Abstract
Intrinsic structural disorder is a prevalent feature of proteins with chaperone activity. Using a complementary set of techniques, we have structurally characterized LjIDP1 (intrinsically disordered protein 1) from the model legume Lotus japonicus, and our results provide the first structural characterization of a member of the Lea5 protein family (PF03242). Contrary to in silico predictions, we show that LjIDP1 is intrinsically disordered and probably exists as an ensemble of conformations with limited residual β-sheet, turn/loop, and polyproline II secondary structure. Furthermore, we show that LjIDP1 has an inherent propensity to undergo a large conformational shift, adopting a largely α-helical structure when it is dehydrated and in the presence of different detergents and alcohols. This is consistent with an overrepresentation of order-promoting residues in LjIDP1 compared with the average of intrinsically disordered proteins. In line with functioning as a chaperone, we show that LjIDP1 effectively prevents inactivation of two model enzymes under conditions that promote protein misfolding and aggregation. The LjIdp1 gene is expressed in all L. japonicus tissues tested. A higher expression level was found in the root tip proximal zone, in roots inoculated with compatible endosymbiotic M. loti, and in functional nitrogen-fixing root nodules. We suggest that the ability of LjIDP1 to prevent protein misfolding and aggregation may play a significant role in tissues, such as symbiotic root nodules, which are characterized by high metabolic activity.
Intrinsically disordered (or natively unfolded) proteins (IDPs)5 that are disordered along the entire amino acid chain or contain long disordered regions have recently attracted attention due to their role in important human diseases and their distinct functional features (1). In plants, a loosely defined group of stress-related proteins known as the late embryogenesis-abundant (LEA) proteins are characterized by being almost exclusively intrinsically disordered (reviewed by Tunnacliffe and Wise (2)). Several lines of evidence suggest a role of LEA proteins in the protection against damage caused by different types of stress, in particular desiccation, salt, and cold stress. The exact protection mechanism is unknown; however, the demonstrated ability of some (and probably most) LEA proteins to adopt a more well ordered conformation upon the addition of structure perturbing chemicals and when dried may contribute (2). Such structural plasticity is also known from other IDPs, which undergo disorder-to-order transitions upon ligand binding (3). However, specific ligand interactions have not been described for any LEA protein, and the mechanism by which LEA proteins assert their functions remains elusive.
We report here the biochemical characterization of the LjIDP1 protein encoded by the LjIdp1 gene that is up-regulated during nodule development and in functional nitrogen-fixing nodules of the model legume Lotus japonicus. Leguminous plants are unique in their ability to engage in symbiosis with Rhizobium and allied bacteria. This interaction triggers the development of a new plant organ, the root nodule that hosts the nitrogen fixing rhizobia in intracellular organelle-like symbiosomes. In contrast to most root tissues, nodules are highly metabolically active, and hundreds of genes involved in metabolic processes have higher expression levels in nodules compared with roots (4). A high respiration rate of nodules is needed to provide the large energy input required for nitrogen fixation, and it was estimated that up to 70% of total root respiration is associated with nitrogen fixation (5). The high respiration rate of nodules inevitably leads to a large production of reactive oxygen species (ROS) that have been detected in association with both cellular and functional processes in rhizobium-legume interaction and symbiosis (see Pauly et al. (6) for a review). Several studies have documented increased ROS production during symbiosis and suggest that ROS may influence development and function of symbiotic root nodules. Roles in signal transduction, in hypersensitive responses controlling the infection process, in cross-linking of proteins in the infection thread matrix, and in senescence have been suggested (7-9).
Using real time PCR and microarray analysis, we show that LjIdp1 mRNA levels are high in nodules and propose that LjIDP1 may play a role in oxidative stress tolerance. We show that LjIDP1 has chaperone-like activity and protects two model enzymes subjected to stresses that are known to destabilize protein structure and promote misfolding. The molecular characterization of LjIDP1 presented here provides new information on the hitherto largely uncharacterized Lea5 protein family and focuses attention to the role of intrinsic structural disorder in protein chaperones.
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification of Recombinant LjIDP1—LjIDP1 was expressed with an N-terminal extension (MAHHHHHHVDDDDK = His6 tag + enterokinase (Ek) cleavage site). The following primers were used to PCR-amplify the coding sequence of LjIdp1 flanked by LIC overhangs (underlined): LjIdp1-LIC-forward, 5′-GACGACGACAAGATGGCTCGCTCTTTCACCACCATCAAG-3′; LjIdp1-LIC-stop-reverse, 5′-GAGGAGAAGCCCGGTTCAATTTTTTCCCAGAACCGCAGATCG-3′.
The fragment was subsequently cloned into pRSF-2 Ek-LIC according to the manufacturer's protocol (Novagen; user protocol TB163 Rev.F 0904). Plasmid identity was confirmed by sequencing. The resulting vector, pRSF:LjIdp1, was transformed into Escherichia coli Bl21(DE3). A single BL21(DE3)::LjIDP1 colony was inoculated in 50 ml of LB medium containing 50 μg/ml kanamycin and grown overnight at 37 °C. 10 ml was inoculated in 500 ml of Terrific Broth medium containing 50 μg/ml kanamycin in 5-liter baffled Erlenmeyer flasks at 37 °C. At A600 = 0.8, expression of recombinant LjIDP1 was induced by adding isopropyl β-d-1-thiogalactopyranoside to a final concentration of 1 mm, and after 4 h at A600 = 1.55, cells were harvested and washed in 0.9% (w/v) NaCl and subsequently frozen at -20 °C.
For purification of recombinant LjIDP1, we have employed two different strategies. For strategy 1, cells were opened in ∼5 ml of binding buffer (50 mm potassium phosphate, pH 7.6, 500 mm NaCl, 10 mm β-mercaptoethanol, 1 mm phenylmethanesulfonyl fluoride)/g using a high pressure homogenizer (Emusi-Flex-05; Avestin). The crude extract was cleared by ultracentrifugation (75 min at 50,000 × g; Beckman LB-70 M equipped with a Type 60 Ti rotor) at 4 °C and filtered through a 45-μm filter before loading on a ∼10-ml nickel-charged Fastflow chelating Sepharose (Amersham Biosciences) column pre-equilibrated with washing buffer (50 mm potassium phosphate, pH 7.6, 500 mm NaCl, 0.1 mm phenylmethanesulfonyl fluoride, 45 mm imidazole), at a flow rate of 0.5 ml/min. The column was washed with washing buffer until stable base line. Bound protein was eluted with a gradient of imidazole in washing buffer going from 50 to 300 mm. Fractions containing LjIDP1 were concentrated on a 2-ml Centricon 10 column (Millipore). The concentrated samples were purified to homogeneity by gel filtration chromatography on a Superdex 75 10/300 GL column (GE Healthcare) in 50 mm HEPES, pH 6.5, 100 mm KCl, 5% glycerol. Eluted fractions were analyzed by SDS-PAGE. For strategy 2, cells (2 g) were resuspended in 30 ml of lysis buffer (50 mm Hepes, pH 7.5, 250 mm NaCl, 10 mm imidazole, 6 mm β-mercaptoethanol, one tablet of complete protease inhibitor mixture EDTA-free (Roche Applied Science)). The resuspended cells were heated in a small glass flask for 5 min in a boiling water bath and stirred continually using a small magnet. The flask was subsequently cooled for 5 min in an ice-ethanol bath and sonicated three times for 1 min each. Cell debris and denatured proteins were pelleted by centrifugation, 20,000 rpm for 15 min. The supernatant was loaded onto a 1-ml Histrap FF Crude column (GE Healthcare) pre-equilibrated in wash buffer (50 mm Hepes, 250 mm NaCl, 25 mm imidazole), flow rate 0.5 ml/min. The column was washed in wash buffer until stable base line. Bound proteins were eluted using elution buffer (50 mm Hepes, pH 7.5, 250 mm NaCl, 300 mm imidazole, 5% glycerol) fraction size 0.5 ml. The purity of the eluted protein was analyzed by SDS-PAGE.
Identification of Purified LjIDP1 by Mass Spectrometry—The identity of the purified protein was confirmed by peptide mass fingerprinting of peptides derived from an in-gel trypsin (Promega)-catalyzed proteolytic cleavage. Lyophilized gel pieces were incubated for 45 min on ice with trypsin solution (12.5 ng/μl trypsin from porcine (Promega) in 50 mm NH4CO3). Any remaining liquid was removed, and 60 μl of 50 mm NH4CO3 was added and incubated at 37 °C overnight.
