Abstract
Glucocorticoids (GCs) play important roles in numerous cellular processes, including growth, development, homeostasis, inhibition of inflammation, and immunosuppression. Here we found that GC-treated human lung carcinoma A549 cells exhibited the enhanced formation of the thick stress fibers and focal adhesions, resulting in suppression of cell migration. In a screen for GC-responsive genes encoding actin-interacting proteins, we identified caldesmon (CaD), which is specifically up-regulated in response to GCs. CaD is a regulatory protein involved in actomyosin-based contraction and the stability of actin filaments. We further demonstrated that the up-regulation of CaD expression was controlled by glucocorticoid receptor (GR). An activated form of GR directly bound to the two glucocorticoid-response element-like sequences in the human CALD1 promoter and transactivated the CALD1 gene, thereby up-regulating the CaD protein. Forced expression of CaD, without GC treatment, also enhanced the formation of thick stress fibers and focal adhesions and suppressed cell migration. Conversely, depletion of CaD abrogated the GC-induced phenotypes. The results of this study suggest that the GR-dependent up-regulation of CaD plays a pivotal role in regulating cell migration via the reorganization of the actin cytoskeleton.
Glucocorticoids (GCs)2 strongly affect numerous biological processes, including cell growth, development, homeostasis, inhibition of inflammation, and immunosuppression (1-4). Because of their pharmacological properties, they have been widely used to treat inflammatory and autoimmune diseases (5, 6). They are also potent chemotherapeutic agents for malignant lymphoma, advanced prostate cancer, and some other solid cancers (7-9). Since their discovery, our insight into the molecular mechanisms of GC functions has grown considerably. Glucocorticoid receptor (GR) is critical for transduction of the GC signal (10, 11) and is mainly located in the cytosol when not bound by ligands. Upon ligand binding, GR translocates into the nucleus, binds to the cognate DNA element (GC response element, GRE) within the promoter regions of target genes, and then activates or represses their transcription (10). Despite the many reports on the biological effects of GCs, their effect on cell motility has been little studied.
Cell migration is an essential cellular process for embryogenesis, organogenesis, immune response, and wound healing (12). It is also involved in various pathological events, such as cancer progression (13, 14). During invasion and metastasis, cancer cells dissociate from the primary tumor foci and invade distant target tissues (13). These processes depend on cancer-cell motility. Cell migration is regulated by complex changes in the cytoskeleton, particularly in the actin cytoskeleton, which plays key roles in controlling cell migration and morphology (12, 15, 16).
Caldesmon (CaD) is an actin-, tropomyosin-, and calmodulin-binding protein (17, 18). Two isoforms with different molecular weights (Mr), generated from a single gene by alternative splicing, have been identified as follows: high Mr CaD (h-CaD; 120-150 kDa) and low Mr CaD (l-CaD; 70-80 kDa) (18, 19). h-CaD is exclusively expressed in smooth muscle cells (SMCs), but l-CaD is widely expressed in non-muscle cells (18). h-CaD is a component of smooth muscle thin filaments and regulates their contraction via inhibition of the actin-myosin interaction (20, 21). l-CaD has an alternative function for stabilizing actin filaments in non-muscle cells, in addition to regulating contraction (22, 23). In non-muscle cells, l-CaD is located along stress fibers, membrane ruffles, and lamellipodial extensions (24-26). Overexpression of l-CaD leads to the reorganization of the actin cytoskeleton that correlates with decreased cell motility in several cell lines (26-28). In SMCs and some fibroblasts, transcription of the caldesmon (CALD1) gene is mainly dependent on serum-response factor (SRF) (29-31). It had been reported previously that CaD expression is induced by GC stimulation (32, 33). However, the molecular mechanism underlying the GC-induced expression of CaD remained unknown for a decade.
Here we showed that GCs suppress the migration of lung carcinoma cells, which showed marked enhancement of thick stress fiber formation and focal adhesion assembly, in a Rho-independent manner. By screening for GC-responsive genes that encode actin-interacting proteins, we identified CaD as being specifically responsive to GCs. We found that the CALD1 gene is transcribed GR-dependently. Furthermore, increased expression of CaD was closely correlated with the suppression of cell motility. These results indicate that CaD plays a pivotal role in cell migration in response to GCs, and will shed light on the mechanisms underlying the effects of GCs.
EXPERIMENTAL PROCEDURES
Materials—Phenol red-free Dulbecco's modified Eagle's medium (DMEM), dexamethasone (DEX), cortisol (hydrocortisone), progesterone, β-estradiol, and aldosterone were purchased from Sigma. Fetal bovine serum (FBS) and geneticin were obtained from Invitrogen. To remove endogenous steroids from FBS, it was treated with charcoal-dextran (Sigma) before use (34). Cycloheximide (CHX), cytochalasin D (cytD), and blebbistatin were obtained from Merck. Testosterone, RU486, RU28318 potassium salt, and mitomycin C were obtained from TCI, Cayla Tocris, and Nacalai Tesque, respectively. Anti-CaD antibody was prepared and purified as described previously (35). The following primary antibodies were purchased: mouse anti-α-tubulin (T9026), mouse and rabbit anti-FLAG (F1804, F7425), and mouse anti-vinculin (V9131) were from Sigma; mouse anti-RhoA (sc-418), rabbit anti-GAPDH, (sc-25778), rabbit anti-GR (sc-8992), rabbit anti-SRF (sc-335), goat anti-ROCK2 (sc-1851), and rabbit anti-p-cofilin1 (Ser-3) (sc-12912-R) were from Santa Cruz Biotechnology; mouse and rabbit anti-GFP (A-11120, A-11122) were from Invitrogen; rabbit anti-ROCK1 (4035S), rabbit anti-phosphomyosin light chain 2 (Ser-19) (3671), rabbit anti-myosin light chain 2 (3672), and rabbit anti-profilin1 (3237) were from Cell Signaling Technology; mouse anti-mDia1 (610848) and mouse anti-cofilin1 (612144) were from BD Transduction Laboratories; and mouse anti-non-muscle heavy chain myosin (ab684) were from Abcam. The secondary antibodies, horseradish peroxidase-conjugated donkey anti-rabbit IgG (NA934) and horseradish peroxidase-conjugated sheep anti-mouse IgG (NA931), were purchased from GE Healthcare.
Cell Culture—The human lung carcinoma cell line A549 (National Institutes of Health Science) was cultured in phenol red-free DMEM supplemented with 10% FBS and antibiotics in a 37 °C incubator with 5% CO2.
Expression Vectors and Transfection—The coding region of human glucocorticoid receptor α (GRα) was amplified by PCR using cDNA from A549 cells as a template. To prepare a constitutively active form of GRα, the ligand-binding domain of the carboxyl terminus (525-777) was deleted (GRΔC) (36). The FLAG tag sequence was fused to the 3′ end of the coding sequences of GR and GRΔC by PCR. The fragments were inserted into the mammalian expression vector pCS2(+) (Invitrogen). A549 cells were transiently transfected with these expression vectors using the Lipofectamine 2000 or Lipofectamine LTX transfection reagent (Invitrogen). To establish stable cell lines expressing GFP, GFP-CaD, or GRΔC-cFLAG, A549 cells were transfected by retroviral infection, using a retroviral gene delivery and expression system. The pLNCX2-GFP, pLNCX2-GFP-CaD, and pLNCX2-GRΔC-cFLAG plasmids were constructed by inserting the coding sequence of enhanced GFP, GFP-CaD (37), or GRΔC-cFLAG into the pLNCX2 plasmid (Clontech). Using the BD Retro-X Universal Retroviral Expression System (Clontech), amphotropic retroviral particles were produced by GP2-293 packaging cells. A549 cells were infected with the retrovirus in the presence of 4 μg/ml Polybrene for 24 h. The infected cells were cultured with 1000 μg/ml Geneticin (Invitrogen), and the drug-resistant cells were selected.
RNA Interference—Small interference RNAs (siRNA) for human CaD (CaD siRNA1, sequence was not supplied, sc-29880, Santa Cruz Biotechnology; and CaD siRNA2, target sequence, CAGATAGGTATCAATATGTTT, Qiagen), human GR (target sequence, GAGGATCATGACTACGCTCAA, Qiagen), human SRF (target sequence, CCGCGTGAAGATCAAGATGGA, Qiagen), and a control (sc-37007, Santa Cruz Biotechnology) were purchased. The cells were transfected with the siRNAs using Lipofectamine RNAiMAX transfection reagent (Invitrogen), cultured for 1 day, and then used in subsequent assays.
