Abstract
We have previously proposed that Euglena gracilis possesses a pathway for the production of ascorbate (AsA) through d-galacturonate/l-galactonate as representative intermediates (Shigeoka, S., Nakano, Y., and Kitaoka, S. (1979) J. Nutr. Sci. Vitaminol. 25 ,299 -307). However, genetic evidence proving that the pathway exists has not been obtained yet. We report here the identification of a gene encoding aldonolactonase, which catalyzes a penultimate step of the biosynthesis of AsA in Euglena. By a BLAST search, we identified one candidate for the enzyme having significant sequence identity with rat gluconolactonase, a key enzyme for the production of AsA via d-glucuronate in animals. The purified recombinant aldonolactonase expressed in Escherichia coli catalyzed the reversible reaction of l-galactonate and l-galactono-1,4-lactone with zinc ion as a cofactor. The apparent Km values for l-galactonate and l-galactono-1,4-lactone were 1.55 ± 0.3 and 1.67 ± 0.39 mm, respectively. The cell growth of Euglena was arrested by silencing the expression of aldonolactonase through RNA interference and then restored to the normal state by supplementation with l-galactono-1,4-lactone. Euglena cells accumulated more AsA on supplementation with d-galacturonate than d-glucuronate. The present results indicate that aldonolactonase is significant for the biosynthesis of AsA in Euglena cells, which predominantly utilize the pathwayviad-galacturonate/l-galactonate. The identification of aldonolactonase provides the first insight into the biosynthesis of AsA via uronic acids as the intermediate in photosynthetic algae including Euglena.
Vitamin C (l-ascorbic acid (AsA)2) represents a group of essential nutrients for humans and is a hydrophilic antioxidant synthesized by all animals except primates and photosynthetic organisms, including plants and algae (2). AsA has a pivotal role both as an antioxidant and as an enzyme cofactor in animals. In plants, AsA has been documented to play multiple roles in the control of photosynthesis, cell expansion and growth, and transmembrane electron transport in addition to its well characterized role as an antioxidant (3, 4).
With regard to the biosynthesis of AsA in living organisms, the major pathways differ between animals and plants. In animals, the committed step in the biosynthesis utilizes d-glucuronate (d-GlcUA), l-gulonate (l-GulA), and l-gulono-1,4-lactone (l-GulL) as a direct precursor of AsA (5). This pathway is a branch of the d-GlcUA pathway via the pentose phosphate pathway. A microsome-localized enzyme, l-GulL oxidase, catalyzes the terminal step to produce AsA. Genetic evidence that l-GulL is synthesized directly from l-GulA, not mediated by d-glucuronolactone, has been provided by the identification of SMP30 in rats as gluconolactonase (6).
In contrast to animals, plants utilize l-galactono-1,4-lactone (l-GalL) as the terminal precursor for the biosynthesis of AsA, which is produced from l-galactose via GDP-d-mannose and GDP-l-galactose as the intermediates (Man/Gal pathway) (4, 7, 8). The l-GalL is finally oxidized by the action of l-GalL dehydrogenase located in the mitochondrial inner membrane, resulting in the production of AsA (9, 10). Recently, an overall picture of the pathway was completed with the identification and analysis of a causal gene in low-AsA vtc mutants in Arabidopsis thaliana (11, 12). Genetic evidence indicates that the Man/Gal pathway is of critical importance. A null Arabidopsis mutant of the CYT1 gene, which encodes the same protein as VTC1 (GDP-Man pyrophosphorylase, an enzyme required for production of GDP-Man from Man 1-P), showed lethality in seedlings as well as caused reduced AsA levels (13). Dowdle et al. (14) also reported that VTC2 encodes a GDP-l-Gal phosphorylase enzyme that breaks down GDP-l-Gal phosphorolytically, producing l-Gal 1-P, and the knock-out mutant of two VTC2 genes shows growth arrest immediately upon germination and the cotyledons subsequently bleach. These findings indicated that the Man/Gal pathway is the only physiologically significant source of AsA in Arabidopsis plants.
Although the Man/Gal pathway appears to be the predominant pathway, several other possible biosynthetic pathways have been proposed via uronic acid intermediates (15, 16). The isolation of an aldo-keto reductase specific for d-galacturonate (d-GalUA) from ripening strawberry fruits and the generation of transgenic plants overexpressing its gene in Arabidopsis indicated the contribution of the alternative pathway via d-GalUA to the production of AsA (15). In the pathway, although the conversion of l-galactonate (l-GalA), the product derived form the reduction of d-GalUA, to l-GalL is indispensable, aldonolactonase (ALase), a key catalyzing enzyme, is still missing. In addition to the pathway via d-GalUA, other possible pathways via d-GlcUA or l-gulose are also proposed, although most of the enzymes related to these pathways have not been identified yet. Therefore, it is still unclear how these pathways contribute to the biosynthesis of AsA in planta.
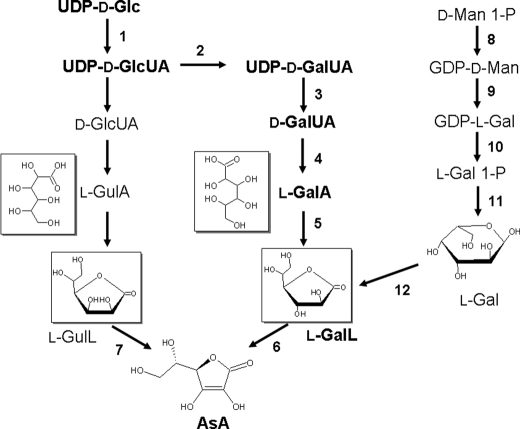
In contrast to animals and plants, although some eukaryotic microorganisms also contain significant concentrations of cellular AsA, only scarce data are available on the way of biosynthesis. Chlorella and Prototheca contained appreciable amounts of AsA, and adding Man, l-GalA and l-GalL to the Prototheca medium resulted in significantly increased levels of AsA, suggesting the presence of a Man/Gal pathway similar to that in plants (17, 18). It was recently identified that a protozoan parasite, Trypanosoma brucei, possesses a functional gene encoding d-arabinono-1,4-lactone oxidase that can also oxidize l-GalL, although the exact pathway arriving at the final precursor has not been clarified yet (19). In Euglena gracilis containing high concentrations of AsA, the biosynthetic pathway was first proposed by Shigeoka et al. (1). Radioisotope tracer studies revealed that the biosynthesis of AsA in Euglena proceeded via the conversion of UDP-d-GlcUA to UDP-d-GalUA, with production of d-GalUA and reduction to l-GalA (Fig. 1). l-GalL was the final precursor in the production of AsA. These findings indicate that Euglena has developed a pathway analogous to the alternative pathway of AsA biosynthesis via d-GalUA, which is unclear in plants. Detection of l-GalL dehydrogenase activity in the Euglena cells (20) and characterization of purified d-GalUA reductase (21) strongly supported the contribution of the pathway via d-GalUA/l-GalA in promoting AsA biosynthesis in this alga.
