Abstract
The crystal structure of glucooligosaccharide oxidase from Acremonium strictum was demonstrated to contain a bicovalent flavinylation, with the 6- and 8α-positions of the flavin isoalloxazine ring cross-linked to Cys130 and His70, respectively. The H70A and C130A single mutants still retain the covalent FAD, indicating that flavinylation at these two residues is independent. Both mutants exhibit a decreased midpoint potential of ∼+69 and +61 mV, respectively, compared with +126 mV for the wild type, and possess lower activities with kcat values reduced to ∼2 and 5%, and the flavin reduction rate reduced to 0.6 and 14%. This indicates that both covalent linkages increase the flavin redox potential and alter the redox properties to promote catalytic efficiency. In addition, the isolated H70A/C130A double mutant does not contain FAD, and addition of exogenous FAD was not able to restore any detectable activity. This demonstrates that the covalent attachment is essential for the binding of the oxidized cofactor. Furthermore, the crystal structure of the C130A mutant displays conformational changes in several cofactor and substrate-interacting residues and hence provides direct evidence for novel functions of flavinylation in assistance of cofactor and substrate binding. Finally, the wild-type enzyme is more heat and guanidine HCl-resistant than the mutants. Therefore, the bicovalent flavin linkage not only tunes the redox potential and contributes to cofactor and substrate binding but also increases structural stability.
Flavoenzymes catalyze a wide range of biological redox reactions and are ubiquitously found in all organisms. Hundreds of flavoproteins have been reported, and the majority binds the flavin cofactor noncovalently. However, there is a subset in which the flavin moiety is covalently linked to the protein (1). Covalent flavinylation is found to proceed through an autocatalytic process. To date, five different types of single covalent linkages have been identified: four types via the 8α-methyl group of the isoalloxazine ring cross-linking to a histidine, cysteine, or tyrosine and one type via the C-6 atom to a cysteine. It has been shown that the roles of the covalent linkage include modulation of the flavin redox properties (2-9), stabilization of the protein structures (10), facilitation of electron transfer (11), promotion of tight association of different subunits (6, 12), avoidance of the 6-hydroxyl modification (13), and prevention of loss of the weakly bound oxidized FAD (7).
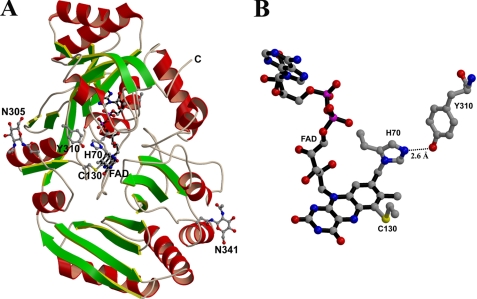
Glucooligosaccharide oxidase (GOOX)2 from Acremonium strictum has been screened for potential application in alternative carbohydrate assays and oligosaccharide acid production (14). This covalent flavoenzyme catalyzes the oxidation of glucose, lactose, cellobiose, maltose, cello-oligosaccharides, and malto-oligosaccharides (15). GOOX contains a 25-residue leader signal, and hence the residues are numbered according to the mature protein in A. strictum. Recently, we have determined the crystal structure of GOOX, demonstrating that this enzyme harbors a bicovalent flavinylation, namely a 6-S-cysteinyl, 8α-N1-histidyl FAD (see Fig. 1) (16). The FAD cofactor is covalently attached to two residues, His70 and Cys130. The same type of bicovalent FAD is also observed in the recently reported structure of aclacinomycin oxidase from Streptomyces galilaeus (17). In addition, biochemical studies have revealed the same type of bicovalent linkage in Eschscholzia californica berberine bridge enzyme (EcBBE), Fusarium graminearum chitooligo-saccharide oxidase (FgChitO), hexose oxidase from Chondrus crispus (CcHEOX), and Dbv29 involved in the glycopeptide A40926 maturation (8, 9, 18, 19). Based on the structure fold, both GOOX and S. galilaeus aclacinomycin oxidase are grouped into the vanillyl-alcohol oxidase (VAO) superfamily (20, 21). Although members of this family share a similar FAD-binding domain, they contain three distinct FAD-binding modes with four different flavin linkages: a double-covalent flavinylation as mentioned above, three types of single-covalent flavinylation including 8α-N1-histidylation, 8α-N3-histidylation, and 8α-O-tyrosylation, and noncovalent binding.
FIGURE 1.
A, the GOOX structure. Like other VAO members, the protein consists of two separate domains responsible for FAD and substrate binding. The helices are colored in red, whereas the strands are in green. The cofactor FAD, His70, Cys130, Asn305, Tyr310, and Asn341 are displayed as ball-and-stick representations. The loop from Val312 to Glu314 is omitted for clarity. B, a bicovalent flavinylation of 6-S-cysteinyl, 8α-N1-histidyl FAD. Tyr310 hydrogen bonds with His70 and hence may modulate the covalent flavinylation at this residue. However, mutation analysis displays that unlike His61 in PsVAO, Tyr310 has little effect on flavinylation and enzyme catalysis.
