Abstract
SepF (Septum Forming) protein has been recently identified through genetic studies, and it has been suggested to be involved in the division of Bacillus subtilis cells. We have purified functional B. subtilis SepF from the inclusion bodies overexpressed in Escherichia coli. Far-UV circular dichroism and fluorescence spectroscopic analysis involving the extrinsic fluorescent probe 1-anilinonaphthalene-8-sulfonic acid suggested that the purified SepF had characteristics of folded proteins. SepF was found to promote the assembly and bundling of FtsZ protofilaments using three complimentary techniques, namely 90° light scattering, sedimentation, and transmission electron microscopy. SepF also decreased the critical concentration of FtsZ assembly, prevented the dilution-induced disassembly of FtsZ protofilaments, and suppressed the GTPase activity of FtsZ. Further, thick bundles of FtsZ protofilaments were observed using fluorescein isothiocyanate-labeled SepF (FITC-SepF). Interestingly, FITC-SepF was found to be uniformly distributed along the length of the FtsZ protofilaments, suggesting that SepF copolymerizes with FtsZ. SepF formed a stable complex with FtsZ, as evident from the gel filtration analysis. Using a C-terminal tail truncated FtsZ (FtsZΔ16) and a C-terminal synthetic peptide of B. subtilis FtsZ (366-382); we provided evidence indicating that SepF binds primarily to the C-terminal tail of FtsZ. The present work in concert with the available in vivo data support a model in which SepF plays an important role in regulating the assembly dynamics of the divisome complex; therefore, it may have an important role in bacterial cell division.
The formation of transverse septum is the primary requirement during binary fission in the rod-shaped bacteria (1, 2). According to Hamoen et al. (3), the process of septum formation can be thought to result from three sequential events: (i) Z-ring formation, (ii) divisome assembly, and (iii) septum synthesis. The septal ring formation is initiated by the assembly of the FtsZ monomers into a ring like structure called a “Z-ring” (4). The Z-ring functions as a scaffold for accessory cell division proteins, and several proteins play important roles in spatial and temporal regulation of the Z-ring formation (5).
The negative regulatory proteins (MinCDE, EzrA, SulA, nucleoid occlusion, and chaperon ClpX) are crucial for the destabilization and positioning of the Z-ring (6-10). For instance, deletion mutants of negative-acting factors, MinC system (MinC, MinD), DivIVA, and EzrA, show abnormal cell morphology and multiple Z-rings at polar and midcell positions in Bacillus subtilis (1, 7, 11).
The positive regulatory proteins, namely ZapA, ZipA, and FtsA, on the other hand, play key roles in the stabilization and the positioning of the Z-ring (12-14). The cytoplasmic ZapA protein induces FtsZ polymerization (12), and the actin-like protein FtsA is recruited to potential division site in the earliest stage of the Z-ring formation in B. subtilis (15). FtsA has been shown to interact directly with FtsZ, and the disruption of ftsA leads to severe deficiency in the division of B. subtilis cells (16, 17).
Recent studies showed that the SepF protein (renamed for the product of ylmF gene) complements for the lack of FtsA protein (3). Whereas the overexpression of ylmF in FtsA-depleted cells suppressed the division defects, the deletion of ylmF gene in FtsA depleted B. subtilis cells resulted in a complete inhibition of the Z-ring formation and cell division (18). The overlapping functional roles of SepF and FtsA proteins in the formation of the Z-ring indicate that SepF may be involved during early stages of cell division (18). In contrast, Hamoen et al. (3) have argued that the main function of SepF may not be the initiation of the assembly of the divisome complex, but it is required for the proper execution of the septum synthesis as a late division protein.
SepF is well conserved almost in all Gram-positive bacteria (18). The disruption of the ylmF gene resulted in partial impairment of cell division in B. subtilis (18). Also, ylmF inactivated cells of Streptococcus pneuomoniae were morphologically different (19). SepF interacts with FtsZ as suggested by the yeast two-hybrid analysis, and it has been found to localize to cell division sites in a FtsZ-dependent manner (3). The focus of our present work is to elucidate the effects of B. subtilis SepF on the assembly of FtsZ in vitro.
We have purified functional SepF from the inclusion bodies overexpressed in Escherichia coli. In vitro, SepF promoted the assembly of FtsZ, reduced the critical concentration of FtsZ assembly, induced bundling of FtsZ protofilaments, stabilized FtsZ polymers, and suppressed the GTPase activity of FtsZ. Further, SepF formed a stable complex with FtsZ monomers and was found to be recruited in the FtsZ bundles. We also obtained evidence suggesting that the conserved C-terminal tail of FtsZ is the primary site of interaction with SepF. The results indicated that SepF may play an important role during cytokinesis in B. subtilis.
EXPERIMENTAL PROCEDURES
Materials—Pipes,2 phenylmethylsulfonyl fluoride, bovine serum albumin (BSA), β-mercaptoethanol, GTP, FITC, and Sephadex G-100 were purchased from Sigma. Isopropyl β-d-1-thiogalactopyranoside was purchased from Calbiochem. 1-Anilinonaphthalene-8-sulfonic acid (ANS) was purchased from Molecular Probes. Bio-Gel® P-6 resin was purchased from Bio-Rad; nickel(II)-nitrilotriacetic acid coupled to Sepharose® was purchased from Qiagen. Nickel(II)-iminodiacetic acid coupled to Sepharose® was purchased from Amersham Biosciences. All of the other chemicals used were of analytical grade. Pfu Turbo DNA polymerase was obtained from Stratagene. E. coli BL21(DE3) (B F- dcm ompT hsdS (rB-, mB-) gal λ(DE3)) and plasmid pET16b(+) were from Novagen.
Cloning and Overexpression of SepF—The genomic DNA of B. subtilis ATCC6633 was used for the PCR amplifications of the desired orf using Pfu Turbo DNA polymerase. The primers 5′-ACAGCAATGCATATGAAAGATAAACTG-3′ (forward) and 5′-TCATGGATCCTTACCACCTCTGATGTTCG-3′ (reverse) were used to amplify the coding sequence of sepF. The PCR product was cloned into NdeI and BamHI site of pET16b(+) vector (pET16-sepF). E. coli XL1 Blue was used as the cloning host. The recombinant plasmid was subsequently transformed into E. coli BL21(DE3) for the overexpression of SepF. The nucleotide sequences of the chromosomal sepFas well that of pET16-sepF construct were confirmed by complete sequencing of sepF orf using an automated DNA sequencer (ABI-PRISM 377). The deduced amino acid sequence of expressed SepF matched with B. subtilis strain 168 (Gen-Bank™ accession number Z9912) except for two changes (V29E and D86V). However, the sequence of our construct matched exactly with the genomic sepF of B. subtilis ATCC6633 strain used for cloning.