FIGURE 1.
The pathway of AsA biosynthesis in Euglena and plants. A radio-tracer experiment provides evidence for the biosynthesis of AsA via uronic acids in Euglena. The unnumbered reactions are uncertain; however, enzyme 4 catalyzes the conversion of d-GlcUA to l-GulA (21). The pathway from d-Man 1-P to l-GalL (Man/Gal pathway, reactions 8-12) mainly operates in higher plants. Enzymes catalyzing the numbered reactions are: 1, UDP-Glc dehydrogenase; 2, UDP-Glc-4-epimerase; 3, UDP- d-GalUA pyrophosphatase/phosphorylase; 4, d-GalUA reductase; 5, ALase; 6, l-GalL dehydrogenase; 7, l-GulL oxidase/dehydrogenase; 8, GDP-d-Man pyrophosphorylase (VTC1); 9, GDP-Man-3′,5′-epimerase; 10, GDP-l-Gal phosphorylase (VTC2/VTC5); 11, l-Gal 1-P phosphatase (VTC4); 12, l-Gal dehydrogenase.
In this study, to understand the biosynthesis of AsA in Euglena in detail, we have identified and characterized ALase, catalyzing the penultimate step from l-GalA to l-GalL. We report that Euglena cells silenced of the expression of ALase by RNAi are unable to grow unless supplemented with l-GalL. Based on the present findings, we discuss the significance of AsA biosynthesis via d-GalUA/l-GalA and the physiological role of AsA in this organism.
EXPERIMENTAL PROCEDURES
Materials—l-GalL, l-GulL, and d-GulL were purchased from Sigma-Aldrich. d-GalL and d-glucono-δ-lactone were purchased from Wako Pure Chemical (Osaka, Japan). l-GalA and l-GulA were prepared by the hydrolysis of l-GalL and l-GulL, respectively, under basic conditions (21). Twenty milliliters of 0.3 m NaOH was added to 100 μl of 10 mm l-GalL or l-GulL, the mixture was vigorously agitated by vortexing for 20 s, and 20 μl of 0.3 m hydrochloric acid was added to neutralize the solution.
Strain and Culture—E. gracilis strain Z was grown in Koren-Hutner medium under continuous light at a photosynthetic photon flux density of 24 μmol m-2 s-1 at 26 °C for 6 days, by which time the stationary phase was reached (22).
Enzyme Assay—The activity of lactonase was assayed based on the change in absorbance of a p-nitrophenol pH indicator using a modified version of a previously described method (23). The reaction mixture contained 10 mm PIPES buffer, pH 6.5, 250 μm p-nitrophenol, 5 mm d-glucono-δ-lactone, 75 μm ZnCl2, and an enzyme in a total volume of 1 ml. Decolorization of p-nitrophenol by acidification was monitored at 405 nm and quantified by titration with known amounts of HCl.
Cloning of ALase cDNA and Expression of Recombinant Protein in Escherichia coli—Based on the Euglena EST data base from the Protest EST Program, a gene-specific primer was designed to amplify the missing 3′ end of the ALase EST (ELL00003818). The primer was 5′-TGACTGCCAGTCTGGGGACC-3′ and was used sequentially for 3′-rapid amplification of cDNA ends (3′-RACE) with a 5′/3′-RACE kit (Roche Applied Science). The complete sequence has been deposited with the DDBJ data bank under accession number AB306917. Then the full-length coding sequence of Euglena ALase was amplified by PCR with rALase-F (5′-CTCGAGATGCGAACGCTGGCCACCGTC-3′) and rALase-R (5′-AAGCTTACACGAGTTCAGCACAGCC-3′). The upstream and downstream primers contained XhoI and HindIII restriction sites, respectively. PCR amplification was carried out using Blend Taq (Toyobo, Osaka, Japan). The template was from a single-strand cDNA pool that was made from 6-day-old Euglena cells. The amplified product was cloned in the vector pGEM-T (Promega, Madison, MI), and the sequence was verified by DNA sequencing. The XhoI/HindIII fragment was extracted and subcloned into the corresponding restriction site of the expression vector pCold TF (Takara, Shiga, Japan). E. coli cells (BL21-CodonPlus (DE3)-RIL, Stratagene) were transformed with the resulting constructs. The cells were grown in LB medium supplemented with 34 μgml-1 chloramphenicol and 50 μgml-1 ampicillin at 37 °C. When the culture reached an absorbance at 0.4-0.5 at 600 nm, 1 mm isopropyl β-d-thiogalactopyranoside was added, and the cells were grown further at 15 °C for 20 h.
Site-directed Mutagenesis—The Euglena ALase mutants, N116A, N161A, D219A, and S262A, were constructed using ALase cDNA as a template with a two-step PCR by KOD-Plus polymerase (Toyobo). The oligonucleotides used were: N116A-F 5′-GAAAGGTTCATACCGGAGCGCAGATGGAAAATGTG-3′, N116A-R 5′-CACATTTTCCATCTGCGCTCCGGTATGAACCTTTC-3′, N161A-F 5′-GTGTCACCATCGGAGCCGGTCTTGCCTGGAG-3′, N161A-R 5′-CTCCAGGCAAGACCGGCTCCGATGGTGACAC-3′, D219A-F 5′-GAGGCGGCCCCCGCTGGGATGACCATCGAC-3′, D219A-R 5′-GTCGATGGTCATCCCAGCGGGGGCCGCCTC-3′, S262A-F 5′-GCGAAGTACACGACCGCCGTGGCGCTCGGG-3′, and S262A-R 5′-CCCGAGCGCCACGGCGGTCGTGTACTTCGC-3′. Changed nucleotides are underlined. Successful mutagenesis was confirmed by DNA sequencing. The resultant mutants were subcloned into the pCold TF expression vector (Takara) and then introduced into E. coli cells as described above.
SDS-PAGE—SDS-PAGE was performed in a 12.5% polyacrylamide slab gel using a Tris/Gly buffer system. The gels were stained with Coomassie Blue G-250.