The tryptic peptides were recovered by adsorption on a C18 ZipTip (Millipore) and dispensed on the MALDI target plate in 1.5 μl of MALDI matrix solution (4 mg/ml α-cyano-4-hydroxycinnamic acid in 70% acetonitrile, 30% 0.1% trifluoroacetic acid). The recovered peptides were analyzed by MALDI-mass spectrometry on a quadrupole/time-of-flight Ultima Global mass spectrometer (Micromass).
Circular Dichroism Spectroscopy—All CD spectra were recorded
at the UV1 beamline at the Institute for Storage Ring Facilities at Aarhus
University, Denmark, using synchrotron radiation provided by the ASTRID
storage ring. Samples were recorded in a closed 0.1-mm Suprasil quartz cell
(Hellma, GmbH). The sample and base-line spectra were acquired using three
consecutive scans, in 1-nm increments and a 2.5-s dwell time. The temperature
dependence of the CD signal was determined using spectra recorded from 20 to
80 °C in steps of 5 °C. Samples were allowed to equilibrate at each
temperature for 10 min before data acquisition. The spectra were averaged,
base line-subtracted, and mildly smoothed with a Savitzky-Golay filter using
the CDtool software (10). For
the dehydration experiment, the sample cell containing LjIDP1 was
left open in the cell chamber, which was continually purged with dry nitrogen
gas. Dehydration occurs from the top through the holes that are used to fill
the cell. All spectra were recorded in 10 mm potassium phosphate
(pH 7.0), 75 mm NaF except for the studies on the dehydrated
protein, which were done in 10 mm potassium phosphate (pH 7.0), 100
mm NaF. Concentration of each of the protein samples was determined
using a double quantitative amino acid analysis or by measuring
A280 on an ND-1000 spectrophotometer (NanoDrop). The
average of five measurements was used using (His-Ek-LjIDP1)
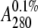 = 0.854 as determined using the
ProtParam software on the ExPASy server (available on the World Wide Web).
= 0.854 as determined using the
ProtParam software on the ExPASy server (available on the World Wide Web).
Quantification of Secondary Structure Content—The relative content of secondary structure elements was determined from the CD data using the program CDSSTR (11) and the SP175 (12) reference data set at the Dichroweb Server (12).
Fourier Transform Infrared Spectroscopy—FTIR spectroscopy was carried out using a Tensor 27 (Bruker) FTIR spectrophotometer equipped with a DTGS Midinfrared detector and a Golden Gate single reflection diamond attenuated total reflectance cell (Specac). Approximately 6 μg of purified LjIDP1 was dried on the crystal in a flow of dry nitrogen. LjIDP1 had been lyophilized and reconstituted in D2O. Spectra were recorded from 4000 to 1000 cm-1 using a nominal resolution of 2 cm-1 and 64 accumulations. Lorentzian curve fitting was done in the OPUS 5.5 system (Bruker).
Analytical Gel Filtration Chromatography—Analytical gel filtration chromatography was performed on a Superdex™ 75 10/300 GL (GE Healthcare) using an Acta purifier. The column was calibrated with bovine serum albumin (Rs = 35.7 Å, molecular mass 66 kDa), carbonic anhydrase (Rs 25.1 Å, mass 29 kDa), and cytochrome c (Rs 17.4 Å, mass 12.4 kDa) included in (MW-GF200) purchased from Sigma. All proteins were analyzed in the same buffer (100 mm HEPES, pH 7, 250 mm NaCl, 20 mm imidazole, 5% (v/v) glycerol) and a flow rate of 0.35 ml/min at 4 °C.
NMR Spectroscopy of LjIDP1—One-dimensional 1H NMR data were acquired on a Bruker Avance 700 WB spectrometer with a TBI probe head at 293.2 K with 128 transients. The water signal was suppressed by 3-9-19 WATERGATE. The concentration of LjIDP1 was 0.4 mm. Urea was added to a concentration of 6 m, and the solution was concentrated back to a protein concentration of 0.4 mm on a 500-μl Vivaspin column (Sartorius) before repeating the NMR experiment.
Citrate Synthase (CS) Dehydration Assay—The effect of dehydration on the activity of CS (EC 4.1.3.7) from porcine heart (Sigma) was determined as follows. CS (250 μg/ml) was dehydrated in the absence or presence of protectants (His6-Ek-LjIDP1 in 10 mm HEPES, pH 7.5, bovine serum albumin (BSA) (Sigma) in water or trehalose in water. CS and BSA had been dialyzed against water overnight in 3× 100 ml of water. The tested concentrations of His6-Ek-LjIDP1 and BSA were 200 μg/ml, whereas for trehalose, it was 75.6 mg/ml (0.2 m). The total volume was 30 μl (adjusted with distilled water). Dehydration was performed in a Speedvac 100 (SAVANT) equipped with an Edwards E8M2 high vacuum pump (Edwards). The dehydrated samples were rehydrated in 30 μl of distilled water after the first round of dehydration in 28 μl after the second, 26 μl after the third, and so on. Aggregated protein was pelleted by centrifugation for 3 min at 14,000 rpm, and the protein remaining in solution was measured spectrophotometrically at 280 nm on a ND-1000 spectrophotometer (NanoDrop).
CS Cryoprotection Assay—The effect of freezing on the activity of CS (250 μg/ml) was determined using the same concentrations and volumes as described for the CS dehydration assay. Samples were flash frozen in liquid nitrogen for 1 min and subsequently thawed at ambient temperature. Residual activity was assayed as described below.
CS Activity Assay—The activity of CS was determined essentially according to Srere (13). 2 μl of CS with or without protectant from the cryoprotection and dehydration assay was added to 500 μl of Act buffer (50 mm Tris-HCl, pH 8.0, 2 mm EDTA, 100 μm oxaloacetic acid, 100 μm 5,5′-dithiobis-2-nitrobenzoic acid, 95 μm acetyl-CoA (disodium salt)). A412 was measured every 0.5 s for 90 s in a UV-2101 PC spectrophotometer equipped with a thermostatically controlled CPS-260 cell positioner (Shimadzu). Measurements were performed at 25 °C.
LHD Dehydration Assay—The effect of dehydration on the activity of lactate dehydrogenase (LDH) (from rabbit muscle; Sigma; EC 1.1.1.27) was performed essentially as described for CS, except the LDH concentration was 100 μg/ml.
LDH Cryoprotection Assay—The effect of freezing on the activity of LDH (100 μg/ml) was determined as described for CS.
LDH Activity Assay—To determine the LDH activity, 2 μl of each sample were added to a 500-μl quartz cuvette containing 500 μl of sodium phosphate buffer (pH 6), 200 μm NADH, and 2 mm pyruvate. Change in A340 was monitored every 0.1 s for 1 min in a UV-2101 PC spectrophotometer equipped with a thermostatically controlled CPS-260 cell positioner (Shimadzu). Measurements were performed at 25 °C.
Bioinformatics on LjIdp1—The data set called “28IDP1” comprising LjIDP1 and a set of 27 homologous proteins was collected by BLAST searching the amino acid sequence of LjIDP1 against the nonredundant uniref100 data base, and hits with E-values less than 10-3 were selected. Amino acid profiling was performed using the World Wide Web-based tool Composition Profiler, taking advantage of the standard amino acid data sets, Disprot 3.4 and Swissprot51, provided by the program. For the prediction of intrinsic structural disorder in LjIDP1 using PONDR (available on the World Wide Web), the average of the VLXT, XL1_XT, CAN_XT, and VL3-BA was used. The cumulative distribution function analysis of LjIDP1 PONDR VL-XT scores was also done using the PONDR software (available on the World Wide Web).
Protease Sensitivity Assay—25 μg of His6-Ek-LjIDP1, lysozyme (Sigma), and α-synuclein in 50 mm NH4CO3 were incubated at 37 °C for 1 h with decreasing amounts of trypsin (Promega). The following ratios were tested ([His-LjIDP1/lysozyme/α-synuclein]/[trypsin]): 1:10-2, 1:10-3. 1:10-4, and 1:10-5. Total reaction volume was 32 μl. After incubation, 8 μl of 6× SDS loading buffer were added to each digest, vortexed, boiled 2 min, vortexed, boiled 2 min, and analyzed on a 10-20% acrylamide gradient gel.
