Abstract
Myb family transcription factors are important in regulating cell proliferation, differentiation, and cell cycle progression. Giardia lamblia differentiates into infectious cysts to survive outside of the host. During encystation, genes encoding cyst wall proteins (CWPs) are coordinately induced. We have identified an encystation-induced Myb2 protein, which binds to the promoter regions of the cwp genes and myb2 itself in vitro. To elucidate the role of Myb2 in G. lamblia, we tested the hypothesis that Myb2 can activate encystation-induced genes. We found that overexpression of Myb2 resulted in an increase of expression of CWP1 at both protein and mRNA levels. Interestingly, the Myb2-overexpressing trophozoites had increased capability to differentiate into cysts. In cotransfection assays, Myb2 was able to transactivate the cwp promoters and its own promoter in vivo, suggesting that its gene can be positively autoregulated. Moreover, deletion of the N- or C-terminal domain resulted in a decrease of transactivation and autoregulation function of Myb2. We also found that the promoter of a newly identified encystation-induced gene, the giardial myeloid leukemia factor-like gene, has the Myb2 binding sites and that its mRNA levels were increased by Myb2 overexpression. Chromatin immunoprecipitation assays confirmed that Myb2 was bound to the promoters with its binding sites. Transfection of the myb2 antisense construct reduced the levels of the cwp1 transcripts and cyst formation. Our results suggest that Myb2 is a potent transactivator of the cwp genes and other endogenous genes and plays an important role in G. lamblia differentiation into cysts.
Numerous parasitic protozoa have developmental stages in their life cycle that are essential for disease transmission (1). We used Giardia lamblia, which has a relatively simple two-stage life cycle that can be completed in vitro as a model for studying the evolution of cellular differentiation (2, 3). G. lamblia is an important human pathogen that causes outbreaks of water-borne diarrhea (4, 5). During encystation, G. lamblia synthesizes a resistant extracellular wall, which is composed of proteins and polysaccharide (2, 3), protecting the parasite from hypotonic lysis by fresh water and from gastric acid during infection of the new host.
Despite the importance of cyst wall biogenesis during giardial encystation, the molecular mechanisms governing transcriptional regulation remain poorly understood. Expression of genes encoding three cyst wall structural proteins (Cyst Wall Protein 1 (CWP1), CWP2, and CWP3)3 (6-8) and an enzyme in the cyst wall polysaccharide biosynthetic pathway (glucosamine-6-phosphate isomerase-B, G6PI-B) increases with similar kinetics (9-10), suggesting the importance of regulation at transcriptional level.
In addition to its medical importance, G. lamblia is of biological interest in understanding the mechanisms of eukaryotic evolution (11-13). It has fewer cellular components for DNA synthesis, transcription, and RNA processing (13). The lack of clear giardial homologs to these proteins suggests their divergence or their functional redundancy with other proteins in some pathways. G. lamblia has a highly divergent TATA-binding protein and lacks eight of the twelve general transcription initiation factors (14, 15). It also has many unusual features with regard to transcription. Unusually short 5′-flanking regions (<65 bp) with no consensus TATA boxes or other cis-acting elements identified in late-branching eukaryotic promoters are sufficient for the expression of some genes (6, 7, 10, 16-19). Instead, AT-rich sequences have been found around the transcription start sites of many genes, functionally similar to the initiator element in late-branching eukaryotes (6, 7, 10, 16-19).
Few transcription factors that have been characterized to date are involved in cwp gene regulation (20-22). The GARP-like protein 1 (named from the maize GOLDEN2, Arabidopsis response-regulator proteins and the Chlamydomonas Psr1 protein) and the AT-rich interaction domain-family transcription factors may be involved in transcriptional regulation of many different genes including the encystation-induced cwp1 gene (21, 22). Interestingly, we have identified an encystation-induced Myb2 protein, which binds to the promoters of four key encystation-induced genes, cwp1, cwp2, cwp3, g6pi-b, and myb2 itself, suggesting that Myb2 may be involved in co-ordinating their differential expression (20).
In late-branching eukaryotes, Myb proteins are DNA-binding transcription factors that regulate specific gene expression in differentiation of different cell types (23). Myb family transcription factors are important in regulating physiological processes in organisms as diverse as fungi, plants, and mammals (24-29). Myb proteins function as transcriptional activators or repressors via association with some cofactors on the promoter context of specific target genes to regulate development, cell differentiation, cell cycle, apoptosis, and cancer (26, 30-34). For example, mammalian c-Myb is required for development of mature B and T cells (31). Several c-Myb target genes have been identified, such as promyelocytic-specific mim-1, T cell receptor gamma and delta chains, and c-myb itself (35-42).
In the previous study, we have identified an encystation-induced Myb2 protein, which binds to the promoters of the cwp genes in vitro (20). In this study, we found that the constitutively overexpressed Myb2 can increase the expression of endogenous CWP1 at both protein and mRNA levels. Interestingly, the Myb2 overexpressing trophozoites had increased capability to differentiate into cysts. We also found that the cwp1, cwp3, myb2, and g6pi-b promoters can be activated to differing degrees by Myb2, suggesting that Myb2 can transactivate these genes and that the myb2 gene can be positively autoregulated. Deletion of the N- or C-terminal domain resulted in a decrease of transactivation and autoregulation function of Myb2. Transfection of the myb2 antisense construct reduced the levels of the cwp1 transcripts and cyst formation. In the previous studies, we have found that stable transfection systems can increase the levels of cwp1-3 transcripts, CWP2 protein and cyst formation during vegetative growth, indicating that stable transfection systems may trigger an encystation-like physiologic response in G. lamblia trophozoites (43). Interestingly, we have found that the expression of the gene encoding orf 16424 was also up-regulated by ∼1.4-4.8-fold in both encysting cells and stably transfected cells (43). The function of the orf 16424 is unknown. Blast searches suggest that it is similar to Drosophila and mouse myeloid leukemia factor, which can reprogram erythroleukemic cells (data not shown) (43). Therefore, we named it giardial myeloid leukemia factor-like (MLFL) protein. In the present study, we found that the mlfl gene has the Myb2 binding sites and that the levels of mlfl mRNA, like those of cwp mRNA, also increased in the Myb2 overexpressing cell line. We used chromatin immunoprecipitation (ChIP) assays to confirm the binding of Myb2 to these gene promoters. Our results suggest that Myb2 could function as a transactivator of the cwp and other endogenous genes to regulate G. lamblia differentiation.
EXPERIMENTAL PROCEDURES
G. lamblia Culture—Trophozoites of G. lamblia WB (ATCC 30957), clone C6, were cultured in modified TYI-S33 medium (44) and encysted as previously described (8). Cyst count was performed on vegetative cultures as previously described (43).