Isolation and Purification of SepF—Recombinant SepF protein of B. subtilis was overexpressed and purified from E. coli BL21(DE3) cells. The expressed SepF protein carried pET16 His tag fused at its N terminus. The cells containing the desired gene were grown at 37 °C in LB medium containing 100 μg/ml ampicillin. The cells were induced at the late log phase (A600 = ∼0.8) by the addition of 0.5 mm isopropyl β-d-1-thiogalactopyranoside and incubated for an additional 4 h. Subsequently, the cells were harvested by centrifugation at 10,000 × g at 4 °C for 10 min. The cell pellet was resuspended in lysis buffer (50 mm Tris-HCl, pH 8.0, and 250 mm NaCl) containing 0.1% β-mercaptoethanol and 2 mm phenylmethylsulfonyl fluoride and was homogenized for 5 min on ice. The cell suspension was incubated with 1 mg/ml lysozyme for 1 h on ice. The cells were disrupted by ultrasonication with 30 pulses at 30-s intervals 10 times. The cell lysate was cleared by centrifugation at 10,000 × g for 10 min at 4 °C. SepF was found only in the inclusion bodies. The inclusion bodies were separated and washed with lysis buffer containing 0.05% β-mercaptoethanol and 1 mm phenylmethylsulfonyl fluoride for 5 times at 10,000 × g for 10 min at 4 °C and were subsequently dissolved in denaturing buffer (lysis buffer containing 8 m urea). The denatured SepF was refolded in the stabilizing buffer (50 mm Tris-HCl, pH 8.0, containing 50 mm NaCl, 10% glycerol, 0.4 m l-arginine, and 2% glycine) by dilution method (20). During the refolding step, drops containing solubilized inclusion bodies were diluted to a large volume (100 ml) of stabilizing buffer. The refolded SepF protein was recovered in the soluble supernatant after centrifugation at 120,000 × g for 30 min at 4 °C. The supernatant was dialyzed against 25 mm Tris-HCl and 150 mm NaCl for overnight at 4 °C. The cleared supernatant containing SepF protein was adjusted for final concentration of 10 mm imidazole by the addition of imidazole (pH 8.0) and was loaded onto a nickel(II)-iminodiacetic acid-Sepharose affinity column. The column was washed extensively with a buffer containing 25 mm Pipes, pH 6.8, 250 mm NaCl, and 25 mm imidazole. Bound proteins were eluted with a discontinuous gradient of imidazole (50, 100, 250, and 500 mm). SepF was found to elute at 250 and 500 mm imidazole concentrations as adjudged from the Coomassie Blue-stained 12% SDS-PAGE. The fractions containing SepF were pooled and desalted by using P-6 resin. SepF was transferred to 42 °C for 10 h to remove misfolded SepF by precipitation method. The protein was concentrated using Amicon® Ultra PL-10 (Millipore) at 4 °C. The purity level of refolded SepF was determined by scanning the Coomassie Blue-stained 12% SDS-PAGE (≥95% pure), using ImageJ software. The concentration of purified protein was measured by the Bradford method using BSA as a standard (21). The purified protein was aliquoted and stored at -80 °C.
The molecular mass of the His-tag fused SepF was determined to be 19.6 kDa using Axima-CFR matrix-assisted laser desorption ionization time-of-flight mass spectrometry (Kratos Analytical, Manchester, UK). The measured molecular weight matched with the calculated molecular weight of the His tag fused SepF protein sequence deduced from the DNA sequence.
The SepF protein was incubated with Factor Xa (Novagen) (50:1 w/w) for 24 h at 25 °C as described in the manufacturer's protocol. Factor Xa was removed with Xarrest Agarose (capture kit; Novagen) after cleavage of the His tag from SepF. The Factor Xa-treated SepF protein solution was incubated with spin column containing Xarrest Agarose slurry. His tag-removed SepF protein was collected after centrifugation at 2,000 × g for 2 min. The purified SepF without His tag was characterized using different spectroscopic analysis and was used for the subsequent experiments. The amino acid sequence of the SepF protein was analyzed for the prediction of transmembrane domain(s) that could anchor the protein to the lipid bilayer.
Isolation and Purification of B. subtilis FtsZ—Recombinant B. subtilis full-length FtsZ and C-terminal tail 16-residues truncated FtsZ (FtsZΔC16) were overexpressed and purified from E. coli (BL21)pLysS clones individually by affinity chromatography using chelating Sepharose (nickel(II) nitrilotriacetic acid) matrix as described earlier (22). FtsZ concentration was measured by the Bradford method using BSA as a standard and adjusting the final concentration of FtsZ using a correction factor of 0.80 for the FtsZ/BSA ratio (23). The purified proteins were frozen and stored at -80 °C.
CD and Spectrofluorometer Spectra Analysis—SepF (25 μm) was incubated for 30 min on ice in the absence and presence of 8 m urea prepared in the 50 mm phosphate buffer, pH 6.8. The spectra of SepF with and without urea were recorded at 25 °C using a JASCO Spectropolarimeter (model J-810) equipped with a JASCO PTC 423S Peltier temperature control system. A quartz cuvette of 0.1-cm path length was used for the CD measurements. Each spectrum was the average of five scans. Deconvolution and statistical analysis of the CD spectra were performed using CDNN and SSE software from JASCO and OriginPro 7.5 software, respectively.
Fluorescence spectroscopic studies of the SepF protein were performed using a JASCO FP-6500 spectrofluorometer with an appropriate blank spectrum subtracted from the respective protein spectrum. SepF (2 μm) solution was prepared in the 25 mm Pipes buffer (pH 6.8) and incubated in the absence and presence of 8 m urea for 1 h at 25 °C. The ANS (100 μm) was added to the refolded and unfolded solutions of SepF and further incubated for 30 min at 25 °C. The ANS fluorescence spectra were recorded using 360 nm as an excitation wavelength, and the emission spectra were recorded over the range of 460-560 nm, with the excitation and emission bandwidths fixed at 10 nm each.
Light Scattering Signal—SepF, FtsZ, and FtsZΔC16 proteins were precleared by spinning at 280,000 × g for 10 min at 4 °C for all of the experiments. The effect of SepF on the assembly of full-length FtsZ and FtsZΔC16 were monitored by 90° light scattering signals using a JASCO 6500 spectrofluorometer. FtsZ (3.4 μm) or FtsZΔC16 (3.5 μm) was incubated without or with different concentrations (1, 2 and 3 μm) of SepF in 25 mm Pipes buffer, pH 6.8, for 30 min on ice. After incubation, 10 mm MgCl2 and 1 mm GTP were added to the reaction mixture. The reaction mixture was placed immediately into a quartz cuvette (Sterna Cells) for monitoring the light scattering signal at 37 °C for 10 min using 400 nm for excitation and emission spectra with slits width of 1 and 5 nm, respectively.
