Abstract
Overexpression of the ped/pea-15 gene in mice impairs glucose tolerance and leads to diabetes in conjunction with high fat diet treatment. PED/PEA-15 is also overexpressed in type 2 diabetics as well as in euglycemic offspring from these subjects. The cause(s) of this abnormality remains unclear. In the present work we have cloned and localized the promoter region of the human PED/PEA-15 gene within the first 230 bp of the 5®-flanking region. A cis-acting regulatory element located between -320 and -335 bps upstream the PED/PEA-15 gene transcriptional start site (+1) is recognized by both the hepatocyte nuclear factor 4α (HNF-4α) and the chicken ovalbumin upstream promoter transcription factor II (COUP-TFII), two members of the steroid/thyroid superfamily of transcription factors, both of which are involved in the control of lipid and glucose homeostasis. HNF-4α represses PED/PEA-15 expression in HeLa cells, whereas COUP-TFII activates its expression. In hepatocytes, the activation of PED/PEA-15 gene transcription is paralleled by the establishment of a partially dedifferentiated phenotype accompanied by a reduction in mRNA levels encoded by genes normally expressed during liver development. Cotransfection of HeLa cells with a reporter construct containing the PED/PEA-15 response element and various combinations of HNF-4α and COUP-TFII expression vectors indicated that COUP-TFII antagonizes the repression of the PED/PEA-15 gene by HNF-4α. Thus, at least in part, transcription of the PED/PEA-15 gene in vivo is dependent upon the intracellular balance of these positive and negative regulatory factors. Abnormalities in HNF-4α and COUP-TFII balance might have important consequences on glucose tolerance in humans.
Phosphoprotein enriched in diabetes/phosphoprotein enriched in astrocytes (PED/PEA-15)3 is a cytosolic phosphoprotein widely expressed in different tissues and highly conserved in mammals (1-4). It binds to and modulates the function of a number of signaling proteins and effectors. PED/PEA-15 binds several pro- and anti-apoptotic proteins thereby exerting a broad anti-apoptotic function (5-9). It also controls mitogenic signaling by binding extracellular-regulated kinases (ERKs) and anchoring ERKs to the cytoplasm (10). Indeed, changes in PED/PEA-15 expression play an important role in tumor development and sensitivity to anti-neoplastic agents (11, 12). PED/PEA-15 binds to phospholipase D, enhancing its stability and increasing intracellular diacylglycerol levels (13, 14). This effect, in turn, activates classical protein kinase C isoforms and generates resistance to insulin action on glucose metabolism in peripheral tissues. Protein kinase C dysregulation by PED/PEA-15 also impairs glucose-stimulated insulin secretion in β cells in mice (14, 15).
PED/PEA-15 gene maps on human chromosome 1q21-22 (4) and is overexpressed in type 2 diabetics as well as in the euglycemic offspring from these individuals. Interestingly, in these same subjects, PED/PEA-15 levels correlate with insulin resistance (4, 16). PED/PEA-15 cellular levels are regulated by ubiquitinylation and proteasomal degradation (17). However, run-on experiments in cultured cells from type 2 diabetic subjects demonstrated that, at least in part, the overexpression observed in these subjects is caused by transcriptional abnormalities (4). The molecular details responsible for these abnormalities and the mechanisms responsible for PED/PEA-15 gene regulation are still unclear.
Hepatocyte nuclear factor-4α (HNF-4α) and the chicken ovalbumin upstream promoter transcription factor II (COUP-TFII) are two members of the steroid/thyroid superfamily of transcription factors involved in the control of glucose homeostasis (18-20). Studies in mice in which the early lethal phenotype is circumvented have revealed that HNF-4α is essential for hepatocyte differentiation both at the morphological and the functional levels (21) and for accumulation of hepatic glycogen stores and generation of normal hepatic epithelium (22). Point mutations in HNF-4α impair liver and pancreatic regulation of glucose homeostasis and cause Maturity Onset Diabetes of the Young type 1 (MODY1). More recently, genetic and biochemical evidence has been generated, indicating that HNF-4α may also have a role in the development of more common forms of type 2 diabetes (23-25).
Most of the promoter elements interacting with HNF-4α can also recognize the COUP-TFs (26-28), one of the most extensively studied orphan receptors. COUP-TFs regulate a number of biological processes including embryonic development (29) and neural cell fate determination (30). COUP-TFs may also affect glucose homeostasis. Indeed, in vitro studies indicate that COUP-TFII, also termed Arp-1, regulates several genes involved in glucose and lipid metabolism including insulin gene expression in pancreatic β -cells (31, 32). Functionally, COUP-TFII has been identified as a transcriptional repressor of genes activated by HNF-4α. However, evidence is also present in the literature indicating that, at least in certain circumstances, COUP-TFII activates gene expression (33, 34). The specific function of COUP-TFII likely depends upon the repertoire of coregulatory proteins interacting with COUP-TFII and HNF-4α in each specific context (35-43).
In the present paper we have cloned and characterized the promoter region of the human PED/PEA-15 gene. We obtained evidence that HNF-α and COUP-TFII compete for binding to PED/PEA-15 promoter. Although HNF-4α represses PED/PEA-15 transcription, COUP-TFII functions as transcriptional activator. HNF-4α/COUP-TFII control of PED/PEA-15 transcriptional activity may determine hepatocyte differentiation and gluco-regulatory functions.
EXPERIMENTAL PROCEDURES
Materials—Media, sera, and antibiotics for cell culture and the Lipofectamine reagent were purchased from Invitrogen. Goat polyclonal HNF-4α and rabbit polyclonal COUP-TFII antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA). PED/PEA-15 antibody has been previously described (4, 9). The mouse monoclonal HA antibody was from Roche Applied Science. The protein A- and G-Sepharose beads were from Pierce. The pCDNA3/HNF-4α expression vector, which contains the HNF-4α coding region, was a generous gift from Dr. Graeme Bell (Dept. of Medicine, University of Chicago, Chicago, IL). The pCMV6-XL5COUP-TFII expression plasmid and the pCMV6-XL5 vector were from OriGene Technologies, Inc. (Rockville, MD). Radiochemicals and Western blot and ECL reagents were purchased from Amersham Biosciences. All other reagents were from Sigma.
Plasmid Constructs—A 2.0-kilobase KpnI-XhoI genomic fragment containing the 5′-flanking region of the human PED/PEA-15 gene was amplified (PCR) and subcloned between the corresponding restriction sites of the luciferase expression vector pGL3 Basic (Promega, Madison, WI). The resulting plasmid, termed pPED2000, was used as a template for PCR generation of progressively deleted fragments of the PED/PEA-15 5′-flanking region. These were also subcloned in the luciferase cloning vector. To minimize the possibility of introducing errors during the PCR, the Expand Long Template PCR System (Roche Applied Science) was used, and DNA was amplified according to the manufacturer's instructions. The identity and orientation of the PCR fragments were then assessed by restriction enzyme analysis and sequencing.
The HNF-4α binding site on PED/PEA-15 promoter (TCATCCAAAGGTCA) was mutagenized by PCR to TCATCCCCCGGTCA. Two mutated inner primers (pPED477mutFor CGTGGTCATCCCCCGGTCAAAAG and pPED477mutRev CTTTTGACCGGGGGATGACCAGG) and two outer primers (Ped477 KpnI and Ped (XhoI) antisense; see supplemental Table 1) were used. Two mutated products were amplified using the pPED477 construct as a template and the Expand Long Template PCR System (Roche Applied Science) to minimize the possibility of introducing errors during the PCR. In the first reaction the mutated inner sense primer was used together with the Ped (XhoI) antisense, whereas the Ped477 KpnI primer was used in the second reaction together with the mutated inner antisense. The mutant HNF-4α response element (HRE) was then reconstituted with the Expand Long Template PCR System (Roche Applied Science) using the outer primers. The mutant fragment (477mut) obtained was then inserted into the pGL3 Basic Vector and completely sequenced.