RNA Extraction and Northern Blot Analysis—Total RNA was extracted from G. lamblia clones C6 at the indicated differentiation stages in the legends of Figs. 1, 5B, and 8C using TRIzol reagent (Invitrogen). For Northern blot analysis, 10 μg of total RNA was fractionated and transferred to charged nylon membranes (Biodyne B membrane, Pall). Full-length coding region probes of myb2 (GenBank™ accession number AY082882), cwp1 (GenBank™ accession number U09330), cwp2 (GenBank™ accession number U28965), ran (GenBank™ accession number U02589), mlfl (orf 16424, GenBank™ accession number for genomic DNA: XM_764168; orf 16424 in G. lamblia genome data base)(13), phosphoglycerate kinase (pgk, GenBank™ accession number for genomic DNA: XM_762975), and bip (GenBank™ accession number for genomic DNA: XM_766560, for protein: XP_771653) genes were prepared by PCR amplification of genomic DNA using primers: myb2F (ATGTTACCGGTACCTTCT) and myb2R (TCAGGGTAGCTTCTCACG), cwp1F (ATGATGCTCGCTCTCCTT) and cwp1R (TCAAGGCGGGGTGAGGCA), cwp2F (ATGATCGCAGCCCTTGTT) and cwp2R (TCACCTTCTGCGGACAAT), ranF (ATGTCTGACCCAATCAGC) and ranR (TCAATCATCGTCGGGAAG), mlflF (CACCATGAGTAGAACGCCAAAC) and mlflR (GTAGCGACGATTACCGGA), pgkF (ATGTCCTTAGCGAAGCTCTCC) and pgkR (CTTCTTGTCAGACAGTCTGAT), and bipF (ATGACGTCTAGTCACGTTAA) and bipR (GAGTTCATCTTTTTCTGCAT), respectively. Radiolabeled probes were prepared using the Rediprime II kit (Amersham Biosciences). The membranes were hybridized and washed as previously described (10). Equal loading was confirmed by reprobing the Northern blots with radiolabeled ribosomal DNA. The ribosomal DNA fragment for large subunit ribosomal RNA (GenBank™ accession number X05397) was amplified by PCR using primers RIBOF (GGCCTGCCCCTCGCCCGC) and RIBOR (CCCCTCAGTCCTCCGGGG) and a genomic DNA template. Radiolabeled ribosomal DNA probes were prepared as described above. Hybridization signals were imaged and quantified using a Storm system (Molecular Dynamics). Two independently generated stably transfected lines were made from each construct and each of these lines was assayed three separate times. The results are expressed as relative expression level over control. Student's t-tests were used to determine statistical significance of differences between samples.
FIGURE 1.
Induction of cwp1 and cwp2 gene expression in the Myb2-overexpressing cell line. A, diagrams of the 5′-Δ5N-Pac and pPTMyb2 plasmid. The pac gene (open box) is under the control of the 5′- and 3′-flanking regions of the gdh gene (striated box). In construct pPTMyb2, the myb2 gene is under the control of the constitutively expressed α2-tubulin promoter (striped box) and 3′-flanking region of the ran gene (dotted box). The filled box indicates the coding sequence of the AU1 epitope tag. B, Northern blot analysis of gene expression in the Myb2-overexpressing cell line. Total RNA blots made from 5′-Δ5N-Pac or pPTMyb2 cell cultures during vegetative growth were hybridized with specific gene probes as indicated (upper panels). Ribosomal RNA loading controls are in the bottom panel. Representative results are shown. The numbers show the relative activity, which reflects expression relative to that in controls.
FIGURE 5.
Mapping of the Myb2 transactivation domain. A, analysis of Myb2 deletion mutants. The specific transfectants were cultured in encystation medium for 24 h (Enc) and then subjected to Western blot analysis, using anti-AU1 antibody for detection. Coomassie Blue-stained total protein loading control is shown below. B, analysis of cwp1 gene expression. Total RNA was harvested from specific transfectants cultured in encystation medium for 24 h. Northern blots were hybridized with the cwp1 and myb2 gene probes (upper panels). Ribosomal RNA loading controls are in the bottom panels. Representative results are shown. The numbers show the relative activity, which reflects expression relative to that in controls. Full-length myb2 transcripts (shown by arrows) and deletion mutant transcripts (shown by arrowheads) were detected in the pNMybΔC and pNMybΔN transfectants. Only the relative activity of the full-length myb2 transcripts (shown by arrows) was shown for the pNMybΔC and pNMybΔN transfectants.
FIGURE 8.
Transfection of the myb2 antisense construct reduced the levels of the cwp1 transcripts and cyst formation. A, diagrams of the myb2 antisense construct. A 1590-bp PCR fragment of the myb2 gene was cloned in the opposite (antisense) orientation to its own promoter into the pNM5 (Fig. 6). The resulting construct, pNMyb2as, contains full-length coding sequences complementary to the mRNA sequences of the myb2 gene. B, transfection of the myb2 antisense construct reduced the levels of cyst formation. Wild-type non-transfected WB C6 cells, pNM5 and pNMyb2as stable transfectants were cultured in encystation medium for 48 h and then subjected to cyst count. The sum of total cysts is expressed as relative expression level over control. Values are shown as means ± S.E. C, transfection of the myb2 antisense construct reduced the cwp1 transcript levels. Total RNA blots made from vegetative non-transfected WB C6 cells, pNM5 or pNMyb2as stable transfectants were hybridized with specific probes as indicated (upper panels). Ribosomal RNA loading controls are in the bottom panel. Representative results are shown. The numbers show the relative activity, which reflects expression relative to that in controls.
Plasmid Construction—All constructs were verified by DNA sequencing with BigDye Terminator 3.1 DNA Sequencing kit and an ABI 3100 DNA Analyzer (Applied Biosystems). Plasmid 5′-Δ5N-Pac was a gift from Dr. Steven Singer and Dr. Theodore Nash (45). Plasmid pRANneo, pPW1, pNM5, 4s/-42/+3, -42/+3, pNMybΔ1, and pNMybΔC have been previously described (20, 21, 46). The myb2 gene was amplified with oligonucleotides Myb2BF (GGCGTCATGATACCGGTACCTTCTCAGCCA) and Myb2AUER (GGCGGAATTCTCAGATGTATCGATACGTATCGGGTAGCTTCTCACGGGGAAG), digested with BspHI/EcoRI, and ligated in place of the NcoI/EcoRI-excised luciferase gene in pNT5 (20). The resulting plasmid, pNTMyb2, contained the myb2 gene controlled by the α2-tubulin promoter with an AU1 tag fused at its C terminus. A NheI/PstI fragment containing the luciferase gene, 32-bp ran promoter and two copies of a 19-bp tet operator sequence from pPop2N (47) was replaced by the NheI/PstI-excised myb2 gene and α2-tubulin promoter from pNTMyb2, resulting in pPTMyb2. A NheI/ClaI fragment containing the luciferase gene, 32-bp ran promoter, and two copies of a 19-bp tet operator sequence from pPop2N (47) was replaced by the NheI/ClaI-excised luciferase gene and cwp3 promoter from pNC35 (8), resulting in pPC35. The 5′-flanking region of the genomic myb2 gene was amplified with oligonucleotides Myb25NF (GGCGGCTAGCTGATTCAAAAGAATTAGACGGACTG) and Myb25m1NR (GGCGCCATGGTAGCAGTACAGAGTAATTATTATTTTAGTAAGGACATCAAGAAGAAAAAATACAT, mutated nucleotides are underlined), digested with NheI/NcoI, and ligated in place of the NheI/NcoI-excised 32-bp ran promoter and two copies of a 19-bp tet operator sequence in pNLop2-1 (20). The resulting plasmid, pNM5m, contained the luciferase gene under the control of the myb2 promoter with a mutation on the Myb2 binding site. For constructing pNMybΔ2, a PCR with oligonucleotide Myb2D2F (GGCGCTGCAGCACGACTATGGAGAGTACTACCAT) and Myb2AUER generated a 0.7-kb product that was digested with PstI and EcoRI. Another PCR with primers Myb2D2R (GGCGCTGCAGCTCCATGGGATCGTGAGTAATAAC) and Myb25NF generated a 1-kb PCR product that was digested with PstI and NheI and cloned into NheI/EcoRI-digested pNLop2-1 (20) with the 0.7-kb PstI/EcoRI fragment. The resulting pNMybΔ2 contains a myb2 gene lacking a region with some similarity to Pseudomonas putida dehRI protein (residues 157-212) (Fig. 4). For constructing pNMybΔN, a PCR with oligonucleotide Myb2D4F (GGCGCTGCAGATGACGAACTGGGCACCAGAAGAAGAC) and Myb2AUER generated a 0.4-kb product that was digested with PstI and EcoRI. Another PCR with primers Myb2D4R (GGCGCTGCAGTACAGAGTAATTATTATTTTAGTAAGC) and Myb25NF generated a 0.3-kb PCR product that was digested with PstI and NheI and cloned into NheI/EcoRI-digested pNLop2-1 (20) with the 0.4-kb PstI/EcoRI fragment. The resulting pNMybΔN contains a myb2 gene lacking the N-terminal 343 amino acids (residues 2-410) and leaves both Myb repeats and the C-terminal 10 amino acids (residues 410-530)(Fig. 4, A and B). For constructing pNMyb2as, a PCR with primers Myb2BF (GGCGTCATGATCAGGGTAGCTTCTCACGGGGAAG) and Myb2EF (GGCGGAATTCATGTTACCGGTACCTTCTCAGC) generated a 1590-bp PCR product that was digested with BspHI and EcoRI and ligated in place of the NcoI/EcoRI-excised luciferase gene in pNM5 (20).