Real Time PCR Analysis of LjIdp1—Poly(A)+ mRNA was isolated using the Dynabeads® mRNA direct kit (Invitrogen) according to the manufacturer's instructions. DynaBeads RNA samples were tested for DNA contamination by PCR using primers specific for the untranscribed Nin gene promoter (5′-GTTTTCAAGAATGGGAGGGG-3′, 5′-CTCCTCTGGTTTCATTGGTG-3′). Samples containing DNA were incubated with DNase I, treated with phenol/chloroform, and retested for contaminating DNA. First strand cDNA was synthesized using reverse transcriptase (Roche Applied Science) and oligo(dT) primer. Transcript levels were determined by semiquantitative real time PCR on a Lightcycler (Roche Applied Science) using the FastStart DNA master SYBR greenI kit (Roche Applied Science). The relative quantification software (Roche Applied Science) was used to determine the efficiency-corrected relative transcript concentration, normalized to a calibrator sample (a pool of mRNA from the different tissues and conditions). ATP synthase was used as a reference gene (AW719841). Primer sequences and conditions were as follows: ATP synthase, annealing 60 °C, measurement 80 °C, 5′-CAATGTCGCCAAGGCCCATGGTG-3′ and 5′-AACACCACTCTCGATCATTTCTCTG-3′; LjIdp1, annealing 64 °C, measurement 85 °C, 5′-GATGTCGCGGACCTGCG-3′ and 5′-CCCATATATGTCCGAATCCCC-3′.
RESULTS
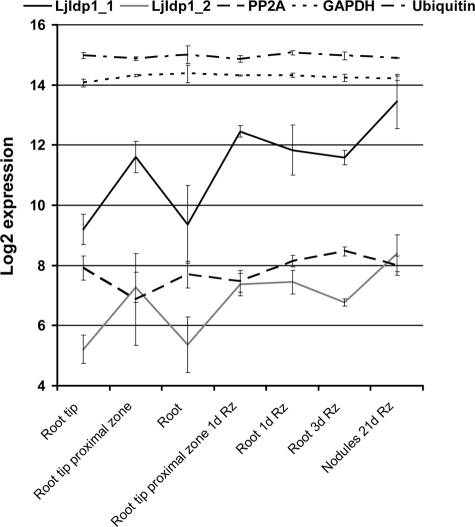
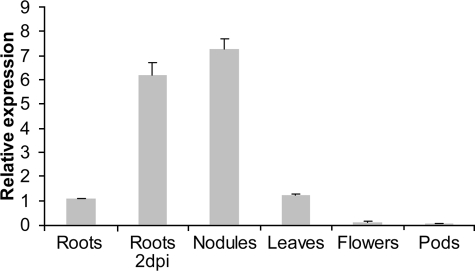
LjIdp1 Is Highly Transcribed in Inoculated Roots and Mature Root Nodules—Taking advantage of the newly available 61K Lotus japonicus GeneChip from Affymetrix, expression of the LjIdp1 gene was profiled at various time points postinoculation and in different organs and tissues.6 Expression of LjIdp1 was up-regulated in roots upon inoculation with the Lotus microsymbiont Mesorhizobium loti. After only 24 h postinoculation, expression had increased and reached a maximum in mature nodules, where LjIdp1 transcript levels were ∼8-16-fold higher than in uninoculated roots (Fig. 1). Interestingly, in uninoculated roots, significantly higher LjIdp1 mRNA levels were detected in the root tip proximal zone encompassing the invasion zone where legume roots are competent for rhizobial infection (∼1-cm segment of the root located just above the root tip). Furthermore, the expression is increased in the root tip proximal zone upon inoculation with M. loti. Since the tissue sample “root” includes the “root tip proximal zone,” the observed increase in LjIdp1 mRNA levels in the root 24 h after inoculation could at least partly originate from increased LjIdp1 mRNA levels in the root tip proximal zone. To confirm and further characterize the expression pattern of LjIdp1, the expression profile was analyzed by quantitative PCR. Real time PCR analysis revealed high LjIdp1 mRNA levels in roots 2 days postinoculation with M. loti and in mature nodules (Fig. 2). Intermediate levels were found in leaves and uninoculated roots, whereas low levels were found in pods and flowers.
FIGURE 1.
Steady state level of LjIdp1 mRNA determined by microarray analysis. Expression of LjIdp1 in different root tissues, inoculated roots, and mature nodules. LjIdp1 is represented on the chip with two probe sets (LjIdp1_1 and LjIdp1_2) with different efficiencies. Legend code example, “Root 1d Rz” is roots 1 day postinoculation with Rhizobium. PP2A, protein phosphatase 2A; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
FIGURE 2.
Relative expression level of LjIdp1 in different plant organs and roots 2 days post inoculation with Rhizobium as determined by real time PCR. Error bars, S.D. values.
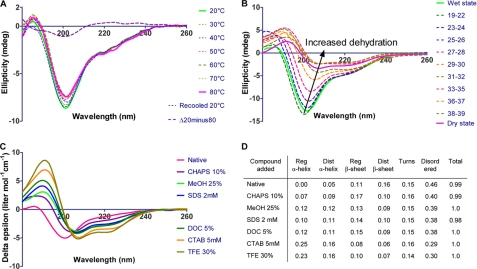
LjIDP1 Is Intrinsically Disordered—To gain information about the conformation and physiochemical properties of LjIDP1, the protein was expressed with an N-terminal, 14-residue extension containing a polyhistidine tag suitable for affinity purification and an enterokinase protease site. Pure (>95% homogeneity) recombinant LjIDP1 protein was purified from E. coli BL21(DE3) extracts using a combination of either immobilized metal ion affinity chromatography (IMAC) followed by size exclusion chromatography or IMAC combined with an initial heating step. Neither the presence of the N-terminal extension nor the purification procedure had any influence on the structure as determined by far UV CD spectroscopy, and therefore His-tagged LjIDP1 was used (not shown). The secondary structure content of recombinant LjIDP1 was analyzed by synchrotron radiation CD spectroscopy (SR-CD) (Fig. 3A). The high light intensity of the synchrotron beam allows recording of CD spectra down to 180 nm, which makes subsequent quantification of the relative secondary structure content more reliable (14). Like unfolded polypeptides, the CD spectrum of LjIDP1 shows a large negative peak at ∼200 nm (Fig. 3A). However, the small negative ellipticity at ∼220 nm (unlike the zero or positive values shown by unfolded polypeptides (15)) indicates the presence of some β-sheet structure. This was supported by secondary structure quantification of the CD spectra, estimating the β-sheet content of LjIDP1 to be ∼25% (Fig. 3D). The predicted high fraction of distorted β-sheet indicates the presence of many short β-strands or alternatively that the β-sheet fraction is transient and fluctuating in nature. The analysis also indicates the presence of ∼15% turn structures. Note that LjIDP1 was predicted to be essentially devoid of α-helical structure.
FIGURE 3.
SR-CD spectroscopy of LjIDP1. A, the SR-CD spectra of LjIDP1 showing the effect of temperature on the secondary structure. The spectrum of LjIDP1 has a large negative peak at ∼200 nm typical for an unfolded polypeptide and a small negative ellipticity at ∼220 nm due to β-sheet. Heating the sample from 20 to 80 °C has little effect on the CD spectrum. Note the small intensity and shape of the difference spectrum Δ(20-80 °C) that is similar to the CD spectrum of PPII. B, 70 consecutive SR-CD scans of LjIDP1 undergoing dehydration were collected. Virtually no changes were observed during the first 18 scans, so these were averaged and denoted “wet state.” All major structural changes occurred after 39 scans; therefore, spectra 40-70 were averaged and denoted “dry state.” The spectrum of LjIDP1 in the dry state is characterized by a positive peak at ∼190 nm and negative ellipticity at ∼208 nm and ∼222 nm typical for α-helix. C, SR-CD spectroscopy of LjIDP1 in the presence of different detergents and alcohols. Intriguingly, all agents tested induced α-helix at the expense of the β-sheet and disordered fractions. D, the table shows the quantified relative amounts of secondary structure elements.
To test whether the β-sheet content in LjIDP1 is only of local character or is sufficiently well organized to lead to cooperative unfolding transitions; we investigated the thermal stability of LjIDP1 by SR-CD. Remarkably, the small changes in the CD spectrum resulting from a temperature increase of 20-80 °C, in combination with the absence of a clear isodichroic point in the spectra, strongly suggest that LjIDP1 lacks any well defined structure (Fig. 3A). The difference spectrum Δ(20-80 °C) is positive above 210 nm and has a negative peak at ∼200 nm and crosses zero at ∼210 nm (Fig. 3A). This shape is similar to the CD spectrum of polyproline II secondary structure, which indicates that native LjIDP1 has limited polyproline II structure that is lost upon heating.