To elucidate the functional roles of the bicovalent FAD, mutant proteins of EcBBE, FgChitO, CcHEOX, and Dbv29 have been generated and characterized (8, 9, 18, 19). GOOX shares 45, 25, 25, and 23% sequence identity to FgChitO, CcHEOX, Dbv29, and EcBBE, respectively. So far, except for the FgChitO H94A mutant, previous studies on several VAO and non-VAO members clearly demonstrated that elimination of the FAD covalent bond markedly decreases the flavin redox potential (2-9). Unexpectedly, the FgChitO H94A mutant displays an increased redox potential of +164 mV from +131 mV (9). In addition, the Dbv29 H91A/C151A mutant contains a noncovalent FMN with 10% of the wild-type activity (19), whereas expression trials of the EcBBE and FgChitO double mutants failed (8, 9). Moreover, in Penicillium simplicissimum VAO (PsVAO), mutational analysis displayed an essential role of His61 for autoflavinylation, perhaps through hydrogen bonding between this residue and the attachment site His422 to stabilize the histidine orientation (22). Similarly, in GOOX Tyr310 is the only residue hydrogen bonding to His70 (Fig. 1B), and hence it may be involved in covalent flavinylation. To elucidate the functional roles of the double covalent FAD in GOOX, the single mutants H70A, C130A, and Y310A as well as the H70A/C130A double mutant were generated and characterized.
EXPERIMENTAL PROCEDURES
Protein Characterization—Site-directed mutagenesis, protein isolation, crystallization, and enzyme activity assays were carried out as previously described (15, 16). Because expression in Escherichia coli was not successful, all the recombinant proteins were expressed in Pichia pastoris KM71 in a bioreactor with automatic delivery of methanol. Periodic acid-Schiff staining was utilized for glycoprotein detection (23). UV-visible absorption spectra were recorded with a Beckman DU640B spectrophotometer. Circular dichroism spectra were measured with an Aviv 202 Spectrometer equipped with a thermoelectric cell holder. The spectra were measured over the range of 200-250 nm using three scans for each sample, and the results were expressed in terms of mean residue ellipticity (Φ) in degree cm2 dmol-1. All of the spectra were corrected by subtracting the buffer base line. Recombinant proteins were incubated with gradient concentrations of freshly prepared guanidine HCl for 1 h or exposed to high temperature, and the unfolding process was monitored by circular dichroism spectral change at 222 nm.
Yellow protein crystals of the C130A mutant were grown in 25% polyethylene glycol monomethyl ether 550, 10 mm ZnSO4, and 0.1 m MES (pH 6.5) similar to the wild-type enzyme (16). The diffraction data were collected using the synchrotron radiation at the beamline BL13B1 at NSRRC, Taiwan. The structure was solved by molecular replacement using the refined coordinates of the wild-type GOOX. The cofactor FAD and the mutated residue were omitted from the initial model. The structure was refined using the CNS program (24) with the refinement parameters listed in Tables 1 and 2. Figs. 1 and 7B were generated by MolScript (25), and Fig. 7A was generated by BobScript (25).
TABLE 1.
Parameters of wild-type and mutant GOOX
Steady state kinetic data are the means ± standard errors of at least three independent experiments. The values of the transient kinetic data were extracted from the hyperbolic fit to raw data of typically three to five independent measurements. In the case of kred, the standard deviation of the measurements at substrate saturating conditions is used as an error indicator. E°′ is the mid-point potential for the flavin cofactor determined at pH 7.0 and 25 °C shown as the means ± standard deviation of three independent experimental set-ups. Because of the unusual observations upon evaluation of the redox potentials for H70A, only an approximate value is presented.
| Wild type | H70A | C130A | |
|---|---|---|---|
| kcat (s−1) | 11.90 ± 0.2 | 0.22 ± 0.01 | 0.58 ± 0.025 |
| Km (mm) | 0.061 ± 0.005 | 0.22 ± 0.01 | 0.40 ± 0.02 |
| kcat/Km (s−1m m−1) | 194.3 ± 12.2 | 1.01 ± 0.07 | 1.50 ± 0.03 |
| Kd (mm) | 0.21 ± 0.03 | 0.32 ± 0.02 | 6.4 ± 0.6 |
| kred (s−1) | 170 ± 20 | 1.0 ± 0.1 | 23 ± 2 |
| E°′ (mV) | 126 ± 2 | ∼69 | 61 ± 1 |
| Tm (°C) | 56 | 48 | 52 |
| [Guanidine HCl]½ (m) | 3.2 | 1.9 | 2.4 |
TABLE 2.
Statistics for data collection and structural refinement of the C130A mutant
The values in parentheses are for the highest resolution shell.
| Data collection | |
| Space group | P21 21 21 |
| Cell dimension (Å) | 58.5, 97.6, 98.2 |
| Resolution range (Å) | 50−1.69 (1.75−1.69) |
| Total observations | 845,432 (47,758) |
| Unique reflections | 62,403 (5,427) |
| Completeness (%) | 98.6 (87.2) |
| I/σ<I> | 42.7 (2.2) |
| Rmerge (%) | 6.1 (73.4) |
| Refinement | |
| Resolution range (Å) | 50−1.69 (1.70−1.69) |
| Reflections (F > 0 σF) | 62,403 (636) |
| Rcryst (%) for 90% data | 19.7 (31.8) |
| Rfree (%) for 10% data | 22.4 (31.2) |
| Root mean square deviations | |
| Bond lengths (°) | 0.005 |
| Bond angles (°) | 1.31 |
| Mean B value (Å2) | |
| 3,721 protein atoms | 23.3 |
| 53 FAD atoms | 17.8 |
| 423 water molecules | 33.8 |
FIGURE 7.