FIGURE 4.
Localization of Myb2 and its deletion derivatives. A, diagrams of the plasmids for Myb2 deletion mapping. Each plasmid contains a neo gene (open box) under the control of the 5′- and 3′-flanking regions of the ran gene (dotted box). Plasmid pNMyb contains the myb2 gene (open boxes) under the control of the myb2 its own promoter (striped box) and 3′-flanking region of the ran gene (dotted box). The filled box indicates the coding sequence of the AU1 epitope tag. The Myb repeats are indicated as open boxes labeled Myb. The specific transfectants were cultured in growth medium (Veg) or encystation medium for 24 h (Enc) and then subjected to immunofluorescence analysis, using anti-AU1 antibody for detection. The localization of Myb2 in nuclei (N) or cytosol (C) is summarized in the right panel. “-” denotes negative staining. B, alignment of the amino acid sequences of the giardial Myb2 and P. putida dehRI or S. cerevisiae Rlr1p. Specific sequence similarity search was performed against the GenBank™ data base on the NCBI Web site using the BLASTP algorithm (50). This search identified similarity of giardial Myb2 to P. putida dehRI or S. cerevisiae Rlr1p in the GenBank™ data base. Numbers indicate positions of the residues relative to the first amino acid. Plus (+) and hyphens indicate conserved amino acids and gaps in the respective proteins, respectively.
Transfection, Luciferase Assay, and Western Blot Analysis—Cells transfected with pN series plasmid were selected with G418 as previously described (46). Stable transfectants were maintained at 150 μg/ml G418. Cells transfected with pP series plasmid containing the pac gene were selected and maintained with 54 μg/ml puromycin. For co-transfection assays (see Fig. 3), G. lamblia cells were first transfected with pP series plasmids and selected in 54 μg/ml puromycin. The stable transfectants were transfected with pN series plasmids, and then the cells were doubly selected in both 150 μg/ml G418 and 54 μg/ml puromycin. After stable transfection with specific constructs, luciferase activity was determined in vegetative cells at late log/stationary phase (1.5 × 106 cells/ml) or in 24 h encysting cells as described (10) and was measured with an Optocomp I luminometer (MGM Instruments). Two independently generated stably transfected lines were made from each construct and each of these lines was assayed three separate times. Western blots were probed with anti-AU1 monoclonal antibody (Covance, Princeton, 1/5000 in blocking buffer), and detected with peroxidase-conjugated goat anti-mouse IgG (Pierce, 1/5000) and enhanced chemiluminescence (GE Healthcare).
FIGURE 3.
Transactivation by Myb2 through its own target sequence. In construct pNTMyb2, the myb2 gene is under the control of the constitutively expressed α2-tubulin promoter (striped box) and 3′-flanking region of the ran gene (dotted box). The filled box indicates the coding sequence of the AU1 epitope tag. In the pPW1, pPC35, or pPM5 reporter construct, the luciferase gene (luc+, open box) is under the control of the cwp1, cwp3, or myb2 promoter as shown. In the -42/+3 reporter construct, the luciferase gene (luc, open box) is under the control of the g6pi-b promoter deletion lacking the Myb2 binding site. In the 4s/-42/+3 reporter construct, four Myb2 binding sequences (open boxes) were inserted upstream of the g6pi-b promoter deletion lacking the Myb2 binding site. The neo or pac gene (open box) is under the control of the 5′- and 3′-flanking regions of the ran (dotted box) or gdh gene (striated box). Specific cell lines were produced by the stable co-transfection of reporter construct and the effector construct pNTMyb2. The corresponding control cell lines were produced by the stable co-transfection of reporter construct and the pRANneo construct. Luciferase activity was measured in vegetative cells as described under “Experimental Procedures.” The activity of co-transfectants (+pNTMyb2) relative to the corresponding control cell line (+pRANneo) is presented. Values are shown as means ± S.E. in the right panel.
Generation of anti-CWP1 Antibody—The genomic cwp1 gene was amplified using oligonucleotides W1F (ATGATGCTCGCTCTCCTTGCTCTTG) and W1R (AGGCGGGGTGAGGCAGTACTCTCCG). The product was cloned into the expression vector pCRT7/CT-TOPO (Invitrogen) in-frame with the C-terminal His and V5 tags to generate plasmid pCWP1. The pCWP1 plasmid was freshly transformed into Escherichia coli BL21(DE3)pLysS (QIAexpressionist, Qiagen). An overnight preculture was used to start a 250-ml culture. E. coli cells were grown to an A600 of 0.5, and then induced with 1 mm isopropyl-d-thiogalactopyranoside (Promega) for 2 h. Bacteria were harvested by centrifugation and sonicated in 10 ml of buffer A (100 mm sodium phosphate, 10 mm Tris-Cl, 6 m guanidine hydrochloride, pH 8.0) containing 10 mm imidazole and complete protease inhibitor mixture (Roche Applied Science). The samples were centrifuged, and the supernatant was mixed with 1 ml of a 50% slurry of Ni-NTA superflow (Qiagen). The resin was washed with buffer B (100 mm sodium phosphate, 10 mm Tris-Cl, 8 m urea, pH 8.0) and buffer C (100 mm sodium phosphate, 10 mm Tris-Cl, 8 m urea, pH 6.3) and eluted with buffer E (100 mm sodium phosphate, 10 mm Tris-Cl, 8 m urea, pH 4.5). Fractions containing CWP1 were pooled, dialyzed in 25 mm HEPES pH 7.9, 40 mm KCl, 0.1 mm EDTA, and 15% glycerol, and stored at -70 °C. Protein purity and concentration were estimated by Coomassie Blue and silver staining compared with bovine serum albumin. CWP1 was purified to apparent homogeneity (>95%). Purified CWP1 protein was used to generate rabbit polyclonal antibodies through a commercial vendor (Angene, Taipei, Taiwan).