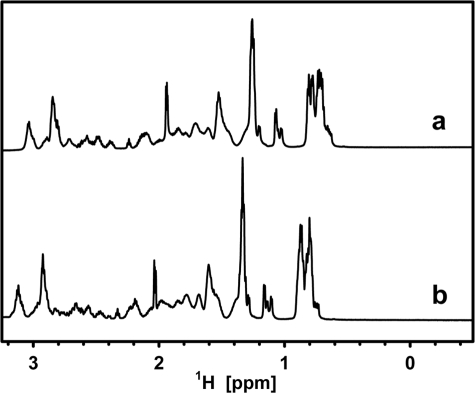
To examine the existence of heat-stable cooperatively folded segments, the NMR spectrum of native LjIDP1 was compared with that of LjIDP1 in a 6 m concentration of the chemical denaturant urea, which is expected to destroy most of the organized structure. The 1H spectrum of LjIDP1 shows no signal dispersion typical of globular proteins, and the similarity of the spectra in the aliphatic region with and without urea indicates that no major structural transitions occurred and supports the notion that LjIDP1 is intrinsically disordered (Fig. 4).
FIGURE 4.
Aliphatic/methyl region of one-dimensional NMR spectra of LjIDP1 in buffer (a) and in buffer plus 6 m urea (b). The low dispersion in a indicates that LjIDP1 lacks well defined structure, and the high similarity between the two spectra indicates that no significant structural rearrangements have occurred.
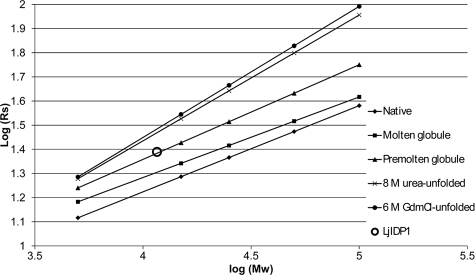
The different protein conformational classes (native, premolten globule, molten globule, urea- and GdmCl-unfolded) have characteristic dependences of the hydrodynamic dimensions, on molecular weight, and we analyzed this aspect of LjIDP1 by analytical gel filtration (16). The His6-tagged LjIDP1 preparation (molecular mass ∼11.6 kDa) has an elution volume that is similar to a globular protein of ∼28 kDa, which in turn corresponds to a Stokes radius of 24.52 Å. As seen in Fig. 5, this combination of molecular mass and Stokes radius suggests that LjIDP1 is in the so-called premolten globule state, which is a state characterized by moderate secondary structure and lack of well defined tertiary structure. This shows that LjIDP1 is a highly disordered and elongated protein that belongs to the “native premolten globule” class of IDPs.
FIGURE 5.
Size exclusion chromatography suggests that LjIDP1 belongs to the native premolten globule class of IDPs. The Stokes radius (Rs) of LjIDP1 was determined by gel filtration chromatography on a Superdex 75 HR 10/300 GL column. LjIDP1 elutes at 11.3 ml, which corresponds to a Stokes radius of 24.5 Å. The empirical equations (trend lines) representing the dependences of the hydrodynamic radii (Rs) on the molecular mass for different protein conformational states is from Uversky (16).
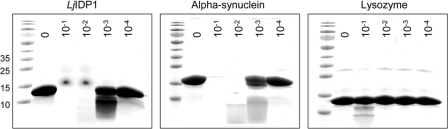
Another well established technique to probe the flexibility of a protein is to investigate the sensitivity to proteolytic degradation (17, 18). Consequently, LjIDP1 was incubated with decreasing amounts of the serine protease trypsin that predominantly cleaves proteins after basic lysine and arginine residues. Undigested LjIDP1 was visualized by SDS-PAGE. Fig. 6 shows that LjIDP1 protease sensitivity is comparable with the intrinsically disordered α-synuclein, whereas the globular protein lysozyme remains intact under the same conditions. This suggests that the amino acid backbone of LjIDP1 is largely accessible to proteolytic attack and in combination with the spectroscopic data unequivocally shows that LjIDP1 is largely disordered along the entire length.
FIGURE 6.
Protease sensitivity assay. LjIDP1, α-synuclein, and lysozyme were incubated with decreasing concentrations of trypsin for 1 h at 37 °C and analyzed by SDS-PAGE on a 10-20% polyacrylamide gradient gel. The numbers on the gels represent the relative fraction of trypsin (w/w).
LjIDP1 Is Prone to Adopt an α-Helix—Many IDPs undergo large conformational changes toward a more ordered conformation upon interaction with other biological components (3, 15, 19-21). If LjIDP1 undergoes such a functionally relevant disorder-to-order transition, the ordered state might be promoted upon the addition of structure-inducing agents. This hypothesis was tested using a number of additives, some of which are known to induce structure (trifluoroethanol (TFE), SDS, and methanol (MeOH)) and some that are not generally known to induce structure (CHAPS, deoxycholate, and cetyltrimethylammoniumbromide (CTAB)) (22-27). Interestingly, all of the tested compounds promoted formation of SR-CD-measurable α-helix structures in LjIDP1. In particular, the detergent CTAB and TFE were effective; both induced ∼40% α-helical structure in LjIDP1 (Fig. 3C). A comparison of the near UV CD spectrum of LjIDP1 in PBS buffer with the spectrum in the presence of CTAB (5 mm) or TFE (30%) revealed only minute differences in the spectra, suggesting that there was very little organized tertiary structure to lose/gain, just as one would expect from a fluctuating structure (not shown).
We further probed the structural propensities of LjIDP1 by analyzing the structure of the dehydrated protein. By dehydrating a protein, the hydrophobic driving force, which is dependent on the presence of bulk water, is lost, and this is expected to destabilize globular proteins. However, at the same time, the major destabilizing force, namely the entropic cost of folding, is expected to decrease, because conformational freedom of the polypeptide chain decreases as water is lost. These two effects of dehydration counteract each other. Since IDPs lack a well defined hydrophobic core, they are only affected by the decrease in the entropic cost of folding, which promotes the formation of a folded structure. Furthermore, most, if not all, backbone polar groups (CO and NH groups) form either intrapeptide hydrogen bonds (between polar groups in the protein) or interpeptide hydrogen bonds (between backbone CO/NH groups and solvent water). IDPs have many interpeptide hydrogen bonds, and dehydration drives the equilibrium between intra- and intermolecular hydrogen bonds toward intramolecular hydrogen bonds as found in α-helix and β-sheets. We monitored structural rearrangements occurring in LjIDP1 during dehydration by leaving the sample cell open in the dry nitrogen-purged sample chamber while continuously collecting CD spectra. Fig. 3B shows the CD spectra of a dehydration experiment of LjIDP1. It is clear that large structural rearrangements occur during dehydration and that the initial spectra are typical of an unfolded peptide, whereas the later spectra resemble that of an α-helical protein. As dehydration proceeds, the concentration of protein and buffer in the remaining liquid will gradually increase. In the experimental setup used, the sample cell is in a vertical position, and because evaporation occurs from the top, most protein will end up in the lower part of the cell (below the “beam window” where the CD signal is measured), which explains the gradual decrease in the intensity (amplitude) of the spectra. The data obtained are qualitative. Quantification of the structural changes is not possible, since the protein concentration in the sample cell changes during the experiment. The presence and shape of the intermediate spectra (spectra 19-39) could be taken as evidence of a gradual folding event between the disordered (wet) state and the ordered (dry) state. However, this is unlikely. From the high voltage signal, we can see that the period from spectrum ∼19 to ∼39 corresponds to the period where the liquid front passes the “beam window” (not shown). This means that any spectrum obtained within this time span includes contributions from both the dry and the wet state.
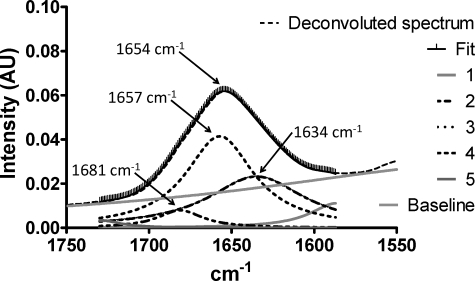
To exclude the possibility that the changes observed are caused by a systematic alignment of the protein during dehydration, which could influence the CD signal, the linear dichroism spectrum of the sample was collected between each CD scan. Linear dichroism is the difference in absorbance of light polarized parallel and perpendicular to the alignment axis. Thus, the greater the alignment of the sample, the greater the linear dichroism signal. The small magnitude of the linear dichroism signal (ΔAbs in the range of 10-4) spectra point toward a limited alignment of the dehydrated sample, and the effect on the CD signal is therefore unlikely to be significant. Similar results were obtained by FTIR spectroscopy of dry LjIDP1 (Fig. 7). Interestingly, the demonstrated propensity of LjIDP1 to adopt α-helical structure is supported by a good correlation between in silico secondary structure predictions and the experimentally determined secondary structure content. Using two different predictors of secondary structure content, PSIpred and Predictprotein, it is estimated that LjIDP1 contains between 30 and 40% α-helix and between 4 and 15% β-sheet, which is in the same range as the experimentally derived values in the presence of CTAB and TFE (28, 29). Obviously, CTAB and TFE are not physiological compounds, but they may mimic the structural changes induced by binding to naturally occurring ligands. The credibility of this approach is supported by a recent study of 24 IDPs characterized by undergoing a disorder-to-order transition upon functioning, in which the authors found a good correlation between the in silico predicted secondary structure content and the structure of the ordered state (30).