Immunofluorescence Microscopic and Time-lapse Image Analyses—Cells grown on coverslips were fixed with 4% formaldehyde and 4% sucrose in phosphate-buffered saline (PBS) for 30 min at room temperature and then permeabilized in blocking solution (0.2% Triton X-100, 0.2% bovine serum albumin, 10% normal goat serum in PBS). The cells were incubated with diluted primary antibody in Can Get Signal Immunostain (Toyobo), followed by the secondary antibody as indicated. To visualize actin filaments, Alexa488-, Alexa568-, or Alexa350-conjugated phalloidin (Invitrogen) was added to the secondary antibody solution. For nuclear staining, Hoechst 33342 or propidium iodide (Invitrogen) was used. For cytochalasin D treatment, cells were treated with 0.2 μm cytD for 20 min. For Triton extraction, cells were treated with 0.1% Triton X-100 in PBS for 3 min at room temperature before fixation. Samples were observed under the IX70 inverted fluorescence microscope (Olympus). Three-dimensional images were obtained using the LSM 5 PASCAL laser scanning microscope (Carl Zeiss). For time-lapse image analysis, A549 cells were observed under an Axiovert 200 M microscope (Carl Zeiss). For cell motility assay, the images of migrating cells were captured over a 5-h span with intervals of 5 min. For cell contraction assay, the images were captured over a 1.5-h span with intervals of 3 min. The frames showing migrating cells were captured manually, and the parameters were analyzed using the DIAS (Soll Technologies, Inc) and ImageJ software.
Migration Assay—The migration assay was performed in 24-well Transwell chambers (BD Biosciences) with a polyethylene terephthalate membrane (8.0-μm pore size). The cells were treated with DEX or other steroids for 48 h. Dimethyl sulfoxide (DMSO) was used as the vehicle. To avoid artifactual data from cell proliferation, the cells were pretreated with 10 μg/ml mitomycin C for 2 h. The cells were suspended in serum-free DMEM and plated in a Transwell chamber (2.5 × 104 cells/well). The lower chamber was filled with DMEM containing 10% FBS. The indicated drugs were added to both the upper and lower chambers. After 24 h to allow migration to occur, the cells on the upper side of the filters were mechanically removed. The cells that had migrated to the lower side were fixed with 4% formaldehyde, stained with hematoxylin-eosin, and counted under a light microscope.
Cell Contraction Assay—Cells were starved with serum-free DMEM for 18 h, and then stimulated with an equal volume of pre-warmed DMEM containing 10% FBS (final concentration: 5%) under phase-contrast microscope. For blebbistatin treatment, 10 μm blebbistatin was treated for 30 min prior to serum stimulation. Cell areas at the indicated time points were calculated, and reduction in cell area over time was used as the criterion for cell contraction (38, 39).
Quantitative Real Time PCR (Real Time qPCR)—The total RNA was extracted from A549 cells using TRIzol reagent (Invitrogen) and reverse-transcribed with MultiScribe reverse transcriptase (Applied Biosystems). The cDNA was amplified with gene-specific primer pairs using SYBR GreenER qPCR SuperMix Universal reagent (Invitrogen). The primer sequences used in this study are listed in supplemental Table S1. The quantities measured by real time PCR were normalized to the GAPDH expression level in each sample.
Western Blot Analysis—The cells were rinsed with PBS followed by lysis in SDS sample buffer. The lysates were separated by electrophoresis in 10-15% polyacrylamide gels, and the separated proteins were transferred onto a nitrocellulose membrane. The membrane was incubated with primary antibody diluted in 5% nonfat dry milk in Tris-buffered saline containing 0.1% Tween 20 (TBS-T), followed by the appropriate horseradish peroxidase-conjugated secondary antibody, as indicated. ImageJ software was used to quantify the band intensity, and the value was normalized to the α-tubulin expression level in each sample. To determine Rho activity, Rho binding domain (RBD)-agarose pulldown assay was performed using a Rho activation kit (Upstate Biotechnology, Inc.), according to the manufacturer's instructions. A549 cells were cultured with vehicle or 1 μm DEX for 24 h, quickly washed with ice-cold PBS, and then lysed in lysis buffer. Equal volumes of cell lysates were incubated with 30 μl of GST-RBD beads for 45 min at 4 °C. The bound Rho protein was eluted by SDS-sample buffer. The proteins in the samples from the RBD beads, and the total cell lysates were separated by SDS-PAGE and transferred onto a polyvinylidene difluoride membrane. The amount of RhoA protein was analyzed by Western blot using anti-RhoA antibody.
Reporter Assays—The HeLa-type promoter region of the human CALD1 gene (-1908 to +207) was amplified by PCR and cloned into pGL3-basic. The reporter construct for the fibroblast-type promoter region of the human CALD1 gene was described previously (31). For the mutant constructs, several substitutions were introduced into the two GRE-like sequences in the fibroblast-type promoter as follows: for GRE-like 1, TGTTCACTTAGCATGGA→ TTTTAACTTAACATGGA; for GRE-like 2, AGAGCAGTGTGTATTC→ AAAGAAGTGTATATTC. A549 cells were transfected with these constructs and pGL3-β-gal for normalization of the transfection efficiency. In some experiments, cells were co-transfected with pCS2(+)hGRΔC-cFLAG. Twenty four h after the transfection, the cells were lysed with Passive Lysis buffer (Promega), and the luciferase and β-galactosidase activities were measured using the luciferase assay system (Promega) and luminescent β-galactosidase detection kit II (Clontech), respectively.
Chromatin Immunoprecipitation (ChIP) Assay—The ChIP assay was performed using a chromatin immunoprecipitation kit (Upstate Biotechnology, Inc.) according to the manufacturer's instructions. A549 cells were cultured with vehicle or 1 μm DEX for 2 h. The cells were fixed with formaldehyde, and the DNA and bound proteins were cross-linked. ChIP assays were performed by co-precipitating the DNA-protein complexes with anti-GRα antibodies or rabbit control IgG (Santa Cruz Biotechnology). The promoter region (-94 to +47) of the human CALD1 gene, which contains the two GRE-like sequences, was amplified from the prepared DNA samples using the primer pair TCCCGACTGTAAACATAGGGGATA and ACAGCCAGAGAGCAAGCAG (fragment size, 141 bp). The GRE-containing promoter region (-1233 to -1073) of the GC-responsive serum and glucocorticoid-inducible kinase 1 (SGK1) gene was amplified using the primer pair CTCACGTGTTCTTGGCATGG and GGAGGGGGCGGAAATAAAAG (fragment size, 160 bp) as a positive control (40). The promoter region (-777 to -516) of the GC-unresponsive GAPDH gene, which does not contain GREs, was also amplified, using the primer pair GATTGTCTGCCCTAATTATC and CAGGCAAAGGCCTAGGAG (fragment size, 261 bp). The amplified products were run on an agarose gel, and the ethidium bromide-stained DNA bands were photographed.
DNA-binding Assay—Human GRα-c-FLAG was synthesized using the TnT high yield in vitro transcription/translation system (Promega) and precleared with Dynal Dynabeads M280 streptavidin (Invitrogen). The oligonucleotides for CaDGRE1 (biotin-GGGATATGTGTTCACTTAGCATGGACTTCT and AGAAGTCCATGCTAAGTGAACACATATCCC), CaDGRE1mut (biotin-GGGATATGTTTTAACTTAACATGGACTTCT and AGAAGTCCATGTTAAGTTAAAACATATCCC), CaDGRE2 (biotin-TATTGAATAGAGCAGTGTGTATTCGGCTGC and GCAGCCGAATACACACTGCTCTATTCAATA), and CaDGRE2mut (biotin-TATTGAATAAAGAAGTGTATATTCGGCTGC and GCAGCCGAATATACACTTCTTTATTCAATA) were annealed and used as affinity probes. The synthesized proteins were incubated with 1 μg of biotinylated DNA fragment in DNA-binding buffer (20 mm HEPES-KOH, pH 7.9, 80 mm KCl, 1 mm MgCl2, 0.2 mm EDTA, 0.5 mm dithiothreitol, 10% glycerol, 0.1% Triton X-100, 50 μg/ml herring sperm DNA, 1% protease inhibitor mixture (Nacalai Tesque)) at 4 °C for 1.5 h. The Dynabeads were then added to the reaction mixtures, which were incubated at room temperature for 1 h. After three washes with DNA-binding buffer, SDS-sample buffer was added to the Dynabeads mixture for SDS-PAGE.
RESULTS
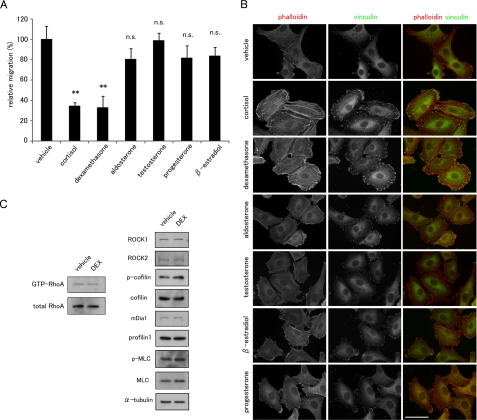
Glucocorticoids Suppress Cell Migration and Reorganize the Actin Cytoskeleton in a Rho-independent Manner—We first examined the effects of GCs on cell migration and morphology. To exclude the effects of any endogenous steroids in FBS, we removed them by charcoal treatment (34) and used phenol red-free DMEM. The cells were treated with each steroid at 1 μm for 48 h and exposed to mitomycin C for 2 h before the assay to prevent the effect of steroids on cell proliferation. In our preliminary experiments, we confirmed that mitomycin C inhibits cell proliferation but not cell viability during this assay. Among the steroids examined, cortisol and synthetic glucocorticoid DEX potently suppressed the motility of human non-small cell lung carcinoma A549 cells. Mineralocorticoid, aldosterone, and sex steroids, including testosterone, progesterone, and β-estradiol, did not induce significant changes in cell migration (Fig. 1A). Phalloidin staining revealed the formation of prominent, straight, thick stress fibers in cortisol- and DEX-treated A549 cells, but no such actin-dependent cytoskeletal changes were seen in cells treated with the other steroids (Fig. 1B). Cortisol and DEX also enhanced the formation of focal adhesions, which were peripherally located at both ends of the stress fiber arrays, as visualized by anti-vinculin antibody staining (Fig. 1B). Because out of the cell lines examined, A549 cells showed the most prominent changes in the actin cytoskeleton in response to GC treatment (data not shown), we used A549 cells in the following experiments and chose DEX as a representative GC.