Purification of Recombinant Euglena ALase—All the procedures for the recombinant proteins were performed at 4 °C. E. coli cells were resuspended in 50 mm potassium phosphate buffer, pH 7.0, containing 0.3 m NaCl and 10 mm imidazole and lysed by sonication. The His-tagged recombinant ALase was purified on a column packed with TALON Metal Affinity resin (Clontech). The trigger factor with a His tag was removed from recombinant ALase by a 20-h digestion with HRV 3C protease (Takara) at 4 °C. To obtain the truncated recombinant ALase, the reaction mixture was applied to the TALON Metal Affinity resin column, and the flow-through protein was collected. The active fraction was stored at -20 °C after the addition of 20% (w/v) glycerol until used.
RNAi Experiments—Silencing of ALase by RNAi was performed as described by Iseki et al. (24). An ∼450-bp length of partial ALase cDNA was PCR-amplified with the addition of the T7 RNA polymerase promoter sequence at one end. The primer sequences were: EgALase RNAi-F 5′-TAATACGACTCACTATAGGGGAGCTGCAGCCAGAGCTTCCGAAAGG-3′ and EgALase RNAi-R 5′-GTTCCTTCTCCACCACACGGGGATATCACTCAGCATAAT-3′ (T7 RNA polymerase promoter sites are underlined). Then the sense and antisense RNAs were synthesized using the PCR products as templates (MEGAscript RNAi kit, Ambion). After purification of transcribed RNA with DNase I digestion followed by phenol/chloroform extraction and ethanol precipitation, double-stranded RNA (dsRNA) was made by annealing equimolar amounts of the sense and antisense RNAs. Euglena cells of 2-day-old cultures were collected and resuspended in culture medium containing 4.2 mm Ca(NO3)2, 3.7 mm KH2PO4, and 2.1 mm MgSO4. Forty-five microliters of the cell suspension (∼1 × 106 cells) was transferred to a 0.4-cm-gap cuvette and electroporated with 5 μl of RNA solution (25 μg of ALase-dsRNA in 50 mm Tris-HCl, pH 7.5, 1 mm EDTA) using Gene Pulser II (Bio-Rad) at 1.2 kV and 25 microfarads. After a 30-min incubation at room temperature, the cell suspension was diluted with fresh Koren-Hutner medium (22) or the medium supplemented with precursors related to AsA biosynthesis and cultured at 27 °C for restoration.
Northern Analysis—Total RNA was extracted from the cells using RNAiso (Takara). The total RNA (10 μg/lane) was separated by electrophoresis through a 1.2% agarose/formaldehyde gel, blotted onto a nylon membrane (Hybond-N+, Amersham Biosciences), and hybridized with a 32P-labeled ALase cDNA prepared by random priming.
Measurement of AsA Concentration—Euglena cells were homogenized in 0.1 m HCl and 1 mm EDTA (∼0.1 g flesh weight ml-1) and centrifuged at 12,000 × g for 5 min. Total AsA content was measured by iron(III) reduction (25).
Data Analysis—The significance of differences between data sets was evaluated by t test. Calculations were carried out with Microsoft Excel software.
RESULTS
The Presence of a Homologue of the Rat Gluconolactonase Gene in Euglena Cells—Our previous identification and characterization of d-GalUA reductase from Euglena indicated that the occurrence of ALase is essential for the conversion of l-GalA, the resultant product of d-GalUA reduction, to l-GalL in this organism (21). Therefore, a BLASTp search was performed against the Euglena EST data base of the Protest EST Program using the amino acid sequence of the rat gluconolactonase involved in the biosynthesis of AsA in mammals (6) as a query. An EST clone encoding the putative Euglena ALase had the cluster ID ELL00003818. The corresponding full-length cDNA was cloned by the rapid amplification of cDNA ends method and submitted to the DDBJ/GenBank/EMBL data base (accession number AB306917). The predicted protein product, designated EgALase, displayed significant sequence identity (30.5%) and similarity (70.2%) with the rat gluconolactonase (Fig. 2).
FIGURE 2.
Sequence alignment of Euglena ALase with SMP30 from rat and human and Drp35 from Staphylococcus and Zymomonas. Conserved amino acid residues for both calcium binding of Staphylococcus Drp35 and zinc binding of Euglena ALase are highlighted in white letters with a black background. The accession numbers are: Euglena ALase (AB306917), rat SMP30 (X69021), human SMP30 (BC050371), Staphylococcus aureus Drp35 (AB030228), and Zymomonas mobilis (AE008692).
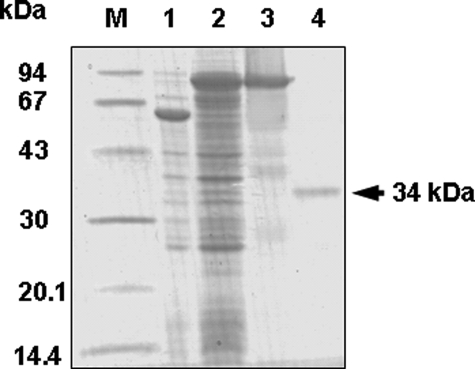
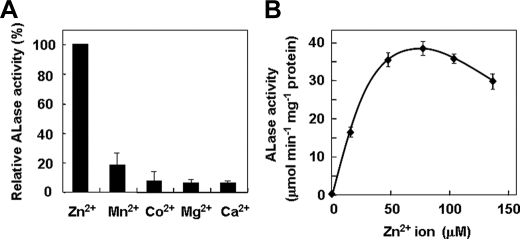
The EgALase Protein Has Lactonase Activity—To clarify the function of the EgALase isolated from the single-strand cDNA pool in 6-day-old Euglena cells, the recombinant EgALase was expressed in E. coli with the addition of N-terminal His tags followed by the trigger factor. The recombinant protein was purified by nickel-affinity chromatography and treated with HRV 3C protease to remove the additional tag proteins. The final product migrated in the SDS-polyacrylamide gel as a single band with an apparent molecular mass of ∼34 kDa (Fig. 3). The recombinant enzyme non-stereospecifically hydrolyzed aldonate lactones, such as d-glucono-δ-lactone, d-/l-GalL, and d-/l-GulL, indicating that the isolated cDNA encodes an aldonolactonase (Table 1). The enzyme required a divalent metal ion for maximum activity which was obtained by the addition of Zn2+ at a concentration of 75 μm (Fig. 4). Other divalent ions, Mn2+, Co2+, Mg2+, and Ca2+, had a significant low effect on the activity to 19, 9, 8, and 7%, respectively, of the enzyme activity given by the case of Zn2+. Therefore, Zn2+ was used at 75 μm for the subsequent experiments to study the catalytic properties of the Euglena lactonase.