Immunofluorescence Assay—Stably transfected cells were harvested from growth medium under drug selection, washed in PBS and attached to glass coverslips (2×106 cells/coverslip), then fixed as previously described (10). Cells were reacted with anti-AU1 monoclonal antibody (BAbCO, 1/300 in blocking buffer) and anti-CWP1 (1/300 in blocking buffer). Anti-mouse ALEXA 568 (Molecular Probes, 1/500 in blocking buffer) and anti-rabbit ALEXA 488 (Molecular Probes, 1/500 in blocking buffer) was used as the detector. Detected proteins were visualized using a Leica TCS SP2 Spectral Confocal System.
ChIP Assay—The pNMyb stable cell line and non-transfected WB cells were inoculated into encystation medium (5 × 107 cells in 45 ml of medium) and harvested after 24 h in encystation medium under drug selection and washed in PBS. Formaldehyde was then added to the cells in PBS at a final concentration of 1%. Cells were incubated at room temperature for 15 min and reactions were stopped by incubation in 125 mm glycine for 5 min. After PBS washes, cells were lysed in luciferase lysis buffer (Promega) and protease inhibitor (Sigma) and then vortexed with glass beads. The cell lysate was sonicated on ice and then centrifuged. Chromatin extract was incubated with anti-AU1 antibody conjugated to beads (Bethyl Laboratories Inc.). The beads were washed twice with luciferase lysis buffer and twice with PBS. The beads were resuspended in elution buffer containing 50 mm Tris-HCl, pH 8.0, 1% SDS and 10 mm EDTA at 65 °C for 4 h. To prepare DNA representing input DNA, 2.5% of chromatin extract without incubation with anti-AU1 beads was combined with elution buffer. Eluted DNA was purified by the QIAquick PCR purification kit (Qiagen). Purified DNA was subjected to PCR followed by agarose gel electrophoresis. The following primers were used to amplify different gene promoters: myb2chF (TGATTCAAAAGAATTAGACGGACTG), myb2chR (AAGAAGAAAAAATACATAGCCATAAAGCG), cwp1chF (AGTAGTTCTCTCTTCTGGCGA) cwp1chR (CCCTGATATTTTATTTCTGTG), cwp2chF (AAAGGATACCCCTTGGGCC), cwp2chR (TTTATTTTCCCAGCCACTGTTGAGCTGCTG), ranchF (AGTGCGCCGTCTGAGCCGCT), ranchR (GCTACTCTCGGTTCCTGGGTTAAAGTTTTA), mlflchF (TGTGTAAGCAACACGTAGTTC), mlflchR (TTTTATTTTTTGCGTTGCATCATGGTCATCATGA), pgkchF (AAGTCGAATGAGAATGCG), pgkchR (TTTTTGCTAAAATCTAAT), and bipchF (GGCTGTGAATGCTAGGCAGCCAGA), and bipchR (TAAAATTTGCTTGTAGACATACGC).
RESULTS
Overexpression of Myb2 Induced the Expression of cwp1/2 Genes—To study the role of Myb2 in G. lamblia, we expressed myb2 constitutively under the control of the α2-tubulin gene promoter (pPTMyb2, Fig. 1A) and observed its gene expression in vegetative cells. Northern blot analysis showed that the mRNA levels of the endogenous myb2 plus vector expressed myb2 in the Myb2-overexpressing cell line increased by ∼2.7-fold (p < 0.05) relative to those in the control cell line which expressed only the puromycin selection marker (5′Δ5N-Pac, Fig. 1B). The endogenous cwp1 and cwp2 genes were up-regulated by ∼3.6-3.8-fold (p < 0.05) in the Myb2 overexpressing cell line (Fig. 1B), suggesting that the overexpressed Myb2 can transactivate the cwp1 and cwp2 genes. The levels of endogenous ran mRNA decreased by ∼70% (p < 0.05) in the Myb2-overexpressing cell line compared with those in the control cell line (Fig. 1B).
In previous studies, we have found that the expression of the cwp1, cwp2, and myb2 genes were up-regulated in stable cell line with drug selection (43). Interestingly, we have found that the expression of the gene encoding orf 16424 was also up-regulated by ∼1.4-4.8-fold in both encysting cells and stably transfected cells (43). The orf 16426 is a myeloid leukemia factor-like (MLFL) protein, and its function is unknown. We have also found that the expression of the phosphoglycerate kinase (pgk) and bip genes was up-regulated in stably transfected cells but not changed in encysting cells (43). We wished to understand the importance of Myb2 for expression of these genes. We found that the mRNA levels of the mlfl gene also increased ∼1.4-fold (p < 0.05) in the Myb2-overexpressing cell line and that the mRNA levels of the pgk and bip gene did not change in the Myb2-overexpressing cell line (Fig. 1B).
Overexpression of Myb2 Increased the Levels of Cyst Wall Protein 1 and Cyst Formation—We further investigated the effect of giardial Myb2 on cyst formation. In previous studies, we have found that some G. lamblia trophozoites may undergo spontaneous differentiation (43). We obtained consistent cyst count data for vegetative G. lamblia cultures during growth to stationary phase (∼4800 cysts/ml for 5′Δ5N-Pac cell line) (43). In this study, we found that the cyst number in the Myb2 overexpressing cell line increased ∼3.2-fold (p < 0.05) relative to the control levels in the 5′Δ5N-Pac cell line (Fig. 2A), indicating that the overexpressed Myb2 can increase the cyst formation.
FIGURE 2.
Localization of CWP1 in the Myb2-overexpressing cell line. A, overexpression of Myb2 increased the levels of cyst formation. 5′-Δ5N-Pac and pPTMyb2 stable transfectants were cultured in growth medium to late log/early stationary phase. Cyst count was performed on these late log/early stationary phase cultures (1.5 × 106 cells/ml). The sum of total cysts is expressed as relative expression level over control. Values are shown as means ± S.E. B, overexpression of Myb2 increased the numbers of CWP1 positive staining cells. The 5′Δ5N-Pac and pPTMyb2 stable transfectants were cultured in growth medium to late log/early stationary phase and then subjected to immunofluorescence assays. The endogenous CWP1 protein was detected by anti-CWP1 antibody. The number of CWP1-positive staining cells were counted and expressed as relative expression level over control. Values are shown as means ± S.E. C and D, localization of CWP1 and Myb2 in the same cell. The endogenous CWP1 protein and vector-expressed Myb2-AU1 protein were detected by anti-CWP1 and anti-AU1 antibodies. The left panel shows that the CWP1 protein is localized to the ESVs. The middle panel shows that the Myb2-AU1 protein is localized to the nuclei and slightly in the cytoplasm. The right panel shows the merged image. The CWP1 protein was stained in the ESVs of all of Myb2-AU1-positive stained cells (data not shown), and two cells are shown in C and D.