FIGURE 1.
Effects of steroid hormones on cell migration and the actin cytoskeleton in A549 cells. A, effects of steroid hormones on cell migration. A549 cells were cultured with the indicated steroid hormones for 48 h and treated with mitomycin C for 2 h preceding the assay. Migration toward FBS in the presence of the steroids was measured in a Transwell migration chamber (mean ± S.D., **, p < 0.01 compared with vehicle; n.s., not significant). B, effect of steroid hormones on the actin cytoskeleton. A549 cells were treated with 1 μm of each steroid for 48 h and stained with phalloidin (red in merged image) and anti-vinculin antibody (green in merged image). Bar, 50 μm. C, effect of DEX on Rho activity. The Rho activity was determined by RBD-beads pulldown assay as described under “Experimental Procedures.” The amounts of total and GTP-RhoA protein were detected by Western blot with anti-RhoA antibody. The amounts of Rho downstream effectors, ROCK1, ROCK2, mDia1, cofilin 1, phospho(Ser-3)-cofilin 1, MLC2, phospho(Ser-19)-MLC2, and profilin were also analyzed by Western blot analysis using their corresponding antibodies.
Small GTPases act as a molecular switch by shuttling between the GDP-bound inactive and the GTP-bound active forms, and they regulate cell motility and adhesion through the remodeling of the actin cytoskeleton (41, 42). In particular, the Rho pathway positively controls stress fiber formation and focal adhesion assembly (41, 42). We examined the possible involvement of Rho and its downstream molecules in response to DEX. However, DEX treatment did not affect the Rho activity as measured by a pulldown assay using RBD (Rho binding domain) beads, by examining the expression levels of the Rho downstream effectors ROCK1, ROCK2, and mDia1, or by evaluating the phosphorylation levels of cofilin and myosin light chain (MLC) (Fig. 1C), suggesting that the DEX-induced enhancement of thick stress fiber formation and focal adhesion assembly is independent of the Rho signaling pathway.
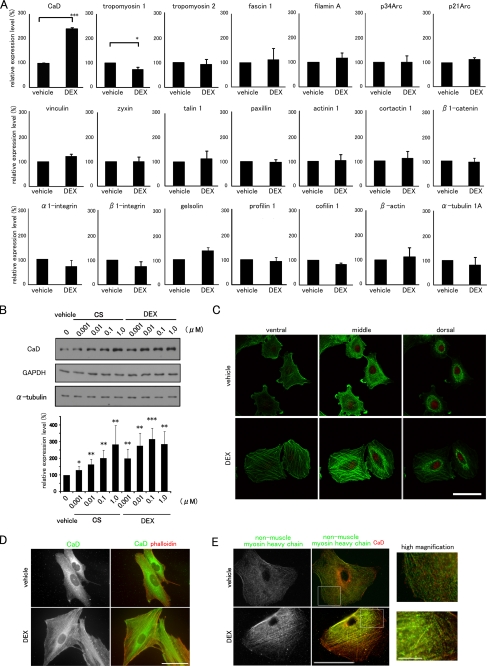
Glucocorticoids Specifically Enhance the Expression of Caldesmon—We then screened the mRNA expression profiles of major actin-interacting proteins in response to DEX using real time qPCR and found that only the CaD mRNA increased (Fig. 2A). Furthermore, cortisol and DEX increased the expression of CaD protein dose-dependently (about 3.0-fold, maximum) (Fig. 2B). Because DEX has a higher affinity for GR than cortisol (43), DEX induced CaD expression more strongly.
FIGURE 2.
CaD is specifically up-regulated by GCs and localizes along thick stress fibers. A, effects of DEX on the expression profiles of actin cytoskeletal genes. Real time qPCR of cDNAs made from A549 cells. The mRNA expression levels of actin-interacting proteins (CaD, tropomyosin 1, tropomyosin 2, fascin 1, filamin A, p34Arc, p21Arc, vinculin, zyxin, talin 1, paxillin, α-actinin 1, cortactin 1, β1-catenin, α1-integrin, β1-integrin, gelsolin, profilin 1, cofilin 1, β-actin, and tubulin α1a) are shown. The data were obtained from three independent experiments (mean ± S.D., *, p < 0.05; ***, p < 0.001). B, GCs induced the up-regulation of the CaD protein in A549 cells. The cells were incubated with cortisol (CS) or dexamethasone (DEX) at the indicated concentrations for 48 h. The expression levels of CaD, α-tubulin, and GAPDH were determined by Western blot analysis. The CaD protein levels were quantified using ImageJ and normalized to α-tubulin expression. (mean ± S.D., *, p < 0.05; **, p < 0.01; ***, p < 0.001 compared with vehicle). C, confocal images of DEX-induced reorganization of the actin cytoskeleton. Vehicle- or DEX-treated A549 cells were stained with phalloidin (green) and propidium iodide (PI) (red). Bar, 50 μm. D, DEX-induced reorganization of the actin cytoskeleton. Vehicle- or DEX-treated A549 cells were stained with anti-CaD antibody (green in merged image) and phalloidin (red in merged image). Bar, 50 μm. E, DEX-induced stress fiber formation. Vehicle- or DEX-treated A549 cells were pretreated with 0.1% Triton X-100 in PBS and fixed, followed by staining with anti-CaD (red in merged image) and anti-non-muscle myosin heavy chain (green in merged image) antibodies. Bar, 50 μm. Areas within the insets are shown at higher magnification (right panels, Bar, 10 μm).
Whereas the untreated control cells showed only the thin stress fibers located at the cell periphery and the localization of actin filaments within the lamellipodia, GC treatment induced formation of the straight, thick stress fibers across the cell bodies (Fig. 2C). CaD was diffusely distributed in the cytoplasm and localized to the lamellipodia in the control cells, but showed dense accumulations along the thick stress fibers in the DEX-treated cells (Fig. 2D). The stress fibers are composed of contractile elements and α-actinin, which are responsible for cell contraction and for exerting tension on the stress fibers (44-46). The non-muscle myosin heavy chain was distributed along the thin actin fibers in control cells, but accumulated along the DEX-induced thick stress fibers in a periodic pattern (Fig. 2E). Compared with CaD, which was distributed entirely along the thick stress fibers, the non-muscle myosin heavy chain in DEX-treated cells was located only along the central portion of the fibers and was absent at the periphery, so the co-localization of CaD and non-muscle myosin heavy chain were restricted within the central portion of the fibers. Together, these results suggest that the suppression of cell migration by GCs is closely associated with a marked enhancement of thick stress fiber formation and focal adhesion assembly via the up-regulation of CaD expression.
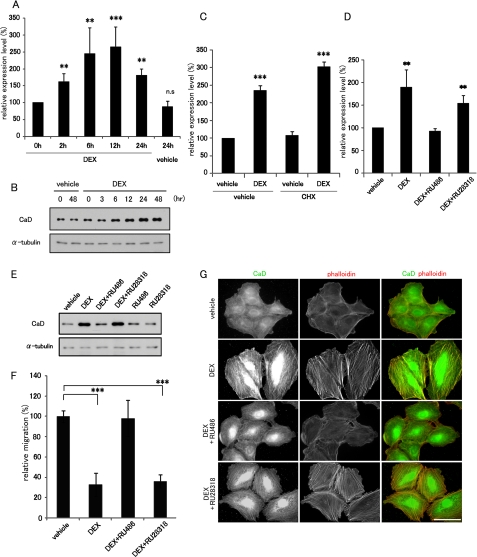
Glucocorticoid Receptor Directly Activates Caldesmon Expression—We next investigated the molecular mechanism underlying the GC-dependent CaD expression. Using real time qPCR, we performed a time course analysis of CaD mRNA expression during DEX treatment and found that CaD mRNA increased after DEX treatment with a peak at 6-12 h, and then gradually decreased thereafter (Fig. 3A). Increased levels of CaD protein were first detected 6 h after DEX treatment, and the up-regulated expression was sustained for more than 48 h (Fig. 3B). To determine whether the DEX-induced up-regulation of CaD mRNA requires de novo protein synthesis, the cells were treated with a potent translation inhibitor, CHX. The DEX-dependent increase of CaD mRNA was apparent even in the presence of CHX (Fig. 3C), indicating that novel protein synthesis was not required.