Sedimentation Assay—FtsZ (3.4 μm) or FtsZΔC16 (3.5 μm) was incubated without or with different concentrations (1, 2, 3, and 6 μm) of SepF in 25 mm Pipes buffer, pH 6.8, for 30 min on ice. Subsequently, 10 mm MgCl2 and 1 mm GTP were added to the reaction milieu and incubated for an additional 20 min at 37 °C. FtsZ polymers were sedimented at 200,000 × g for 25 min at 30 °C. The supernatant was decanted carefully without disturbing the pellet. The pellets were dissolved in 25 μl of SDS loading buffer. The protein concentration in the pellet was estimated from the Coomassie Blue staining of the 12% SDS-PAGE, and the intensity of FtsZ band on SDS-PAGE was measured using ImageJ software.
Effect of SepF on the Critical Concentration of FtsZ Polymerization—Different concentrations of FtsZ were incubated without or with 3 μm SepF in 25 mm Pipes buffer, pH 6.8, for 30 min on ice. After incubation, 10 mm MgCl2 and 1 mm GTP were added to the reaction mixtures and were polymerized at 37 °C for 10 min. FtsZ polymers were sedimented at 280,000 × g at 30 °C for 30 min. The amount of FtsZ in the pellets was quantified by the densitometric analysis of Coomassie Blue-stained 12% SDS-PAGE (22).
Transmission Electron Microscopic Analysis—FtsZ (3.4 μm) or FtsZΔC16 (3.5 μm) was polymerized without or with different concentrations (1, 3, and 6 μm) of SepF as described earlier (24). The polymers were negatively stained and examined by transmission electron microscopy. Briefly, 50 μl of the polymeric suspension was transferred onto carbon-coated copper grids (300 mesh) and blotted dry. The grids were subsequently negatively stained with a 50 μl of 2% uranyl acetate solution and air-dried. The samples were examined using 120v TECNAI G2 12 FEI TEM.
Stability Assay of FtsZ Polymers in the Presence of SepF—FtsZ (22.5 μm) was polymerized in 25 mm Pipes buffer, pH 6.8, containing 10 mm MgCl2 and 1 mm GTP at 37 °C for 20 min. The polymeric suspension was diluted 20-fold in warm 25 mm Pipes buffer, pH 6.8, containing 10 mm MgCl2 and 1 mm GTP in the absence or presence of different concentrations (1, 3, and 6 μm) of SepF and incubated for another 20 min at 37 °C. The polymers were collected by centrifugation at 280,000 × g for 30 min at 30 °C. The supernatant was decanted carefully, and the pellet was dissolved in SDS loading buffer. The samples were analyzed by Coomassie Blue-stained 12% SDS-PAGE. The FtsZ band intensity was analyzed using ImageJ software.
Effect of SepF on the GTPase Activity of FtsZ—FtsZ (3.4 μm) in the absence or presence of SepF (1 and 3 μm) were incubated in 25 mm Pipes buffer, pH 6.8, on ice for 30 min. Subsequently, 10 mm MgCl2 and 1 mm GTP were added to the reaction mixtures, and the kinetics of GTP hydrolysis was measured at 37 °C using the standard malachite green assay (24, 25). The initial rate of GTP hydrolysis of FtsZ assembly was calculated using the data points of the first 10 min.
Chemical Modification of SepF Protein by FITC Labeling—SepF has eight lysine residues, and a fluorescent probe can be introduced in the protein by covalent modification of one of the lysine residues. SepF (20 μm) was incubated with 150 μm FITC in 50 mm sodium phosphate buffer, pH 8, for 4 h on ice. The labeling reaction was quenched by adding 5 mm Tris-HCl, pH 8.0, on ice for 30 min and centrifuged at 50,000 × g for 10 min to remove any aggregates. The unbound FITC molecules were removed from FITC-labeled SepF (FITC-SepF) solution by dialyzing the reaction mixture against 50 mm phosphate buffer, pH 6.8, and followed by size exclusion chromatography using a pre-equilibrated (Bio-Gel® P-4 resin) column at 4 °C (26). The column was equilibrated with 50 mm phosphate buffer, pH 6.8, at 4 °C. The concentration of FITC-SepF was determined from the absorbance at 495 nm using a molar extinction coefficient of 77,000 m-1 cm-1. The concentration of SepF was determined by the method of Bradford. The incorporation ratio of FITC per mol of SepF was determined by dividing the bound FITC concentration by the SepF concentration.
Copolymerization of SepF with FtsZ Polymers—FtsZ (3.4 μm) was incubated without or with 3 and 6 μm SepF in 25 mm Pipes buffer, pH 6.8, for 30 min on ice and then polymerized at 37 °C for 10 min in the presence of 10 mm MgCl2 and 1 mm GTP. The protein sample (15 μl) was observed using a fluorescence microscope (Nikon ECLIPSE TE2000-U) with a 40× objective. The images were captured using a CoolSNAP-Pro camera and processed using ImageJ software.
Monitoring FtsZ-SepF Interactions by Size Exclusion Chromatography—FtsZ (6.8 μm) was incubated with SepF (6 μm) in buffer A (25 mm Pipes buffer, pH 6.8, and 250 mm NaCl) for 30 min on ice. The protein mixture was loaded onto a Sephadex G-100 column (1 × 50 cm) pre-equilibrated with buffer A. The proteins were eluted in 0.5-ml fractions at the flow rate of 0.5 ml/min maintained by peristaltic pump (Amersham Biosciences). The protein concentration of each fraction was measured using the Bradford method (21). The elution profiles of FtsZ and SepF proteins were also individually monitored using the same column. The elution profiles of the FtsZ-SepF complex, SepF and FtsZ were found to be different. The eluted proteins were analyzed on 12% SDS-PAGE by Coomassie Blue staining.
Effects of CTP17 on FtsZ Assembly—CTP17 (a 17-mer peptide corresponding to 366-382 amino acids of B. subtilis FtsZ) was purchased from USV-Custom (Mumbai, India), and the synthesis and the characterization of the peptide was described recently (22). The purity level of the peptide was found to be ≥95%. The peptide was dissolved in 25 mm Pipes buffer, pH 6.8. SepF (3 μm) was incubated without and with different concentrations (10-100 μm) of CTP17 in 25 mm Pipes buffer, pH 6.8, for 30 min on ice. FtsZ (3.4 μm) was added to each of the reaction mixtures and incubated for an additional 30 min on ice. Then 10 mm MgCl2 and 1 mm GTP were added to each of the reaction mixtures, and the assembly kinetics of FtsZ was monitored by light scattering at 37 °C.
RESULTS
Characterization of the Refolded SepF Protein—The B. subtilis SepF protein was refolded from the inclusion bodies overexpressed in E. coli. The conformational state of the purified SepF was monitored using Far-UV CD and spectrofluorometric analysis and compared with that of the denatured SepF.