Animal Studies, Cell Culture, Transfections, RT-PCR, and Western Blot Assay—The PED/PEA-15 transgenic mice have been previously described (15). HeLa, human embryonic kidney 293 (HEK 293), and HepG2 cells were cultured in Dulbec-co's modified Eagle's medium supplemented with 10% fetal bovine serum, penicillin (200 IU/ml), streptomycin (100 μg/ml), and 2% l-glutamine in a humidified CO2 incubator. Stable expression of the HNF-4α short hairpin RNA (shRNA) clone and wild-type PED/PEA-15 cDNA in HepG2 cells was achieved as reported by Condorelli et al. (6). Transient transfection of plasmid DNAs in HeLa, HEK 293, and HepG2 cells were carried out by the calcium phosphate coprecipitation method (42). Briefly, the cells were plated in 60-mm dishes before transfection at a confluence of 1 × 105 cells/dish. 3 μg of the indicated PED/PEA-15 promoter-luciferase construct and 1 μg of the pRSVβ-gal vector (to correct for the variable transfection efficiencies) were then added.
To examine the effect of HNF-4α and COUP-TFII on PED/PEA-15 promoter, HeLa and HepG2 cells were cotransfected with 2 μg of PED/PEA-15 promoter-luciferase together with different amounts of HNF-4α and COUP-TFII expression vectors. Total DNA content (up to 4 μg/plate) was normalized to the empty vector devoid of HNF-4α and COUP-TFII coding sequence. 48 h after transfection the cells were harvested and lysed as described previously (43). Luciferase activity was measured by a commercial luciferase assay kit (Promega). Values were normalized for β-galactosidase. Statistical significance was evaluated by t test analysis.
Total RNAs were prepared by extraction with the RNeasy kit (Qiagen Sciences) according to the manufacturer's instructions. For real time RT-PCR analysis, 1 μg RNA was reverse-transcribed using the Superscript II reverse transcriptase (Invitrogen). Reactions were performed using Platinum SYBR Green qPCR Super-UDG in a iCycler IQ multicolor Real Time PCR detection system (Bio-Rad). All reactions were performed in triplicate, and β-actin was used as the internal standard.
For Western blot analysis, cells were solubilized in lysis buffer (50 mm HEPES (pH 7.5), 150 mm NaCl, 10 mm EDTA, 10 mm Na2P2O7, 2 mm Na3VO4, 100 mm NaF, 10% glycerol, 1% Triton X-100, 1 mm phenylmethylsulfonyl fluoride, 10 μg/ml aprotinin) for 2 h at 4 °C. Cell lysates were clarified by centrifugation at 5000 × g for 20 min, separated by SDS-PAGE, and transferred into 0.45-μm Immobilon-P membranes (Millipore, Bedford, MA). Upon incubation with primary and secondary antibodies, immunoreactive bands were detected by ECL according to the manufacturer's instructions.
shRNA-mediated Knockdown of HNF-4α—To interfere with endogenous HNF-4α expression in HepG2 cells, we use short-hairpin RNAs with the following sequences: clone a (Cl.a) CCGGCCATCACCAAGCAGGAAGTTACTCGAGTAACTTCCTGCTTGGTGATGGTTTTT; clone b (Cl.b) CCGGACCACCCTGGAATTTGAGAATCTCGAGATTCTCAAATTCCAGGGTGGTTTTTT. The cells were transfected with 1 μg of each clone using the Lipofectamine reagent according to manufacturer's recommendations and analyzed 48 h after transfection.
Electrophoretic Mobility Shift Assay—Cells were transfected with 1 μg of expression plasmids as described above. Whole cell extracts (10-15 μg of protein) were incubated for 30 min on ice in binding buffer 5× (50 mm Tris-HCl (pH 8.0), 750 mm KCl, 2.5 mm EDTA, 0.5% Triton X-100 (v/v), 62.5% glycerol (v/v), 1 mm dithiothreitol) containing 600 ng of poly (dI®dC) (GE Healthcare) in the presence of the 32P-labeled probe. Samples were subsequently resolved on 6% polyacrylamide gels (37.5:1 acrylamide/bisacrylamide) in 22.5 mm Tris borate, 0.5 mm EDTA (0.25 × Tris borate EDTA) at 120 V for 2 h at 4 °C. The gel was dried, and the labeled DNA-protein complexes were visualized by autoradiography. Oligonucleotides corresponding to the wild-type, and mutated HREs were synthesized (supplemental Table 1). Radioactive probes were generated by incubation of the appropriate annealed oligonucleotides (5 pmol) in the presence of 20 mCi of [γ-32P]ATP and T4-polynucleotide kinase according to the manufacturer's instructions. The radio-labeled probes were separated from free nucleotides by spin dialysis using Sephadex G-50 (Roche Applied Science). Specific activity obtained was typically ∼105 cpm/ml.
Chromatin Immunoprecipitation Assay—The cross-linking solution, containing 1% formaldehyde, was added directly to cell culture media. The fixation proceeded for 10 min and was stopped by the addition of glycine to a final concentration of 125 mm. Primary human hepatocytes (Lonza Walkersville), HeLa, and HepG2 cells were rinsed twice with cold phosphate-buffered saline plus 1 mm phenylmethylsulfonyl fluoride and then (HeLa and HepG2) scraped. Cells were collected by centrifugation at 800 × g for 5 min at 4 °C. Cells were swelled in cold cell lysis buffer containing 5 mm PIPES (pH 8.0), 85 mm KCl, 0.5% Nonidet P-40, 1 mm phenylmethylsulfonyl fluoride, and inhibitors mixture (Sigma) and incubated on ice for 10 min. Nuclei were precipitated by microcentrifugation at 2000 × g for 5 min at 4 °C, resuspended in nuclear lysis buffer containing 50 mm Tris-HCl (pH 8.0), 10 mm EDTA, 0.8% SDS, 1 mm phenyl-methylsulfonyl fluoride and inhibitors mixture (Sigma), and then incubated on ice for 10 min. Samples were broken by sonication into chromatin fragments of an average length of 500/1000 bp and then microcentrifuged at 16,000 × g for 10 min at 4 °C. The sonicated cell supernatant was diluted 8-fold in chromatin immunoprecipitation (ChIP) dilution buffer containing 0.01% SDS, 1.1% Triton X-100, 1.2 mm EDTA, 16.7 mm Tris-HCl (pH 8.0), and 167 mm NaCl and precleared by adding salmon sperm and conjugating protein at equimolar concentration for 90 min at 4 °C. Precleared chromatin from 1 × 106 cells was incubated with 1 μg of polyclonal antibody (anti-HNF-4α and an unrelated one) or no antibody and rotated at 4 °C for 16 h. Immunoprecipitates were washed five times with radio-immune precipitation assay buffer containing 10 mm Tris-HCl (pH 8.0), 1 mm EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS, 140 mm NaCl, and 1 mm phenylmethylsulfonyl fluoride, twice with LiCl buffer containing 0.25 m LiCl, 1% Nonidet P-40, 1% sodium deoxycholate, 1 mm EDTA, 10 mm Tris-HCl (pH 8.0), and then 3 times with TE (10 mm Tris-HCl (pH 8.0), 1 mm EDTA). Before the first wash, the supernatant from the reaction lacking primary antibody was saved as total input of chromatin and was processed with the eluted immunoprecipitates beginning at the cross-link reversal step. Immunoprecipitates were eluted by adding 1% SDS, 0.1 m NaHCO3, and reverse cross-linked by the addition of NaCl to a final concentration of 200 mm and by heating at 65 °C for at least 4 h. Recovered material was treated with proteinase K, extracted with phenol-chloroform-isoamyl alcohol (25:24:1), and precipitated. The pellets were resuspended in 30 μl of TE and analyzed by PCR using specific primers for the analyzed regions. The input sample was resuspended in 30 μl of TE and diluted 1:10 before PCR.