FIGURE 3.
GR antagonist, RU486, blocks DEX-induced up-regulation of CaD expression. A549 cells were incubated with vehicle or 1 μm DEX for the indicated times, and the expression levels of CaD mRNA (from real time qPCR) (A) and CaD protein (from Western blot) (B) are shown (mean ± S.D., **, p < 0.01, ***, p < 0.001; n.s., not significant). C, A549 cells were pretreated with CHX for 2 h and incubated with vehicle or 1 μm DEX for 24 h in the presence of CHX. CaD mRNA was measured by real time qPCR. D and E, A549 cells incubated with 1 μm DEX in the presence of 10 μm RU486 or 10 μm RU28318. The expression of CaD mRNA (after 24 h (D); from real time qPCR) and CaD protein (after 48 h (E); from Western blot) are shown (mean ± S.D., ***, p < 0.001). F, effect of RU486 or RU28318 on cell migration assayed in Transwell migration chambers. A549 cells were incubated with a combination of 1 μm DEX and 10 μm RU486 or RU28318 (mean ± S.D., ***, p < 0.001). G, A549 cells were incubated with a combination of 1 μm DEX and 10 μm RU486 or 10 μm RU28318 for 48 h and stained with anti-CaD antibody (green in merged image) and phalloidin (red in merged image). Bar, 50 μm.
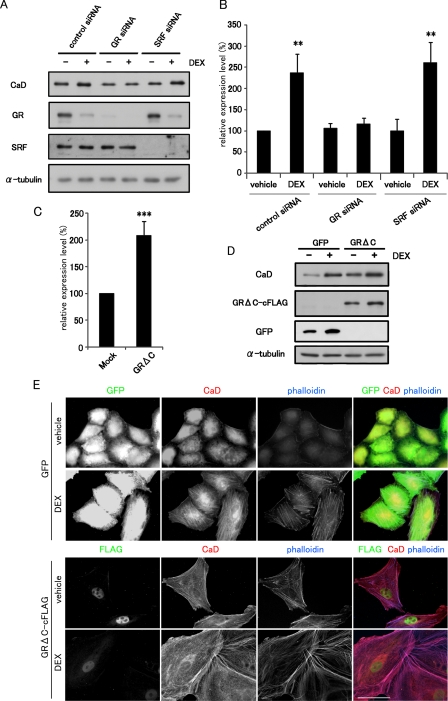
We then examined the involvement of GR in the DEX-induced up-regulation of CaD. A potent GR antagonist, RU486, but not a selective mineralocorticoid receptor (MR) antagonist, RU28318, completely blocked the DEX-induced up-regulation of CaD (Fig. 3, D and E). RU486 also abolished both the DEX-dependent thick stress fiber formation and its suppression of cell migration (Fig. 3, F and G). These results suggest that GR is a critical regulator of CaD expression. We confirmed this finding by depleting GR in A549 cells transfected with GR siRNA. In cells depleted of GR, the DEX-induced up-regulation of CaD was markedly reduced (Fig. 4, A and B). We previously reported that the CALD1 gene is transactivated by the myocardin or myocardin-related transcription factor (MRTF)/SRF pathway in SMCs and fibroblasts (29, 31). However, depletion of SRF did not alter the DEX-induced up-regulation of CaD in A549 cells (Fig. 4, A and B), indicating that this pathway is not involved in the CaD response to DEX. On the other hand, forced expression of a constitutively active GR, GRΔC (36), up-regulated the CaD mRNA levels (Fig. 4C). Furthermore, A549 cells, which stably express GRΔC under control of the cytomegalovirus-immediate early promoter, showed increased expression of the CaD protein compared with GFP-expressing control cells (Fig. 4D). Because of up-regulation of the cytomegalovirus promoter activity in the retrovirus vector by GC (supplemental Fig. S1), the expression levels of both GFP and GRΔC increased in response to DEX. Because GRΔC is a truncated form of GR that lacks its carboxyl terminus, we confirmed that it translocated into the nucleus without its ligand (Fig. 4E). The GRΔC-expressing cells exhibited CaD-linked thick stress fiber formation without DEX as compared with GFP-expressing cells (Fig. 4E). Taken together, these results suggest that the DEX-induced up-regulation of CaD expression is mediated through GR.
FIGURE 4.
GR up-regulates the expression of CaD in A549 cells. A, effect of GR or SRF depletion on the DEX-induced up-regulation of CaD expression. A549 cells transfected with control siRNA, GR siRNA, or SRF siRNA were incubated with vehicle or 1 μm DEX for 48 h. The expression levels of GR, SRF, and CaD were measured by Western blot analysis. B, CaD mRNA expression in the GR- or SRF-depleted cells was measured by real time qPCR (mean ± S.D., **, p < 0.01). C, effect of the expression of GRΔC on the CaD mRNA level in A549 cells transiently transfected with pCS2+GRΔC-cFLAG or the empty vector. The cells were incubated for 24 h. The CaD mRNA levels were measured by real time qPCR (mean ± S.D., ***, p < 0.001). D, expression levels of CaD and GRΔC in GFP- or GRΔC-cFLAG-expressing A549 cells after incubation with vehicle or 1 μm DEX for 48 h. E, cell morphology and CaD localization in GFP- and GRΔC-expressing A549 cells stained with anti-GFP or anti-FLAG (green in merged image), anti-CaD antibodies (red in merged image), and phalloidin (blue in merged image). Bar, 50 μm.
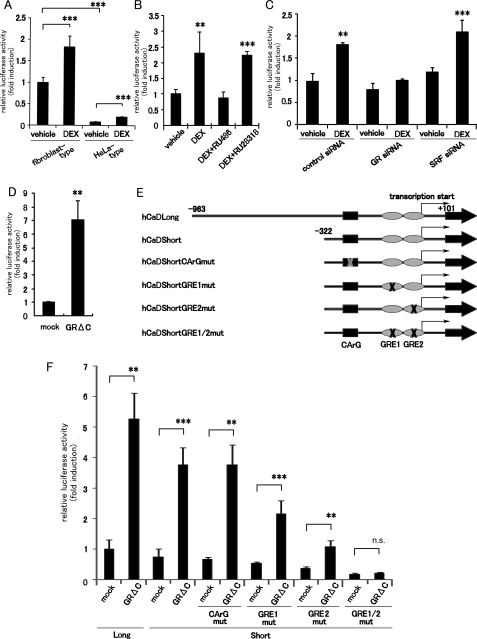
GCs Transactivate the CALD1 Gene via the Two GRE-like Sequences within Its Promoter Region—To elucidate the detailed transcriptional regulation of the CALD1 gene by GR, we analyzed the human CALD1 promoter in a luciferase reporter assay. We previously reported that the human CALD1 gene has two alternative promoter regions, referred to as the HeLa-type and fibroblast-type promoters (47). We compared the DEX responsiveness of the two promoter regions. Although both the promoter constructs responded to DEX stimulation (Fig. 5A), the activity of the fibroblast-type promoter in A549 cells was stronger than that of the HeLa-type one. We also compared the expression levels of the two species of CaD mRNA by reverse transcription-PCR, using three primer sets as follows: one each for the fibroblast- and HeLa-type CaD mRNAs and one to assay the total CaD mRNAs. We found the fibroblast-type transcripts to be more abundant than the HeLa-type ones (supplemental Fig. S2). There was also a correlation between the levels of fibroblast-type and total CaD transcripts with respect to GC responsiveness and the CaD expression pattern after DEX treatment. These results suggest that the reporter assay correctly reflects the activities of the two endogenous CALD1 promoters and that the fibroblast-type CALD1 promoter is responsible for the GC-induced up-regulation of CaD.
FIGURE 5.
Identification of the GC-responsive region in the human CALD1 promoter. A, reporter assay was performed with the human fibroblast-type (-964 to +101) and HeLa-type (-1908 to +207) CALD1 promoters in A549 cells co-transfected with the luciferase reporter plasmid and pGL3β-gal plasmid. The cells were incubated with vehicle or 1 μm DEX for 24 h. The luciferase activities were normalized to the β-galactosidase activity (mean ± S.D., ***, p < 0.001). B, effect of GR or MR antagonists on the fibroblast-type CALD1 promoter activity (mean ± S.D., **, p < 0.01, ***, p < 0.001). C, effect of GR or SRF depletion on the fibroblast-type CALD1 promoter activity (mean ± S.D., **, p < 0.01; ***, p < 0.001). D, luciferase activities in A549 cells co-transfected with reporter plasmids and the pCS2+GRΔC-cFLAG plasmid were measured after a 24 h incubation (mean ± S.D., **, p < 0.01). E, schematic diagram of deletion and mutation constructs of the CArG element or GRE-like sequences in the human fibroblast-type CALD1 promoter. F, GRΔC-responsiveness of the promoter activities of a series of reporter constructs (mean ± S.D., **, p < 0.01; ***, p < 0.001; n.s., not significant).