An analysis of the CD spectra of purified SepF protein revealed a good agreement with those of folded proteins with distinct secondary structures. Deconvolution of the averaged data points (200-260 nm) using CDNN algorithm (27) indicated that SepF contains 44.0% helix, 12.5% β-sheet, 15.1% β-turn, and 28.7% random coil. In comparison, the CD spectrum of SepF in the presence of 8 m urea showed characteristics of denatured proteins. An analysis of the unfolded protein spectrum indicated a strong reduction in the α-helical content and an increase in the random coil content (data not shown).
We also monitored the folding status of SepF protein using an environment-sensitive hydrophobic fluorescence probe ANS. The emission spectra of ANS-SepF complex in the presence of 8 m urea showed a maximum at 520 nm, whereas the fluorescence emission spectra of ANS-SepF complex in the absence of urea showed a maximum at 505 nm (data not shown). In addition, the fluorescence intensity of ANS-SepF complex was found to be 12 ± 2% higher in the presence of 8 m urea than in the absence of urea. The results of CD and spectrofluorometric analysis together suggested that we were able to purify folded SepF.
Effects of SepF on the Assembly of FtsZ in Vitro—The effects of SepF on FtsZ polymerization were assessed by three complimentary techniques, namely 90° light scattering, sedimentation and transmission electron microscopy. SepF enhanced the light scattering signal of FtsZ polymerization in a concentration-dependent manner (Fig. 1A). For example the light scattering signal was increased by 27 ± 11, 53 ± 12, and 78 ± 6% in the presence of 1, 2, and 3 μm SepF, respectively, as compared with that of the control (without SepF). The SepF-mediated increase in the light scattering signal was not due to its oligomerization, because the SepF protein alone did not scatter light appreciably in the absence of FtsZ (data not shown).
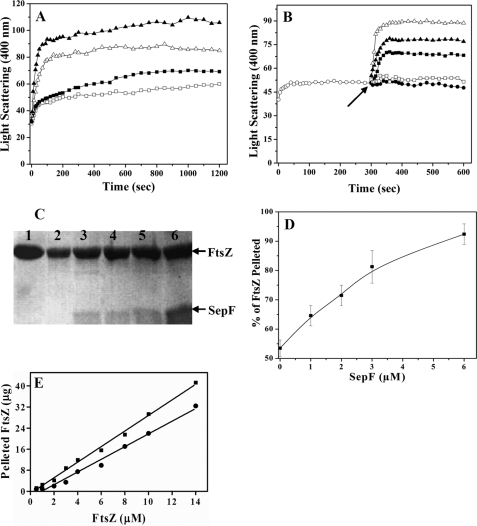
FIGURE 1.
SepF promoted the assembly of FtsZ. The effects of SepF on the assembly kinetics of FtsZ in 25 mm Pipes buffer, pH 6.8, containing 10 mm MgCl2 and 1 mm GTP were monitored by 90° light scattering (Panel A). FtsZ (3.4 μm) was polymerized in the absence (□) or presence of different concentrations of 1 μm (▪), 2 μm (▵), and 3 μm (▴) SepF (A). The experiment was performed three times. FtsZ (3.4 μm, ○) was polymerized in 25 mm Pipes buffer, pH 6.8, containing 10 mm MgCl2 and 1 mm GTP at 37 °C for 5 min (B). Then either different concentrations (1 μm, ▪;3 μm, ▴;6 μm, ▵) of SepF, (□) 6 μm BSA, or (•) 25 mm Pipes buffer, pH 6.8, were added to the cuvette, and the assembly of FtsZ was monitored for an additional 5 min. The arrow indicates the time of the addition of SepF and BSA to the assembly mixtures. The experiment was performed three times. SepF increased the sedimentable polymeric mass of FtsZ (C and D). FtsZ (3.4 μm) was polymerized in the absence and presence of different concentrations of SepF as described under “Experimental Procedures.” The polymers were collected by centrifugation, the pellets were dissolved, and the protein content in the pellets was estimated by Coomassie Blue staining of the 12% SDS-PAGE as described under “Experimental Procedures” (C). Lane 1 represents the total (polymeric plus soluble) FtsZ, and lanes 2-6 represent the polymeric FtsZ in the absence or presence of 1, 2, 3, and 6 μm SepF, respectively. The effects of SepF on the polymeric mass of FtsZ were estimated by densitometric analysis of the Coomassie Blue-stained SDS-PAGE (D). The data are the averages of five independent experiments. SepF reduced the critical concentration of FtsZ assembly (E). The symbols (• and ▪) represent the polymeric mass of FtsZ in the absence and presence of 3 μm SepF, respectively. The experiment was performed four times.
To further analyze the effects of SepF on FtsZ assembly, FtsZ was first polymerized for 5 min in the absence of SepF (Fig. 1B). After 5 min of polymerization, different concentrations of SepF were added into the cuvette, and the polymerization reaction was monitored for an additional 5 min. The addition of SepF to the assembly mixture significantly increased the light scattering signal, indicating that it promoted FtsZ assembly (Fig. 1B). The addition of BSA or Pipes buffer had no detectable effect on the assembly of FtsZ (Fig. 1B).
Under the conditions used for the assembly reaction, ∼53 ± 3% of the total FtsZ was sedimented in the absence of SepF. SepF increased the polymerized mass of FtsZ in a concentration-dependent manner (Fig. 1, C and D). For example, the polymerized mass of FtsZ was increased by 21 ± 5, 34 ± 10, 52 ± 11, and 72 ± 5% in the presence of 1, 2, 3, and 6 μm of SepF, respectively, as compared with the control (without SepF).
The critical concentrations of FtsZ assembly were determined to be 1.1 ± 0.12 and 0.4 ± 0.11 μm, respectively, in the absence and presence of 3 μm SepF, indicating that SepF promoted the assembly of FtsZ (Fig. 1E).
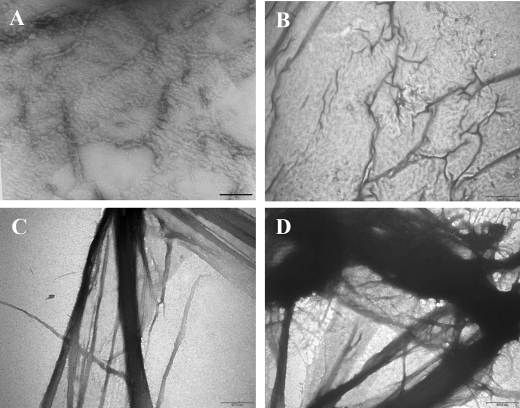
Thin and small protofilaments of FtsZ polymers were seen in the absence of SepF (Fig. 2A). SepF induced bundling of FtsZ protofilaments (Fig. 2, B-D). Thick bundles of FtsZ polymers were abundant in the presence of 3 and 6 μm SepF (Fig. 2, C and D). Formation of such thick and bulky polymer networks was reminiscent of the effects of agents known to cause bundling of FtsZ protofilaments, such as Ca2+, monosodium glutamate, ruthenium red (24, 28, 29), and proteins like ZapA, ZipA, and FtsA (13, 30, 31).