FIGURE 7.
FTIR spectrum of dehydrated LjIDP1. Curve fitting with five Lorentzian peaks produced an excellent fit. Curve 3 (1657 cm-1) was assigned to α-helix, curve 2 (1634 cm-1) to β-sheet, and curve 4 (1681 cm-1) to turns.
The demonstrated ability of LjIDP1 to readily undergo large structural rearrangements in part reflects the highly malleable nature of IDPs. However, the high propensity to adopt α-helix under various conditions suggests that this is an inherent and functionally relevant conformation of LjIDP1.
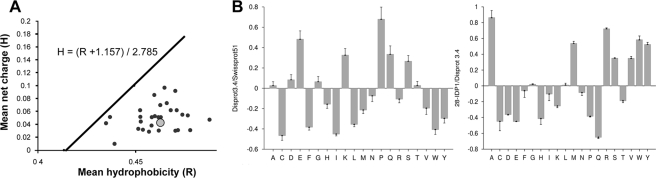
LjIDP1 Has an Unusual Amino Acid Profile—IDPs, compared with globular proteins, have distinct sequence profiles as evidenced by the existence of several programs that can successfully predict intrinsic disorder based on amino acid sequence. Having revealed a high α-helical propensity of LjIDP1, we analyzed whether this is reflected in a higher frequency of “order-promoting residues” in a data set called 28IDP1 containing LjIDP1 and 27 homologous proteins (BLAST search against uniref100 with a 10-3 cut-off) compared with an unbiased data set of IDPs called “Disprot 3.4,” containing 460 verified IDP entries and 1103 verified disordered regions from the Disprot data base (31). Order-promoting residues are here defined as residues overrepresented in IDPs, whereas residues underrepresented in IDPs are defined as “disorder-promoting residues.” An initial comparison of the amino acid composition of Disprot 3.4 with a data set, “Swissprot 51,” which has an amino acid distribution resembling that found in nature, was used to classify residues as either disorder- or order-promoting (Fig. 8B, first panel). The analysis was done using the World Wide Web-based tool “Composition Profiler,” which provides the Disprot 3.4 and Swissprot 51 data sets (32). This analysis revealed that disorder-promoting residues are mainly hydrophilic, whereas order-promoting residues are mainly hydrophobic. We then compared the sequence profile of 28IDP1 with Disprot 3.4 and discovered that all but four amino acids had significantly different representations in the two data sets. Out of the eight amino acids significantly depleted in 28IDP1, five (Gln, Glu, Pro, Asp, and Lys) are disorder-promoting (of which Lys, Gln, Glu, and Pro are the top disorder-promoting residues), two (Cys and His) are order-promoting, and one (Thr) is neutral. Among amino acids significantly enriched in 28IDP1, only one (Ser) is disorder-promoting, five (Val, Tyr, Met, Trp, and Arg) are classified as order-promoting, and one (Ala) is neutral. Furthermore, 28IDP1 is depleted in proline residues compared with Disprot 3.4. It is worth noting that the only three proline residues found in LjIDP1 are located in a short conserved proline-rich domain 64WVPDPVTGYYRP76 that also contains three residues (Thr, Tyr, and Tyr) capable of being phosphorylated. Proline is often found in regions engaging in transient protein-protein interactions, regulated by phosphorylation, and is known to prevent protein aggregation (33, 34).
FIGURE 8.
Composition profiling of LjIDP1 and homologous proteins (28IDP1). A, plot of mean hydrophobicity (H) against mean net charge (R) of 28IDP1 (LjIDP1 (gray circle) and 27 homologous proteins). This group has very similar R and H values, and interestingly, all are located below the boundary line separating small globular proteins (below the line) from IDPs (above the line). The boundary line satisfies the equation, H = (R + 1.157)/2.785. B, amino acid composition profiles of Disprot 3.4 versus Swissprot (first table) and 28IDP1 versus Disprot 3.4 (second table). Residues overrepresented in Disprot 3.4 compared with Swissprot 51 are defined as disorder-promoting, and residues underrepresented are defined as order-promoting. It is evident that disorder-promoting residues are mainly hydrophilic, whereas order-promoting residues are mainly hydrophobic. The subsequent profiling of 28IDP1 versus Disprot 3.4 reveals that all except five residues occur with significantly different frequencies in these data sets and that 28IDP1 has less disorder-promoting and more order-promoting residues than DisProt 3.4.
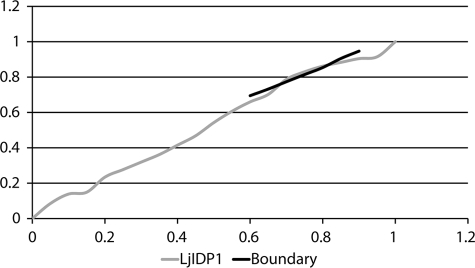
As mentioned, IDPs have distinct sequence profiles and are generally characterized by a high net charge and a high hydrophilicity compared with globular proteins (35, 36). We elaborated our sequence characterization by evaluating these aspects of the individual members of 28IDP1. For this purpose, the mean average hydrophobicity (R) was plotted against the mean net charge (H) for all 28 protein sequences in 28IDP1. Fig. 8A shows that members of 28IDP1 have very similar hydrophobicity and mean net charge, indicating that these parameters are structurally and/or functionally important. Interestingly, all members of 28IDP1 are found below the boundary line separating IDPs (above the boundary) from small globular proteins (below the boundary) as determined by Uversky et al. (36) (Fig. 8A). This result is consistent with predictions from two different predictors of protein disorder, Foldindex and PONDR, which predict LjIDP1 to be ∼60% ordered and ∼40% disordered, respectively (37-39). Proteins with R/H values below the boundary line can be either well ordered rigid structures or molten globules with substantial secondary structure and limited tertiary structure. Cumulative distribution function analysis of PONDR VL-XT scores in combination with R/H analysis has previously been used to distinguish these alternatives (40, 41). As seen in Fig. 9, the cumulative distribution function curve of LjIDP1 nearly superimposes on the boundary line separating ordered proteins (above) from disordered proteins (below), indicating that LjIDP1 is well ordered and flexible without falling into either of the archetypical classes.
FIGURE 9.
The cumulative distribution function curve of LjIDP1 PONDR VL-XT scores superimposes on the boundary line separating ordered proteins (above) from disordered proteins (below).
In conclusion, the amino acid profiling shows that members of 28IDP1 have an unusual amino acid composition compared with the average of IDPs with more order-promoting residues and less disorder-promoting residues. Furthermore, all members of 28IDP1 cluster in a small region of the R/H space that overlaps not with IDPs but with small globular proteins. The high frequency of order-promoting residues in 28IDP1 is consistent with its experimentally determined inherent propensity to adopt a largely α-helical conformation.
LjIDP1 Protects Enzymes from Desiccation-induced Inactivation—Structural disorder has been related to chaperone function and cellular protection against damage induced by various stresses, including heat, freezing, dehydration, salt, and osmotic and oxidative stress. When intrinsically disordered regions engage in interaction with ligands/partners, there is a large entropic cost, which lowers the binding affinity and makes IDPs suitable for transient interactions. Two elegant models have been proposed to explain the chaperone action of IDPs, whose salient feature is multiple rounds of transient interactions between the misfolded region and the IDP (42, 43). A common consequence of the above-mentioned stressors is protein misfolding leading to inactivation and aggregation.