RESULTS
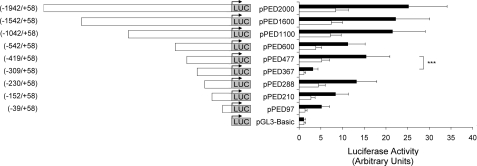
Localization of the Promoter Activity within the 5′-Flanking Region of the PED/PEA-15 Gene—To verify whether the proximal 5′-flanking region of the PED/PEA-15 gene contains a functional promoter, reporter gene assays were performed in HEK 293 and cervical carcinoma (HeLa) cell lines. A 2-kilobase PCR product containing the proximal 5′-flanking region, the transcription start site, and 58 bp of exon 1 was cloned upstream to a promoterless luciferase reporter gene (fragment -1942/+58, pPED2000). Three independent clones were tested showing similar promoter activity both in the HeLa and HEK 293 cells (data not shown). Next the pPED2000 plasmid was used as a template to generate progressive deletion fragments of the PED/PEA-15 5′-flanking region, which were analyzed for promoter activity (Fig. 1). The constructs were transfected in HeLa and HEK 293 cell lines, and luciferase activity was measured after 48 h. Significant transcriptional activity compared with the basal luciferase activity was obtained in both cell lines with all of the deletion constructs encompassing the PED/PEA-15 promoter region -1942/-39. Indeed, the shortest -39/+58 fragment still has a significantly higher luciferase activity compared with the empty vector, suggesting that all the basal promoter activity is contained in the very proximal 5′-flanking region. Because luciferase activity significantly declines from -230, it is possible that the minimal promoter is located within this sequence (Fig. 1). Truncation of promoter at an upstream region between -1042 and -542 and between -419 and -309 produced a decrease in the PED/PEA-15 transcriptional activity, indicating that these regions contain positive regulatory elements of the basal promoter function. A further deletion between -309 and -230 revealed the presence of a potential transcription silencer.
FIGURE 1.
Characterization of the promoter activity of the PED/PEA-15 5®-flanking region. The PED/PEA-15 5′-flanking fragments were cloned upstream to a promoterless luciferase reporter gene in the pGL3 Basic vector as described under “Experimental Procedures”. HeLa (black bars) and HEK 293 cells (gray bars) were then cotransfected with 3 μg of the construct DNAs (or 3 μg of the promoterless pGL3 Basic vector DNA) and 1 μg of the pRSVβ-gal vector DNA. Luciferase activity was assayed as described under “Experimental Procedures” and is presented as the increase above the activity measured with the pGL3 Basic vector. The results are presented as the means (normalized for β-galactosidase activity) ± S.D. of four independent experiments each performed in quadruplicate. Asterisks denote statistically significant differences (p < 0.001).
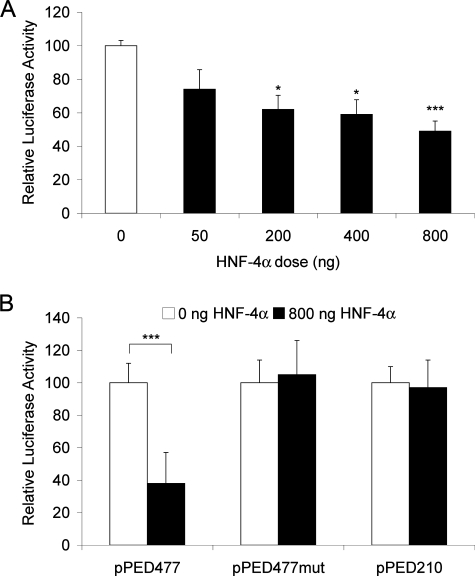
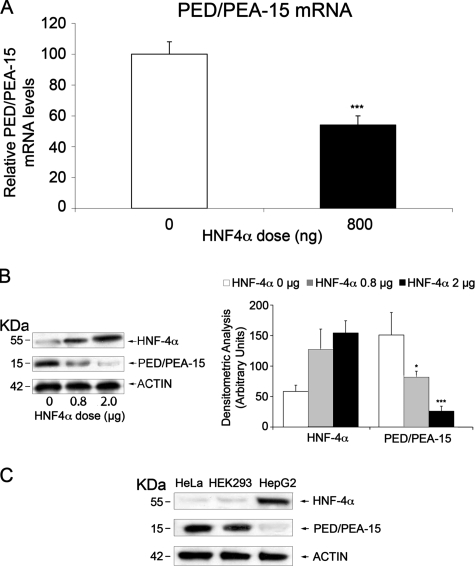
Regulation of the PED/PEA-15 Transcriptional Activity by HNF-4α—In silico analysis of the PED/PEA-15 promoter region revealed the presence of a 5′-GTCATCCAAAGGTCAAA-3′ sequence located between the -419/+58 and -309/+58 fragments. This sequence closely resembles the HRE (44, 45), suggesting the presence of an HNF-4α binding site in the promoter of the PED/PEA-15 gene. To address the role of HNF-4α in PED/PEA-15 promoter transcriptional activity, we co-transfected the -419/+58 PED/PEA-15 promoter-luciferase construct (pPED477) together with an HNF-4α expression vector (pCDNA3/HNF-4α) into HeLa cells, which feature low levels of the endogenous HNF-4α. As shown in Fig. 2A, HNF-4α reduces the reporter gene activity in a dose-dependent manner. This effect is specific, as it is lost when the cells are transfected with a vector containing the mutagenized HNF-4α binding sequence (pPED477mut) and a 5′ deletion construct lacking the HNF-4α site (pPED210) (Fig. 2B). Consistent with these luciferase assays, real time PCR assay on total RNA extracted from HNF-4α-transfected HeLa cells confirmed a 2-fold decrease in PED/PEA-15 expression levels (Fig. 3A). Same analysis was performed by Western blot with similar results; as shown in Fig. 3B, the levels of PED/PEA-15 protein decreases in HNF-4α-transfected HeLa cells. Moreover, we observed an inverse correlation between PED/PEA-15 and HNF-4α protein levels in different cell types. In fact, HepG2 cells present well detectable levels of endogenous HNF-4α and low levels of PED/PEA-15, whereas the opposite is observed in the HeLa and HEK 293 cells, both of which show low levels of HNF-4α and high levels of PED/PEA-15 (Fig. 3C). Overall, our results indicate that HNF-4α negatively regulates PED/PEA-15 gene expression.
FIGURE 2.
Regulation of PED/PEA-15 promoter activity by HNF-4α. A, HeLa cells were co-transfected with 2 μg of the pPED477 PED/PEA-15 promoter-luciferase construct and the indicated amounts of the pCDNA3/HNF-4α expression vector. B, alternatively, the cells were co-transfected with the pPED477, the pPED477mut, or the pPED210 PED/PEA-15 promoter luciferase constructs, and 0.8 μg of the pCDNA3/HNF-4α expression vector. Luciferase activities were normalized for those of β-galactosidase and are presented as the means ± S.D. of four independent experiments each performed in quadruplicate. Asterisks denote statistically significant differences (*, p < 0.05; ***, p < 0.001).
FIGURE 3.
Correlation between PED/PEA-15 and HNF-4α levels. A, HeLa cells were transfected with 0.8 μg of the pCDNA3/HNF-4α expression vector. PED/PEA-15 mRNA levels were then quantitated by RT-PCR. Data were normalized for β-actin mRNA and are expressed as % decrease versus control (untransfected cells). B, HeLa cells were transfected with the indicated amounts of the pCDNA3/HNF-4α vector DNA. The cells were then solubilized and Western-blotted with PED/PEA-15 antibodies followed by reblotting with HNF-4α antibodies. C, lysates from HeLa, HEK 293, and HepG2 cells (40 μg protein/sample) were analyzed by Western blotting with PED/PEA-15 antibodies followed by reblotting with HNF-4α and actin antibodies. All filters were revealed by ECL and autoradiography and quantitated by laser densitometry of the autoradiographs. Bars represent the means ± S.D. of three (B) and four (A) independent experiments. Asterisks denote statistically significant differences (*, p < 0.05; ***, p < 0.001). Representative blots are also shown.