We next asked whether the levels of cyst wall protein 1 increased with the increase of the cwp1 transcripts in the Myb2-overexpressing cell line. The CWP1 protein was not detected in Western blots, which could be due to its low expression during vegetative growth (43). In immunofluorescence assays, the AU1-tagged Myb2 was detected in the nuclei and slightly in the cytoplasm during vegetative growth (Fig. 2, C and D). This finding was similar to that in our previous study (20). Expression was ∼8% positive in vegetative cells. The endogenous CWP1 protein was stained in encystation-secretory vesicles (ESVs) in ∼2% of the control cell line 5′Δ5N-Pac (data not shown) (43). A ∼10-fold increase in numbers (∼20%) of cells positively stained for CWP1 protein was found in the Myb2-overexpressing cell line (Fig. 2B). Interestingly, the CWP1 protein was stained in the ESVs of all of Myb2-AU1 positive stained cells (Fig. 2, C and D and data not shown). The appearance of discrete CWP1-localized ESVs could be due to the presence of the constitutively overexpressed Myb2 in the trophozoites. The results suggest that Myb2 may function in inducing the ESV/cyst formation and in activation of the cwp1 gene.
Myb2 Can Transactivate the cwp Genes Through Its Target Sequence—To determine if Myb2 has the capacity to activate transcription, we co-transfected the Myb2 expression plasmid pNTMyb2 together with constructs in which the luciferase reporter gene is under the control of the cwp1, cwp3, myb2, or g6pi-b synthetic promoter with four Myb2 binding sites (Fig. 3, pPW1, pPC3, pPM5, or 4s/-42/+3). The Myb2-AU1 protein was expressed in the pNTMyb2 cell line as detected in immunofluorescence assays (data not shown). Cotransfection of the pPW1 reporter construct with pNTMyb2 resulted in a ∼3.2-fold enhancement of activity in vegetative cells relative to the control cell line (pPW1+pRANneo, Fig. 3), indicating that Myb2 can transactivate the cwp1 promoter. We also tested other promoter-luciferase constructs and found that the cwp3 and myb2 promoter, and g6pi-b synthetic gene promoter with four Myb2 binding sites can be activated ∼2.1- to ∼4.3-fold by Myb2 (Fig. 3). The results indicate that Myb2 can transactivate the promoters containing its binding sites and it can autoregulate its own promoter. We also found that deletion of the Myb2 binding site in the g6pi-b promoter abolishes transactivation by Myb2 (-42/+3, Fig. 3), indicating that Myb2 can transactivate the g6pi-b promoter through its binding sites.
Mapping of the Myb2 Transactivation Domain—To identify the transactivation domain of the Myb2 protein, we established a pNMyb cell line expressing the myb2 gene controlled by its own promoter with sequences encoding an AU1 tag at its C terminus (Fig. 4A) (20). Using this parental system, we constructed four deletion mutants of Myb2: MybΔ1 that deletes the first Myb repeat (residues 410-468), MybΔ2 that deletes a region with some similarity to P. putida dehRI protein (residues 157-212), MybΔC that deletes both Myb repeats and the C-terminal 10 amino acids (residues 410-530), and MybΔN that deletes the N-terminal 343 amino acids (residues 2-410) and leaves both Myb repeats and the C-terminal 10 amino acids (residues 410-530)(Fig. 4, A and B). MybΔN that deletes the N-terminal 343 amino acids (residues 2-410) also lacks the region with some similarity to Saccharomyces cerevisiae Rlr1p (residues 340-403) (Fig. 4, A and B). These constructs were transfected into G. lamblia and the cellular locations of Myb2 in the transfectants were also examined by immunofluorescence assays. The wild-type Myb2, MybΔ1, and MybΔ2 were localized to nuclei, but MybΔC was localized to cytosol in both vegetative and encysting cells (Fig. 4A, additional data not shown) (20). The MybΔN protein was not detected in vegetative cells, but it was detected in nuclei during encystation (Fig. 4A, additional data not shown).
We further asked whether the protein levels of the Myb2 deletions were changed. As shown by Western blot analysis, the levels of MybΔ1 or MybΔ2 increased significantly compared with those of wild-type Myb2 during encystation (Fig. 5A). However, the levels of MybΔC or MybΔN decreased significantly (Fig. 5A). To understand the effect of different Myb2 deletion mutants, we observed the expression of endogenous cwp1 gene in encysting cells. We found that the cwp1 gene was up-regulated by ∼2-3-fold (p < 0.05) in the pNMybΔ1 and pNMybΔ2 cell lines relative to the pNMyb cell line (Fig. 5B). Higher levels of the MybΔ1 or MybΔ2 protein might account for the higher transactivation activity observed in the pNMybΔ1 and pNMybΔ2 cell lines (Fig. 5A). However, the cwp1 gene was down-regulated by ∼2-fold (p < 0.05) in the pNMybΔCor pNMybΔN cell line relative to the pNMyb cell line (Fig. 5B). The results suggest that the lower levels of the MybΔCor MybΔN protein may be correlated with loss of transactivation. In addition, the inactivity of the MybΔC protein could be due to its inability to enter nucleus (Fig. 4A). The inactivity of the MybΔN protein could be due to the lack of transactivation domain as the MybΔN protein can enter nuclei (Fig. 4A).
Myb2 Gene May Be Autoregulated—We further asked whether the mRNA levels of the Myb2 deletions were changed. As shown by Northern blot analysis, the levels of endogenous myb2 plus mybΔ1 or mybΔ2 mRNA increased by 2-5-fold (p < 0.05) in the pNMybΔ1or pNMybΔ2 cell line relative to the pNMyb cell line (Fig. 5B). This result suggests that the increase in the protein levels of the pNMybΔ1 or pNMybΔ2 could be due to the increased mRNA levels. However, the levels of endogenous myb2, mybΔc, or mybΔn mRNA decreased significantly in the pNMybΔC or pNMybΔN cell line relative to the pNMyb cell line (Fig. 5B). The results suggest that deletion of the N- or C-terminal domain may result in a decrease of the transactivation and autoregulation function of Myb2.
Because the myb2 promoter contains a Myb2 binding site (20), we would like to know whether this Myb2 binding site is required for the myb2 promoter function. To address this, we constructed a myb2 promoter mutant construct (pNM5m, Fig. 6) with an altered Myb2 binding site in the myb2 promoter-reporter construct pNM5 (20). We found that this mutation decreased promoter activity to ∼72 and ∼15% of the wild-type promoter activity during vegetative growth and encystation, respectively (Fig. 6). The induction ratio (luciferase expression in encysting cells relative to that in vegetative cells) of the myb2 promoter mutant construct was ∼2.6. This is much lower than that of the wild-type myb2 promoter construct (∼12.4), indicating that the Myb2 binding site is important for the myb2 promoter activity and that the myb2 gene could be positively autoregulated.
FIGURE 6.