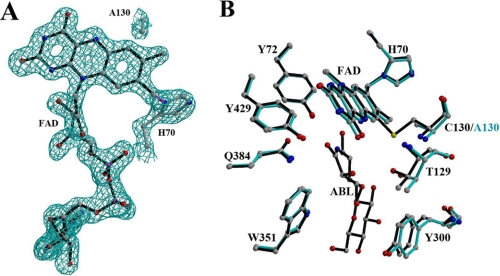
A, the Fo - Fc simulated annealing omit map for the FAD cofactor contoured at the 2 σ level. This density map clearly displays that the C130A mutant still retains the 8α-N1-histidyl bond but not the 6-S-cysteinyl bond. B, superposition of the active-site residues in the wild-type (black) and C130A (cyan) structures. The noncovalent contacts between the flavin C-6 and the Ala130 Cβ atoms result in Thr129 and Tyr300 shifting away from the substrate-binding site.
Determination of Reductive Rates—Kinetic traces of flavin reduction were determined using a stopped flow device from TgK Scientific (SF-61DX2). Oxygen-free conditions were provided in a glove box from Belle technology (0.8 ppm oxygen). Changes in flavin absorbance were followed with a PM-61s photomultiplier, and all concentrations stated in this context were final values after mixing in the flow cell. The samples were rendered anaerobic by flushing with oxygen free nitrogen followed by incubation in the glove box environment. Apparent rate constants for the reductive half-reaction were measured at six or more different concentrations of cellobiose. The term “reductive rates” in the remaining text refers to the apparent rate constant under substrate saturating conditions (obtained from a hyperbolic fit to all data). The actual substrate concentrations varied between the different mutant proteins because of significant changes in the Kd values. Protein concentration for a typical experiment was ∼15 μm. Fitting of the obtained transients at 445 nm was performed with SpecFit 32 (Spectrum Software Associates) using a function of two exponentials. The rate constants of the faster phase (contributing the major part to the observed decrease in flavin absorbance) showed a hyperbolic dependence on the substrate concentration. The contribution of the slower phase (usually more than 10 times slower than the fast rate) decreased with increasing substrate concentrations, but the obtained rate constants also showed a hyperbolic dependence. Up to now, no specific process could be assigned to this latter process.
Redox Potential Determination—Determination of the redox potential was carried out by the dye equilibration method using the xanthine/xanthine oxidase system as described by Massey (26). The reactions were carried out in 50 mm potassium phosphate buffer (pH 7.0) at 25 °C containing benzyl viologen (5 μm) as a mediator, 300 μm xanthine, and xanthine oxidase in catalytic amounts (∼5 nm). To maintain anaerobic conditions throughout the experiment, the reduction was carried out in a glove box (0.8 ppm oxygen; Belle Technology) after rendering all solutions anaerobic by repeated evacuation and flushing with nitrogen. Spectra during the course of reduction were recorded with a KinetaScanT diode array detector (MG-6560) attached to a stopped flow device (SF-61DX2, TgK Scientific) and equipped with an auto-shutter to reduce photochemical effects during the course of the experiment. A typical reduction lasted for 60-90 min to ensure equilibration between dye and protein. Dyes used for analysis and their corresponding redox potentials were toluylene blue (+115 mV) and thionin acetate (+64 mV). The redox potentials were calculated from plots of log([ox]/[red]) of protein versus log([ox]/[red]) of the dye according to Minnaert (27).
RESULTS
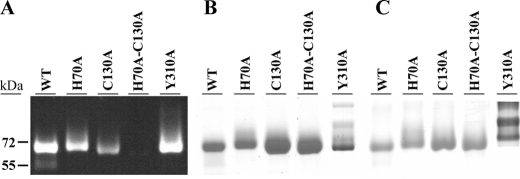
SDS-PAGE Analysis—Wild-type and the single mutant proteins were purified and showed a bright yellow coloration. These proteins were precipitated with 5% trichloroacetic acid, and no free flavin was detected in the supernatant. In-gel fluorescence analysis revealed that the fluorescent bands of the H70A and C130A mutants were similar to the wild-type protein (Fig. 2A). Thus both mutants still retained a covalent FAD, as did the wild-type enzyme. In contrast, the H70A/C130A mutant did not contain any detectable FAD, i.e. the protein solution was colorless lacking flavin absorbance, and no fluorescent band was observed by in-gel fluorescence analysis.
FIGURE 2.
SDS-PAGE analysis of purified wild-type (WT) and mutant enzymes. The gel was visualized by transillumination at 365 nm (A), with Coomassie Brilliant Blue 250 (B), or periodic acid-Schiff staining (C). This analysis demonstrated that FAD is covalently attached in wild type and single mutants but not in the H70A/C130A mutant.
SDS-PAGE analysis with Coomassie Blue R-250 staining and periodic acid-Schiff staining showed in the Y310A mutant a trace amount of proteins with a higher molecular weight containing a larger amount of carbohydrates (Fig. 2, B and C). Weaker fluorescent signals were observed for these protein bands with a longer exposure time. Sequencing by Edman degradation analysis showed that all of the isolated recombinant proteins had similar N termini.