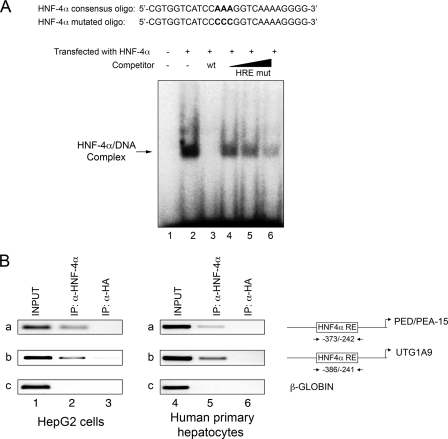
HNF-4α Binds the Cis-acting Element at Position -335 to -320 in the PED/PEA-15 Promoter—We next tested the ability of HNF-4α to bind the putative binding sequence in the PED/PEA-15 gene promoter. Electrophoretic mobility shift assay was performed using a probe containing the putative HNF-4 -binding site, designated HNF-4α RE element (Fig. 4A). Whole cell extracts were obtained from HeLa cells transfected either with an HNF-4α expression plasmid or a control vector. Incubation of HNF-4α-transfected HeLa cell extracts with the radiolabeled probe resulted in a strong band that was absent when the cells were transfected with the empty vector (lanes 1 and 2). Competition with unlabeled HNF-4α RE oligonucleotide abolished this band (lane 3). In contrast, increasing amounts of the mutated HNF-4α RE probe added to the binding reactions competed for HNF-4α binding to the probe encompassing the wild-type sequence much less efficiently (lanes 4-6). These data indicate that in vitro HNF-4α binds to the PED/PEA-15 HNF-4α RE site at -335 to -320 position.
FIGURE 4.
HNF-4α binding to the PED/PEA-15 gene promoter. A, electrophoretic mobility shift assay. Whole cell extracts from HeLa cells transfected with either the empty plasmid (lane 1) or the HNF-4α expression vector (lanes 2-6) were incubated with the 32P-labeled HNF-4α RE probe. Incubation occurred in the absence (lanes 1-2) or the presence of either a 4-fold molar excess of unlabeled HNF-4α RE probe (HRE wt; lane 3) or a 2-, 4-, 10-fold molar excess of unlabeled HNF-4α RE mutated oligonucleotides (HRE mut; lanes 4-6). Proteins were separated on a non-denaturing polyacrylamide gel and revealed by autoradiography. The autoradiograph shown is representative of four independent experiments. B, ChIP assay. Soluble chromatin was prepared from HepG2 cells and human primary hepatocytes as described under “Experimental Procedures” and immunoprecipitated (IP) with either HNF-4α (lanes 2 and 5) or HA antibodies (lanes 3 and 6). Total (INPUT, lanes 1 and 4) and immunoprecipitated DNAs were then amplified using primer pairs covering HNF-4α RE on the PED/PEA-15 (a) and the UGT1A9 promoters (b, positive control) or the β-GLOBIN (c, negative control). The photographs shown are each representative of three independent experiments.
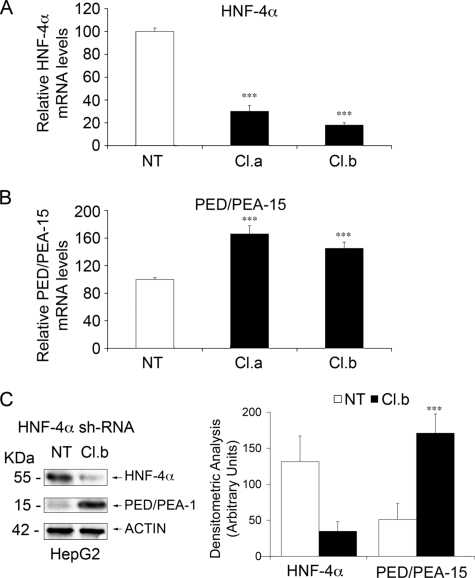
Occupancy of the PED/PEA-15 promoter by HNF-4α in living cells was further analyzed by ChIP experiment. Wild-type HepG2 cells, which show well detectable levels of this transcription factor, were used. Chromatin DNA was precipitated using either the anti-HNF-4α or anti-HA antibodies as the negative control, and the sequence encompassing the HNF-4α consensus binding site was amplified using specific primers. As shown in Fig. 4B, precipitation with HNF-4α (lane 2a) but not HA (lane 3a) antibodies enabled amplification, indicating occupation of the PED/PEA-15 promoter by HNF-4α. As a positive control, a 145-bp fragment containing the HNF-4α response element of the UGT1A9 promoter was also amplified from genomic DNA precipitated with the HNF-4α antibody (lane 2b) (46). Consistently, PCR amplification with oligonucleotides for β-GLOBIN (negative control) did not show any signal. Same results were also obtained with primary human hepatocytes (Fig. 4B). To further assess the direct role of HNF-4α on PED/PEA-15 expression, we used an RNA-mediated interference-mediated approach. Transfection of HepG2 cells with HNF-4α specific shRNA clones caused a decrease of HNF-4α mRNA to 20% that of the endogenous levels (Fig. 5A). In the same cells, a parallel increase of PED/PEA-15 mRNA and protein levels was observed (Fig. 5, B and C), supporting the important role of HNF-4α in controlling PED/PEA-15 expression.
FIGURE 5.
The effect of RNA-mediated interference-mediated silencing of HNF-4α on PED/PEA-15 gene expression in HepG2 cells. HepG2 cells were transiently transfected with two HNF-4α-specific shRNA clones (Cl.a, Cl.b). 48 h upon transfection total RNAs were extracted form transfected and nontransfected cells (NT). HNF-4α (A) and PED/PEA-15 (B) mRNA levels were then quantitated by RT-PCR. Bars represent values normalized for β-ACTIN mRNA. C, HepG2 cells were transiently transfected with the HNF-4α shRNA (Cl.b). After 48 h transfected and non-transfected cells were solubilized, and lysates were Western-blotted with antibodies toward HNF-4α, PED/PEA-15, or β-actin. Filters were revealed by ECL and autoradiography followed by densitometric analysis of the autoradiographs. Bars represent the means ± S.D. of four (A and B) and three (C) independent experiments. Asterisks denote statistically significant differences. A representative blot is shown in the left panel of C.
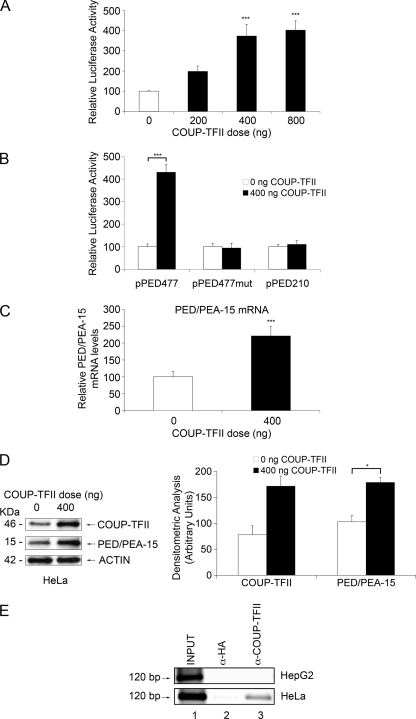
PED/PEA-15 Gene Transcription Is Activated by COUP-TFII—COUP-TF antagonizes HNF-4α-dependent gene expression (27, 28, 47-49). We have, therefore, hypothesized that COUP-TFs may also regulate PED/PEA-15 transcription. To this aim, the pPED477 luciferase construct containing the HRE was transfected in HeLa cells together with the COUP-TFII expression vector (pCMV6-XL5COUP-TFII). As shown in Fig. 6A, COUP-TFII produced an activation of the reporter gene in a dose-dependent manner (Fig. 6A). This effect did not occur in cells transfected with a pPED477mut containing the mutagenized HNF-4α binding site and the 5′ deletion construct lacking this site (pPED210) (Fig. 6B), indicating that COUP-TFII activates the human PED/PEA-15 promoter through the HNF-4α-responsive element. Real time PCR assay and Western blot analysis on COUP-TFII-transfected HeLa cells also confirmed the increase in PED/PEA-15 expression level (Fig. 6, C and D).