Mutation analysis of the Myb2 binding site in the myb2 promoter region. In the pNM5 construct, the luciferase gene (luc+, open box) is under the control of the myb2 promoter and 3′-flanking region of the ran gene (dotted box). The neo gene is under the control of the 5′- and 3′-flanking regions of the ran gene (dotted box). In the pNM5m construct, the mutated Myb2 binding site is indicated by a filled box. After stable transfection with these constructs, luciferase activity was measured in vegetative cells and 24 h encysting cells as described under “Experimental Procedures.” Values are shown as means ± S.E. in the right panel. The induction ratio was obtained by dividing the activity in the encysting cells by the activity in the vegetative cells of each construct.
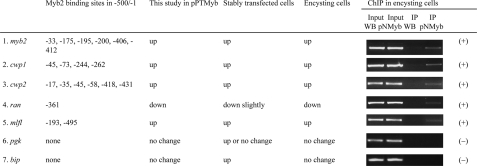
Recruitment of Myb2 to the cwp Promoters—We searched the 500-bp 5′-flanking regions of these giardial genes, and found that myb2, cwp1, cwp2, ran, and mlfl contain Myb2 binding sequences, including CTACAG, CAACAG, CTACTG, or CTACAC on the coding strand or noncoding strand (Fig. 7) (20). The myb2, cwp1, or cwp2 gene has 4-6 upstream Myb2 binding sequences. However, the ran or mlfl gene has 1-2 upstream Myb2 binding sequences. We further used ChIP assays to study association of Myb2 with these promoters in the Myb2-overexpressing cell line. We found that Myb2 was associated with the myb2 its own promoter and the cwp1, cwp2, ran, and mlfl promoters in the pNMyb cell line during encystation (Fig. 7). However, Myb2 was not associated with the pgk and bip gene promoter which have no Myb2 binding site (-500/-1 of the 5′-flanking region, Fig. 7).
FIGURE 7.
Summary of the results from search of Myb2 binding sites and ChIP assays. The 500 bp of the 5′-flanking regions of endogenous genes were searched for Myb2 binding sites CTACAG, CAACAG, CTACTG, CTACAC on the coding strand or noncoding strand. The 3′-ends of the Myb2 binding sites are numbered relative to the translation initiation site (+1). The pNMyb cell line expressing the AU1-tagged Myb2 (Fig. 4) and non-transfected WB cells as the control cell line were cultured in encystation medium for 24 h and then subjected to ChIP assays. Anti-AU1 antibody conjugated to beads was used to assess binding of Myb2-AU1 to endogenous gene promoters. Immunoprecipitated chromatin was analyzed by PCR using primers that amplify the 5′-flanking region of specific genes. Approximately the same amount of PCR product was obtained from input DNA in the pNMyb cell line and the control cell line. At least three independent experiments were performed. Representative results are shown. Immunoprecipitated products of Myb2-AU1 yielded more PCR products of myb2, cwp1, cwp2, ran, and mlfl promoters in the pNMyb cell line relative to the control cell line, indicating that Myb2-AU1 was bound to these promoters (+).
Transfection of the myb2 Antisense Construct Reduced the Levels of the cwp1 Transcripts and Cyst Formation—We next asked whether Myb2 is important for encystation. We found that, when a myb2 antisense-expressing construct (pNMyb2as) was transfected into G. lamblia, the levels of cyst formation decreased significantly in comparison to that in cells transfected with luciferase expression construct (pNM5) or the nontransfected WB C6 cells (Fig. 8, A and B). Northern blot analysis showed that the levels of the myb2 mRNA decreased by ∼60% (p < 0.05) in the pNMyb2as transfectants relative to those in the WB C6 control (Fig. 8C). The levels of the cwp1 mRNA decreased by ∼70% (p < 0.05) in the pNMyb2as transfectants relative to those in the WB C6 control (Fig. 8C). The levels of the ran mRNA increased ∼1.3-fold (p < 0.05) in the pNMyb2as transfectants relative to those in the WB C6 control (Fig. 8C). Similar mRNA levels of the bip gene were found in the pNMyb2 transfectants compared with those in the WB C6 control (Fig. 8C). The results suggest an important role of Myb2 during encystation. In the previous studies, we have found that the cyst formation and the expression of the cwp1 and myb2 genes increased significantly in stable cell lines with drug selection (43). The increased cyst numbers and expression of the cwp1 and myb2 genes were also found in the pNM5 stable transfectants relative to those in the non-transfected WB C6 cells (Fig. 8, B and C).
DISCUSSION
In the previous studies, we have identified a myb2 gene whose expression increased during encystation (20). The presence of the Myb2 binding sites in the proximal 5′-flanking regions of key encystation-induced genes, cwp1, cwp2, cwp3, g6pi-b, and myb2 itself suggests that Myb2 may be involved in co-ordinating their differential expression. To gain insight into the function of Myb2 in cell differentiation, we tested the hypothesis that Myb2 can activate transcription of the endogenous encystation-induced genes. Our results showed that the constitutively overexpressed Myb2 increased the levels of the cwp1 and cwp2 mRNA in vegetative trophozoites. The levels of the CWP1 protein and cyst formation also increased in the Myb2-overexpressing cell line. The overexpressed Myb2 also can transactivate the cwp1, cwp3, myb2, and g6pi-b promoters that contain Myb2 binding sites in vivo. We also found that deletion of the Myb2 binding site abolished transactivation by Myb2, indicating that Myb2 can transactivate the encystation-induced genes through its target sequence. In addition, deletion of the N- or C-terminal domain resulted in a decrease of transactivation function of Myb2. Transfection of the myb2 antisense construct reduced the levels of the cwp1 transcripts and cyst formation. The results suggest that Myb2 may play an important role in induction of encystation.
Many important transcription factors involved in developmental regulation have an autoregulation mechanism, including mammalian c-Myb (36, 48). The presence of a Myb2 binding site upstream of myb2 itself raises the possibility of myb2-positive autoregulation in G. lamblia. We also addressed this by mutation of this Myb2 binding site and found a significant decrease of Myb2 promoter activity. The overexpressed Myb2 also can transactivate myb2 its own promoter. In addition, we also found that deletion of the Nor C-terminal domain resulted in a decrease of autoregulation function of Myb2. ChIP assays confirmed the association of Myb2 with its own promoter. Our results suggest that the myb2 gene may be positively autoregulated, and this may help attain a higher level of Myb2 for induction of cwp genes during encystation.
The Myb2-overexpressing system could be used to increase some possible encystation-induced genes in vegetative cells. In the previous studies, we have found that the expression of the cwp1, cwp2, myb2, and mlfl (orf 16424) genes were up-regulated in both encysting cells and stably transfected cells (43). In the present study, we found that the levels of mlfl mRNA, like those of cwp mRNA, also increased in the Myb2-overexpressing cell line. It is interesting that 4-6 Myb2 binding sequences are present in the 500-bp 5′-flanking regions of the myb2, cwp1, or cwp2 genes. One to two Myb2 binding sequences are present in the 500-bp 5′-flanking regions of the mlfl, or ran genes. Interestingly, ChIP assays confirmed the association of Myb2 with these promoters containing the Myb2 binding sites. Therefore, the ability of Myb2 to transactivate the encystation-induced cwp or mlfl genes may require the binding of Myb2 to its binding sequences. In the previous studies, we have also found that the expression of the phosphoglycerate kinase (pgk) and bip genes was up-regulated in stably transfected cells but not changed in encysting cells (43). In this study, we found that the levels of pgk and bip mRNA did not change in the Myb2-overexpressing cell line. In addition, these two promoters have no Myb2 binding site (-500/-1 of the 5′-flanking region, Fig. 7) and Myb2 was not associated with them. These findings suggest that there might be different signal transduction pathways or different transcriptional mechanisms regulating the expression of the pgk and bip genes in stably transfected cells and Myb2-overexpressing cells.