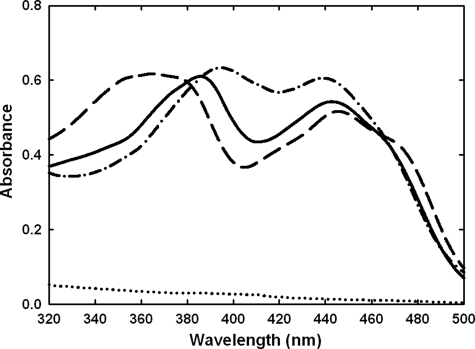
Flavin Absorption Spectra—Wild-type enzyme and the Y310A mutant share similar flavin absorbance spectra with two maxima, ∼385 and 440 nm. The H70A and C130A mutants display a similar absorbance at 440 nm but have distinct spectral characteristics at lower wavelengths (Fig. 3). In the H70A mutant, the shift from 385 to 395 nm may indicate a lack of the 8α-histidyl bond, because removal of this linkage is expected to result in a bathochromic shift of this absorbance maximum (28). Both PsVAO H422A and FgChitO H94A mutants have also been shown to result in a similar shift (3, 9). In contrast, in the C130A mutant the maximum wavelength was shifted to 360-370 nm. The flavin absorbance spectrum of the C130A mutant was similar to that of the EcBBE C166A and FgChitO C154A mutant proteins reported recently (8, 9) but was still apparently different from the unique absorbance spectrum of the 6-hydroxy-FAD (29). This indicates that the FAD C-6 atom is not hydroxylated in these three cysteine variants.
FIGURE 3.
Flavin absorption spectra. The spectra were recorded with a protein concentration of 2 mg/ml in 10 mm Tris-HCl (pH 7.6). Wild-type, solid line; H70A,—·—; C130A,———; and H70A/C130A, ······. These different spectra indicated a substantial change in the FAD microenvironment upon elimination of the 6-S-cysteinyl or 8α-N1-histidyl bond.
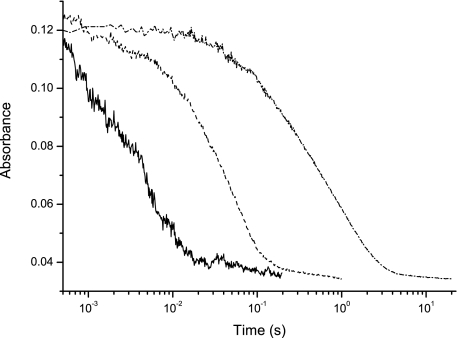
Flavin Reductive Rates and Redox Potentials—Wild-type enzyme and the H70A and C130A mutants were rapidly mixed with various concentrations of cellobiose, and the time course of flavin reduction was followed at 445 nm. Traces close to substrate saturating conditions are shown in Fig. 4. Fitting of the obtained transients resulted in observed rates that showed a hyperbolic dependence with respect to substrate concentration. A summary of the obtained kinetic parameters accounting for cofactor reduction by cellobiose is presented in Table 1. Most notable is the strong shift in the affinity of cellobiose to the C130A mutant protein with a 30-fold higher Kd value.
FIGURE 4.
Flavin reduction in the presence of cellobiose. The kinetic traces shown are representative for each protein under saturating conditions of substrate. Transients from left to right correspond to wild type (solid line), C130A (———), and H70A (—·—).
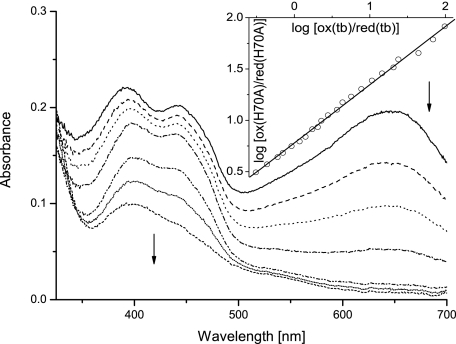
For the determination of the redox potentials, the xanthine/xanthine oxidase system with appropriate reference dyes was employed. For both wild-type and the C130A mutant protein a simultaneous two electron reduction of cofactor and dye was observed. Evaluation of the data according to Minnaert (27) resulted in slopes of 0.90 and 0.98, respectively, indicating simultaneous two electron reduction of the GOOX bound FAD and the reference dye. The calculated redox potentials are 126 ± 2 and 61 ± 1 mV for wild-type and the C130A mutant protein using toluylene blue and thionin acetate, respectively, as reporter dyes. On the other hand, analysis of the redox titrations for the H70A mutant protein revealed a deviation of the slopes from unity varying between 0.43 and 0.67 using different reporter dyes for the experiment. These results indicate that only one electron is transferred to FAD, whereas two are transferred to the reporter dye. Because we do not observe a flavin radical during redox titrations (Fig. 5), we assume that the two-electron transfer to FAD in the H70A mutant is kinetically inhibited and not a result of thermodynamic stabilization of the flavin radical over the hydroquinone form. From the obtained Nernst plots (Fig. 5, inset), a midpoint potential of ∼70 mV was estimated for the H70A mutant using the Nernst equation for a process resulting in a slope of 0.5 and toluylene blue as reference. To confirm this result we also used thionin acetate in a control experiment and obtained a value of ∼67 mV with a slope of 0.45. Another possible explanation for the observed discrepancies would be the lack of equilibrium between the reduction of the cofactor and the reporter dye during the measurement. However, this was ruled out by performing the experiment at different xanthine oxidase concentrations, resulting in experimental set-ups that required between 1 and 6 h for complete reduction of both species. Evaluation of all reductions showed no significant dependence of the slope in the Nernst plots upon the time course of the titration, suggesting that equilibrium was reached in all cases. Taken together these controls indicate that the redox potential calculation is reproducible but should be interpreted with some care because a kinetic barrier for parallel two electron reduction of cofactor and dye appears to exist. The value of 69 mV given in Table 1 for the redox potential of the H70A mutant protein represents an average obtained from titrations with the two reporter dyes utilized in our study.