FIGURE 6.
COUP-TFII action on PED/PEA-15 expression. A, HeLa cells were transiently transfected with 2 μg of the pPED477 PED/PEA-15 promoter-luciferase construct, the indicated amounts of the pCMV6-XL5COUP-TFII plasmid, and 1 μg of the pRSV-βgal plasmid (transfection efficiency control). Alternatively (B), the cells were transfected with the pPED477, the pPED477mut, or the pPED210 PED/PEA-15 promoter luciferase constructs together with 0.4 μg of the COUP-TFII expression vector and 1 μg of the pRSV-βgal expression plasmid. Luciferase activity was assayed as described under “Experimental Procedures” and normalized for β-galactosidase. C, HeLa cells were transfected with 0.4 μg of the COUP-TFII expression vector. At 48 h, PED/PEA-15 mRNA levels were quantitated by RT-PCR and normalized for β-ACTIN mRNA. Bars represent the means ± S.D. of three (A and B) and four (C) independent experiments, each in quadruplicate. All data are expressed as increases above values in control cells. D, HeLa cells were transfected with 0.4 μg of the COUP-TFII expression plasmid and solubilized, and proteins (40 μg/sample) were separated by PAGE and analyzed by Western blotting with COUP-TFII and PED/PEA-15 antibodies as indicated. Filters were revealed by ECL and autoradiography, and autoradiographs were subjected to densitometry. Bars represent the means ± S.D. of three independent experiments. A representative experiment is shown in the left panel. Asterisks denote statistically significant differences (*, p < 0.05; ***, p < 0.001). E, HepG2 and HeLa cells were cross-linked with 1% formaldehyde and subjected to Chip assays using COUP-TFII (lanes 3) and HA antibodies (lanes 2; negative control). Total (lane 1; INPUT) and immunoprecipitated DNAs were amplified using primers covering the HNF-4α RE in the PED/PEA-15 gene. The photographs shown are representative of at least three independent experiments.
As shown in Fig. 6E, COUP-TFII specifically bound the HRE on PED/PEA-15 promoter in native HeLa cells. At variance, COUP-TFII binding to the HRE was almost undetectable in the HepG2 cell lines, which express HNF-4α at higher levels.
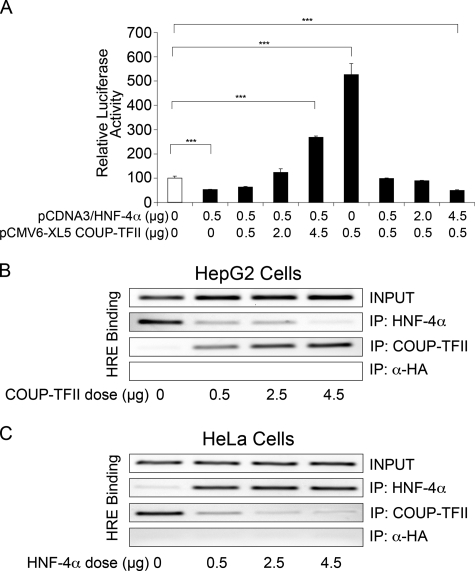
COUP-TFII Opposes HNF-4α Repression of PED/PEA-15 Gene Expression—Because HNF-4α and COUP-TFII appear to bind the same response element on the promoter of PED/PEA-15 gene, we hypothesized that these two proteins compete with each other for binding. Therefore, transient transfection assays were performed with the pPED477 reporter construct and various combinations of HNF-4α and COUP-TFII expression vectors. As shown in Fig. 7A, HNF-4α repression of the pPED477 construct expression in HeLa cells can be completely overcome by increasing amounts of COUP-TFII. Conversely, transactivation by COUP-TFII was repressed by increasing amounts of HNF-4α, indicating that COUP-TFII antagonizes repression of the PED/PEA-15 gene by HNF-4α. Moreover, ChIP experiments in HepG2 cells demonstrate that HNF-4α binding to PED/PEA-15 promoter is reduced by increasing amounts of COUP-TFII (Fig. 7B), whereas the opposite was obtained in HeLa cells in which COUP-TFII binding to PED/PEA-15 promoter is antagonized by increasing amount of HNF-4α (Fig. 7C).
FIGURE 7.
Antagonistic effects of HNF-4α and COUP-TFII on PED/PEA-15 promoter. A, HeLa cells were transiently transfected with 2 μg of the pPED477 PED/PEA-15 promoter-luciferase construct alone (open bar) or in combination with the indicated amounts of the pCMV6-XL5COUP-TFII and the pCDNA3/HNF-4α plasmids. Luciferase activity was assayed and normalized as described under “Experimental Procedures” Bars represent the mean ± S.D. of three independent experiments. Asterisks denote statistically significant differences (p < 0.001). B and C, ChIP assays. Soluble chromatin was prepared from HepG2 (B) and HeLa (C) cell lines as described under “Experimental Procedures” and immunoprecipitated (IP) with HNF-4α, COUP-TFII, and HA antibodies. Total (INPUT) and immunoprecipitated DNAs were amplified using primer pairs covering HNF-4α RE on the PED/PEA-15 promoter. The photographs shown are representative of three independent experiments.
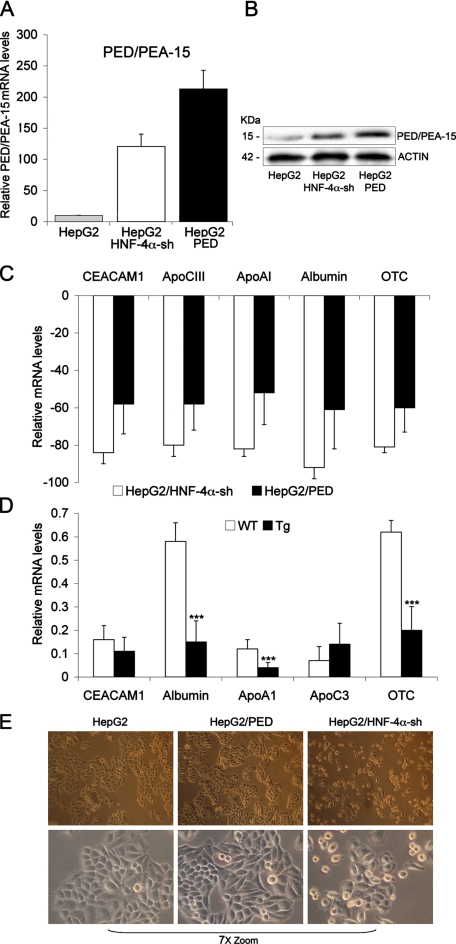
PED/PEA-15 Overexpression Decreases Hepatocyte Marker Gene Expression—Fetal livers obtained from HNF-4α-/- mice fail to express a large array of genes encoding several apolipoproteins, metabolic proteins, and serum factors whose expression is essential for the function of a differentiated parenchyma (21). Indeed, HNF-4α acts as hepatocyte differentiation factor during mammalian liver development. Consistently, HepG2 cells stably transfected with an HNF-4α-specific shRNA clone show about an 80% reduction in mRNA levels of several genes expressed in differentiated hepatocytes (CEACAM1, apoCIII, apoAI, albumin, OTC (ornithine transcarbamyolase), Fig. 8C) compared with wild-type cells (p < 0.001). Because these cells feature high levels of PED/PEA-15 mRNA and protein (Fig. 8, A and B), we investigated whether PED/PEA-15 overexpression may affect hepatocyte cell differentiation. To this aim we have stably transfected HepG2 cells with a vector driving expression of the human PED/PEA-15 cDNA, increasing the PED/PEA-15 mRNA and protein levels (Fig. 8, A and B, third lane). As shown in Fig. 8C, the abundance of CEACAM1, apoCIII, apoAI, albumin, and OTC RNAs was reduced by about 60% in the HepG2/Ped compared with the wild-type cells (p < 0.001). To further address this issue, we analyzed livers from ped/pea-15 transgenic mice. These animals have been previously characterized and reported (15). The abundance of the CEACAM1, apoAI, albumin, and OTC mRNA was reduced by 20-50% in their livers (p < 0.001, Fig. 8D). The HepG2/Ped cells also displayed an intermediate morphology compared with both wild-type and HNF-4α shRNA-transfected cells with a less polygonal cell shape and a more disorganized monolayer formation with less looser cell-cell contacts compared with wild-type cells (Fig. 8E).
