Abstract
We previously reported that the heparan sulfate 3-O-sulfotransferase (3OST)-5 produces a novel component of heparan sulfate, i.e. the tetrasulfated disaccharide (Di-tetraS) unit (Mochizuki, H., Yoshida, K., Gotoh, M., Sugioka, S., Kikuchi, N., Kwon, Y.-D., Tawada, A., Maeyama, K., Inaba, N., Hiruma, T., Kimata, K., and Narimatsu, H. (2003) J. Biol. Chem. 278 ,26780 -26787). In the present study, we investigated the potential of other 3OST isoforms to produce Di-tetraS with heparan sulfate and heparin as acceptor substrates. 3OST-2, 3OST-3, and 3OST-4 produce Di-tetraS units as a major product from both substrates. 3OST-5 showed the same specificity for heparin, but the production from heparan sulfate was very low. Di-tetraS production by 3OST-1 was negligible. We then investigated the presence of Di-tetraS units in heparan sulfates from various rat tissues. Di-tetraS was detected in all of the tissues analyzed. Liver and spleen contain relatively high levels of Di-tetraS, 1.6 and 0.95%, respectively. However, the content of this unit in heart, large intestine, ileum, and lung is low, less than 0.2%. We further determined the expression levels of 3OST transcripts by quantitative real time PCR. The 3OST-3 transcripts are highly expressed in spleen and liver. The 3OST-2 and -4 are specifically expressed in brain. These results indicate that the Di-tetraS unit is widely distributed throughout the body as a rare and unique component of heparan sulfate and is synthesized by tissue-specific 3OST isoforms specific for Di-tetraS production.
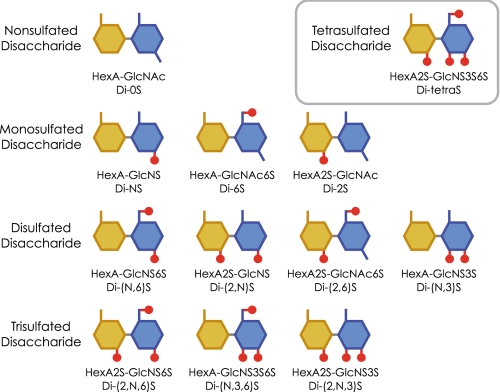
Heparan sulfate is synthesized by most mammalian cells as a component of proteoglycans and interacts with numerous proteins such as growth factors, morphogens, receptors, and extracellular matrix proteins to regulate various physiological processes (1-5). The heparan sulfate molecule is composed of densely sulfated regions or sulfated domains connected by mostly nonsulfated and N-acetyl-rich regions (6-9). Most of the interactions between heparan sulfate and functional proteins are thought to occur at the sulfated domains having specific arrangements of disaccharide isoforms (10-13). Heparan sulfate is initially synthesized as a polymer of the repeating disaccharide, -glucuronic acid-β1,4-N-acetylglucosamine-α1,4-. This polymer is then partially N-deacetylated/N-sulfated and subsequently undergoes epimerization of glucuronic acid to iduronic acid, 2-O-sulfation of hexuronic acid, and 6-O-sulfation of glucosamine residues. Additionally, a rare but functionally important modification, 3-O-sulfation of the glucosamine residue, also occurs. These modification processes are generally not uniform and result in a variety of disaccharide isoforms (14). There are 12 sulfation patterns of disaccharide units (Fig. 1), and some of these units have isoforms of glucuronic acid or iduronic acid. N-Unsubstituted glucosamine residues are also reported as minor components (15, 16). Combinations of these sulfated isoforms enable the formation of disaccharide sequences specific for individual ligand proteins. Rare and heavily sulfated disaccharides in particular may be critical components for the formation of such specific sequences.
FIGURE 1.
Disaccharide units of heparan sulfate. Twelve sulfation patterns of disaccharide units are illustrated. The abbreviations are shown under the substance.
Most of the enzymes involved in the biosynthesis of heparan sulfate have been cloned and characterized. Some of them are present as isoforms and may play important roles in generating the diverse and specific arrangements of disaccharide units (17, 18). Six isoforms of human 3-O-sulfotransferase (3OST)2 have been reported (19-21) and thought to produce the key structure for the functional domain. 3OST-1 transfers sulfate to GlcA-GlcNS±6S and produces the GlcA-GlcNS3S±6S unit essential to the antithrombin (AT)-binding domain (19, 22). 3OST-2 transfers sulfate to GlcA2S/IdoA2S-GlcNS units (23). 3OST-3A and -3B, having identical catalytic domains, transfer sulfate to N-unsubstituted glucosamine residues and form IdoA2S-GlcNH23S±6S units (24). 3OST-3-modified heparan sulfate specifically binds to glycoprotein D (gD) of herpes simplex virus type-1 (HSV-1) and also makes cells susceptible to HSV-1 entry (25, 26). The reaction products of 3OST-5 have been determined to be HexA-GlcNS3S6S (Di-(N,3,6)S), HexA2S-GlcNS3S (Di-(2,N,3)S), and HexA2S-GlcNS3S6S (Di-tetraS) in our previous report (27). 3OST-5-modified heparan sulfate binds to both AT and gD (21). Recently, the Di-tetraS unit was also detected from the products of 3OST-2, -3, and -4 by mass spectrometry (28-30). However, the proportion of Di-tetraS units in the product of each isoform is still unknown.