FIGURE 2.
Transmission electron microscopic analysis of the effects of SepF on FtsZ assembly. FtsZ (3.4 μm) was polymerized in the absence and presence of different concentrations of SepF at 37 °C for 20 min in 25 mm Pipes buffer, pH 6.8, containing 1 mm GTP and 10 mm MgCl2. The samples were prepared for transmission electron microscopy as described under “Experimental Procedures.” A, B, C, and D show FtsZ polymers in the absence or presence of 1, 3, and 6 μm of SepF, respectively. The scale bars show 1 μm.
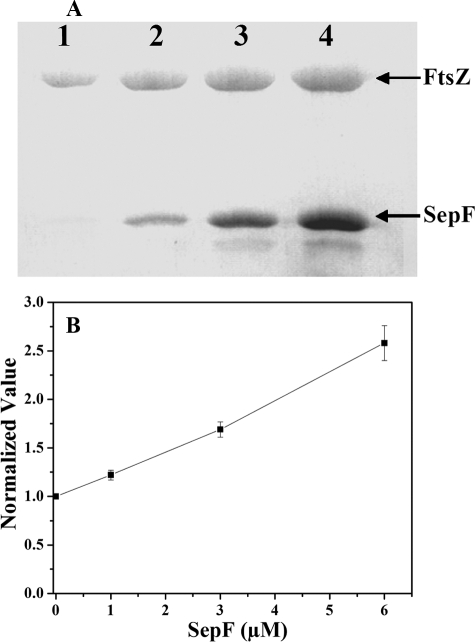
SepF Prevented Dilution-induced Disassembly of FtsZ Polymers—Preformed FtsZ polymers were diluted 20 times to a final concentration of 1.1 μm FtsZ into warm Pipes buffer containing 1 mm GTP, 10 mm MgCl2. These mixtures were incubated without or with different concentrations of SepF at 37 °C for an additional 20 min. The polymers were then collected by sedimentation. The amount of FtsZ pelleted was found to increase with increasing concentration of SepF (Fig. 3A). For example, the amount of FtsZ pelleted was increased by 22 ± 1, 70 ± 8, and 158 ± 19% in the presence of 1, 3, and 6 μm SepF, respectively, as compared with that of the control (Fig. 3B). A similar increase in the pelleted FtsZ was also observed when the preformed FtsZ polymers were diluted to a final concentration of 0.4 μm FtsZ in the presence of SepF, suggesting that SepF prevented the disassembly of the FtsZ protofilaments.
FIGURE 3.
SepF prevented dilution-induced disassembly of FtsZ polymers. Preformed FtsZ polymers were diluted 20 times in warm 25 mm Pipes buffer, pH 6.8, containing 1 mm GTP, 10 mm MgCl2 without or with different concentrations of SepF. The reaction mixtures were incubated for an additional 20 min at 37 °C (A). The samples were centrifuged, dissolved, and adjudged on a Coomassie Blue-stained 12% SDS-PAGE. The FtsZ band intensities were determined by ImageJ software, and the normalized value of the FtsZ band intensity was plotted against SepF concentrations as shown in B. The experiment was performed three times.
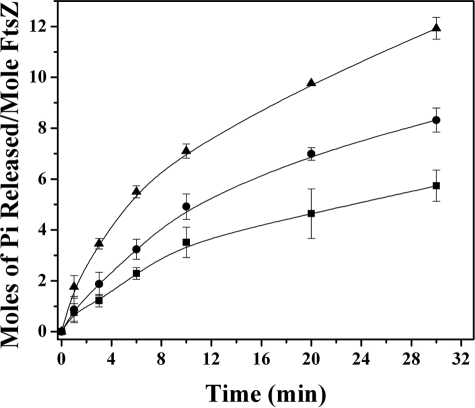
SepF Reduced the GTPase Activity of FtsZ—SepF suppressed the initial rate of GTP hydrolysis of FtsZ from the value of 0.7 ± 0.04 min-1 in the absence of SepF to 0.5 ± 0.03 and 0.34 ± 0.06 min-1 in the presence of 1 and 3 μm SepF, respectively. Concomitantly, a significant reduction in the extent of GTPase activity of FtsZ was also observed in the presence of SepF (Fig. 4). For example, 1 and 3 μm SepF reduced the extent of GTP hydrolysis by 30 ± 7 and 51 ± 6%, respectively, as compared with the control after 30 min of hydrolysis.
FIGURE 4.
Effects of SepF on the GTPase activity of FtsZ. FtsZ (3.4 μm) was polymerized in 25 mm Pipes buffer, pH 6.8, containing 1 mm GTP and 10 mm MgCl2 in the absence (▴) and presence of 1 μm (•) and 3 μm (▪) SepF. The experiment was performed three times.
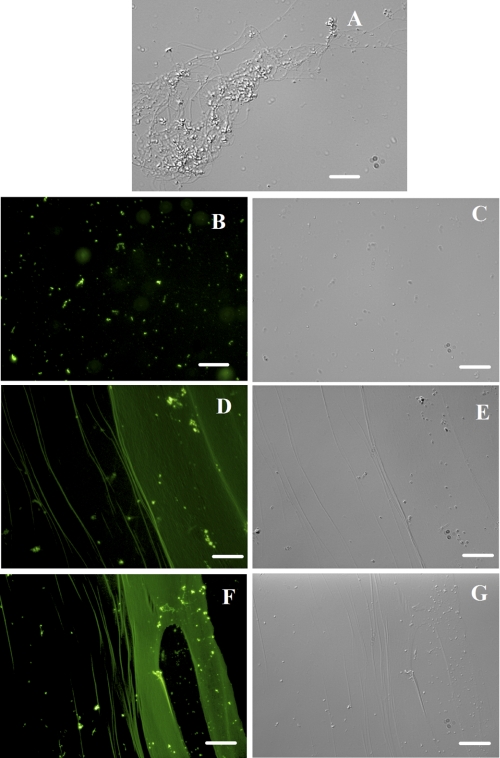
SepF Copolymerized with FtsZ—The stoichiometry of incorporation of FITC per SepF molecule was determined to be 0.6 ± 0.14. FtsZ was polymerized in the absence and presence of different concentrations of FITC-SepF to examine whether SepF could copolymerize with FtsZ. The polymer bound FITC-SepF was visualized by fluorescence microscopy. FITC-SepF was clearly visualized on FtsZ protofilaments (Fig. 5). Although individual protofilaments were visible at low concentration of FITC-SepF (Fig. 5, D and E), thick bundles of FtsZ were seen at high concentrations of FITC-SepF (Fig. 5, F and G). These results are in agreement with the light scattering and electron microscopic analysis presented here, emphasizing further that SepF induced FtsZ bundling. The uniform recruitment of FITC-SepF in FtsZ bundles also provided clues that SepF might form stable complexes with FtsZ.