FIGURE 8.
PED/PEA-15 action on the expression of hepatocyte differentiation markers. A, total RNA preparations were obtained from HepG2 wild-type cells and from cells stably expressing either the HNF-4α-specific shRNA (HepG2/HNF-4α-sh) or the human PED/PEA-15 gene (HepG2/PED). B, lysates from HepG2 wild-type cells and from cells stably expressing either the HNF-4α-specific shRNA (HepG2/HNF-4α-sh) or the human PED/PEA-15 gene (HepG2/PED) (40 mg of protein/sample) were analyzed by Western blotting with PED/PEA-15 antibody followed by reblotting with actin antibody. The levels of PED/PEA-15 (A) CEACAM1, apoCIII, apoAI, albumin, and OTC mRNAs (C) were then quantitated by RT-PCR and normalized for β-actin mRNA. mRNA levels shown in the figure are relative to those in control (HepG2 wild type) cells. Bars represent the means ± S.D. of data from four independent experiments. D, RNA preparations were obtained from livers of ped/pea-15 transgenic and control mice (non-transgenic littermates). The indicated mRNAs were subsequently quantitated by RT-PCR and normalized for β-actin. Bars represent the means ± S.D. of four independent experiments, where reactions were performed in triplicate using pooled total RNA preparations from six mice/genotype. Asterisks denote statistically significant differences (p < 0.001). E, phase contrast microphotographs of HepG2, HepG2/PED, and HepG2/HNF-4α-sh cells (upper panel, 100× magnification; lower panel, 7×-zoomed microphotographs of the above fields).
DISCUSSION
Several lines of evidence indicate that a multitude of concurrent alterations contribute to type 2 diabetes onset and progression (50-52). Among these defects, the PED/PEA-15 gene has been found to be overexpressed in about 30% of individuals affected by type 2 diabetes and in their first-degree relatives. Studies in cellular and animal models have shown a cause-effect relationship between the overexpression of PED/PEA-15 and impaired insulin action (4, 14-16, 53). Thus, clarifying the mechanism of PED/PEA-15 transcriptional regulation will help in understanding its abnormalities in type 2 diabetes. In the present study we have isolated and further characterized the PED/PEA-15 promoter region, and we have identified cis elements involved in the regulation of PED/PEA-15 human gene expression.
The transcription start site of the PED/PEA-15 gene has already been mapped (54). Analysis of the 5′ genomic sequence revealed that this gene lacks a TATA box and the first 600 bp flanking the transcription start region have a high GC content (64%), including several consensus Sp1 sites. These data are consistent with our results showing that the shortest 5′-flanking fragment spanning nucleotides -39/+58 contains all the elements necessary to achieve basal promoter activity.
Deletion analyses showed that HNF-4α, a member of the steroid receptor class of transcription factors, binds to the -419/-309 fragment and represses PED/PEA-15 transcription through this region (Figs. 1 and 2). Indeed transfection experiments of both an HNF-4α expression vector in HeLa cells and an HNF-4α-specific shRNA clone in HepG2 cells confirmed that this transcription factor is capable of repressing PED/PEA-15 expression both at the RNA and at the protein levels (Figs. 3 and 5). Electrophoretic mobility shift assay and ChIP assays have further shown that the promoter site recognized by HNF-4α is located between nucleotides -335 and -320 (Fig. 4). This element closely resembles the HNF-4α REs found in other genes and the GGGGCAAAGGTCA consensus HNF-4α binding site (55). In addition we have shown that the same responsive element is recognized by COUP-TFII, another member of the orphan receptor family (Fig. 6). We found that COUP-TFII activates PED/PEA-15 gene expression by binding to the HRE (Fig. 6), whereas HNF-4α functions as a transcriptional repressor. The observation that these two transcription factors bind to the HRE in a mutually exclusive manner (Fig. 7) suggests that their mechanism of action involves direct competition at the level of DNA binding. Thus, transcription of the PED/PEA-15 gene is dependent upon the intracellular balance of these positive and negative regulatory factors. Competition for DNA binding has been described for other members of the steroid receptor superfamily, which because of their highly related zinc finger DNA binding domains, can interact with overlapping or identical DNA elements. For example, estrogen and thyroid hormone receptors exert opposite regulatory effects via their competitive binding to the estrogen response element (56).
In certain cell types where COUP-TFII occupies the HRE, transcription of PED/PEA-15 gene is enhanced, whereas in other cell types, such as the hepatocytes, where HNF-4α is present in relatively abundant amounts, COUP-TFII is replaced by HNF-4α, and PED/PEA-15 transcription is repressed. At variance from the PED/PEA-15 gene, COUP-TFII most often serves as a transcriptional silencer (33), whereas HNF-4α is an activator (27). There is also evidence, however, indicating that the function of many nuclear hormone receptors is dependent upon the actions of distinct receptor binding cofactors that differentially recognize occupied and unoccupied receptors (34-41). Indeed, nuclear receptors often interact with co-regulators affecting their target genes in a tissue- and gene-specific manner (57). For instance, it has been reported that PGC-1α co-activates HNF-4α and induces CYP7A1 gene transcription during starvation in mice (58). But Song et al. (59) also demonstrated that Prox1, an early specific marker for liver and pancreas development from the foregut endoderm, interacts with HNF-4α and suppresses CYP7A1 gene transcription. These authors have generated evidence suggesting that Prox1 competes with PGC-1α for HNF-4α binding and, thus, inhibits PGC-1α co-activating activity. The identification of ligands cooperating with HNF-4α and COUP-TFII in the regulation of PED/PEA-15 gene is currently in progress in the laboratory and is expected to help clarify the mechanism linking PED/PEA-15 overexpression and the genes controlling its expression. Indeed, PED/PEA-15 overexpression likely represents a molecular abnormality downstream of distinct diabetes risk genes and serves as an effector of these genes.
Recently, Parviz et al. (22) demonstrated the importance of HNF-4α in the formation of a hepatic epithelium and liver morphogenesis (22). Moreover, targeted disruption of HNF-4α in the adult liver results in impaired lipid homeostasis, abnormal glycogen deposition, and hepatic hypertrophy (18). In fact, individuals with HNF-4α mutations leading to MODY1 exhibit impaired lipid homeostasis before the onset of β cell dysfunction and hyperglycemia, indicating the presence of a primary hepatic lesion (60). Here, we show that the activation of PED/PEA-15 gene expression in hepatocytes is paralleled by the establishment of a partially dedifferentiated phenotype accompanied by reduction in mRNA levels of genes, which are expressed during normal liver development (Fig. 8). Thus, PED/PEA-15 overexpression seems to have similar consequences as HNF-4α haploinsufficiency. These include impaired liver differentiation as well as β cell dysfunction, both of which contribute to the progression toward diabetes.