FIGURE 5.
Redox potential determination of H70A GOOX. Selected spectra of the course of reduction are shown with the inset representing the Nernst plot for calculation of the redox potential. In the example given, the mutant protein with a concentration of 16 μm was reduced with the xanthine/xanthine oxidase system in the presence of toluylene blue. Data points for analysis were extracted at 443 and 645 nm for FAD and the dye, respectively, where no significant contribution of the other chromophore can be observed. The first spectrum is shown with a solid line and followed by consecutive spectra as indicated by the arrows with the short-dashed line representing the fully reduced sample.
Enzyme Kinetics—Kinetic parameters were determined using cellobiose as the substrate (Table 1). The amount of protein used to assay wild-type enzyme and the Y310A mutant protein was 5-10 μg/ml, whereas 150-300 μg/ml was used for the H70A and C130A single mutant proteins. The H70A and C130A mutant proteins showed a substantial reduction in the kcat values decreasing by a factor of 54 and 20, respectively. At the same time, the Km increased by a factor of 4-and 6.5, respectively. In contrast, no activity was detected when the purified H70A/C130A double mutant protein was assayed, apparently caused by the total lack of the essential FAD cofactor (see above). The addition of exogenous FAD to the reaction solution was not able to restore any detectable activity.
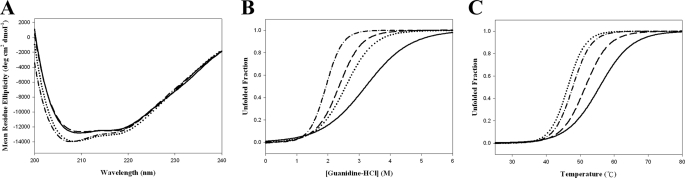
Structural Stability—The similar far-UV circular dichroism spectra of the wild-type and mutant proteins suggested that elimination of either or both the 6-S-cysteinyl and 8α-N1-histidyl bonds, the latter resulting in the loss of FAD binding, did not significantly affect the global tertiary structure (Fig. 6A). The thermal and guanidine HCl-induced unfolding experiments indicated a typical monophasic denaturation curve, a two-state unfolding process (Fig. 6, B and C). The corresponding melting temperatures and [guanidine HCl]½ values for the H70A, C130A, H70A/C130A, and wild-type proteins were estimated to be 48 °C, 1.9 m; 52 °C, 2.4 m; 46 °C, 2.6 m; and 56 °C/3.2 m, respectively.
FIGURE 6.
A, circular dichroism spectra. The spectra were recorded with a protein concentration of 0.2 mg/ml in 20 mm sodium phosphate (pH 7.6). Wild type, solid line; H70A,—·—; C130A,———; and H70A/C130A, ······. The loss of either or both the 6-S-cysteinylation and 8α-N1-histidylation did not significantly affect the overall fold. B and C, guanidine HCl-induced (B) and thermal-induced (C) unfolding process monitored by ellipticity changes at 222 nm. The greater thermal and chemical resistance of the wild-type enzyme than the mutants revealed an enhancement of the structural stability by the bicovalent flavinylation.
The C130A Mutant Structure—The C130A mutant protein crystals only appeared at a protein concentration of more than 50 mg/ml. The first simulated annealing omit maps clearly demonstrate that the mutant protein contains a single covalent 8α-N1-histidyl FAD, whereas the covalent linkage to the 6-position of the isoalloxazine ring is missing because of substitution of Cys130 to alanine (Fig. 7A). The C130A mutant protein and wild-type structure are virtually identical with a root mean square deviation of 0.27 Å for 474 Cα atoms. However, residues 128-130 and 298-300 in the structural vicinity to residue 130 show some differences (Fig. 7B). The distance of the flavin C-6 atom to the Ala130 Cβ is 3.3 Å, whereas it is 2.7 Å to the Cys130 Cβ. The isoalloxazine ring has also undergone subtle changes that might contribute to the observed alteration in the flavin redox properties as reported for PsVAO (3).
DISCUSSION
Independent Formation of the Double Covalent Attachments—The EcBBE C166A, GOOX H70A and C130A, FgChitO H94A, C154A, CcHEOX H79K, and Dbv29 H91A and C151A single mutant proteins all retained a covalent FAD (8, 9, 18, 19). This demonstrates that both modifications can be formed independently. Wild-type enzyme and the Y310A mutant protein share similar enzyme activity and flavin absorbance spectra, suggesting that the Tyr310 mutant protein still retains the double flavinylation. Thus unlike His61 in PsVAO, the hydrogen bond of Tyr310 to His70 is not involved in covalent flavinylation. Therefore, formation of the 6-S-cysteinyl and 8α-N1-histidyl bonds may be through independent processes and seem not to require assistance by specific residues.