FIGURE 3.
Production of recombinant Euglena ALase. The soluble proteins were extracted from E. coli cells transformed with an empty vector pColdTF or the vector containing the Euglena ALase (pColdTF/EgALase) after isopropyl 1-thio-β-d-galactopyranoside induction. The recombinant His- and TF-tag protein was purified in a nickel-nitrilotriacetic acid column. HRV3C protease was used to detach the fused tag. The samples were separated by 12.5% SDS-PAGE and visualized with Coomassie Brilliant Blue. M, molecular marker; lane 1, soluble fraction of cell extract transformed with an empty pColdTF vector; lane 2, soluble fraction of cell extract transformed with pColdTF/EgALase; lane 3, elution fraction from nickel-nitrilotriacetic acid; lane 4, purified recombinant EgALase after treatment with HRV 3C protease.
TABLE 1.
Comparison of substrate specificities of the recombinant Euglena ALase with the homologous enzyme from rat
All substrates were at a concentration of 5 mm. Data are the means of three independent determinations ± S.E.
|
Substrate |
Specific activity |
|
|---|---|---|
| rEgALase | SMP30a | |
| μmol min-1 mg-1 of protein | ||
| d-Glucono-δ-lactone | 39.0 ± 0.8 (100%) | 217 ± 3.6 (100%) |
| l-Glucono-δ-lactone | 5.4 ± 2.2 (2.49%) | |
| l-Galactono-γ-lactone | 24.5 ± 3.3 (62.8%) | 0.8 ± 0.0 (0.37%) |
| d-Galactono-γ-lactone | 27.3 ± 2.3 (70.0%) | 9.4 ± 0.3 (4.33%) |
| l-Gulono-γ-lactone | 40.9 ± 0.1 (105%) | 2.7 ± 0.1 (1.24%) |
| d-Gulono-γ-lactone | 20.1 ± 0.5 (51.5%) | 12.0 ± 0.9 (5.53%) |
Kondo et al. (6).
FIGURE 4.
Effect of divalent metal ions on Euglena ALase activity. A, ALase activity of the purified recombinant enzyme was determined in the presence of 75 μm concentrations of various divalent metal ions. B, ALase activity was determined at the indicated concentrations of ZnCl2. Values are expressed as the mean ± S.E. of three independent experiments.
Catalytic and Other Properties of the Recombinant Euglena ALase—Normal hyperbolic kinetics were obtained for all substrates listed in Table 2. The Km and Vmax values for d-glucono-δ-lactone were 2.08 ± 0.45 mm and 120 ± 22.7 μmol min-1 mg-1 protein, respectively, and the optimum pH was 6.5 (data not shown). The Km values for l-GalL and l-GulL were 1.67 ± 0.39 and 3.05 ± 0.75 mm, respectively, whereas the Km values for l-GalA and l-GulA were 1.55 ± 0.3 and 4.55 ± 0.23 mm, respectively. Although the Km values for l-GulL and l-GulA were two to three times higher than those for l-GalL and l-GalA, the Vmax values for them were approximately two times higher than those for l-GalL and l-GalA. These findings indicated the catalytic efficiency (Kcat/Km) of Euglena ALase for these substrates to be almost equal and that the reaction between uronic acid and lactone is reversible.
TABLE 2.
Kinetic parameters of purified recombinant ALase
The values are the mean ± S.E. for three independent experiments.
| ALase/substrate | Km | Vmax | kcat | kcat/Km |
|---|---|---|---|---|
| mm | μmol min−1 mg−1 protein | s−1 | s−1 mm−1 | |
| rEgALase | ||||
| l-Galatono-γ-lactone | 1.67 ± 0.39 | 333 ± 23.3 | 450 ± 32 | 270 |
| l-Galactonic acid | 1.55 ± 0.30 | 455 ± 212 | 615 ± 287 | 397 |
| l-Gulono-γ-lactone | 3.05 ± 0.75 | 769 ± 140 | 1040 ± 189 | 341 |
| l-Gulonic acid | 4.55 ± 0.23 | 1111 ± 226 | 1503 ± 306 | 330 |
| d-Glucono-σ-lactone | 2.08 ± 0.45 | 120 ± 22.7 | 160 ± 31 | 77 |
| SMP30a | ||||
| d-Glucono-σ-lactone | 9.40 | 345 | 192 | 20 |
Kondo et al. (6).
Various compounds listed in Table 3 were examined as to their effects on the ALase activity, with l-GalL and l-GalA as substrates. EDTA, a chelating reagent, at 1 mm strongly inhibited the activity in both directions. Reducing reagents, such as AsA and dithiothreitol, had no effect on the enzyme. On the other hand, the activity was inhibited by treatment with N-ethylmaleimide, a sulfhydryl reagent. Interestingly, the activity toward l-GalL as a substrate was completely inhibited by 0.4 mm N-ethylmaleimide, whereas that toward l-GalA was only inhibited 47.5% at the same concentration compared with activity levels without N-ethylmaleimide. These results suggest that a Cys residue is involved in the recognition of substrates by the Euglena ALase. In contrast to the results for N-ethylmaleimide, 0.5 mm H2O2 caused a 27 and 32% activation with l-GalL and l-GalA as the substrate, respectively.
TABLE 3.
Effect of various compounds on purified recombinant ALase from Euglena
The values are the mean ± S.E. for three independent experiments.