FIGURE 5.
Copolymerization of FITC-SepF with FtsZ. FtsZ (3.4 μm) was polymerized in the presence of different concentrations of FITC-SepF. FtsZ polymers and FITC-SepF was observed under differential interference contrast microscopy shown in A (3.4 μm FtsZ alone), C (6 μm FITC-SepF alone), E (3.4 μm FtsZ plus 3 μm FITC-SepF), and G (3.4 μm FtsZ plus 6 μm FITC-SepF). These were also seen under fluorescence microscopy as in B (6 μm FITC-SepF alone), D (3.4 μm FtsZ plus 3 μm FITC-SepF), and F (3.4 μm FtsZ plus 6 μm FITC-SepF). The scale bars show 20 μm.
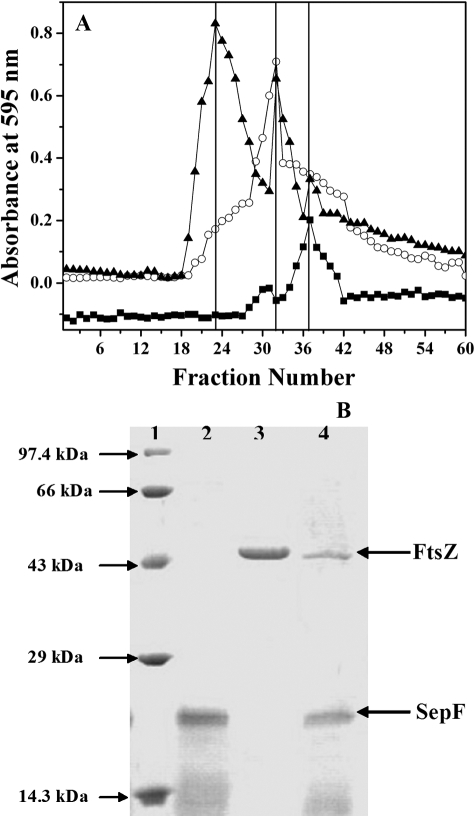
SepF Formed a Stable Complex with FtsZ—The interaction between SepF and FtsZ was examined by size exclusion column chromatography. Distinct elution profiles of FtsZ, SepF, and FtsZ-SepF complex were observed, and they were eluted at fractions 38, 32, and 24, respectively (Fig. 6). The elution profiles of the FtsZ and SepF mixture also showed the presence of uncomplexed FtsZ and SepF, and their elution peaks matched with the distinguishing elution peaks of the individual proteins. The results suggested that SepF directly interacted with FtsZ and formed a stable complex with FtsZ in vitro.
FIGURE 6.
SepF directly interacts with FtsZ. FtsZ (6.8 μm) was incubated with SepF (6 μm) for 30 min on ice and passed through a gel filtration (Sephadex G-100) column (as described under “Experimental Procedures”). A shows the elution profiles of FtsZ (○), SepF (▪), and FtsZ in the presence of SepF (▴). B shows the Coomassie Blue staining of the 12% SDS-PAGE of the Marker (lane 1), SepF alone (lane 2), FtsZ alone (lane 3), and FtsZ-SepF complex (lane 4). One of two similar experiments is shown.
Effects of SepF on the Assembly of C-terminal Truncated FtsZ (FtsZΔC16)—Under the conditions used for the assembly, FtsZΔC16 was found to assemble more efficiently than the full-length FtsZ (Figs. 1 and 7). For example, ∼72 ± 3% of the FtsZΔC16 proteins were pelleted as polymers, whereas ∼53 ± 3% of FtsZ proteins were pelleted as polymers in the absence of SepF. In contrast to the strong effects of SepF on the light scattering signal of the assembly of the full-length FtsZ, SepF was found to exert modest effects on the light scattering signal of the assembly of FtsZΔC16, indicating that it does not induce strong bundling of the FtsZΔC16 polymers (Fig. 7A). SepF also displayed a modest effect on the polymerized mass of FtsZΔC16 (Fig. 7, B and C).
FIGURE 7.
Effects of SepF on the assembly of FtsZΔC 16. The kinetics of FtsZΔC 16 (3.5 μm) assembly in the absence (□) or presence 1 μm (▪), 2 μm (▵), and 3 μm (▴) of SepF are shown in A. FtsZΔC16 (3.5 μm) was polymerized in the absence and presence of different concentrations of SepF, and the polymers were collected by centrifugation. The protein content in the pellets was estimated by Coomassie Blue staining of the 12% SDS-PAGE as described under “Experimental Procedures” (Panel B). Lane 1 represents the total FtsZΔC16, and lanes 2, 3, 4, 5, and 6 represent the polymeric FtsZΔC16 in the absence or presence of 1, 2, 3, and 6 μm SepF, respectively. The effects of SepF on the polymeric mass of FtsZΔC16 were estimated by densitometric analysis of the Coomassie Blue-stained SDS-PAGE (C). The data are the averages of three experiments. D shows the transmission electron microscopic analysis of FtsZΔC16 (3.5 μm) polymers in the absence and presence of different concentrations of SepF. The scale bars show 1 μm.
In the absence of SepF, FtsZΔC16 produced thin filaments as well as small bundles (Fig. 7D). SepF did not induce a large change in the size of FtsZΔC16 bundles (Fig. 7D), whereas it strongly induced bundling of FtsZ prototofilaments (Fig. 2). In the presence of 3 and 6 μm SepF, the width of the FtsZΔC16 bundles was found to be much smaller as compared with that of the full-length FtsZ (Figs. 2 and 7D). The modest effect of SepF on the assembly of FtsZΔC16 could be due to the loss of its primary binding site on FtsZ because of the deletion of 16 C-terminal tail residues from the full-length FtsZ. Alternatively, the efficient assembly of FtsZΔC16 could partly mask some of the effects of SepF on the assembly of FtsZΔC16.
To examine whether SepF exerts its assembly promoting effects primarily by binding at the C-terminal tail of FtsZ, we used a 17-mer C-terminal tail (366-382) FtsZ peptide (22) having the amino acid sequence DDTLDIPTFLRNRNKRG with the premise that the synthetic peptide should be able to compete with FtsZ for its binding to SepF. CTP17 had no detectable effect on the assembly kinetics of FtsZ (22). The peptide inhibited the assembly promoting activity of SepF in a concentration-dependent manner (Fig. 8). For example, SepF (3 μm) increased the light scattering signal of FtsZ assembly by 73 ± 6, 30 ± 6, and 14 ± 4% in the absence and presence of 50 and 100 μm CTP17, respectively. The results suggested that the peptide inhibited the binding of SepF to FtsZ, and the C-terminal tail of FtsZ is the primary site of interaction with SepF.