Additive Enhancement of the Flavin Redox Potential by Bicovalent Linkage—The bicovalent FAD is also the first example of 6-S-cysteinyl FAD. The 6-S-cysteinyl FMN in trimethylamine dehydrogenase has been shown to prevent enzyme inactivation by hydroxylation at the flavin C-6 atom (13). In the GOOX C130A, EcBBE C166A, and FgChitO C154A mutant proteins, a lack of the unique features in the 6-hydroxyl flavin absorbance spectrum indicates that the 6-S-cysteinyl bond is not for protection against 6-hydroxylation. Instead, in these three enzymes, elimination of the 6-S-cysteinylation leads to a similar significant drop of the redox potential, from +126 to +61 mV, +132 to +53 mV (8), and +131 to +70 mV (9), respectively. This is consistent with several flavin derivative studies, demonstrating that both 6-S-cysteinyl and 8α-histidyl substitutions are able to cause an increase in redox potential by ∼50 mV (30, 31).
Previous studies on several single covalent flavoenzymes and flavin derivatives have demonstrated that elimination of the 8α-histidyl modification markedly reduces the redox potential (2-9, 31). In this study we have confirmed a lower redox potential for the H70A mutant protein, although the determined potential of ∼+69 mV, must be considered with care because the Nernst slopes deviate from unity. However, we are confident that the observed deviation is not due to the lack of equilibrium during the reductive titration nor due to the loss of electrons because both processes would show some dependence upon the rate of reduction. Because this is not the case, and we did not detect any flavin radicals during redox titrations, we assume that the observed Nernst slope of ∼0.5 is due to kinetic inhibition of the electron transfer to FAD in the H70A mutant. It should be noted here that also for FgChitO, the redox potential determination of this protein variant (H94A) did not proceed as smoothly as for the other proteins studied (9). As mentioned by these authors (9), the mutant protein precipitated during reduction and could not be reoxidized thereafter. The extended time necessary for reduction might also reflect the kinetic barrier for electron transfer to FAD as observed in our case. In combination these studies suggest that the missing covalent linkage to position 8-α of the cofactor has a significant effect upon the properties of FAD; however, the precise function still has to be demonstrated. Unlike for the FgChitO H94A mutant protein with an increased potential but similar to other previously reported flavoenzymes, we show that the GOOX H70A mutant has a decreased redox potential, from +126 mV to ∼+69 mV. Taken together, our studies suggest that in GOOX both flavin linkages contribute to an additive increase of the redox potential. In fact, GOOX, EcBBE, and FgChitO, possess a very similar redox potential, the highest potential determined for any flavin-dependent enzyme.
Distinct Impacts of Covalent FAD in Enzyme Catalysis—With no exceptions so far, elimination of the covalent flavin attachment to the protein results in a lower level of the rate of flavin reduction (3, 8, 9) and the kcat value. The EcBBE C166A, FgChitO C154A, and GOOX H70A mutant proteins exhibit a good correlation: The decrease of the redox potential (-79, -61, and -56 mV, respectively) correlates directly with the rate of flavin reduction (370-, 375-750-, and 170-fold lower rates). However, the flavin reduction rate of the GOOX C130A mutant protein is only slowed down ∼7-fold with a lower redox potential of 65 mV, suggesting that other factors in the active site compensate for the lower oxidative power of the FAD. Similarly, even though the PsVAO H422A mutant protein exhibits a large drop of the redox potential from +55 to -65 mV, the flavin reduction rate slowed down only 11-fold (3). In this context, it is interesting that the FgChitO H94A mutant displays an increased potential of 33 mV but a lower reduction rate by 3 orders of magnitude, again indicating that other parameters bear on the rate of reduction of the cofactor.
The EcBBE C166A, GOOX H70A and C130A, and FgChitO H94A and C154A mutant proteins showed a reduction in the kcat values, of ∼17-, 54-, 20-, 8-, and 3-fold, respectively (8, 9). 8α-N1-Histidylation seems to contribute more to kcat than 6-S-cysteinylation, even though the latter is near the target N5 atom. On the other hand, the Km values for the GOOX H70A and C130A, and FgChitO H94A and C154A mutant proteins increase by factors of 4, 6.5, 34, and 75, respectively. 6-S-Cysteinylation seems to affect the substrate binding more than 8α-N1-histidylation. The GOOX C130A mutant protein structure provides direct evidence for a novel function of 6-S-cysteinylation in assistance of substrate binding (Fig. 7B). Replacement of Cys130 with alanine eliminates the 6-S-cysteinyl bond and causes some conformational differences in surrounding residues including residues 128-130 and 298-300. Among them, Thr129 and Tyr300 directly interact with the substrate (16), and hence their shift in position in the C130A mutant protein appears to weaken substrate binding, resulting in increased Km and Kd values (Table 1). Similarly, deletion of the 8α-N1-histidylation at His70 may induce spatial changes of another substrate-interacting residue, Tyr72. Therefore, we suspect that the marked increased Km values in the FgChitO H94A and C154A mutants are due to Tyr96, Thr153, and Phe319 shifting away from the substrate-binding site upon deletion of the covalent flavinylation.