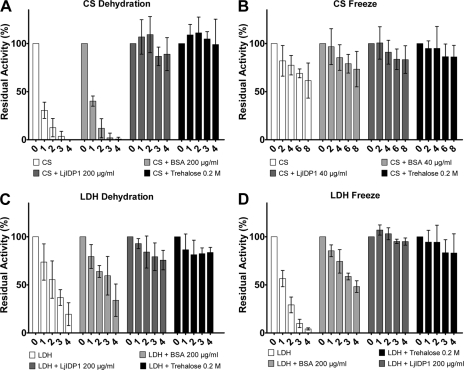
For this reason, we tested the ability of LjIDP1 to prevent the loss of enzymatic activity of two well established test enzymes, LDH and CS, subjected to freezing and dehydration stress, which is known to induce the formation of insoluble protein aggregates. When CS alone was subjected to repeated cycles of dehydration and rehydration, it formed insoluble aggregates accompanied by the loss of enzyme activity (Fig. 10A). After one round, less than a third of the original activity remained, and after three rounds, virtually no activity could be measured. As controls, the effect of well known cryoprotective agents and widely used additives bovine serum albumin (BSA) and trehalose was measured (44-46). Interestingly, the addition of 200 μg/ml LjIDP1 effectively prevented dehydration-induced inactivation of CS, and even after four cycles of dehydration/rehydration, the activity was retained. In contrast, at the same concentration, BSA had only a limited ability to protect CS activity. Protein concentrations were measured before and after the experiment, and as expected, in samples devoid of activity, the protein concentration was very low, whereas in samples with full activity, the initial concentration was retained, confirming that LjIDP1 prevented CS inactivation by preventing the formation of dehydration-induced insoluble protein aggregates (not shown).
FIGURE 10.
LjIDP1 prevents dehydration- and freezing-induced enzyme inactivation. A, residual activity of CS (250 μg/ml) before and after repeated cycles of dehydration/rehydration either alone of in the presence of stabilizing agents. C, similar experiment with LDH (100 μg/ml). LjIDP1 effectively prevents inactivation of CS and LDH even after four cycles of dehydration/rehydration. B, residual activity of CS (250 μg/ml) before and after repeated cycles of freezing/thawing, either alone or in the presence of stabilizing agents. D, similar experiment with LDH 100 μg/ml. CS alone is only slightly affected by freezing, and after eight cycles of freezing/thawing, it retains ∼60% activity. In contrast, LDH alone quickly loses activity upon freezing, but adding LjIDP1 effectively prevents this. Numbers on the x axis refer to the number of cycles. The activity at the start (cycle 0) of the experiments is set to 100%. Error bars, 95% confidence intervals.
In order to investigate if the ability of LjIDP1 to prevent dehydration-induced aggregation is a general feature, the same experiment was performed with LDH. As seen in Fig. 10C, LDH activity decreased upon dehydration although less pronouncedly than CS. This inactivation was almost completely abolished by the addition of 200 μg/ml LjIDP1, which was similar to the effect of adding high concentrations (0.2 m) of trehalose.
LjIDP1 Protects Enzymes against Freeze-induced Inactivation—Freezing is another type of stress that can induce protein misfolding and aggregation (46). In order to further characterize the ability of LjIDP1 to prevent stress induced inactivation, we tested the effect of LjIDP1 in preventing inactivation of LDH and CS undergoing repeated cycles of freezing and thawing. Fig. 10D shows that LDH alone quickly loses activity upon freezing, having only ∼10% residual activity after four freeze/thaw cycles. However, the addition of 200 μg/ml LjIDP1 efficiently prevents inactivation better than BSA.
The effect of freezing on the activity of CS is limited, and after eight freeze/thaw cycles, the activity is nearly 60% (Fig. 10B). The addition of LjIDP1 at a low concentration (40 μg/ml) offers protection against inactivation to an extent similar to BSA and slightly less than trehalose at high concentration.
DISCUSSION
The structural investigations presented here show that LjIDP1 is largely disordered and lacks any cooperatively folded regions, thereby placing LjIDP1 in the subgroup of IDPs that are disordered along the entire polypeptide chain. This is in agreement with LjIDP1 existing as an ensemble of ill defined, largely unordered conformers with limited fluctuating β-strand, turn/loop, and polyproline II structure.
It is noteworthy that in silico structure predictions of LjIDP1 are consistent with the experimentally determined structure of LjIDP1 in the induced folded state rather than the native unfolded state. A similar correlation between the in silico predictions and structure of the folded ligand-bound state opposed to the native disordered state was found for 24 IDPs (30). Furthermore, Prilusky et al. (37) compared the efficacy of four predictors and found Foldindex and PONDR to have a prediction efficiency of 77 and 72%, respectively, for IDPs and 88 and 93%, respectively, for folded proteins. The different efficiency in predicting IDPs and folded proteins is in part a consequence of these algorithms being optimized to minimize the rate of false prediction of disordered residues (48). The conservative estimates of disorder content by Foldindex and PONDR in combination with our results, which show that LjIDP1 is enriched in order-promoting and depleted in disorder-promoting residues compared with the average of IDPs, provide a possible explanation to the inaccurate prediction of the folding status of LjIDP1. Together, these studies uncover a need for modifications of the bioinformatics tools designed for predicting intrinsic structural disorder, possibly through a careful selection of the protein sequences used to train algorithms like PONDR VL-XT.
It is interesting to speculate about the role of the high α-helical propensity of LjIDP1. One possibility is that it may be mechanistically important, since it is well known that many IDPs undergo a disorder-to-order transition upon ligand binding.
Alternatively, being intrinsically disordered, LjIDP1 is predisposed to aggregate, and it is possible that the high α-helical propensity of LjIDP1 evolved to prevent aggregation, which is consistent with Chiti et al. (49) demonstrating an inverse correlation between α-helix propensity and aggregation propensity. Similarly, it is worth noting that other natively disordered LEA proteins with chaperone-like activities also display high α-helical propensities (50-54).
We have demonstrated that LjIDP1 effectively protects two model enzymes against stress-induced inactivation and aggregation. Similar chaperone-like activities have been reported for other IDPs α-synuclein, Tau, α- and β-casein, minichaperones (catalytically active fragments of larger chaperones), and a number of LEA proteins from different protein families (51, 55-60). Tompa and Csermely (42) presented an elegant mechanistic model, “the entropy transfer model,” which intimately relates intrinsic disorder to protein and RNA chaperone activities. This model includes several mechanistic elements that may apply to the protein-stabilizing effect of LjIDP1 and possibly other LEA proteins. One of the suggested mechanisms and the hallmark of this model is that transient interactions between the misfolded region and the IDP result in unfolding of the misfolded region, allowing it to retain the native conformation. The unfolding reaction is driven by an entropy transfer event, in which the loss of conformational entropy in the IDP upon interaction with the substrate can pay the thermodynamic cost of breaking the nonnative contacts in the misfolded region.
Alternatively, LjIDP1 might stabilize proteins by a mechanism involving transient hydrophobic interactions, a mechanism that was recently suggested to explain the effect of “small cageless chaperones” exemplified by α-casein and mini-GroEL, by two different groups using molecular dynamics simulations (43, 61). The “small cageless chaperones” were found to increase the folding yields of a model substrate through a combination of shielding misfolded aggregation prone surfaces and an increased refolding rate. Interestingly both the molecular dynamics studies and the “entropy transfer model” support a model for LjIDP1 action, which is dependent on rapid and transient interactions. A peculiar feature of LjIDP1 is that it contains only three proline residues, which is in contrast to the observation that IDPs in general are highly enriched in proline residues. It is noteworthy that these three residues are found in a conserved 12-amino acid domain. The biophysical properties of proline make it suitable for transient interactions in protein-protein interaction interfaces, and it is possible that this domain is implicated in the transient interactions that are a hallmark of the mechanistic models presented above (33, 62). It follows that it would be obvious to investigate the protein-stabilizing effect of a truncated LjIDP1 lacking the conserved domain and to analyze the protein-stabilizing effect, including the ability to prevent fibrillation, of this domain alone. Two short peptides, iAb11 (RDLPFFPVPID) and YiAb11 (RDLPFYPVPID), have been shown to effectively inhibit Aβ fibrillation (implicated in Alzheimer disease), and interestingly these peptides share sequence similarities with the proline-rich (64WVPDPVTGYYRP76) domain of LjIDP1 (63). Potentially, a small peptide, like the 12-residue conserved domain from LjIDP1, that effectively prevents fibrillation of, for example, α-synuclein (implicated in Parkinson and Alzheimer disease) could be of therapeutic value.
Based on the results presented here, we suggest that LjIDP1 functions as an integral member of the complement of factors dedicated to maintain cellular integrity. These results focus attention to the role of intrinsic disorder for the function of chaperones, and we suggest that “disordered chaperone-like proteins” exemplified by LjIDP1 could provide several advantages compared with classical chaperones. 1) Disordered chaperone-like proteins do not require an energy input in the form of ATP for functioning. 2) By functioning as molecular shields, disordered chaperone-like proteins can prevent aggregation of misfolded proteins. 3) Disordered chaperone-like proteins are much smaller than classical chaperones, thereby providing an energetic advantage in terms of cost of synthesis and contributing less to molecular crowding.