|
Compound |
Concentration |
Relative activity |
|
|---|---|---|---|
| l-GalA | l-GalL | ||
| mm | % | ||
| None | 100 | 100 | |
| EDTA | 1 | 1.2 | 0.0 |
| AsA | 1 | 94.7 | 98.8 |
| Dithiothreitol | 1 | 97.3 | 105 |
| H2O2 | 0.1 | 123 | 118 |
| 0.5 | 132 | 127 | |
| N-Ethylmaleimide | 0.4 | 52.5 | 0.0 |
| 4.0 | 33.3 | 0.0 | |
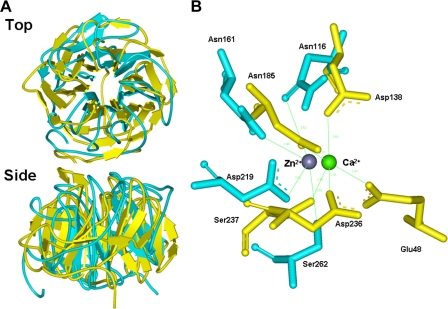
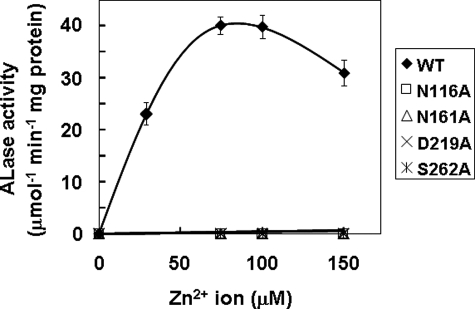
Identification of Zn2+ Binding Residues in Euglena ALase—A homology model of the Euglena lactonase was generated using the molecular graphics package Molecular Operating Environment (Chemical Computing Group Inc., Montreal, Canada) on the basis of the crystal structure of Drp35, a Ca2+-dependent lactonase hydrolyzing aromatic lactones from Staphylococcus aureus (PDB codes 2DG1) as a template (26). Drp35 has a six-bladed β-propeller structure with two calcium ions. One calcium ion binds at the center of the β-propeller structure and is essential for the lactonase activity (26). Interestingly, although the primary structure of Euglena ALase had comparatively little homology (11% identity) with that of Staphylococcus Drp35 (Fig. 2), the calculated topology of Euglena ALase was quite similar to that of Drp35 (Fig. 5A). Comparison of the center of the β-propeller structure of Euglena ALase with that of Staphylococcus Drp35 suggests the presence of four residues coordinating with the zinc ion (Fig. 5B). Among them, Asn-116, Asn-161, and Asp-219 in Euglena ALase corresponded to Asp-138, Asn-185, and Asp-236, respectively, in Staphylococcus Drp35. Additionally, Ser-262 of Euglena ALase was estimated to be involved in coordinating with the zinc ion. To test the significance of the corresponding residues in Euglena ALase, these four residues were individually substituted with Ala, then the activity of each purified recombinant mutant was determined. As a result, none of mutants showed activity even at up to 150 μm concentrations of zinc ion (Fig. 6), indicating that these four residues, including the novel Ser-262, are essential for the ALase activity and participate in the binding of zinc ion.
FIGURE 5.
Structural comparison of Staphylococcus Drp35 and predicted model of Euglena ALase. A, superim-position of Staphylococcus Drp35 and Euglena ALase. The protein structure of Drp35 and ALase is shown in yellow-green and pale blue, respectively. B, Ca2+ binding center in Drp35 and prediction of Zn2+-binding residues in Euglena ALase. The residues in Drp35 and ALase are shown in yellow-green and pale blue, respectively.
FIGURE 6.
Effect of amino acid substitution on Euglena ALase activity. ALase activities of affinity-purified mutant enzymes were determined at the indicated concentrations of ZnCl2. Values are expressed as the mean ± S.E. for three independent experiments. WT, wild type.
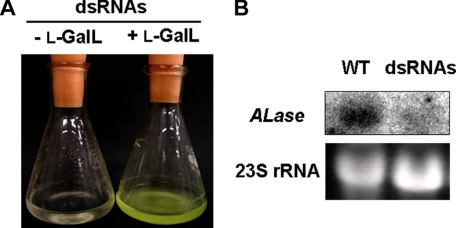
Silencing of ALase Expression Results in Growth Defects—To examine the physiological significance of the ALase to the biosynthesis of AsA in Euglena, we transiently silenced the expression of ALase by RNA-mediated interference. A dsRNA synthesized from part of the Euglena ALase sequence was introduced into Euglena cells by electroporation. Interestingly, the cells grew in medium supplemented with l-GalL (an intermediate that is downstream of ALase) 1 week after the introduction of the dsRNA (Fig. 7A). The AsA level in the dsRNA-containing cells after supplementation with l-GalL was 25 μmol of 10-9 cells. The cells did not grow in KH medium without any supplements at least within the study period. Northern analysis and an assay of ALase activity of the dsRNA-containing cells showed no significant detectable signal and activity of ALase, respectively, confirming the complete suppression of endogenous ALase by the introduction of dsRNA (Fig. 7B). These results clearly indicate that the expression of ALase is essential for the biosynthesis of AsA in Euglena, and AsA plays a pivotal role in cell growth.
FIGURE 7.
Phenotypic characterization of ALase-silenced Euglena cells. A, growth phenotype of dsRNA-introduced cells in a medium supplemented with l-GalL. Cells were restored and grown for 1 week in the presence of 1 mm l-GalL after the introduction of dsRNAs for ALase. Cultures of representative cells grown in a medium supplemented with 1 mm l-GalL were photographed 1 week after the dsRNA was introduced. B, Northern blot analysis. Each lane was loaded with 10 μg of total RNA from 1-week-old wild type (WT) cells and dsRNA-introduced cells. The RNA blot was hybridized with a 32P-labeled ALase cDNA probe. The bottom shows ethidium bromide-stained rRNA as a RNA-loading control.
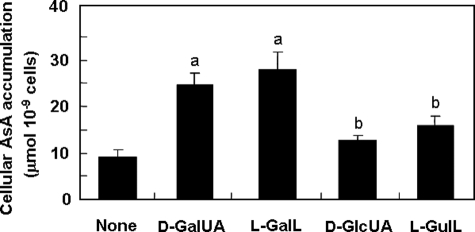
Effect of Treatment with Precursors on the Total AsA Level in Euglena Cells—To evaluate the effect of intermediates involved in the two possible pathways via d-GalUA/l-GalA or d-GlcUA/l-GulA on cellular AsA accumulation, Euglena cells were treated with 5 mm d-GalUA, l-GalL, d-GlcUA, or l-GulL under light at 55 μmol m-2 s-1 for 6 h. As shown in Fig. 8, d-GalUA and l-GalL, the precursors for the pathway via d-GalUA/l-GalA, were converted to AsA even more effectively than d-GlcUA and l-GulL for the pathway via d-GlcUA/l-GulA. The result strongly confirms the possibility that the pathway via d-GalUA/l-GalA is an effective route to highly maintain the cellular total AsA level in Euglena cells.
FIGURE 8.
Effect of precursors on AsA formation in Euglena. Euglena cells were incubated with 5 mm concentrations of each compound under light (55 μmol m-2 s-1) at 25 °C for 6 h. Values are expressed as the mean ± S.E. for three independent experiments. Values that were significantly different between control and treated cells are indicated; a, p < 0.05; b, p < 0.01.