Previous studies in β cells evidenced that PED/PEA-15 restrains the expression of HNF-3β (Foxa2) (53), a master transcription factor controlling several differentiation regulatory genes, including HNF-4α (61). One might speculate that PED/PEA-15 loops back on HNF-4α, thereby causing dedifferentiation by silencing the expression of HNF-4α itself. Indeed, HepG2 cells overexpressing a PED/PEA-15 cDNA feature reduced HNF-4α levels (data not shown). Further studies are in progress in the laboratory to address this issue and clarify the significance of PED/PEA-15 levels to hepatocyte differentiation.
In conclusion, in this study we have cloned and characterized the PED/PEA-15 promoter. We have further shown that two transcription factors, HNF-4α and COUP-TFII, compete for binding to the HRE site on PED/PEA-15 gene promoter. Although COUP-TFII increases PED/PEA-15 gene expression, HNF-4α antagonizes PED/PEA-15 transactivation. Polymorphisms in the consensus binding sequence for these two proteins may affect PED/PEA-15 expression and increase the susceptibility to type 2 diabetes.
Supplementary Material
Acknowledgments
We thank Dr. Graeme Bell (University of Chicago, Chicago, IL) for kindly providing the pCDNA3/HNF4a plasmid.
This work was supported, in part, by European Community FP6 EUGENE2 Contract LSHM-CT-2004-512013, FP7 PREPROBEDIA Grant 201681, and by grants from the European Foundation for the Study of Diabetes (EFSD)/Lilly, the Associazione Italiana per la Ricerca sul Cancro), and the Ministero dell'Università e della Ricerca Scientifica. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1.
Footnotes
The abbreviations used are: PED/PEA-15, phosphoprotein enriched in diabetes/phosphoprotein enriched in astrocytes; HNF-4α, hepatocyte nuclear factor 4α; COUP-TFII, chicken ovalbumin upstream promoter transcription factor II; RT, reverse transcription; HRE, HNF-4α response element; shRNA, short hairpin RNA; PIPES, piperazine-N,N′-bis(2-ethanesulfonic acid); ChIP, chromatin immunoprecipitation; RE, response element.
References
- 1.Danziger, N., Yokoyama, M., Jay, T., Cordier, J., Glowinski, J., and Chneiweiss, H. (1995) J. Neurochem. 64 1016-1025 [DOI] [PubMed] [Google Scholar]
- 2.Araujo, H., Danziger, N., Cordier, J., Glowinski, J., and Chneiweiss, H. (1993) J. Biol. Chem. 268 5911-5920 [PubMed] [Google Scholar]
- 3.Estelles, A., Yokoyama, M., Nothias, F., Vincent, J. D., Glowinski, J., Vernier, P., and Chneiweiss, H. (1996) J. Biol. Chem. 271 14800-14806 [DOI] [PubMed] [Google Scholar]
- 4.Condorelli, G., Vigliotta, G., Iavarone, C., Caruso, M., Tocchetti, C. G., Andreozzi, F., Cafieri, A., Tecce, M. F. P. F., Beguinot, L., and Beguinot, F. (1998) EMBO J. 17 3858-3866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Condorelli, G., Vigliotta, G., Cafieri, A., Trencia, A., Andalo, P., Oriente, F., Miele, C., Caruso, M., Formisano, P., and Beguinot, F. (1999) Oncogene 18 4409-4415 [DOI] [PubMed] [Google Scholar]
- 6.Condorelli, G. A., Trencia, A., Vigliotta, G., Perfetti, A., Goglia, U., Cassese, A., Musti, A. M., Miele, C., Santopietro, S., Formisano, P., and Beguinot, F. (2002) J. Biol. Chem. 277 11013-11018 [DOI] [PubMed] [Google Scholar]
- 7.Estelles, A., Charlton, C. A., and Blau, H. M. (1999) Dev. Biol. 216 16-28 [DOI] [PubMed] [Google Scholar]
- 8.Hao, C., Beguinot, G., Condorelli, G., Trencia, A., Van Meir, E. G., Yong, V. W., Parney, I. F., Roa, W. H., and Petruk, K. C. (2001) Cancer Res. 61 1162-1170 [PubMed] [Google Scholar]
- 9.Trencia, A., Perfetti, A., Cassese, A., Vigliotta, G., Miele, C., Oriente, F., Santopietro, S., Giacco, F., Condorelli, G., Formisano, P., and Beguinot, F. (2003) Mol. Cell. Biol. 23 4511-4521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Formstecher, E., Ramos, J., Fauquet, M., Calderwood, D., Hsieh, J., Canton, B., Nguyen, X., Barnier, J., Camonis, J., Ginsberg, M., and Chneiweiss, Y. (2001) Dev. Cell 1 239-250 [DOI] [PubMed] [Google Scholar]
- 11.Xiao, C., Yang, B. F., Asadi, N., Beguinot, F., and Hao, C. (2002) J. Biol. Chem. 277 25020-25025 [DOI] [PubMed] [Google Scholar]
- 12.Formisano, P., Perruolo, G., Libertini, S., Santopietro, S., Troncone, G., Raciti, A., Oriente, F., Portella, G., Miele, C., and Beguinot, F. (2005) Oncogene 24 7012-7021 [DOI] [PubMed] [Google Scholar]
- 13.Zhang, Y., Redina, O., Altshuller, Y. M., Yamazaki, M., Ramos, J., Chneiweiss, Y., Kanaho, Y., and Frohman, M. A. (2000) J. Biol. Chem. 275 35224-35232 [DOI] [PubMed] [Google Scholar]
- 14.Condorelli, G., Vigliotta, G., Trencia, A., Maitan, M. A., Caruso, M., Miele, C., Oriente, F., Santopietro, S., Formisano, P., and Beguinot, F. (2001) Diabetes 50 1244-1252 [DOI] [PubMed] [Google Scholar]
- 15.Vigliotta, G., Miele, C., Santopietro, S., Portella, G., Perfetti, A., Maitan, M. A., Cassese, A., Oriente, F., Trencia, A., Fiory, F., Romano, C., Tiveron, C., Tatangelo, L., Troncone, G., Formisano, P., and Beguinot, F. (2004) Mol. Cell. Biol. 24 5005-5015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Valentino, R., Lupoli, G. A., Raciti, G. A., Oriente, F., Farinaro, E., Della Valle, E., Salomone, M., Riccardi, G., Vaccaro, O., Donnarumma, G., Sesti, G., Hribal, M., Cardellini, M., Miele, C., Formisano, P., and Beguinot, F. (2006) Diabetologia 49 3058-3066 [DOI] [PubMed] [Google Scholar]
- 17.Perfetti, A., Oriente, F., Iovino, S., Alberobello, T., Barbagallo, A. P. M., Esposito, I., Fiory, F., Teperino, R., Ungaro, P., Miele, C., Formisano, P., and Beguinot, F. (2007) J. Biol. Chem. 282 8648-8657 [DOI] [PubMed] [Google Scholar]
- 18.Hayhurst, G., Lee, Y., Lambert, G., Ward, J., and Gonzalez, F. (2001) Mol. Cell. Biol. 21 1393-1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pereira, F., Tsai, M., and Tsai, S. (2000) Cell. Mol. Life Sci. 57 1388-1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xanthopoulos, K., Prezioso, V., Chen, W., Sladek, F., Cortese, R., and Darnell, J. J. (1991) Proc. Natl. Acad. Sci. U. S. A. 88 3807-3811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, J., Ning, G., and Duncan, S. A. (2000) Genes Dev. 14 464-474 [PMC free article] [PubMed] [Google Scholar]
- 22.Parviz, F., Matullo, C., Garrison, W., Savatski, L., Adamson, J., Ning, G., Kaestner, K., Rossi, J., Zaret, K., and Duncan, S. (2003) Nat. Genet. 34 292-296 [DOI] [PubMed] [Google Scholar]