The LjIDP1 protein together with the Lea5 family belongs to a larger group of stress-related proteins, known as the LEA proteins, functionally implicated in the acquisition of tolerance to abiotic stresses (e.g. freezing, desiccation, and oxidative stress). Little is known about Lea5 members, but their expression can for the most part be correlated with high levels of ROS. The most well characterized Lea5 gene is A. thaliana AtLea5, which is induced by drought, by application of the stress hormone abscisic acid, by oxidative agents, and in senescent leaves of A. thaliana. This gene was recently identified in a screen for genes involved in oxidative stress tolerance and has been suggested to play a role in the controlled degradation of the photosynthetic apparatus in the dark and under stress conditions (64-66). Another study showed that transgenic Nicotiana tabacum overexpressing a Lea5 member from the shrub Tamarix androssowii had increased tolerance against desiccation and clear indications of reduced oxidative stress (67). Interestingly, two Lea5 members from poplar were recently shown to be up-regulated in rust (fungus)-infected leaves, which are known to elicit the ROS-associated hypersensitive plant defense response (68). Other Lea5 genes can be implicated with water (69) or water, salt, and heat stress (70), which are stressors usually accompanied by oxidative stress.
With this in mind, one may speculate whether LjIDP1 functions as a chaperone in protecting against ROS-induced protein destabilization. Moreover, we have compared the oxidant radical absorbance capacity of LjIDP1 with BSA and found LjIDP1 to perform ∼40% better than BSA (not shown). This suggests that LjIDP1, in addition to the putative function as a protein chaperone, may function as an antioxidant in vivo. Interestingly, the phenomenon of multiple functions of one protein, also known as “moonlighting,” is predicted to be a feature of IDPs (71).
The expression analysis of LjIdp1 presented here shows that LjIdp1 mRNA levels are high in the root tip proximal zone, inoculated roots, and mature nodules. The root tip proximal zone encompasses tissues undergoing large physiological changes involving differentiation and elongation of root cells and root hairs, which can be expected to be accompanied by a high respiration rate and concomitant high ROS production. Consistently, Ramu et al. (72) found constitutive high superoxide levels in the root tip proximal zone and an increased zone of superoxide production upon inoculation with Rhizobium.
Interestingly, a recent study showed that a Medicago truncatula homolog of LjIdp1 is highly expressed during nodule senescence along with catalase (antioxidant) and two proteases. Nodule senescence leading to death of both nodule cells and bacteroids is characterized by a general degradation of cellular constituents and probably involves ROS generation (47, 73).
Hence, the expression pattern of LjIdp1 presented here could correlate well with high ROS levels, produced either as a consequence of high respiration rates in the root tip proximal zone and nodules and possibly as part of the nodule autoregulation mechanism and nodule senescence. However, it is worth noting that LjIdp1 mRNA was found in all tissue types tested, albeit at different levels, indicating that the function is not nodulation-specific.
Acknowledgments
S.H. is grateful to Morten Dueholm and Steen Vang Petersen for providing expert advice and technical assistance for the FTIR and MS analysis, respectively.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: IDP, intrinsically disordered (or natively unfolded) protein; LEA, late embryogenesis-abundant; ROS, reactive oxygen species; Ek, enterokinase; MALDI, matrix-assisted laser desorption ionization; FTIR, Fourier transform infrared; CS, citrate synthase; BSA, bovine serum albumin; LDH, lactate dehydrogenase; SR-CD, synchrotron radiation CD spectroscopy; TFE, trifluoroethanol; CHAPS, 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid; CTAB, cetyltrimethylammoniumbromide.
N. Jorgensen, S. Radutoiu, L. Krusell, V. Voroshilova, M. Hannah, N. Goffard, D. Sanchez, F. Lippold, M. Udvardi, T. Ott, S. Sato, S. Tabata, P. Liboriussen, G. Vestergaard, L. Schauser, and J. Stougaard, manuscript in preparation.
References
- 1.Uversky, V. N., Oldfield, C. J., and Dunker, A. K. (2008) Annu. Rev. Biophys. 37 215-246 [DOI] [PubMed] [Google Scholar]
- 2.Tunnacliffe, A., and Wise, M. J. (2007) Naturwissenschaften 94 791-812 [DOI] [PubMed] [Google Scholar]
- 3.Dyson, H. J., and Wright, P. E. (2005) Nat. Rev. Mol. Cell Biol. 6 197-208 [DOI] [PubMed] [Google Scholar]
- 4.Colebatch, G., Desbrosses, G., Ott, T., Krusell, L., Montanari, O., Kloska, S., Kopka, J., and Udvardi, M. K. (2004) Plant J. 39 487-512 [DOI] [PubMed] [Google Scholar]
- 5.Walsh, K. B., Vessey, J. K., and Layzell, D. B. (1987) Plant Physiol. 85 137-144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pauly, N., Pucciariello, C., Mandon, K., Innocenti, G., Jamet, A., Baudouin, E., Herouart, D., Frendo, P., and Puppo, A. (2006) J. Exp. Bot. 57 1769-1776 [DOI] [PubMed] [Google Scholar]
- 7.Vasse, J., Billy, F., and Truchet, G. (1993) Plant J. 4 555-566 [Google Scholar]
- 8.Stacey, G., McAlvin, C. B., Kim, S. Y., Olivares, J., and Soto, M. J. (2006) Plant Physiol. 141 1473-1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Santos, R., Herouart, D., Sigaud, S., Touati, D., and Puppo, A. (2001) Mol. Plant Microbe Interact. 14 86-89 [DOI] [PubMed] [Google Scholar]
- 10.Lees, J. G., Smith, B. R., Wien, F., Miles, A. J., and Wallace, B. A. (2004) Anal. Biochem. 332 285-289 [DOI] [PubMed] [Google Scholar]
- 11.Compton, L. A., and Johnson, W. C., Jr. (1986) Anal. Biochem. 155 155-167 [DOI] [PubMed] [Google Scholar]
- 12.Whitmore, L., and Wallace, B. A. (2004) Nucleic Acids Res. 32 W668-73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Srere, P. A. (1966) J. Biol. Chem. 241 2157-2165 [PubMed] [Google Scholar]
- 14.Miles, A. J., and Wallace, B. A. (2006) Chem. Soc. Rev. 35 39-51 [DOI] [PubMed] [Google Scholar]
- 15.Uversky, V. N. (2002) Protein Sci. 11 739-756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uversky, V. N. (2002) Eur. J. Biochem. 269 2-12 [DOI] [PubMed] [Google Scholar]
- 17.Tsai, C. J., Polverino de Laureto, P., Fontana, A., and Nussinov, R. (2002) Protein Sci. 11 1753-1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Otzen, D. E., Lundvig, D. M., Wimmer, R., Nielsen, L. H., Pedersen, J. R., and Jensen, P. H. (2005) Protein Sci. 14 1396-1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sugase, K., Dyson, H. J., and Wright, P. E. (2007) Nature 447 1021-1025 [DOI] [PubMed] [Google Scholar]
- 20.Dyson, H. J., and Wright, P. E. (2002) Curr. Opin. Struct. Biol. 12 54-60 [DOI] [PubMed] [Google Scholar]
- 21.Tompa, P. (2005) FEBS Lett. 579 3346-3354 [DOI] [PubMed] [Google Scholar]
- 22.Karlin, D., Longhi, S., Receveur, V., and Canard, B. (2002) Virology 296 251-262 [DOI] [PubMed] [Google Scholar]
- 23.Longhi, S., Receveur-Brechot, V., Karlin, D., Johansson, K., Darbon, H., Bhella, D., Yeo, R., Finet, S., and Canard, B. (2003) J. Biol. Chem. 278 18638-18648 [DOI] [PubMed] [Google Scholar]