We therefore used the reporter assay to analyze the fibroblast-type promoter region. The fibroblast-type promoter was activated about 2.5-fold by DEX treatment, and RU486 blocked this activation (Fig. 5B). GR depletion also abolished the DEX-dependent promoter activation (Fig. 5C). Transient transfection of GRΔC strongly enhanced the promoter activity by more than 5-fold (Fig. 5D). These results support the hypothesis that the responsiveness of the promoter construct to GRΔC precisely mimics the up-regulation of CaD expression induced by GC.
To identify the GC-responsive cis-elements within the fibroblast-type CALD1 promoter region, we analyzed a series of truncated promoter constructs or constructs bearing point mutations (Fig. 5E). Compared with the long promoter fragment (-982 to +101), a short one (-322 to +101) showed slightly reduced promoter activity, but the activation rate was not compromised (Fig. 5F). Mutation of the CArG element (-297 to -288), which is a pivotal cis-element for SRF-mediated transcription of the CALD1 gene, did not affect the promoter activity. However, mutation of the two GRE-like sequences near the transcriptional start site (GRE1, -68 to -52; GRE2, -6 to +10), which are conserved among mammalian species (supplemental Fig. S3), did alter the reporter activity. Constructs with mutations in either GRE1 or GRE2 showed decreased responsiveness to GRΔC. A double-mutated construct, with point mutations in both of the GRE-like sequences, completely abolished the GRΔC responsiveness (Fig. 5F). A similar result was obtained in response to DEX stimulation (data not shown). These results indicate that GR enhances the CALD1 promoter activity via GRE1 and GRE2.
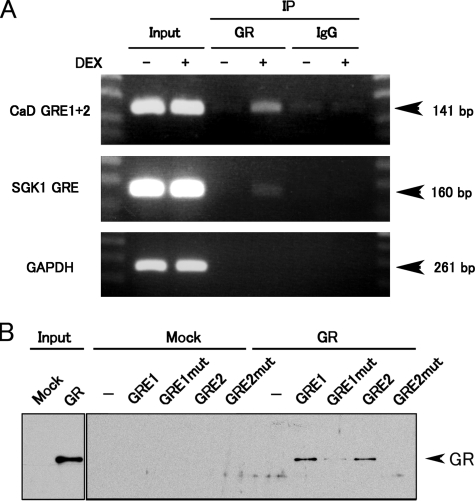
To examine the GR binding to GRE1 and GRE2 in vivo, we performed a chromatin immunoprecipitation (ChIP) assay. Fragments of the CALD1 promoter containing GRE1 or GRE2 were successfully amplified from only DEX-treated, but not untreated, samples (Fig. 6A), suggesting that GR binds to the two GRE-like sequences in response to DEX in vivo. We also confirmed that GR translocated into the nucleus of the A549 cells upon DEX stimulation (data not shown). We further examined GR binding to the promoter by an in vitro DNA-binding assay. Biotinylated GRE1 or GRE2 oligonucleotides were incubated with in vitro-translated GR protein and harvested with streptavidin beads. As shown in Fig. 6B, GR bound to both GRE1 and GRE2. The affinity of GR for mutated GRE1 was greatly reduced, and it was abolished for mutated GRE2. Taken together, these results indicate that GR binds directly to the two GRE-like sequences within the CALD1 promoter and transactivates the CALD1 gene.
FIGURE 6.
GR binds to the GRE-like sequences in the human CALD1 promoter in vivo and in vitro. A, ChIP assay of A549 cells treated with vehicle or 1 μm DEX for 2 h. The cell extracts were subjected to ChIP using the anti-GR antibody. The DNA fragments of the CALD1 promoter region containing the two GRE-like sequences were amplified by PCR. The SGK1 and GAPDH promoter regions were used as the positive and negative controls, respectively. B, DNA-binding assay of GR to GRE-like sequences. Each biotinylated DNA fragment containing a GRE-like sequence was incubated with in vitro-translated GR protein and collected on streptavidin beads. The DNA fragments with mutated GRE-like sequences were also used to ascertain the binding specificity of GR. DNA-bound GR was detected by Western blot with anti-GR antibody.
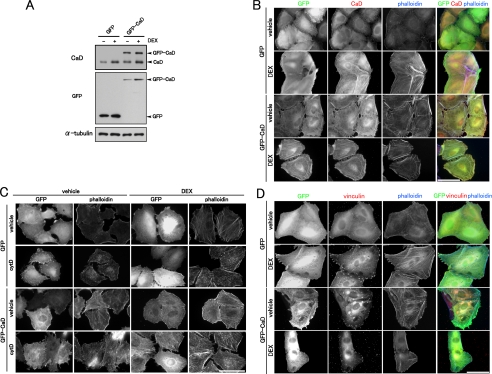
Caldesmon Suppresses Cell Migration via the Enhancement of Thick Stress Fiber Formation and Focal Adhesion Assembly—We examined whether the forced expression of exogenous CaD could mimic the effects of GCs on the reorganization of the actin cytoskeleton and cell migration. We prepared A549 cells stably expressing GFP-CaD (Fig. 7A). The total amounts of CaD (endogenous CaD plus GFP-CaD) in non-DEX-treated GFP-CaD-expressing cells were ∼2.5-fold greater than in the GFP-expressing cells. The expression level of GFP-CaD also increased in response to DEX (about 2.5-fold) (Fig. 7A) as that of GFP-expressing cells, because of up-regulation of the promoter activity in the retrovirus vector by GC (supplemental Fig. S1). In the GFP-CaD-expressing cells, we observed the straight, thick stress fibers that lie across the cell body, as seen with GC treatment. GFP-CaD accumulated along the thick stress fibers regardless of DEX treatment (Fig. 7B). CaD stabilizes actin filaments by protecting them from actin-severing proteins (48, 49) and actin-depolymerizing agents (23). The thick stress fibers formed in both GFP-CaD-expressing and DEX-treated cells were stable against the actin-depolymerizing agent cytochalasin D (Fig. 7C). Focal adhesions were also enhanced in GFP-CaD-expressing cells (Fig. 7D). Consistent with the enhanced stress fiber formation and focal adhesion assembly having a major effect on cell migration, in the GFP-CaD-expressing cells, migration was greatly reduced compared with that of the GFP-expressing cells, even in the absence of DEX (Fig. 8A). We further examined the effect of up-regulated CaD on actomyosin-based cell contraction. The contractile activity was reduced in both DEX-treated and GFP-CaD-expressing cells, similar to that seen in the cells treated with myosin II ATPase inhibitor blebbistatin. These results suggest that up-regulated CaD also inhibits actomyosin-based contractile activity in vivo (Fig. 8B). Thus, the forced expression of exogenous CaD mimicked the effect of DEX treatment on the reorganization of the actin cytoskeleton and on cell migration.
FIGURE 7.
Effect of forced expression of GFP-CaD on the actin cytoskeleton. A, forced expression of GFP-CaD. The expression levels of GFP-CaD, control GFP, and endogenous CaD in A549 transfectants were analyzed by Western blotting with anti-CaD or anti-GFP antibody. B, effect of DEX on the actin cytoskeleton of A549 cells stably expressing GFP or GFP-CaD. The cells were stained with anti-GFP (green in merged image) and anti-CaD (red in merged image) antibodies and phalloidin (blue in merged image). Bar, 50 μm. C, effect of forced expression of GFP-CaD on stress fiber stability to cytochalasin D (cytD). GFP- or GFP-CaD-expressing A549 cells were incubated with vehicle or 1 μm DEX for 48 h. The cells were treated with or without 0.2 μm cytD for 20 min, and stained with anti-GFP antibody and phalloidin. Bar, 50 μm. D, effect of DEX on focal adhesion assembly of A549 cells stably expressing GFP or GFP-CaD. The cells were stained with anti-GFP (green in merged image) and anti-vinculin (red in merged image) antibodies and phalloidin (blue in merged image). Bar, 50 μm.
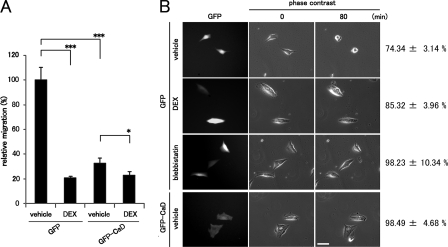
FIGURE 8.
Effects of forced expression of GFP-CaD on cell migration and contraction. A, effect of forced expression of GFP-CaD on cell migration. The migration activity of GFP- or GFP-CaD-expressing A549 cells was measured in a Transwell migration chamber with or without 1 μm DEX. (mean ± S.D., *, p < 0.05; ***, p < 0.001). B, effect of forced expression of CaD on serum-induced cell contraction. GFP- or GFP-CaD-expressing A549 cells were treated with vehicle or 1 μm DEX for 48 h. The cells were starved with serum-free DMEM for 18 h and then stimulated with DMEM containing FBS for 80 min. Cell contraction was determined from reduction in cell area after serum stimulation (mean ± S.E.). (GFP-expressing cells: vehicle, n = 54; DEX, n = 39; blebbistatin, n = 39. GFP-CaD expressing cells: vehicle, n = 36). Bar, 50 μm.