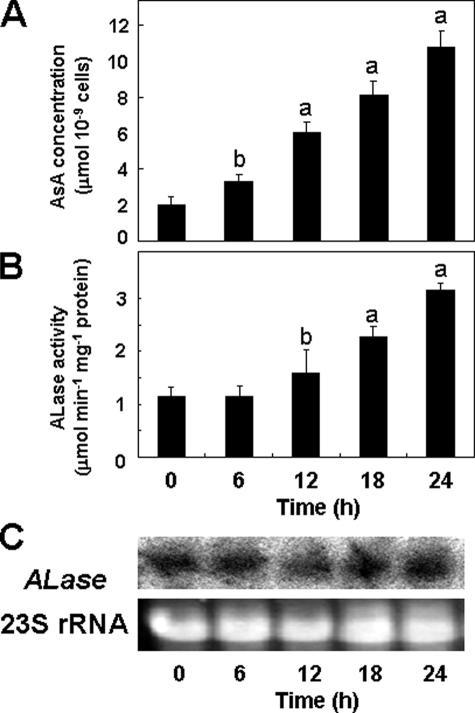
Effect of Illumination on ALase Expression in Euglena—It has been reported that the cellular levels of total AsA in Euglena cells markedly depended on light (1, 27), and in particular, the activity of l-GalL dehydrogenase in dark-grown Euglena was augmented by illumination (20). Therefore, we investigated the relationship between ALase expression and AsA accumulation under light. When Euglena cells grown in the dark were moved into light, cellular AsA levels increased steadily for 24 h, reaching a maximum of 11 μmol 10-9 cells, which was comparable with the level attained by green cells illuminated continuously (Fig. 9). The ALase activity eventually increased ∼3-fold with illumination for 24 h. In contrast to the ALase activity, the transcript levels remained constant, suggesting that ALase is post-transcriptionally regulated in response to light, reflecting the cellular level of AsA in Euglena cells.
FIGURE 9.
AsA level and expression of ALase under light in dark-grown Euglena. The cells were illuminated at an intensity of 55 μmol m-2 s-1. Samples were taken out at the indicated times and used for the determination of AsA, the assay of ALase, and Northern hybridization. Values are expressed as the mean ± S.E. for three experiments. A, AsA level. B, ALase activity. C, Northern blot analysis. Ten micrograms of total RNA extracted from each sample was electrophoresed through a formaldehyde-containing agarose gel, capillary blotted onto a nylon membrane, and hybridized with 32P-labeled cDNA of Euglena ALase. Ethidium bromide staining of the rRNA is shown for the equality of loading. Values that were significantly different between control and illuminated cells are indicated. a, p < 0.05; b, p < 0.01.
DISCUSSION
Our previous analysis of intermediates for AsA biosynthesis with radio-tracer experiments in Euglena has provided evidence that this organism utilizes a pathway via d-GalUA/l-GalA for the biosynthesis (1). In the present study we have successfully identified ALase and demonstrated that it is the bona fide enzyme in the biosynthesis of AsA in Euglena.
Characterization of Euglena ALase—The recombinant Euglena ALase recognized a broad range of substrates including various types of aldonate lactone, with a requirement for zinc ion as a cofactor. Such properties are well consistent with the rat enzyme (6). Zinc ion was the most effective divalent cation and played a critical role in the catalysis (Fig. 4). This was confirmed by the finding that EDTA inhibits the activity (Table 3). The requirement of divalent cation is common to most lactone hydrolases (28). Recent progress in the structural analysis of various lactonases has identified the residues coordinating with the divalent cation (26, 29). For example, N-acyl-l-homoserine lactonase from Erwinia requires zinc ions for its activity, and a structural analysis revealed that it contains two Zn2+ molecules coordinating partly with a conserved Zn2+ binding motif, HXHXDHXnH, at the center of the catalytic site (29). The Euglena ALase had low identity (7.7%) and similarity (43%) with the Erwinia enzyme and lacked a Zn2+ binding motif conserved within the primary sequence. Although the Erwinia enzyme was not suitable as a template for structural modeling of Euglena ALase, the Staphylococcus Drp35, a drug-responsive protein with Ca2+-dependent aromatic lactone hydrolyzing activity (26), provided useful information about the residues coordinating with the zinc ion of the Euglena enzyme. Based on a structural comparison with Drp35 and a mutational analysis, four residues (Asn-116, Asn-161, Asp-219, and Ser-262) in Euglena ALase were identified as significant for the activity (Figs. 5 and 6). These four resides were completely conserved in the rat gluconolactonase (Fig. 2). Three of them (Asn-116, Asn-161, and Asp-219) coordinate with Ca2+ in Drp35 (26). More importantly, the residue corresponding to Glu-48 in Drp35 was not found in Euglena ALase, whereas Ser-262 in Euglena ALase was not conserved in Drp35, assuming that this difference accounts for the coordination with zinc ion, but not calcium ion, in this type of lactonase.
In higher plants, feedback regulation of l-Gal dehydrogenase and GDP-d-Man-3,5-epimerase has been reported to be one of the regulatory mechanisms in AsA biosynthesis (30, 31). Treatment with AsA had no effect on the ALase activity (Table 3), indicating a lack of feedback regulation of the activity in the biosynthesis of AsA in Euglena. On the other hand, Euglena ALase showed redox sensitivity caused by H2O2 treatment (Table 3). Activation of d-GalUA reductase by H2O2 has also been observed in our previous study (21). Because AsA levels in Euglena cells increased under oxidative stress (20, 27) and light (Fig. 8), the activation of the enzymes in response to H2O2 levels seems to be a key regulatory mechanism promoting AsA biosynthesis in Euglena cells under stress.
The Pathway via d-GalUA/l-GalA Is the Significant Route for AsA Biosynthesis in Euglena—Silencing of Euglena ALase resulted in an arrest of cell growth in the normal KH medium, but the cells grew in medium supplemented with l-GalL (Fig. 7). This result provides compelling evidence that ALase is essential for AsA biosynthesis, and the pathway via uronic acid is the major source of AsA in Euglena cells. We previously reported that a d-GalUA reductase purified from Euglena had almost the same catalytic efficiency with two uronic acids, d-GalUA and d-GlcUA (21). Judging from the enzymology parameters of both d-GalUA reductase and ALase, it is difficult to understand which is the more effective route to promote the level of AsA in Euglena cells. However, it is clear from Fig. 8 that l-GalL is the most effective precursor of AsA. l-GulL and d-GlcUA are ∼41 and 48% less effective as precursors in the biosynthesis of AsA than l-GalL and d-GalUA, respectively. These findings are well consistent with the results of the tracer experiment reported by Shigeoka et al. (1). Additionally, crude extracts prepared from Euglena cells had significant dehydrogenase activity with l-GalL, but not l-GulL, as a substrate, whereas they had no oxidase activity with l-GulL or l-GalL.3 Consequently, we conclude that the pathway via d-GalUA/l-GalA is the major route for the biosynthesis of AsA in this organism. It is worth noting that, from a direct comparative tracer experiment, the mean ratio of the conversion of d-[U-14C]glucose to d-GalUA was ∼2-fold higher than that to d-GlcUA (1), suggesting the key point controlling the flux of the pathway is the epimerization of UDP-d-GlcUA to UDP-d-GalUA.