FIGURE 8.
CTP17 inhibited the assembly promoting effect of SepF. SepF (3 μm) was incubated in the absence and presence of different concentrations (10-100 μm) of CTP17 in 25 mm Pipes buffer, pH 6.8, for 30 min on ice. Then FtsZ (3.4 μm) was added to the reaction mixtures and incubated for an additional 30 min on ice. The assembly reaction was started by adding 10 mm MgCl2 and 1 mm GTP and transferring the mixtures to 37 °C for 10 min. The panel shows the kinetics of FtsZ (3.4 μm) assembly in the absence (▪) or presence of 3 μm SepF without (▪) or with 10 μm (▵), 25 μm (•), 50 μm (○), 75 μm (♦), and 100 μm CTP17 (⋄), respectively. The plot (□) shows FtsZ (3.4 μm) assembly in the presence of 100 μm CTP17 alone. The experiment was performed three times.
DISCUSSION
The assembly/disassembly kinetics of the cytokinetic Z-ring in B. subtilis is considered to be regulated by the action of several accessory proteins such as FtsA, ZapA, SepF, MinCD/DivIV, and EzrA (3-6, 18, 32). An alteration in the expression profiles of these regulatory proteins generally impairs bacterial cell division. We present here the first biochemical characterization of B. subtilis SepF and its effects on the assembly of FtsZ in vitro. SepF formed a stable complex with FtsZ in vitro, suggesting that SepF interacts directly with FtsZ. The observed increases in the light scattering signal and the amount of polymeric FtsZ were likely to be due to the bundling of FtsZ protofilaments in the presence of SepF. Bundles of FtsZ protofilaments were seen at low molar ratios of SepF to FtsZ, indicating that SepF stabilizes FtsZ protofilaments. FITC-SepF was found to be incorporated along the length of the FtsZ protofilaments, suggesting that SepF copolymerizes with FtsZ.
How does SepF induce bundling of FtsZ? SepF is known to exist as dimer (18). The data presented here indicate that SepF dimer mediates FtsZ bundling by stabilizing lateral interactions between FtsZ monomers in the protofilaments, like that observed for ZapA (33). This proposal is consistent with the in vivo observations that SepF is directly recruited early on the Z-ring, and its subcellular localization primarily occurs in a FtsZ-dependent manner (3, 18).
The dynamicity of the Z-ring and the GTPase activity of FtsZ are considered to be closely linked (34, 35). On the basis of correlation between slow subunit turnover, GTPase, and GDP-induced disassembly, a polymer recycling model of FtsZ assembly has been advocated (35). According to this model, GTP is hydrolyzed following FtsZ polymerization, and the bound GDP induces disassembly facilitating polymer recycling and dynamics. In the present work, the stabilization of FtsZ protofilaments in the presence of SepF was found to occur in parallel with a reduction in the GTPase activity of FtsZ, indicating that SepF might reduce the GTPase activity of FtsZ by stabilizing the protofilaments.
At low concentration (3.5 μm) FtsZΔC16 assembled more efficiently than the full-length FtsZ (Figs. 1 and 7). However, at high concentration (14.4 μm), FtsZ also polymerized extensively, and the extents of assembly of FtsZ and FtsZΔC16 were not significantly different (22). In the present work, SepF caused a modest increase in the assembly and bundling of FtsZΔC16. Further, a 17-mer C-terminal peptide (366-382) of B. subtilis FtsZ countered the effect of SepF on FtsZ polymerization. The results indicated that the 16 residues at the C-terminal tail of FtsZ may be the primary site of interactions between SepF and FtsZ proteins. This is in-line with the suggestion that SepF may interact with the C-terminal tail residues of FtsZ (18).
SepF has been shown to complement for the depletion of a positive-acting factor, FtsA (18). FtsA has been suggested to bind to the conserved C-terminal tail of FtsZ (36), and most likely, it plays a role in anchoring the Z-ring to the cell wall as well as in the recruitment of downstream cell division proteins (17, 37). The bioinformatic analysis of SepF amino acid sequence did not reveal the presence of a transmembrane helix. However, the presence of an in-plane membrane anchor, spanning N-terminal amino acid residues (KDKLKN7), was predicted in SepF sequence by AmphipaSeek program (38) Analogous to SepF, another cytosolic protein, ZapA, has been shown to promote the assembly and bundling of FtsZ and to colocalize with the Z-ring in vivo (12). The functional overlap of SepF with FtsA and ZapA suggests that SepF can act during the early events of cell division because both ZapA and FtsA support the assembly of the Z-ring during the early stages of the cell division.
It has also been proposed that SepF acts as a late division protein and is required for the proper execution of septum synthesis (3). The disruption of SepF resulted in partial impairment of cell division in B. subtilis (18). However, cells division arrest was observed when the expression of SepF was repressed in the absence of EzrA, though the Z-ring still assembled (3). The synergistic effect of ezrA and sepF double mutants can be rationalized with the present in vitro results in view of the Z-ring assembly dynamics, which is thought to be critical for the constriction of Z-ring and for cytokinesis (39).
In contrast to the assembly promoting effects of SepF, EzrA inhibits the assembly and bundling of FtsZ protofilaments, increases critical concentration of FtsZ assembly, and depolymerizes preformed FtsZ polymers in vitro (22). SepF and EzrA thus show antagonistic effects on the assembly and stability of the FtsZ polymer in vitro. However, similar to SepF, EzrA also concentrates at the cytokinetic ring in an FtsZ-dependent manner (7) and binds to the C-terminal tail of FtsZ (22). It is possible that a fine balance in the activities of SepF and EzrA is critical for the assembly dynamics of the Z-ring and thus for the divisome constriction in Gram-positive bacteria. This is in agreement with our earlier suggestion that the assembly dynamics of FtsZ in the Z-ring is regulated by the competition between positive and negative regulators sharing the same binding site on FtsZ (22).
In conclusion, we suggest that SepF may be a multifunctional protein playing a role in the Z-ring formation during the early events as well as during the late stages of the cell division by regulating the assembly dynamics of the Z-ring. The findings that SepF promotes FtsZ assembly, copolymerizes with FtsZ, and stabilizes the preformed FtsZ polymers indicate that SepF may act antagonistically to a negative-acting factor, EzrA, and that it can act as a functional homolog of positive regulatory factors such as ZapA and FtsA.
