed] [Google Scholar]
- 41.Mitsuda, H., Tsuge, H., Tomozawa, Y., and Kawai, F. (1970) J. Vitaminol. (Kyoto) 16 31-38 [DOI] [PubMed] [Google Scholar]
- 42.Balakrishnan, C. V., Vaidyanathan, C. S., and Rao, N. A. (1977) Eur. J. Biochem. 78 95-102 [DOI] [PubMed] [Google Scholar]
- 43.Kornberg, A., and Pricer, W. E., Jr. (1950) J. Biol. Chem. 186 763-778 [PubMed] [Google Scholar]
- 44.Byrd, J. C., Fearney, F. J., and Kim, Y. S. (1985) J. Biol. Chem. 260 7474-7480 [PubMed] [Google Scholar]
- 45.Lee, R. S., and Ford, H. C. (1988) J. Biol. Chem. 263 14878-14883 [PubMed] [Google Scholar]
- 46.Shin, H. J., and Mego, J. L. (1988) Arch. Biochem. Biophys. 267 95-103 [DOI] [PubMed] [Google Scholar]
- 47.Bradford, M. M. (1976) Anal. Biochem. 72 248-254 [DOI] [PubMed] [Google Scholar]
- 48.Cline, K. (1986) J. Biol. Chem. 261 14804-14810 [PubMed] [Google Scholar]
- 49.Douce, R., Bourguignon, J., Brouquisse, R., and Neuburger, M. (1987) Methods Enzymol. 148 403-415 [Google Scholar]
- 50.Roje, S., Janave, M. T., Ziemak, M. J., and Hanson, A. D. (2002) J. Biol. Chem. 277 42748-42754 [DOI] [PubMed] [Google Scholar]
- 51.Meyer, S. L. (1975) Data Analysis for Scientists and Engineers, pp. 39-48, John Wiley, New York
- 52.Karimi, M., Inzé, D., and Depicker, A. (2002) Trends Plant Sci. 7 193-195 [DOI] [PubMed] [Google Scholar]
- 53.Sheen, J. (2001) Plant Physiol. 127 1466-1475 [PMC free article] [PubMed] [Google Scholar]
- 54.Wirtz, W., Stitt, M., and Heldt, H. W. (1980) Plant Physiol. 66 187-193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Arnon, D. I. (1949) Plant Physiol. 24 1-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Felsenstein, J. (2004) PHYLIP (Phylogeny Inference Package), Version 3.6, University of Washington, Seattle, WA
- 57.Zimmermann, P., Hirsch-Hoffmann, M., Hennig, L., and Gruissem, W. (2004) Plant Physiol. 136 2621-2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fischer, M., and Bacher, A. (2006) Physiol. Plantarum 126 304-318 [Google Scholar]
- 59.Quinlivan, E. P., Roje, S., Basset, G., Shachar-Hill, Y., Gregory, J. F., 3rd, and Hanson, A. D. (2003) J. Biol. Chem. 278 20731-20737 [DOI] [PubMed] [Google Scholar]
- 60.Herz, S., Eberhardt, S., and Bacher, A. (2000) Phytochemistry 53 723-731 [DOI] [PubMed] [Google Scholar]
- 61.Fischer, M., Römisch, W., Saller, S., Illarionov, B., Richter, G., Rohdich, F., Eisenreich, W., and Bacher, A. (2004) J. Biol. Chem. 279 36299-36308 [DOI] [PubMed] [Google Scholar]
- 62.Jordan, D. B., Bacot, K. O., Carlson, T. J., Kessel, M., and Viitanen, P. V. (1999) J. Biol. Chem. 274 22114-22121 [DOI] [PubMed] [Google Scholar]