Physiological Role of AsA in Euglena Cells—Little is known about the physiological significance of AsA in photosynthetic algae including Euglena, except that it is an important antioxidant and essential donor for reactions of AsA peroxidase (EC 1.11.1.1.) (8, 32, 33). In addition to confirming the pathway via d-GalUA/l-GalA as the significant route of AsA biosynthesis, our results showed the importance of AsA in Euglena cells. A delay of cell growth has also demonstrated by an analysis of Trypanosoma mutants with disrupted orthologs encoding plant l-GalL dehydrogenase (19). SMP30, gluconolactonase, knock-out mice showed typical symptom of vitamin C deficiency including scurvy and did not grow well (6). This was due to a lack of collagen biosynthesis caused by a defective activity of AsA-dependent 2-oxoglutarate-dependent dioxygenase that is required for hydroxyproline synthesis. In Arabidopsis plants, Dowdle et al. (14) have reported that the simultaneous disruption of two genes encoding GDP-l-Gal phosphorylase (vtc2/vtc5), a key enzyme of the Man/Gal pathway, resulted in growth arrest upon germination, and the mutants resumed their growth on supplementation with l-Gal or AsA, indicating the function of AsA to be essential for plant growth. They also assumed that the growth arrest of the mutants reflects the role of AsA in growth, possibly due to the decrease in synthesis of extracellular matrix hydroxyproline-rich glycoproteins and/or plant hormones such as ethylene, gibberellic acid, and abscisic acid by suppression of AsA-dependent peptidyl-prolyl hydroxylase and dioxygenase activities.
In the case of Euglena, the reason for the growth arrest caused by the suppression of AsA biosynthesis would be different from that observed in Arabidopsis plants. It is possible that the AsA deficiency affects the regulation of the cell cycle in Euglena. Periodic variations in cAMP levels play a major signaling role in the progression of the Euglena cell cycle and the cAMP and enzymes adenylate cyclase and phosphodiesterase which control the level constitute a relay of the endogenous circadian clock (34). It is also suggested that cGMP serves as an upstream effector that mediates the cAMP oscillation by regulation of the metabolic enzymes (35). It is worth mentioning that AsA augmented cGMP levels in human lymphocytes (36), although the phenomenon is yet to be confirmed in Euglena. It has been reported that the level of AsA in Euglena is well synchronized with circadian rhythms (20, 27), also supporting a positive relationship between AsA biosynthesis and the regulation of the cell cycle. Although further investigations needed to conclude the effect of AsA levels on the control of the cell cycle, the silencing of ALase provides a good model with which to analyze the physiological role of AsA in Euglena cells.
The Possibility That the Pathway via d-GalUA/l-GalA Is Present in Other Photosynthesizing Organisms—The ALase plays a central role in the biosynthesis of AsA in Euglena. This article describes the first example to our knowledge of the biosynthesis of AsA via uronic acid in photosynthetic algae. To detect an ALase in other algae and plants, we performed a BLASTp search using both the Euglena and rat enzymes as a query sequence. The search found no orthologs in the genomes of higher plants, at least in Arabidopsis, rice, and Populus, whereas additional putative orthologs of ALase, which display significant sequence identity (∼30%) with the Euglena and rat ALase proteins, were found in a limited number of eukaryotic algae, including Ostreococcus lucimarinus, Thalassiosira pseudonana, and Phaeodactylum tricornutum (supplemental Fig. S1). A sequence alignment among these homologous proteins showed that four amino acid residues coordinating with the zinc ion in Euglena ALase (Asn-116, Asn-161, Asp-219, and Ser-262) are completely conserved at the appropriate sequence position in some species (O. lucimarinus and T. pseudonana), indicating the possibility that these algae contain an active ALase with properties similar to those of Euglena and the rat. In contrast, no significant orthologs were found in Cyanidioschyzon merolae, Chlamydomonas reinhardtii, Volvox carteri, and Ostreococcus tauri. It is worth noting that all the algae listed above possess predicted orthologs encoding l-GalL dehydrogenase. It is, therefore, assumed that algae have developed diverse pathways for the biosynthesis of AsA, including the pathway via d-GalUA/l-GalA in Euglena and the Man/Gal pathway in higher plants. Because the information available on the biosynthesis of AsA in algae is quite limited at the present moment, further investigation is needed to understand the distribution of these pathways among photosynthesizing algae.
Supplementary Material
The nucleotide sequence(s) reported in this paper has been submitted to the DDBJ/GenBank™/EBI Data Bank with accession number(s) AB306917.
This work was supported in part by The Sumitomo Foundation (to T. I.) and Grant-in-aid for Scientific Research 19208031 from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (to S. S.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
Footnotes
The abbreviations used are: AsA, l-ascorbic acid; ALase, aldonolactonase; l-GalA, l-galactonate; d-GalUA, l-galacturonate; l-GalL, l-galactono-1,4-lactone; l-GlcUA, l-glucuronate; l-GulA, l-gulonate; l-GulL, l-gulono-1,4-lactone; PIPES, 1,4-piperazinediethanesulfonic acid; RNAi, RNA interference; dsRNA, double-stranded RNA.