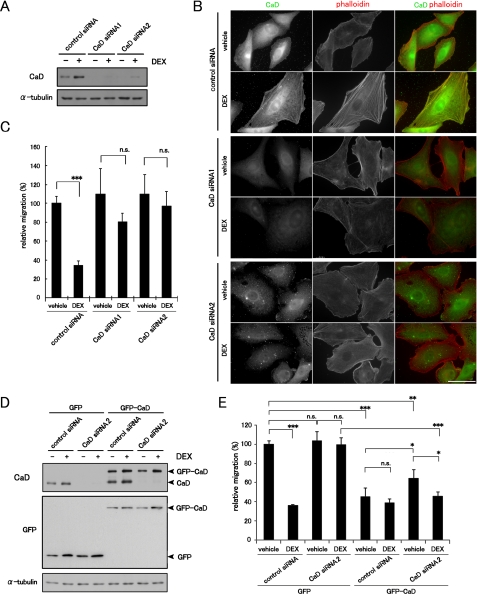
We next examined the effect of CaD depletion on cell migration and the actin cytoskeleton. Two different siRNAs for human CaD effectively depleted the CaD protein in A549 cells (Fig. 9A). The depletion of CaD blocked the DEX-induced thick stress fiber formation (Fig. 9B and supplemental Fig. S4). The depletion of CaD also abolished suppression of cell migration (Fig. 9C), although the CaD-depleted cells showed no obvious changes in cell migration in the absence of DEX. These loss-of-function experiments therefore support the idea of a pivotal role for CaD in DEX-induced thick stress fiber formation and suppression of cell migration. Conversely, expression of GFP-CaD rescued the effects of depleting the endogenous CaD. Because the target sequence of CaD siRNA2 is located in the 3′-untranslated region of the human CaD mRNA, it can deplete only the endogenous CaD, but not GFP-CaD, which lacks the 3′-untranslated region (Fig. 9D). The endogenous CaD-depleted, GFP-CaD-expressing cells showed an ∼1.5-fold increase in the total amounts of CaD (GFP-CaD only) (Fig. 9D), formed the stress fibers (supplemental Fig. S5), and exhibited the modestly reduced migratory activity (Fig. 9E).
FIGURE 9.
Effects of CaD depletion on the actin cytoskeleton and cell migration. A, depletion of CaD expression using siRNA. A549 cells were transfected with CaD siRNAs or control siRNA, and incubated with or without 1 μm DEX for 48 h. The expression levels of CaD protein were detected by Western blot analysis. B, effect of CaD depletion on the formation of stress fibers in response to DEX. The cells were stained with anti-CaD antibody (green in merged image) and phalloidin (red in merged image). Bar, 50 μm. C, cell migration of CaD-depleted cells was measured in a Transwell migration chamber with or without 1 μm DEX (mean ± S.D., ***, p < 0.001). D, GFP-CaD expression counteracts the effects of endogenous CaD depletion. GFP- and GFP-CaD-expressing A549 cells transfected with CaD siRNA2 were incubated with or without 1 μm DEX. CaD siRNA2 repressed the expression of endogenous CaD, but not of exogenous GFP-CaD, which lacked the siRNA target, the 3′-untranslated region. E, cell migration of CaD siRNA2-transfected GFP-CaD- or GFP-expressing cells in a Transwell migration chamber with or without 1 μm DEX. (mean ± S.D., *, p < 0.05; **, p < 0.01; ***, p < 0.001; n.s., not significant).
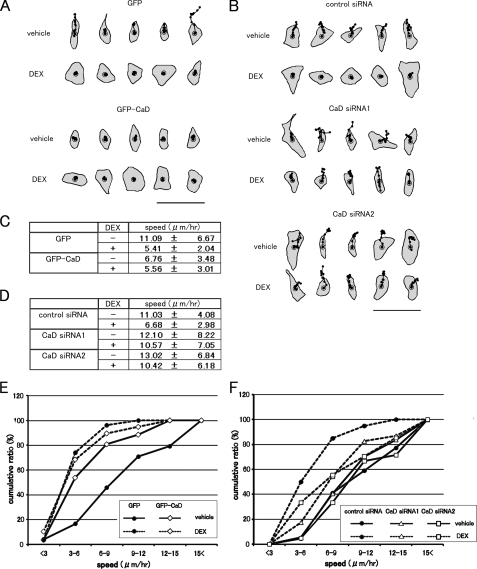
We further analyzed the spontaneous cell motility in these transfectants. DEX-treated or untreated cells were plated on culture dishes and observed with or without GC treatment. The speeds of migrating cells were calculated using DIAS software, by tracing the paths of individual cells in time-lapse movies (Fig. 10, A and B). DEX treatment significantly slowed the migration speeds of cells expressing GFP or transfected with control siRNA (Fig. 10, C and D). The GFP-CaD-expressing cells showed slower migration without DEX (Fig. 10C), but the migration speeds of the CaD-depleted cells were hardly affected by DEX (Fig. 10D). We compared the migration speeds of the various transfectants by cumulative plots, and we found that DEX treatment increased the number of slower migrating cells compared with the control cells (Fig. 10, E and F). The forced expression of GFP-CaD mimicked the DEX-induced increase in slower migrating cells (Fig. 10E). By contrast, CaD depletion abolished the slower migration induced by DEX (Fig. 10F). These results suggest that both the reorganization of the actin cytoskeleton and the migratory activity of A549 cells depend on the expression levels of CaD.
FIGURE 10.
Effect of forced expression or depletion of CaD on cell motility. A, cell motility was analyzed by tracing cell movement in time-lapse images, using DIAS software. GFP- or GFP-CaD-expressing A549 cells were incubated with vehicle or 1 μm DEX for 48 h, and representative centroid tracks of the cells are shown (25-min intervals for 5 h). Bar, 100 μm. B, A549 cells transfected with CaD siRNAs (CaD siRNA1 or CaD siRNA2) or control siRNA were incubated with vehicle or 1 μm DEX for 48 h, and representative centroid tracks of the cells are shown (25-min intervals for 5 h). Bar, 100 μm. C, migration speeds of GFP- or GFP-CaD-expressing A549 cells were calculated from the cell tracks (GFP-expressing cells: vehicle, n = 24; DEX, n = 27. GFP-CaD-expressing cells: vehicle, n = 26; DEX: n = 19) (mean ± S.D.). D, migration speeds of CaD-depleted A549 cells calculated from the cell tracks (control siRNA: vehicle, n = 22; DEX, n = 20. CaD siRNA1: vehicle, n = 37; DEX, n = 46. CaD siRNA2: vehicle, n = 21; DEX, n = 27) (mean ± S.D.). E, cumulative percentage plots of the migration speeds of GFP- and GFP-CaD-expressing A549 cells. GFP-expressing cells (•); GFP-CaD-expressing cells (⋄); -DEX (vehicle), solid lines; +DEX, dashed lines. F, cumulative percentage plots of the migration speeds of CaD-depleted and control A549 cells. Control siRNA-transfected cells (•); CaD siRNA1-transfected cells (Δ); CaD siRNA2-transfected cells (□); -DEX (vehicle), solid lines; and +DEX, dashed lines.
DISCUSSION
GC-dependent Transcription of the CALD1 Gene—GCs are reported to regulate the transcription of numerous GC-responsive genes, such as SGK1 (serum- and glucocorticoid-inducible kinase 1), IL-2Rα (interleukin-2 receptor α), and proopiomelanocortin (10). However, few GC-responsive cytoskeletal genes are known. Although CaD expression was reportedly induced by GC stimulation and involved in the remodeling of the actin cytoskeleton (32, 33), its molecular mechanism and biological function remained largely unclear. In this study, we clearly demonstrate a GC-dependent transcriptional mechanism for the CALD1 gene. The transcriptional regulation of the CALD1 gene has been analyzed mainly in SMCs because of the abundance of CaD in these cells. The SMC-specific transcription of the CALD1 gene is regulated by the coordination of a transcriptional triad composed of the SRF and Nkx homeobox and GATA transcription factors (30). Myocardin is a recently identified co-factor for SRF-mediated transcription of the cardiac and smooth muscle-restricted genes (49), and the Rho-MRTFs-SRF pathway in normal fibroblasts transactivates the actin cytoskeletal genes, including the CALD1 gene (31, 50). GR- and SRF-knockdown experiments show that the GC-induced CaD expression solely depends on GR but not on SRF (Figs. 3, 4, 5). These results indicate that the MRTF/SRF pathway is not required for GC-induced transactivation of the CALD1 gene in A549 cells. Our ChIP and DNA-binding assays reveal that GR directly binds to the two GRE-like sequences in the human fibroblast-type CALD1 promoter (Fig. 6). Because the GRE1 and GRE2 sequences are highly conserved among mammalian species (supplemental Fig. S3), the GC-dependent transcription of the CALD1 gene through the GRE-like sequences may be a common mechanism among mammalian species.