Unlike Myb proteins in many other organisms whose Myb domains are N-terminal (24-26, 49), the Myb domain in giardial Myb2 is near the C terminus. Myb2 contains only two repeats that appear to function like the R2R3 domains in Myb proteins of other organisms. In this study, we found that the first Myb repeat may not be important for nuclear localization and transactivation. Deletion of the first Myb repeat (MybΔ1) or a region with some similarity to P. putida dehRI protein (MybΔ2, residues 157-212) retained higher levels of nuclear Myb2 proteins with transactivation activity. This indicates that these two coding regions may be negative regulatory regions for repression of transcription or mRNA stability. Myb proteins with one repeat have been identified in plants (25). The one repeat Myb protein can bind to DNA as a homodimer (51, 52). Therefore, the one-Myb-repeat MybΔ1 that can enter the nucleus may still be functional. On the other hand, the levels of the myb2 mRNA from the N- or C-terminal deletion mutants (MybΔN or MybΔC) were very low, indicating that both N- and C-terminal coding regions were required for myb2 transcription or mRNA stability. Interestingly, the levels of endogenous myb2 and cwp1 mRNA were also lower in the MybΔN- or MybΔC-overexpressing cell line, indicating that removal of the N- or C-terminal region results in a significant decrease of transactivation and autoregulation ability. The lower transactivation ability of MybΔC protein could be due to its lower levels or its inability to enter nucleus. However, because the MybΔN protein can enter nuclei, the inactivity of the MybΔN protein could be due to its lower levels or the lack of the transactivation domain. This indicates that the N-terminal 343 amino acids (residues 2-410) containing a region with some similarity to S. cerevisiae Rlr1p (residues 340-403) or a stretch of acidic amino acid (residues 225-232, Fig. 1A) has a transactivation ability. It is known that acidic characteristics are associated with the eukaryotic transactivation domains. The existence of distinct Myb transactivation domains may interact with other transcription factors to modulate transcription in vivo.
Our results showed that constitutively expressed Myb2 increased the expression of cwp1 and cwp2 genes by ∼3.6-3.8-fold in vegetative trophozoites. However, the cwp1 promoter could be increased by ∼47-fold during encystation (21). Although Myb2 can also function as a transactivator in vegetative cells, it may still need to cooperate with some other transcription factors that are induced during encystation to transactivate these cyst wall protein genes. In late-branching eukaryotes, Myb proteins regulate specific target genes by interacting with other classes of DNA binding proteins that occupy directly adjacent binding sites within the target promoter region. For, example, c-Myb can cooperate with NF-M/c-EBP (b-ZIP family) or Ets-2 (helix-turn-helix) to activate the promyelocytic-specific mim-1 gene (38, 53). c-Myb and AML1/CBF (Runt family) activate T-cell receptor delta enhancer (39, 40). Therefore, it is possible that giardial Myb2 functions as an activator via association with some encystation-specific cofactors on the promoter context of encystation-induced genes.
Our study provides evidence for the involvement of Myb2 in the differentiation of G. lamblia trophozoites into cysts. Our findings provide new insight into distinct functional domains of Myb2 and suggest testable transcriptional mechanisms in the protozoan G. lamblia.
Acknowledgments
We thank Yi-Li Liu and I-Ching Huang for technical support with DNA sequencing. We also thank the researchers and administrators of the G. lamblia Genome database.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBank™/EBI Data Bank with accession number(s) AY082882.
This work was supported by grants from the National Science Council (NSC 94-2320-B-002-093 and NSC 96-2320-B-002-040-MY3) and the National Health Research Institutes (NHRI-EX96-9510NC and NHRI-EX97-9510NC) in Taiwan and was also supported in part by the Dept. of Medical Research in NTUH. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: CWP, cyst wall protein; PBS, phosphate-buffered saline; G6PI-B, glucosamine-6-phosphate isomerase-B; ChIP, chromatin immunoprecipitation assay.
References
- 1.Marshall, M. M., Naumovitz, D., Ortega, Y., and Sterling, C. R. (1997) Clin. Microbiol. Rev. 10 67-85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gillin, F. D., and Reiner, D. S. (1996) Annu. Rev. Microbiol. 50 679-705 [DOI] [PubMed] [Google Scholar]
- 3.Eichinger, D. (2001) Curr. Opin. Microbiol. 4 421-426 [DOI] [PubMed] [Google Scholar]
- 4.Wolfe, M. S. (1992) Clin. Microbiol. Rev. 5 93-100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adam, R. D. (2001) Clin. Microbiol. Rev. 14 447-475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lujan, H. D., Mowatt, M. R., Conrad, J. T., Bowers, B., and Nash, T. E. (1995) J. Biol. Chem. 270 29307-29313 [DOI] [PubMed] [Google Scholar]
- 7.Mowatt, M. R., Lujan, H. D., Cotton, D. B., Bower, B., Yee, J., Nash, T. E., and Stibbs, H. H. (1995) Mol. Microb. 15 955-963 [DOI] [PubMed] [Google Scholar]
- 8.Sun, C. H., McCaffery, J. M., Reiner, D. S., and Gillin, F. D. (2003) J. Biol. Chem. 278 21701-21708 [DOI] [PubMed] [Google Scholar]
- 9.Van Keulen, H., Steimle, P. A., Bulik, D. A., Borowiak, R. K., and Jarroll, E. L. (1998) J. Eukaryot. Microbiol. 45 637-642 [DOI] [PubMed] [Google Scholar]
- 10.Knodler, L. A., Svard, S. G., Silberman, J. D., Davids, B. J., and Gillin, F. D. (1999) Mol. Microbiol. 34 327-340 [DOI] [PubMed] [Google Scholar]
- 11.Sogin, M. L., Gunderson, J. H., Elwood, H. J., Alonso, R. A., and Peattie, D. A. (1989) Science 243 75-77 [DOI] [PubMed] [Google Scholar]