T. Ishikawa, H. Nishikawa, and S. Shigeoka, unpublished results.
References
- 1.Shigeoka, S., Nakano, Y., and Kitaoka, S. (1979) J. Nutr. Sci. Vitaminol. 25 299-307 [DOI] [PubMed] [Google Scholar]
- 2.Davey, M. W., Van Montagu, M., Inze, D., Sanmartin, M., Kanellis, A., Smirnoff, N., Benzie, I. J. J., Strain, J. J., Favell, D., and Fletcher, J. (2000) J. Sci. Food Agric. 80 825-860 [Google Scholar]
- 3.Smirnoff, N. (2000) Curr. Opin. Plant Biol. 3229 -235 [PubMed] [Google Scholar]
- 4.Ishikawa, T., Dowdle, J., and Smirnoff, N. (2006) Physiol. Plant. 126343 -355 [Google Scholar]
- 5.Nishikimi, M., and Yagi, K. (1996) Subcell. Biochem. 2517 -39 [DOI] [PubMed] [Google Scholar]
- 6.Kondo, Y., Inai, Y., Sato, Y., Handa, S., Kubo, S., Shimokado, K., Goto, S., Nishikimi, M., Maruyama, N., and Ishigami, A. (2006) Proc. Natl. Acad. Sci. U. S. A. 1035723 -5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smirnoff, N., Conklin, P. L., and Loewus, F. A. (2001) Annu. Rev. Plant Physiol. Plant Mol. Biol. 52 437-467 [DOI] [PubMed] [Google Scholar]
- 8.Ishikawa, T., and Shigeoka, S. (2008) Biosci. Biotechnol. Biochem. 721143 -1154 [DOI] [PubMed] [Google Scholar]
- 9.Bartoli, C. G., Pastori, G. M., and Foyer, C. H. (2000) Plant Physiol. 123335 -344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yabuta, Y., Yoshimura, K., Takeda, T., and Shigeoka, S. (2000) Plant Cell Physiol. 41 666-675 [DOI] [PubMed] [Google Scholar]
- 11.Conklin, P. L., Norris, S. R., Wheeler, G. L., Williams, E. H., Smirnoff, N., and Last, R. L. (1999) Proc. Natl. Acad. Sci. U. S. A. 964198 -4203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conklin, P. L., Gatzek, S., Wheeler, G. L., Dowdle, J., Raymond, M. J., Rolinski, S., Isupov, M., Littlechild, J. A, and Smirnoff, N. (2006) J. Biol. Chem. 28115662 -15670 [DOI] [PubMed] [Google Scholar]
- 13.Lukowitz, W., Nickle, T. C., Meinke, D. W., Last. R. L., Conklin, P. L., and Somerville, C. R. (2001) Proc. Natl. Acad. Sci. U. S. A. 982262 -2267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dowdle, J., Ishikawa, T., Gatzek, S., Rolinski, S., and Smirnoff, N. (2007) Plant J. 52 673-689 [DOI] [PubMed] [Google Scholar]
- 15.Agius, F., Gonzalez-Lamothe, R., Caballero, J. L., Munoz-Blanco, J., Botella, M. A., and Valpuesta, V. (2003) Nat. Biotechnol. 21177 -181 [DOI] [PubMed] [Google Scholar]
- 16.Lorence, A., Chevone, B. I., Mendes, P., and Nessler, C. L. (2004) Plant Physiol. 1341200 -1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Running, J. A., Severson, D. K., and Schneider, K. J. (2002) J. Ind. Microbiol. Biotechnol. 29 93-98 [DOI] [PubMed] [Google Scholar]
- 18.Running, J. A., Burlingame, R. P., and Berry, A. (2003) J. Exp. Bot. 541841 -1849 [DOI] [PubMed] [Google Scholar]
- 19.Wilkinson, S. R., Prathalingam, S. R., Taylor, M. C., Horn, D., and Kelly, J. M. (2005) Proc. Natl. Acad. Sci. U. S. A. 10211645 -11650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shigeoka, S., Nakano, Y., and Kitaoka, S. (1979) Agric. Biol. Chem. 432187 -2188 [Google Scholar]
- 21.Ishikawa, T., Masumoto, I., Iwasa, N., Nishikawa, H., Sawa, Y., Shibata, H., Nakamura, A., Yabuta, Y., and Shigeoka, S. (2006) Biosci. Biotechnol. Biochem. 702720 -2726 [DOI] [PubMed] [Google Scholar]
- 22.Koren, L. E., and Hutner, S. H. (1967) J. Protozool. 1417. [DOI] [PubMed] [Google Scholar]
- 23.Hucho, F., and Wallenfels, K. (1972) Biochim. Biophys. Acta 276176 -179 [DOI] [PubMed] [Google Scholar]
- 24.Iseki, M., Matsunaga, S., Murakami, A., Ohno, K., Shiga, K., Yoshida, K., Sugai, M., Takahashi, T., Hori, T., and Watanabe, M. (2002) Nature 4151047 -1051 [DOI] [PubMed] [Google Scholar]
- 25.Kampfenkel, K., Van Montagu, M., and Inze, D. (1995) Anal. Biochem. 225165 -167 [DOI] [PubMed] [Google Scholar]
- 26.Tanaka, Y, Morikawa, K., Ohki, Y., Yao, M., Tsumoto, K., Watanabe, N., Ohta, T., and Tanaka, I. (2007) J. Biol. Chem. 2825770 -5780 [DOI] [PubMed] [Google Scholar]
- 27.Kiyota, M., Numayama, N., and Goto, K. (2006) J. Photochem. Photobiol. B. 84 197-203 [DOI] [PubMed] [Google Scholar]
- 28.Shimizu, S., Kataoka, M., Shimizu, K., Hirakata, M., Sakamoto, K., and Yamada, H. (1992) Eur. J. Biochem. 209383 -390 [DOI] [PubMed] [Google Scholar]
- 29.Kim, M. H., Choi, W. C., Kang, H. O., Lee, J. S., Kang, B. S., Kim, K. J., Derewenda, Z. S., Oh, T. K., Lee, C. H., and Lee, J. K. (2005) Proc. Natl. Acad. Sci. U. S. A. 10217606 -17611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mieda, T., Yabuta, Y., Rapolu, M., Motoki, T., Takeda, T., Yoshimura, K., Ishikawa, T., and Shigeoka, S. (2004) Plant Cell Physiol. 451271 -1279 [DOI] [PubMed] [Google Scholar]
- 31.Wolucka, B. A., and Van Montagu, M. (2003) J. Biol. Chem. 27847483 -47490 [DOI] [PubMed] [Google Scholar]
- 32.Ishikawa, T., Takeda, T., Kohno, H., and Shigeoka, S. (1996) Biochim. Biophys. Acta 1290 69-75 [DOI] [PubMed] [Google Scholar]
- 33.Shigeoka, S., Ishikawa, T., Tamoi, M., Miyagawa, Y., Takeda, T., Yabuta, Y., and Yoshimura, K. (2002) J. Exp. Bot. 531305 -1319 [PubMed] [Google Scholar]
- 34.Carré, I. A., and Edmunds, L. N., Jr. (1993) J. Cell Sci. 1041163 -1173 [DOI] [PubMed] [Google Scholar]
- 35.Tong, J., and Edmunds, L. N., Jr. (1993) Biochem. Pharmacol. 452087 -2091 [DOI] [PubMed] [Google Scholar]
- 36.Atkinson, J. P., Weiss, A., Ito, M., Kelly, J., and Parker, C. W. (1979) J. Cyclic Nucleotide Res. 5 107-123 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.