Formation of a FAD Binding Cavity—So far, among the single covalent flavoenzymes, mutation of the FAD-linking residue results in noncovalently bound FAD and causes only minor conformational changes (3, 5-6, 32, 33). Virtually identical structures of the wild-type PsVAO and the mutant proteins with no or noncovalently bound FAD demonstrate that the apoprotein is fully folded with subsequent formation of a pre-organized FAD-binding site (3, 22, 34).
In contrast to EcBBE and FgChitO, the GOOX double mutant protein H70A/C130A was expressed and isolated successfully. Even though the H70A/C130A mutant protein did not contain any detectable bound FAD, it folds into a similar structure (Fig. 6A). However, unlike the single covalent flavoenzymes and Dbv29, this apoprotein does not possess an appropriate cavity for binding of the oxidized FAD. Residues 128-130 have direct contacts with the isoalloxazine ring, and the change in position observed in the C130A mutant protein may weaken the interaction network between GOOX and the cofactor. For example, in wild-type protein, the Thr129 N hydrogen bonds with the flavin O4 with a distance of 3.0 Å, whereas this distance in the C130A mutant is 3.6 Å. Similarly, residues 65-72 form an extensive interaction network with FAD, particularly with the pyrophosphate moiety (16), and hence disruption of the 8α-N1-histidylation at His70 may affect the local structures and compromise the FAD-binding site. Therefore the GOOX H70A/C130A double mutant protein demonstrates that the covalent attachment is essential for the binding of the oxidized flavin cofactor and hence suggests that the covalent linkage accomplishes the formation of a well organized FAD binding cavity. This is consistent with the hypothesis by Fraaije and co-workers (9). The identified bicovalent flavoenzymes catalyze oxidation of relatively bulky compounds and hence possess an open substrate-binding groove; therefore, the bicovalent linkage might have evolved to anchor the FAD cofactor in a proper orientation for substrate binding and oxidation. In addition, the higher melting temperature and greater guanidine HCl resistance of wild-type GOOX compared with the mutants display that the bicovalent FAD attachment markedly improves the structure stability of the protein (Fig. 6, B and C).
Conclusions—In summary, the results presented here reveal new insights into the functional roles of the 6-S-cysteinyl, 8α-N1-histidyl FAD in GOOX. The bicovalent linkages are needed for efficient catalysis of sugar oxidation through an additive enhancement of the flavin redox potential and the cofactor and substrate binding, facilitation of the reduction rate, and attainment of the active conformation. In addition, the distinct correlations between the redox potentials and the kinetic parameters in the wild-type and mutant EcBBE, FgChitO and GOOX suggest that the bicovalent flavinylation may play versatile roles in various flavoenzymes.
The atomic coordinates and structure factors (code 3E0T) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
This work was supported by National Science Council of Taiwan Grant NSC94-2311-B010-016-MY3 (to S.-H. L.) and by the Austrian Fonds zur För-derung der Wissenschaftlichen Forschung (FWF) through Doktoratskolleg “Molecular Enzymology” Grant W901-B05 (to P. M.). This work was also supported in part by the National Research Program for Genomic Medicine and National Science Council of Taiwan. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: GOOX, glucooligosaccharide oxidase; EcBBE, E. californica berberine bridge enzyme; FgChitO, F. graminearum chitooligosaccharide oxidase; CcHEOX, hexose oxidase from C. crispus; PsVAO, P. simplicissimum vanillyl-alcohol oxidase; MES, 4-morpholineethanesulfonic acid.
References
- 1.Mewies, M., McIntire, W. S., and Scrutton, N. S. (1998) Protein Sci. 7 7-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blaut, M., Whittaker, K., Valdovinos, A., Ackrell, B. A., Gunsalus, R. P., and Cecchini, G. (1989) J. Biol. Chem. 264 13599-13604 [PubMed] [Google Scholar]
- 3.Fraaije, M. W., van den Heuvel, R. H., van Berkel, W. J., and Mattevi, A. (1999) J. Biol. Chem. 274 35514-35520 [DOI] [PubMed] [Google Scholar]
- 4.Nandigama, R. K., and Edmondson, D. E. (2000) J. Biol. Chem. 275 20527-20532 [DOI] [PubMed] [Google Scholar]