- 24.Sonnichsen, F. D., Van Eyk, J. E., Hodges, R. S., and Sykes, B. D. (1992) Biochemistry 31 8790-8798 [DOI] [PubMed] [Google Scholar]
- 25.Lehrman, S. R., Tuls, J. L., and Lund, M. (1990) Biochemistry 29 5590-5596 [DOI] [PubMed] [Google Scholar]
- 26.Nelson, J. W., and Kallenbach, N. R. (1986) Proteins 1 211-217 [DOI] [PubMed] [Google Scholar]
- 27.Jirgenson, B. (1961) Arch. Biochem. Biophys. 94 59-67 [DOI] [PubMed] [Google Scholar]
- 28.Rost, B., Yachdav, G., and Liu, J. (2004) Nucleic Acids Res. 32 W321-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jones, D. T. (1999) J. Mol. Biol. 292 195-202 [DOI] [PubMed] [Google Scholar]
- 30.Fuxreiter, M., Simon, I., Friedrich, P., and Tompa, P. (2004) J. Mol. Biol. 338 1015-1026 [DOI] [PubMed] [Google Scholar]
- 31.Sickmeier, M., Hamilton, J. A., LeGall, T., Vacic, V., Cortese, M. S., Tantos, A., Szabo, B., Tompa, P., Chen, J., Uversky, V. N., Obradovic, Z., and Dunker, A. K. (2007) Nucleic Acids Res. 35 D786-D793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vacic, V., Uversky, V. N., Dunker, A. K., and Lonardi, S. (2007) BMC Bioinformatics 8 211-218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kay, B. K., Williamson, M. P., and Sudol, M. (2000) FASEB J. 14 231-241 [PubMed] [Google Scholar]
- 34.Rousseau, F., Serrano, L., and Schymkowitz, J. W. (2006) J. Mol. Biol. 355 1037-1047 [DOI] [PubMed] [Google Scholar]
- 35.Radivojac, P., Iakoucheva, L. M., Oldfield, C. J., Obradovic, Z., Uversky, V. N., and Dunker, A. K. (2007) Biophys. J. 92 1439-1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uversky, V. N., Gillespie, J. R., and Fink, A. L. (2000) Proteins 41 415-427 [DOI] [PubMed] [Google Scholar]
- 37.Prilusky, J., Felder, C. E., Zeev-Ben-Mordehai, T., Rydberg, E. H., Man, O., Beckmann, J. S., Silman, I., and Sussman, J. L. (2005) Bioinformatics 21 3435-3438 [DOI] [PubMed] [Google Scholar]
- 38.Romero, P., Obradovic, Z., Li, X., Garner, E. C., Brown, C. J., and Dunker, A. K. (2001) Proteins 42 38-48 [DOI] [PubMed] [Google Scholar]
- 39.Obradovic, Z., Peng, K., Vucetic, S., Radivojac, P., and Dunker, A. K. (2005) Proteins 61 Suppl. 7, 176-182 [DOI] [PubMed] [Google Scholar]
- 40.Mohan, A., Sullivan, W. J., Jr., Radivojac, P., Dunker, A. K., and Uversky, V. N. (2008) Mol. Biosyst. 4 328-340 [DOI] [PubMed] [Google Scholar]
- 41.Dunker, A. K., Obradovic, Z., Romero, P., Garner, E. C., and Brown, C. J. (2000) Genome Inform. Ser. Workshop Genome Inform. 11 161-171 [PubMed] [Google Scholar]
- 42.Tompa, P., and Csermely, P. (2004) FASEB J. 18 1169-1175 [DOI] [PubMed] [Google Scholar]
- 43.Jewett, A. I., and Shea, J. E. (2006) J. Mol. Biol. 363 945-957 [DOI] [PubMed] [Google Scholar]
- 44.Santarius, K. A., and Franks, F. (1998) Cryo-letters 19 37-48 [Google Scholar]
- 45.Crowe, J. H., Crowe, L. M., Oliver, A. E., Tsvetkova, N., Wolkers, W., and Tablin, F. (2001) Cryobiology 43 89-105 [DOI] [PubMed] [Google Scholar]
- 46.Tamiya, T., Okahashi, N., Sakuma, R., Aoyama, T., Akahane, T., and Matsumoto, J. J. (1985) Cryobiology 22 446-456 [DOI] [PubMed] [Google Scholar]
- 47.Alesandrini, F., Mathis, R., Van de Sype, G., Herouart, D., and Puppo, A. (2003) New Phytol. 158 131-138 [Google Scholar]
- 48.Oldfield, C. J., Cheng, Y., Cortese, M. S., Brown, C. J., Uversky, V. N., and Dunker, A. K. (2005) Biochemistry 44 1989-2000 [DOI] [PubMed] [Google Scholar]
- 49.Chiti, F., Stefani, M., Taddei, N., Ramponi, G., and Dobson, C. M. (2003) Nature 424 805-808 [DOI] [PubMed] [Google Scholar]
- 50.Goyal, K., Tisi, L., Basran, A., Browne, J., Burnell, A., Zurdo, J., and Tunnacliffe, A. (2003) J. Biol. Chem. 278 12977-12984 [DOI] [PubMed] [Google Scholar]
- 51.Goyal, K., Walton, L. J., and Tunnacliffe, A. (2005) Biochem. J. 388 151-157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grelet, J., Benamar, A., Teyssier, E., Avelange-Macherel, M. H., Grunwald, D., and Macherel, D. (2005) Plant Physiol. 137 157-167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tolleter, D., Jaquinod, M., Mangavel, C., Passirani, C., Saulnier, P., Manon, S., Teyssier, E., Payet, N., Avelange-Macherel, M. H., and Macherel, D. (2007) Plant Cell 19 1580-1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pouchkina-Stantcheva, N. N., McGee, B. M., Boschetti, C., Tolleter, D., Chakrabortee, S., Popova, A. V., Meersman, F., Macherel, D., Hincha, D. K., and Tunnacliffe, A. (2007) Science 318 268-271 [DOI] [PubMed] [Google Scholar]
- 55.Zhang, X., Fu, X., Zhang, H., Liu, C., Jiao, W., and Chang, Z. (2005) Int. J. Biochem. Cell Biol. 37 1232-1240 [DOI] [PubMed] [Google Scholar]
- 56.Bhattacharyya, J., and Das, K. P. (1999) J. Biol. Chem. 274 15505-15509 [DOI] [PubMed] [Google Scholar]
- 57.Bhattacharyya, J., Padmanabha Udupa, E. G., Wang, J., and Sharma, K. K. (2006) Biochemistry 45 3069-3076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tian, R., Nie, C. L., and He, R. Q. (2004) Neurochem. Res. 29 1863-1872 [DOI] [PubMed] [Google Scholar]
- 59.Chatellier, J., Hill, F., Lund, P. A., and Fersht, A. R. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 9861-9866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mouillon, J. M., Gustafsson, P., and Harryson, P. (2006) Plant Physiol. 141 638-650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stan, G., Brooks, B. R., and Thirumalai, D. (2005) J. Mol. Biol. 350 817-829 [DOI] [PubMed] [Google Scholar]
- 62.Petrella, E. C., Machesky, L. M., Kaiser, D. A., and Pollard, T. D. (1996) Biochemistry 35 16535-16543 [DOI] [PubMed] [Google Scholar]
- 63.Poduslo, J. F., Curran, G. L., Kumar, A., Frangione, B., and Soto, C. (1999) J. Neurobiol. 39 371-382 [PubMed] [Google Scholar]
- 64.Mowla, S. B., Cuypers, A., Driscoll, S. P., Kiddle, G., Thomson, J., Foyer, C. H., and Theodoulou, F. L. (2006) Plant J. 48 743-756 [DOI] [PubMed] [Google Scholar]
- 65.Gosti, F., Bertauche, N., Vartanian, N., and Giraudat, J. (1995) Mol. Gen. Genet. 246 10-18 [DOI] [PubMed] [Google Scholar]
- 66.Weaver, L. M., Gan, S., Quirino, B., and Amasino, R. M. (1998) Plant Mol. Biol. 37 455-469 [DOI] [PubMed] [Google Scholar]
- 67.Wang, Y., Jiang, J., Zhao, X., Liu, G., Yang, C., and Zhan, L. (2006) Plant Sci. 171 655-662 [Google Scholar]
- 68.Rinaldi, C., Kohler, A., Frey, P., Duchaussoy, F., Ningre, N., Couloux, A., Wincker, P., Le Thiec, D., Fluch, S., Martin, F., and Duplessis, S. (2007) Plant Physiol. 144 347-366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Galau, G. A., Wang, H. Y., and Hughes, D. W. (1993) Plant Physiol. 101 695-696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Naot, D., Ben-Hayyim, G., Eshdat, Y., and Holland, D. (1995) Plant Mol. Biol. 27 619-622 [DOI] [PubMed] [Google Scholar]
- 71.Tompa, P., Szász, C., and Buday, L. (2005) Trends Biochem. Sci. 30 484-489 [DOI] [PubMed] [Google Scholar]
- 72.Ramu, S. K., Peng, H., and Cook, D. R. (2002) Mol. Plant-Microbe Interact. 15 522-528 [DOI] [PubMed] [Google Scholar]
- 73.Pladys, D., and Vance, C. P. (1993) Plant Physiol. 103 379-384 [DOI] [PMC free article] [PubMed] [Google Scholar]