GC abnormalities cause various dysfunctions (1, 3, 51). GCs are also reported to be essential for the organogenesis of highly GC-responsive tissues, such as brain, lung, spleen, and liver, although the sensitivity to GCs differs substantially among these tissues (52, 53). Indeed, A549 cells, which originated from lung epithelium, exhibit the most potent DEX responsiveness with respect to the up-regulation of CaD of several cell lines examined. The relatively low basal expression of CaD in A549 cells accentuates the DEX-induced up-regulation of CaD expression, but DEX responsiveness is less significant in cells with an abundance of CaD, such as SMCs (data not shown). Therefore, CaD-mediated inhibition of cell migration by GCs might be predominant in cells that express CaD at relatively low levels before stimulation. Our data support the idea that the CaD-mediated suppression of cell migration and the reorganization of the actin cytoskeleton may be involved in both the development of GC-responsive tissues and pathogenic processes within them.
GC-induced Up-regulation of Caldesmon Expression Suppresses Cell Migration via the Reorganization of the Actin Cytoskeleton in a Rho-independent Manner—Although the role of CaD in smooth muscle contraction is well studied so far (18), our understanding with respect to its role in non-muscle cells has been meager. Our present results indicate that the CaD-mediated suppression of cell migration is directly linked to the enhancement of thick stress fiber formation and focal adhesion assembly. The stress fibers include the structural and contractile cytoarchitecture for cell morphology and motility (44-46). The focal adhesions provide attachment sites for the cell to the substratum and link to the stress fibers (54). For example, the disruption of stress fiber formation and/or their maintenance by inhibitors of actin polymerization or myosin II ATPase disrupts the focal adhesions (45). In untreated A549 cells, the actin bundles are present in the cell periphery, and the thin stress fibers are distributed throughout the cell body. In contrast, the stress fibers formed in GC-treated cells are long and thick, and traverse the intracellular space to be anchored at both ends by enhanced focal adhesions (Figs. 1 and 2).
CaD has a dual function in regulating the actin cytoskeleton, via its control of actomyosin contractility (20, 21) and the stabilization of actin filaments (22, 23). CaD is also thought to have an actin filament bundling/cross-linking activity in vitro (17, 55). The straight, thick stress fiber formation mediated by up-regulated CaD may be mainly due to its actin filament-stabilizing and bundling/cross-linking activities. Actually, thick stress fibers formed in CaD up-regulated cells are more stable against cytochalasin D (Fig. 7C and supplemental Fig. S4). The focal adhesion assembly might be promoted by the CaD-linked thick stress fibers, because of the interdependency between stress fibers and focal adhesions.
Cells migrate by lamellipodial extension, and the transient attachment of lamellipodia at the leading edge and their retraction and detachment at the rear require dynamic remodeling of the actin cytoskeleton (12, 15). The stress fibers and focal adhesions are generally assumed to supply the contractile force for the turnover of cell adhesions at the rear (15). However, their roles in cell motility are controversial. It has been well documented that Rho signaling plays a crucial role in cell migration via stress fiber and focal adhesion formation. Stimulation by several chemoattractants such as vascular endothelial growth factor, basic fibroblast growth factor, SDF-1α (stromal cell-derived factor 1α), and endothelin-1 promote cell motility via the activation of Rho and its downstream effectors (56-59). Rho activates mDia1, ROCKs, and their downstream effectors, LIMKs, which phosphorylate and inactivate cofilin, leading to the enhancement of actin polymerization. The Rho/ROCK pathway is also involved in the myosin-linked regulation of force generation. Activated ROCKs phosphorylate MLC and inhibit myosin phosphatase. Thus, the increased phosphorylation of MLC activates the actin-myosin interaction, resulting in the facilitation of cell motility (60, 61). In contrast, we demonstrated in this study that the GC-induced suppression of cell migration is closely linked to the enhancement of thick stress fiber formation and focal adhesion assembly via up-regulated CaD (Figs. 1, 2, and 7, 8, 9). Because the Rho, cofilin, and MLC activities in A549 cells are not altered by DEX treatment, the effects of GC are independent of the Rho/ROCK pathway in this system (Fig. 1). In fact, the forced expression of GFP-CaD mimics the GC-induced reorganization of the actin cytoskeleton and suppression of cell migration without DEX treatment (Figs. 7 and 8). In addition, although CaD stabilizes the stress fibers at the cell periphery of DEX-treated A549 cells, it is also co-localizes with non-muscle myosin along the central region of the thick stress fibers (Fig. 2E). Because CaD negatively regulates actomyosin contractility (20, 21), up-regulated CaD may also play a crucial role in inhibiting the generation of stress fiber-based contractile force (Fig. 8). Taken together, these results suggest that the CaD-induced stabilization of stress fibers and inhibition of contractile force generation by stress fibers lead to the suppression of cell migration through a Rho-independent pathway.
Yemelyanov et al. (62) reported that GR expression is markedly decreased in prostate cancers and that the restoration of GR expression in cancer cells suppresses the progression of the malignancy. We recently reported that CaD negatively regulates the formation of invadopodia/podosomes, which are dynamic cell adhesion structures involved in cancer cell invasion (37). The forced expression of CaD inhibits invadopodium/podosome formation, leading to suppression of the invasive activity of cancer cells (63). Thus, CaD appears to be a potent suppressor of cancer cell invasion. Taken together with the literature, our present results suggest that the increased expression of CaD induced by GCs could provide a novel therapeutic strategy for blocking some cases of cancer cell invasion. Our findings may also contribute to a deeper understanding of the physiological functions of GCs in cell motility.
Supplementary Material
This work was supported by Grant-in-aid for Scientific Research 15GS0312 from the Ministry of Education, Culture, Sports, Science and Technology of Japan (to K. S.) and Grant-in-aid for Scientific Research 20240038 from the Japan Society for the Promotion of Science (to K. S.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1-S5.
Footnotes
The abbreviations used are: GC, glucocorticoid; CaD, caldesmon; GR, glucocorticoid receptor; GRE, GC-response element; DEX, dexamethasone; FBS, fetal bovine serum; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; SRF, serum-response factor; RBD, Rho binding domain; siRNA, small interference RNA; DMEM, Dulbecco's modified Eagle's medium; PBS, phosphate-buffered saline; GFP, green fluorescent protein; MR, mineralocorticoid receptor; ChIP, chromatin immunoprecipitation; qPCR, quantitative PCR; SMC, smooth muscle cell; cytD, cytochalasin D; CHX, cycloheximide; MRTF, myocardin-related transcription factor; MLC, myosin light chain.
References
- 1.Munck, A., Guyre, P. M., and Holbrook, N. J. (1984) Endocr. Rev. 5 25-44 [DOI] [PubMed] [Google Scholar]
- 2.Barnes, P. J. (1998) Clin. Sci. 94 557-572 [DOI] [PubMed] [Google Scholar]
- 3.Turnbull, A. V., and Rivier, C. L. (1999) Physiol. Rev. 79 1-71 [DOI] [PubMed] [Google Scholar]
- 4.Tronche, F., Opherk, C., Moriggl, R., Kellendonk, C., Reimann, A., Schwake, L., Reichardt, H. M., Stangl, K., Gau, D., Hoeflich, A., Beug, H., Schmid, W., and Schütz, G. (2004) Genes Dev. 18 492-497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wick, G., Hu, Y., Schwarz, S., and Kroemer, G. (1993) Endocr. Rev. 14 539-563 [DOI] [PubMed] [Google Scholar]