- 5.Motteran, L., Pilone, M. S., Molla, G., Ghisla, S., and Pollegioni, L. (2001) J. Biol. Chem. 276 18024-18030 [DOI] [PubMed] [Google Scholar]
- 6.Efimov, I., Cronin, C. N., and McIntire, W. S. (2001) Biochemistry 40 2155-2166 [DOI] [PubMed] [Google Scholar]
- 7.Hassan-Abdalah A., Zhao, G., and Jorns, M. S. (2006) Biochemistry 45 9454-9462 [DOI] [PubMed] [Google Scholar]
- 8.Winkler, A., Kutchan, T. M., and Macheroux, P. (2007) J. Biol. Chem. 282 24437-24443 [DOI] [PubMed] [Google Scholar]
- 9.Heuts, D. P., Winter, R. T., Damsma, G. E., Janssen, D. B., and Fraaije, M. W. (2008) Biochem. J. 413 175-183 [DOI] [PubMed] [Google Scholar]
- 10.Caldinelli, L., Iametti, S., Barbiroli, A., Fessas, D., Bonomi, F., Piubelli, L., and Pollegioni, L. (2008) Protein Sci. 17 409-419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim, J., Fuller, J. H., Kuusk, V., Cunane, L., Chen, Z. W., Mathews, F. S., and McIntire, W. S. (1995) J. Biol. Chem. 270 31202-31209 [DOI] [PubMed] [Google Scholar]
- 12.Eschenbrenner, M., Chlumsky, L. J., Khanna, P., Strasser, F., and Jorns, M. S. (2001) Biochemistry 40 5352-5367 [DOI] [PubMed] [Google Scholar]
- 13.Huang, L., Scrutton, N. S., and Hille, R. (1996) J. Biol. Chem. 271 13401-13406 [DOI] [PubMed] [Google Scholar]
- 14.Lin, S. F., Yang, T. Y., Inukai, T., Yamasaki, M., and Tsai, Y. C. (1991) Biochim. Biophys. Acta 1118 41-47 [DOI] [PubMed] [Google Scholar]
- 15.Lee, M. H., Lai, W. L., Lin, S. F., Hsu, C. S., Liaw, S. H., and Tsai, Y. C. (2005) Appl. Environ. Microbiol. 71 8881-8887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang, C. H., Lai, W. L., Lee, M. H., Chen, C. J., Vasella, A., Tsai, Y. C., and Liaw, S. H. (2005) J. Biol. Chem. 280 38831-38838 [DOI] [PubMed] [Google Scholar]
- 17.Alexeev, I., Sultana, A., Mantsala, P., Niemi, J., and Schneider, G. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 6170-6175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rand, T., Qvist, K. B., Walter, C. P., and Poulsen, C. H. (2006) FEBS J. 273 2693-2703 [DOI] [PubMed] [Google Scholar]
- 19.Li, Y. S., Ho, J. Y., Huang, C. C., Lyu, S. Y., Lee, C. Y., Huang, Y. T., Wu, C. J., Chan, H. C., Huang, C. J., Hsu, N. S., Tsai, M. D., and Li, T. L. (2007) J. Am. Chem. Soc. 129 13384-13385 [DOI] [PubMed] [Google Scholar]
- 20.Fraaije, M. W., van Berkel, W. J., Benen, J. A. E., Visser, J., and Mattevi, A. (1998) Trends Biochem. Sci. 23 206-207 [DOI] [PubMed] [Google Scholar]
- 21.Leferink, N. G., Heuts, D. P., Fraaije, M. W., and van Berkel W. J. (2008) Arch. Biochem. Biophys. 474 292-301 [DOI] [PubMed] [Google Scholar]
- 22.Fraaije, M. W., van Den Heuvel, R. H., van Berkel, W. J., and Mattevi, A. (2000) J. Biol. Chem. 275 38654-38658 [DOI] [PubMed] [Google Scholar]
- 23.Zacharius, R. M., Zell, T. E., Morrison, J. H., and Woodlock, J. J. (1969) Anal. Biochem. 30 148-152 [DOI] [PubMed] [Google Scholar]
- 24.Brünger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., Read, R. J., Rice, L. M., Simonson, T., and Warren, G. L. (1998) Acta Crystallogr. Sect. D Biol. Crystallogr. 54 905-921 [DOI] [PubMed] [Google Scholar]
- 25.Esnouf, R. M. (1999) Acta Crystallogr. Sect. D Biol. Crystallogr. 55 938-940 [DOI] [PubMed] [Google Scholar]
- 26.Massey, V. (1991) in Flavins and Flavoproteins (Curti, B., Zanetti, G., and Ronchi, S., eds) pp. 59-66, Walter de Gruyter, Como, Italy
- 27.Minnaert, K. (1965) Biochim. Biophys. Acta 110 42-56 [DOI] [PubMed] [Google Scholar]
- 28.Kenney, W. C., Edmondson, D. E., and Seng, R. L. (1976) J. Biol. Chem. 251 5386-5390 [PubMed] [Google Scholar]
- 29.Igarashi, K., Verhagen, M. F., Samejima, M., Schulein, M., Eriksson, K. E., and Nishino, T. (1999) J. Biol. Chem. 274 3338-3344 [DOI] [PubMed] [Google Scholar]
- 30.Ghisla, S., Kenney, W. C., Knappe, W. R., McIntire, W., and Singer, T. P. (1980) Biochemistry 19 2537-2544 [DOI] [PubMed] [Google Scholar]
- 31.Williamson, G., and Edmondson, D. E. (1985) Biochemistry 24 7790-7797 [DOI] [PubMed] [Google Scholar]
- 32.Mauch, L., Bichler, V., and Brandsch, R. (1989) FEBS Lett. 257 86-88 [DOI] [PubMed] [Google Scholar]
- 33.Lim, L., Molla, G., Guinn, N., Ghisla, S., Pollegioni, L., and Vrielink, A. (2006) Biochem. J. 400 13-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van den Heuvel, R. H., Fraaije, M. W., Mattevi, A., and van Berkel, W. J. (2000) J. Biol. Chem. 275 14799-14808 [DOI] [PubMed] [Google Scholar]