Acknowledgments
We thank Sophisticated Analytical Instrument Facility, Indian Institute of Technology Bombay for providing the transmission electron microscopy facility. We sincerely thank Dr. R. Mukhopadhyaya (Bhabha Atomic Research Centre) for helping with DNA sequencing.
This work is supported by a grant from the Dept. of Science and Technology, Government of India (to D. P.) and a Council of Scientific and Industrial Research doctoral fellowship (to J. K. S.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: Pipes, piperazine-N,N′-bis(2-ethanesulfonic acid); BSA, bovine serum albumin; FITC, fluorescein isothiocyanate; ANS, 1-anilinonaphthalene-8-sulfonic acid.
References
- 1.Edwards, D. H., and Errington, J. (1997) Mol. Microbiol. 24 905-915 [DOI] [PubMed] [Google Scholar]
- 2.Jones, L. J., Carballido-López, R., and Errington, J. (2001) Cell 104 913-922 [DOI] [PubMed] [Google Scholar]
- 3.Hamoen, L. W., Meile, J. C., de Jong, W., Noirot, P., and Errington, J. (2006) Mol. Microbiol. 59 989-999 [DOI] [PubMed] [Google Scholar]
- 4.Romberg, L., and Levin, P. A. (2003) Annu. Rev. Microbiol. 57 125-154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rothfield, L., Taghbalout, A., and Shih, Y. L. (2005) Nat. Rev. Microbiol. 3 959-968 [DOI] [PubMed] [Google Scholar]
- 6.Raskin, D. M., and de Boer, P. A. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 4971-4976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levin, P. A., Kurtser, I. G., and Grossman, A. D. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 9642-9647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Justice, S. S., García-Lara, J., and Rothfield, L. I. (2000) Mol. Microbiol. 2 410-423 [DOI] [PubMed] [Google Scholar]
- 9.Woldringh, C. L., Mulder, E., Valkenburg, J. A., Wientjes, F. B., Zaritsky, A., and Nanninga, N. (1990) Res. Microbiol. 141 39-49 [DOI] [PubMed] [Google Scholar]
- 10.Weart, R. B., Nakano, S., Lane, B. E., Zuber, P., and Levin, P. A. (2005) Mol. Microbiol. 57 238-249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee, S., and Price, C. W. (1993) Mol. Microbiol. 7 601-610 [DOI] [PubMed] [Google Scholar]
- 12.Gueiros-Filho, F. J., and Losick, R. (2002) Genes Dev. 16 2544-2556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geissler, B., Elraheb, D., and Margolin, W. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 4197-4202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pichoff, S., and Lutkenhaus, J. (2002) EMBO J. 21 685-693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feucht, A., Lucet, I., Yudkin, M. D., and Errington, J. (2001) Mol. Microbiol. 40 115-125 [DOI] [PubMed] [Google Scholar]
- 16.Wang, X., Huang, J., Mukherjee, A., Cao, C., and Lutkenhaus, J. (1997) J. Bacteriol. 179 5551-5559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jensen, S. O., Thompson, L. S., and Harry, E. J. (2005) J. Bacteriol. 187 6536-6544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ishikawa, S., Kawai, Y., Hiramatsu, K., Kuwano, M., and Ogasawara, N. (2006) Mol. Microbiol. 60 1364-1380 [DOI] [PubMed] [Google Scholar]
- 19.Fadda, D., Pischedda, C., Caldara, F., Whalen, M. B., Anderluzzi, D., Domenici, E., and Massidda, O. (2003) J. Bacteriol. 185 6209-6214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arakawa, T., Ejima, D., Tsumoto, K., Obeyama, N., Tanaka, Y., Kita, Y., and Timasheff, S. N. (2007) Biophys. Chem. 127 1-8 [DOI] [PubMed] [Google Scholar]
- 21.Bradford, M. M. (1976) Anal. Biochem. 72 248-254 [DOI] [PubMed] [Google Scholar]
- 22.Singh, J. K., Makde, R. D., Kumar, V., and Panda, D. (2007) Biochemistry 46 11013-11022 [DOI] [PubMed] [Google Scholar]
- 23.Lu, C., Stricker, J., and Erickson, H. P. (1998) Cell. Motil. Cytoskeleton 40 71-86 [DOI] [PubMed] [Google Scholar]
- 24.Beuria, T. K., Krishnakumar, S. S., Sahar, S., Singh, N., Gupta, K., Meshram, M., and Panda, D. (2003) J. Biol. Chem. 278 3735-3741 [DOI] [PubMed] [Google Scholar]
- 25.Geladopoulos, T. P., Sotiroudis, T. G., and Evangelopoulos, A. E. (1991) Anal. Biochem. 192 112-116 [DOI] [PubMed] [Google Scholar]
- 26.Santra, M. K., and Panda, D. (2003) J. Biol. Chem. 278 21336-21343 [DOI] [PubMed] [Google Scholar]
- 27.Bohm, G., Muhr, R., and Jaenicke, R. (1992) Protein Eng. 5 191-195 [DOI] [PubMed] [Google Scholar]
- 28.Yu, X. C., and Margolin, W. (1997) EMBO J. 16 5455-5463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Santra, M. K., Beuria, T. K., Banerjee, A., and Panda, D. (2004) J. Biol. Chem. 279 25959-25965 [DOI] [PubMed] [Google Scholar]
- 30.Hale, C. A., and de Boer, P. A. (1997) Cell 88 175-185 [DOI] [PubMed] [Google Scholar]
- 31.Margolin, W. (2003) Curr. Biol. 13 R16-R18 [DOI] [PubMed] [Google Scholar]
- 32.Michie, K. A., and Löwe, J. (2006) Annu. Rev. Biochem. 75 467-492 [DOI] [PubMed] [Google Scholar]
- 33.Low, H. H., Moncrieffe, M. C., and Löwe, J. (2004) J. Mol. Biol. 341 839-852 [DOI] [PubMed] [Google Scholar]
- 34.Scheffers, D. J., and Driessen, A. J. (2002) Mol. Microbiol. 43 1517-1521 [DOI] [PubMed] [Google Scholar]
- 35.Huecas, S., Schaffner-Barbero, C., García, W., Yébenes, H., Palacios, J. M., Díaz, J. F., Menéndez, M., and Andreu, J. M. (2007) J. Biol. Chem. 282 37515-37528 [DOI] [PubMed] [Google Scholar]
- 36.Ma, X., and Margolin, W. (1999) J. Bacteriol. 181 7531-7544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pichoff, S., and Lutkenhaus, J. (2005) Mol. Microbiol. 55 1722-1734 [DOI] [PubMed] [Google Scholar]
- 38.Sapay, N., Guermeur, Y., and Deléage, G. (2006) BMC Bioinformatics 7 255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Osawa, M., Anderson, D. E., and Erickson, H. P. (2008) Science 320 792-794 [DOI] [PMC free article] [PubMed] [Google Scholar]