- 12.Hashimoto, T., Nakamura, Y., Kamaishi, T., Nakamura, F., Adachi, J., Okamoto, K., and Hasegawa, M. (1995) Mol. Biol. Evol. 12 782-793 [DOI] [PubMed] [Google Scholar]
- 13.Morrison, H. G., McArthur, A. G., Gillin, F. D., Aley, S. B., Adam, R. D., Olsen, G. J., Best, A. A., Cande, W. Z., Chen, F., Cipriano, M. J., Davids, B. J., Dawson, S. C., Elmendorf, H. G., Hehl, A. B., Holder, M. E., Huse, S. M., Kim, U. U., Lasek-Nasselquist, E., Manning, G., Nigam, A., Nixon, J. E., Palm, D., Passamaneck, N. E., Prabhu, A., Reich, C. I., Reiner, D. S., Samuelson, J., Svard, S. G., and Sogin, M. L. (2007) Science 317 1921-1926 [DOI] [PubMed] [Google Scholar]
- 14.Seshadri, V., McArthur, A. G., Sogin, M. L., and Adam, R. D. (2003) J. Biol. Chem. 278 27804-27810 [DOI] [PubMed] [Google Scholar]
- 15.Best, A. A., Morrison, H. G., McArthur, A. G., Sogin, M. L., and Olsen, G. J. (2004) Genome Res. 14 1537-1547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holberton, D. V., and Marshall, J. (1995) Nucleic Acids Res. 23 2945-2953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun, C. H., and Tai, J. H. (1999) J. Biol. Chem. 274 19699-19706 [DOI] [PubMed] [Google Scholar]
- 18.Yee, J., Mowatt, M. R., Dennis, P. P., and Nash, T. E. (2000) J. Biol. Chem. 275 11432-11439 [DOI] [PubMed] [Google Scholar]
- 19.Elmendorf, H. G., Singer, S. M., Pierce, J., Cowan, J., and Nash, T. E. (2001) Mol. Biochem. Parasitol. 113 157-169 [DOI] [PubMed] [Google Scholar]
- 20.Sun, C. H., Palm, D., McArthur, A. G., Svard, S. G., and Gillin, F. D. (2002) Mol. Microbiol. 46 971-984 [DOI] [PubMed] [Google Scholar]
- 21.Sun, C. H., Su, L. H., and Gillin, F. D. (2006) Mol. Biochem. Parasitol. 146 45-57 [DOI] [PubMed] [Google Scholar]
- 22.Wang, C. H., Su, L. H., and Sun, C. H. (2007) J. Biol. Chem. 282 8905-8914 [DOI] [PubMed] [Google Scholar]
- 23.Weston, K. (1998) Curr. Opin. Genet. Dev. 8 76-81 [DOI] [PubMed] [Google Scholar]
- 24.Wieser, J., and Adams, T. H. (1995) Genes Dev. 9 491-502 [DOI] [PubMed] [Google Scholar]
- 25.Jin, H., and Martin, C. (1999) Plant Mol. Biol. 41 577-585 [DOI] [PubMed] [Google Scholar]
- 26.Oh, I. H., and Reddy, E. P. (1999) Oncogene 18 3017-3033 [DOI] [PubMed] [Google Scholar]
- 27.Stracke, R., Werber, M., and Weisshaar, B. (2001) Curr. Opin. Plant Biol. 4 447-456 [DOI] [PubMed] [Google Scholar]
- 28.Raffaele, S., Vailleau, F., Léger, A., Joubès, J., Miersch, O., Huard, C., Blée, E., Mongrand, S., Domergue, F., and Roby, D. (2008) Plant Cell 20 752-767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park, J. S., Kim, J. B., Cho, K. J., Cheon, C. I., Sung, M. K., Choung, M. G., and Roh, K. H. (2008) Plant Cell Rep. 27 985-994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Allen, R. D., 3rd, Bender, T. P., and Siu, G. (1999) Genes Dev. 13 1073-1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bender, T. P., Kremer, C. S., Kraus, M., Buch, T., and Rajewsky, K. (2004) Nat. Immunol. 5 721-729 [DOI] [PubMed] [Google Scholar]
- 32.Sala, A. (2005) Eur. J. Cancer. 41 2479-2484 [DOI] [PubMed] [Google Scholar]
- 33.Ramsay, R. G. (2005) Growth Factors 23 253-261 [DOI] [PubMed] [Google Scholar]
- 34.Kolodziejska, K. M., Noyan-Ashraf, M. H., Nagy, A., Bacon, A., Frampton, J., Xin, H. B., Kotlikoff, M. I., and Husain, M. (2008) Circ. Res. 102 554-561 [DOI] [PubMed] [Google Scholar]
- 35.Ness, S. A., Marknell, A., and Graf, T. (1989) Cell. 59 1115-1125 [DOI] [PubMed] [Google Scholar]
- 36.Nicolaides, N. C., Gualdi, R., Casadevall, C., Manzella, L. and Calabretta, B. (1991) Mol. Cell. Biol. 11 6166-6176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ness, S. A., Kowenz-Leutz, E., Casini, T., Graf, T., and Leutz, A. (1993) Genes Dev. 7 749-759 [DOI] [PubMed] [Google Scholar]
- 38.Burk, O., Mink, S., Ringwald, M., and Klempnauer, K. H. (1993) EMBO J. 12 2027-2038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hernandez-Munain, C., and Krangel, M. S. (1994) Mol. Cell. Biol. 14 473-483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hsiang, Y. H., Goldman, J. P., and Raulet, D. H. (1995) J. Immunol. 154 5195-5204 [PubMed] [Google Scholar]
- 41.Evans, J. L., Moore, T. L., Kuehl, W. M., Bender, T., and Ting, J. P. (1990) Mol. Cell. Biol. 10 5747-5752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lidonnici, M. R., Corradini, F., Waldron, T., Bender, T. P., and Calabretta, B. (2008) Blood 111 4771-4779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Su, L. H., Lee, G. A., Huang, Y. C., Chen, Y. H., and Sun, C. H. (2007) Mol. Biochem. Parasitol. 156 124-135 [DOI] [PubMed] [Google Scholar]
- 44.Keister, D. B. (1983) Trans. R. Soc. Trop. Med. Hyg. 77 487-488 [DOI] [PubMed] [Google Scholar]
- 45.Singer, S. M., Yee, J., and Nash, T. E. (1998) Mol. Biochem. Parasitol. 92 59-69 [DOI] [PubMed] [Google Scholar]
- 46.Sun, C. H., Chou, C. F., and Tai, J. H. (1998) Mol. Biochem. Parasitol. 92 123-132 [DOI] [PubMed] [Google Scholar]
- 47.Chen, Y. H., Su, L. H., and Sun, C. H. (2008) Int. J. Parasitol. 38 1305-1317 [DOI] [PubMed] [Google Scholar]
- 48.Nair, M., Bilanchone, V., Ortt, K., Sinha, S., and Dai, X. (2007) Nucleic Acids Res. 35 1687-1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Katzen, A. L., Jackson, J., Harmon, B. P., Fung, S. M., Ramsay, G., and Bishop, J. M. (1998) Genes Dev. 12 831-843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D. J. (1997) Nucleic Acids Res. 25 3389-3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mercy, I. S., Meeley, R. B., Nichols, S. E., and Olsen, O. A. (2003) J. Exp. Bot. 54 1117-1119 [DOI] [PubMed] [Google Scholar]
- 52.Lu, C. A., Ho, T. H., Ho, S. L., and Yu, S. M. (2002) Plant Cell 14 1963-1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dudek, H., Tantravahi, R. V., Rao, V. N., Reddy, E. S., and Reddy, E. P. (1992) Proc. Natl. Acad. Sci. U. S. A. 89 1291-1295 [DOI] [PMC free article] [PubMed] [Google Scholar]