- 23.Black, M. H., Fingerlin, T. E., Allayee, H., Zhang, W., Xiang, A. H., Trigo, E., hartiala, J., Lehtinen, A. B., Haffner, S. M., Bergman, R. N., McEachin, R. C., Kjos, S. L., Lawrence, J. M., Buchanan, T. A., and Watanabe, R. M. (2008) Diabetes 57 1048-1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Menjivar, M., Granados-Silvestre, M. A., Montùfar-Robles, I., Herrera, M., Tusiè-Luna, M. T., Canizales-Quinteros, S., Aguilar-Salinas, C., and Ortiz-Lòpez, M. G. (2008) Clin. Genet. 73 185-187 [DOI] [PubMed] [Google Scholar]
- 25.Johansson, S., Raeder, H., Eide, S. A., Midthjell, K., Hveem, K., Søvik, O., Molven, A., and Njølstad, P. R. (2007) Diabetes 56 3112-3117 [DOI] [PubMed] [Google Scholar]
- 26.Ktistaki, E., Lacorte, J.-M., Katrakili, V., Zannis, I., and Talianidis, I. (1994) Nucleic Acids Res. 22 4689-4696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ladias, J. A. A., Cladaras-Hadzopoulou, M., Kardassis, D., Cardot, P., Cheng, J., Zannis, V. I., and Cladaras, C. (1992) J. Biol. Chem. 267 15849-15860 [PubMed] [Google Scholar]
- 28.Mietus-Snyder, M., Sladek, F. M., Ginsburg, G. S., Kuo, C. K., Ladias, J. A. A., Darnell, J. E., and Karathanasis, S. K. (1992) Mol. Cell. Biol. 12 1708-1718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qiu, Y., Tsai, S. Y., and Tsai, M.-J. (1994) Trends Endocrinol. Metab. 5 234-239 [DOI] [PubMed] [Google Scholar]
- 30.Qiu, Y., Cooney, A. J., Kuratani, S., De Mayo, F. J., Tsai, S. Y., and Tsai, M.-J. (1994) Proc. Natl. Acad. Sci. U. S. A. 91 4451-4455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hwung, Y.-P., Crowe, D., Wang, L.-H., Tsai, S. Y., and Tsai, M.-J. (1988) Mol. Cell. Biol. 8 2070-2077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lou, D., Tannour, M., Selig, L., Thomas, D., Kahn, A., and Vasseur-Cognet, M. (1999) J. Biol. Chem. 274 28385-28394 [DOI] [PubMed] [Google Scholar]
- 33.Park, J., Tsai, S., and Tsai, M. (2002) Keio J. Med. 52 174-181 [DOI] [PubMed] [Google Scholar]
- 34.Chen, J., and Evans, R. (1995) Nature 377 454-457 [DOI] [PubMed] [Google Scholar]
- 35.Horlein, A., Naar, A., Heinzel, T., Torchia, J., Gloss, B., Kurokawa, R., Ryan, A., Kamei, Y., Soderstrom, M., and Glass, C. (1995) Nature 377 397-404 [DOI] [PubMed] [Google Scholar]
- 36.Onate, S., Tsai, S., Tsai, M.-J., and O'Malley, B. (1995) Science 270 1354-1357 [DOI] [PubMed] [Google Scholar]
- 37.Cavailles, V., Dauvois, S., L'Horset, F., Lopez, G., Hoare, S., Kushner, P., and Parker, M. (1995) EMBO J. 14 3741-3751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Le Douarin, B., Zechel, C., Garnier, J., Lutz, Y., Tora, L., Pierrat, P., Heery, D., Gronemeyer, H., Chambon, P., and Losson, R. (1995) EMBO J. 14 2020-2033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee, J., Ryan, F., Swaffield, J., Johnston, S., and Moore, D. (1995) Nature 374 91-94 [DOI] [PubMed] [Google Scholar]
- 40.Kurokawa, R., Soderstrom, M., Horlein, A., Halachmi, S., Brown, M., Rosenfeld, M. G., and Glass, C. K. (1995) Nature 377 451-454 [DOI] [PubMed] [Google Scholar]
- 41.Burris, T., Nawaz, Z., Tsai, M.-J., and O'Malley, B. W. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 9525-9529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wigler, M., Pellicer, A., Silverstein, S., Axel, R., Urlaub, G., and Chasin, L. (1979) Proc. Natl. Acad. Sci. U. S. A. 1979 1373-1376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hatzis, P., and Talianidis, I. (2001) Mol. Cell. Biol. 21 7320-7330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lausen, J., Thomas, H., Lemm, I., Bulman, M., Borgschulze, M., Lingott, A., Hattersley, A., and Ryffel, G. U. (2000) Nucleic Acids Res. 28 430-437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eeckhoute, J., Formstecher, P., and Laine, B. (2001) Mol. Endocrinol. 15 1200-1210 [DOI] [PubMed] [Google Scholar]
- 46.Barbier, O., Girard, H., Inoue, Y., Duez, H., Villeneuve, L., Kamiya, A., Fruchart, J., Guillemette, C., Gonzalez, F., and Staels, B. (2005) Mol. Pharmacol. 67 241-249 [DOI] [PubMed] [Google Scholar]
- 47.Cairns, W., Smith, C., McLaren, A., and Wolf, C. (1996) J. Biol. Chem. 271 25269-25276 [DOI] [PubMed] [Google Scholar]
- 48.Galson, D., Tsuchiya, T., Tendler, D., Huang, L., Ren, Y., Ogura, T., and Bunn, H. (1995) Mol. Cell. Biol. 15 2135-2144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kimura, A., Nishiyory, A., Murakami, T., Tsukamoto, T., Hata, S., Osumi, T., Okamura, R., Mori, M., and Takiguchi, M. (1993) J. Biol. Chem. 274 11125-11133 [PubMed] [Google Scholar]
- 50.Mercado, M. M., McLenithan, J. C., Silver, K. D., and Shuldiner, A. R. (2002) Curr. Diab. Rep. 2 83-95 [DOI] [PubMed] [Google Scholar]
- 51.Bonadonna, R. C., and De Fronzo, R. A. (1991) Diabetes Metab. 17 112-135 [PubMed] [Google Scholar]
- 52.Owen, K. R., and McCarthy, M. I. (2007) Curr. Opin. Genet. Dev. 17 239-244 [DOI] [PubMed] [Google Scholar]
- 53.Miele, C., Raciti, A., Cassese, A., Romano, C., Giacco, F., Oriente, F., Paturzo, F., Andreozzi, F., Zabatta, A., Troncone, G., Bosch, F., Pujol, A., Chneiweiss, H., Formisano, P., and Beguinot, F. (2007) Diabetes 56 622-633 [DOI] [PubMed] [Google Scholar]
- 54.Wolford, J. K., Bogardus, C., Ossowski, V., and Prochazka, M. (2000) Gene (Amst.) 241 143-148 [DOI] [PubMed] [Google Scholar]
- 55.Fraser, J. D., Martinez, V., Straney, R., and Briggs, M. R. (1998) Nucleic Acids Res. 26 2702-2707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Glass, C. K., Holloway, J. M., Devary, O. V., and Rosenfeld, M. G. (1988) Cell 54 313-323 [DOI] [PubMed] [Google Scholar]
- 57.Glass, C. K., and Rosenfeld, M. G. (2000) Genes Dev. 14 121-141 [PubMed] [Google Scholar]
- 58.Shin, D., Campos, J., Gil, G., and Osborne, T. (2003) J. Biol. Chem. 5 50047-50052 [DOI] [PubMed] [Google Scholar]
- 59.Song, K., Tiangang, L., and Chiang, J. (2006) J. Biol. Chem. 281 10081-10088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shih, D. Q., Dansky, H. M., Fleisher, M., Assmann, G., Fajans, S. S., and Stoffel, M. (2000) Diabetes 49 832-837 [DOI] [PubMed] [Google Scholar]
- 61.Wolfrum, C., Besser, D., Luca, E., and Stoffel, M. (2003) Diabetes 52 2989-299514633861 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.