- 6.Adcock, I. M., and Lane, S. J. (2003) J. Endocrinol. 178 347-355 [DOI] [PubMed] [Google Scholar]
- 7.Planey, S. L., and Litwack, G. (2000) Biochem. Biophys. Res. Commun. 279 307-312 [DOI] [PubMed] [Google Scholar]
- 8.Coiffier, B., Lepage, E., Briere, J., Herbrecht, R., Tilly, H., Bouabdallah, R., Morel, P., Van Den Neste, E., Sallesf, G., Gaulardl, P., Reyes, F., Lederlin, P., and Gisselbrecht, C. (2002) N. Engl. J. Med. 346 235-242 [DOI] [PubMed] [Google Scholar]
- 9.Fakih, M., Johnson, C. S., and Trump, D. L. (2002) Urology 60 553-561 [DOI] [PubMed] [Google Scholar]
- 10.Schoneveld, O. J., Gaemers, I. C., and Lamers, W. H. (2004) Biochim. Biophys. Acta 1680 114-128 [DOI] [PubMed] [Google Scholar]
- 11.Zhou, J., and Cidlowski, J. A. (2005) Steroids 70 407-417 [DOI] [PubMed] [Google Scholar]
- 12.Ridley, A. J., Schwartz, M. A., Burridge, K., Firtel, R. A., Ginsberg, M. H., Borisy, G., Parsons, J. T., and Horwitz, A. R. (2003) Science 302 1704-1709 [DOI] [PubMed] [Google Scholar]
- 13.Condeelis, J., and Segall, J. E. (2003) Nat. Rev. Cancer 3 921-930 [DOI] [PubMed] [Google Scholar]
- 14.Wang, W., Goswami, S., Sahai, E., Byskoff, J. B., Segall, J. E., and Condeelis, J. S. (2005) Trends Cell Biol. 15 138-145 [DOI] [PubMed] [Google Scholar]
- 15.Pantaloni, D., Le Clainche, C., and Carlier, M. F. (2001) Science 292 1502-1506 [DOI] [PubMed] [Google Scholar]
- 16.Chhabra, E. S., and Higgs, H. N. (2007) Nat. Cell Biol. 9 1110-1121 [DOI] [PubMed] [Google Scholar]
- 17.Sobue, K., Muramoto, Y., Fujita, M., and Kakiuchi, S. (1981) Proc. Natl. Acad. Sci. U. S. A. 78 5652-5655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sobue, K., and Seller, J. R. (1991) J. Biol. Chem. 266 12115-12118 [PubMed] [Google Scholar]
- 19.Ueki, N., Sobue, K., Kanda, K., Hada, T., and Higashino, K. (1987) Proc. Natl. Acad. Sci. U. S. A. 84 9049-9053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sobue, K., Morimoto, K., Inui, M., Kanda, K., and Kakiuchi, S. (1982) Biomed. Res. 3 188-196 [Google Scholar]
- 21.Ngai, P. K., and Walsh, M. P. (1984) J. Biol. Chem. 259 13656-13659 [PubMed] [Google Scholar]
- 22.Ishikawa, R., Yamashiro, S., and Matsumura, F. (1989) J. Biol. Chem. 264 7490-7497 [PubMed] [Google Scholar]
- 23.Warren, K. S., Shutt, D. C. McDermott, J. P., Lin, J. L. Soll, D. R., and Lin, J. J. (1994) J. Cell Biol. 125 359-368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Owada, M. K., Hakura, A., Iida, K., Yahara, I., Sobue, K., and Kakiuchi, S. (1984) Proc. Natl. Acad. Sci. U. S. A. 81 3133-3137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bretscher, A., and Lynch, W. (1985) J. Cell Biol. 100 1656-1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eppinga, R. D., Li, Y., Lin, J. L., Mak, A. S., and Lin, J. J. (2006) Cell Motil. Cytoskeleton 63 543-562 [DOI] [PubMed] [Google Scholar]
- 27.Li, Y., Lin, L. C., Reiter, R. S., Daniels, K., Soll, D. R., and Lin, J. J. C. (2004) J. Cell Sci. 117 3593-3604 [DOI] [PubMed] [Google Scholar]
- 28.Mirzapoiazova, T., Kolosova, I. A., Romer, L., Garcia, J. G. N., and Verin, A. D. (2005) J. Cell. Physiol. 203 520-528 [DOI] [PubMed] [Google Scholar]
- 29.Yano, H., Hayashi, K., Momiyama, T., Saga, H., Haruna, M., and Sobue, K. (1995) J. Biol. Chem. 270 23661-23666 [DOI] [PubMed] [Google Scholar]
- 30.Nishida, W., Nakamura, M., Mori, S., Takahashi, M., Ohkawa, Y., Tadokoro, S., Yoshida, K., Hiwada, K., Hayashi, K., and Sobue, K. (2002) J. Biol. Chem. 277 7308-7317 [DOI] [PubMed] [Google Scholar]
- 31.Morita, T., Mayanagi, T., and Sobue, K. (2007) Exp. Cell Res. 313 3432-3445 [DOI] [PubMed] [Google Scholar]
- 32.Castellino, F., Heuser, J., Marchetti, S., Bruno, B., and Luini, A. (1992) Proc. Natl. Acad. Sci. U. S. A. 89 3775-3779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Castellino, F., Ono, S., Matsumura, F., and Luini, A. (1995) J. Cell Biol. 131 1223-1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reiner, G. C. A., Katzenellenbogen, B. S., Bindal, R. D., and Katzenellenbogen, J. A. (1984) Cancer Res. 44 2302-2308 [PubMed] [Google Scholar]
- 35.Tanaka, J., Watanabe, T., Nakamura, N., and Sobue, K. (1993) J. Cell Sci. 104 595-606 [DOI] [PubMed] [Google Scholar]
- 36.Meijsing, S. H., Elbi, C., Luecke, H. F., Hager, G. L., and Yamamoto, K. R. (2007) Mol. Cell. Biol. 27 2442-2451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morita, T., Mayanagi, T., Yoshio, T., and Sobue, K. (2007) J. Biol. Chem. 282 8454-8463 [DOI] [PubMed] [Google Scholar]
- 38.Fay, F. S., and Delise, C. M. (1973) Proc. Natl. Acad. Sci. U. S. A. 70 641-645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bodin, P., Richard, S., Travo, C., Berta, P., Stoclet, J. C., Papin, S., and Travo, P. (1991) Arterioscler. Thromb. Vasc. Biol. 260 C151-C158 [DOI] [PubMed] [Google Scholar]
- 40.Itani, O. A., Liu, K. Z., Cornish, K. L., Campbell, J. R., and Thomas, C. P. (2002) Am. J. Physiol. 283 E971-E979 [DOI] [PubMed] [Google Scholar]
- 41.Nobes, C., and Hall, A. (1995) Cell 81 53-62 [DOI] [PubMed] [Google Scholar]
- 42.Van Aelst, L., and D'Souza-Schorey, C. (1997) Genes Dev. 11 2295-2322 [DOI] [PubMed] [Google Scholar]
- 43.Munck, A., and Foley, R. (1976) J. Steroid Biochem. 7 1117-1122 [DOI] [PubMed] [Google Scholar]
- 44.Kreis, T. E., and Birchmeier, W. (1980) Cell 22 555-561 [DOI] [PubMed] [Google Scholar]
- 45.Hotulainen, P., and Lappalainen, P. (2006) J. Cell Biol. 173 383-394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pellegrin, S., and Mellor, H. (2007) J. Cell Sci. 120 3491-3499 [DOI] [PubMed] [Google Scholar]
- 47.Hayashi, K., Yano, H., Hashida, T., Takeuchi, R., Takeda, O., Asada, K., Takahashi, E., Kato, I., and Sobue, K. (1992) Proc. Natl. Acad. Sci. U. S. A. 89 12122-12126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yonezawa, N., Nishida, E., Maekawa, S., and Sakai, H. (1988) Biochem. J. 251 121-127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang, D., Chang, P. S., Wang, Z., Sutherland, L., Richardson, J. A., Small, E., Krieg, P. A., and Olson, E. N. (2001) Cell 105 851-862 [DOI] [PubMed] [Google Scholar]
- 50.Morita, T., Mayanagi, T., and Sobue, K. (2007) J. Cell Biol. 179 1027-1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.De Kloet, E. R., Vreugdenhil, E., Oitzl, M. S., and Joels, M. (1998) Endocr. Rev. 19 269-301 [DOI] [PubMed] [Google Scholar]
- 52.Cole, T. J., Blendy, J. A., Monaghan, A. P., Krieglstein, K., Schmid, W., Aguzzi, A., Fantuzzi, G., Hummler, E., Unsicker, K., and Schutz, G. (1995) Genes Dev. 9 1608-1621 [DOI] [PubMed] [Google Scholar]
- 53.Bamberger, C. M., Schulte, H. M., and Chrousos, G. P. (1996) Endocr. Rev. 17 245-261 [DOI] [PubMed] [Google Scholar]
- 54.Zimerman, B., Volberg, T., and Geiger, B. (2004) Cell Motil. Cytoskeleton 58 143-159 [DOI] [PubMed] [Google Scholar]
- 55.Lynch, W. P., Riseman, V. M., and Bretscher, A. (1987) J. Biol. Chem. 262 7429-7437 [PubMed] [Google Scholar]
- 56.Vicente-Manzanares, M., Cabrero, J. R., Rey, M., Pérez-Martínez, M., Ursa, A., Itoh, K., and Sánchez-Madrid, F. (2002) J. Immunol. 168 400-410 [DOI] [PubMed] [Google Scholar]
- 57.Van Nieuw Amerongen, G. P., Koolwijk, P., Versteilen, A., and Van Hinsbergh, V. W. (2003) Arterioscler. Thromb. Vasc. Biol. 23 211-217 [DOI] [PubMed] [Google Scholar]
- 58.Zheng, R., Iwase, A., Shen, R., Goodman, O. B., Jr., Sugimoto, N., Takuwa, Y., Lerner, D. J., and Nanus, D. M. (2006) Oncogene 25 5942-5952 [DOI] [PubMed] [Google Scholar]
- 59.Abe, M., Sogabe, Y., Syuto, T., Yokoyama, Y., and Ishikawa, O. (2007) J. Cell. Biochem. 102 1290-1299 [DOI] [PubMed] [Google Scholar]
- 60.Fukata, Y., Amano, M., and Kaibuchi, K. (2001) Trends Pharmacol. Sci. 22 32-39 [DOI] [PubMed] [Google Scholar]
- 61.Riento, K., and Ridley, A. J. (2003) Nat. Rev. Mol. Cell Biol. 4 446-456 [DOI] [PubMed] [Google Scholar]
- 62.Yemelyanov, A., Czwornog, J., Chebotaev, D., Karseladze, A., Kulevitch, E., Yang, X., and Budunova, I. (2006) Oncogene 26 1885-1896 [DOI] [PubMed] [Google Scholar]
- 63.Yoshio, T., Morita, T., Kimura, Y., Tsudii, M., Hayashi, N., and Sobue, K. (2007) FEBS Lett. 581 3777-3782 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.