In the present study, we conducted three lines of investigation to expand our knowledge of the Di-tetraS unit and grasp the physiological significance of this unique structure in heparan sulfate. First, we examined the potential of 3OST family enzymes to produce Di-tetraS. Second, we investigated the presence of the Di-tetraS unit in heparan sulfate from various rat tissues. Third, we determined the expression levels of 3OST isoforms in human tissues by quantitative real time PCR analysis.
EXPERIMENTAL PROCEDURES
Materials—The following commercial materials were used. Heparan sulfate from bovine kidney, heparinase, heparitinase I, heparitinase II, and an unsaturated glycosaminoglycan disaccharide kit were purchased from Seikagaku (Tokyo, Japan). [35S]PAPS was obtained from PerkinElmer Life Sciences. Heparin from porcine intestinal mucosa, PAPS, pepstatin A, leupeptin, anti-FLAG M2 agarose affinity gel, and anti-FLAG M2 peroxidase conjugate were obtained from Sigma; the CarboPac PA1 column was from Dionex (Sunnyvale, CA); n-heptyl-β-d-thioglucoside was from Dojindo (Kumamoto, Japan); Marathon-Ready cDNA of various human tissues was from Clontech (Mountain View, CA); Platinum TaqDNA polymerase, pCR2.1-TOPO vector, Escherichia coli competent cells (TOP10 and DH10Bac), pFastBac1 vector, and Sf21 insect cells were from Invitrogen; polyvinylidene difluoride membrane was from Bio-Rad; SuperSignal was from Pierce; and DEAE-Sephacel was from GE Healthcare.
Construction and Purification of 3OST Family Enzymes Fused with FLAG Peptide—Recombinant 3OST family enzymes were expressed as a fusion protein with FLAG peptide and affinity-purified, based on the methods reported previously (27). DNA fragments coding the putative catalytic domain were amplified by PCR using the Marathon-Ready cDNA of human kidney or placenta as a template. The PCR was performed with a Platinum TaqDNA polymerase, according to the manual supplied. The 5′ and 3′ primers for 3OST-1 were ACAGAATTCAATGGCCGCGCTGCTCCTG and ACAGCATGCGTTTCTGAGCTTAGCTTATTGC (corresponding to amino acids 1-307), for 3OST-2 were ACAGAATTCAGACGACCTGGGTCGGAGC and ACAGCATGCGAGAGCCCTTTCCTTTCGTG (corresponding to amino acids 42-367), for 3OST-3 were ACAGAATTCACGCTGCCAGACCCTGTCC and ACAGCATGCGTTATCCATCCCAGCCAAAG (corresponding to amino acids 45-406), for 3OST-4 were ACAGCGGCCGCCAGCGAGATGATCACGGCTC and ACAGGTACCCCTCTGCCTCTCTAGCCTC (corresponding to amino acids 147-456), and for 3OST-5 were ACAGCGGCCGCAGATAGGCTACAACCCATTTGC and ACAGGTACCTTAGGGCCAGTTCAATGTCCTC (corresponding to amino acids 35-346), respectively. At the 5′ end of the oligonucleotide primers, restriction enzyme recognition sites were introduced, a EcoRI or NotI site for the sense primers and a SphI or KpnI site for the antisense primers. The PCR products were subcloned into pCR2.1-TOPO vector and amplified in TOP10 competent cells, according to the manual supplied. The subcloned DNAs of 3OST isoforms were sequenced on both strands by the dideoxy chain termination method using a DNA sequencer (Applied Biosystems 3130xl). The DNA clones having the same nucleotide sequences found in GenBank were used for the following study. The subcloned DNAs were digested with appropriate restriction enzymes and cloned into multicloning sites of pFastBac1 expression plasmid, containing a fragment encoding a signal peptide of honeybee melittin (MKFLVNVALVFMVVYISYIYA) or preprotrypsin (MSALLILALVGAAVA) and the FLAG peptide (DYKDDDDK). DH10Bac competent cells were transformed with the expression vectors and bacmid DNAs prepared from the cells. Recombinant viruses were prepared according to the instruction manual of Bac-to-Bac Baculovirus Expression Systems (Invitrogen). Sf21 insect cells were infected with the recombinant virus at one multiplicity of infection and incubated at 27 °C for 3 days until the survival rate was less than 50% to yield recombinant 3OST proteins fused with the FLAG peptide. The recombinant enzymes were purified using anti-FLAG M2 agarose affinity gels. 3OST-1 and 3OST-2 were purified from cell lysates and other 3OSTs from various culture media. The cells were collected by centrifugation and suspended to 50 mm Tris-HCl, pH 7.4, containing 150 mm NaCl, 0.5 mm EDTA, 5 μm pepstatin A, 25 μm leupeptin, and 0.5% (W/V) n-heptyl-β-d-thioglucoside. After incubation at 0 °C for 30 min, the cell extracts were obtained by centrifugation. To the culture medium, NaCl and n-heptyl-β-d-thioglucoside were added to final concentrations of 150 mm and 0.5%, respectively. The affinity gel was added to the culture media or the cell extracts and stirred gently at 4 °C for 5 h. After centrifugation, the supernatant was aspirated. The affinity gel was washed three times with 20 mm Tris-HCl, pH 7.4, containing 0.5 m NaCl and 0.5% n-heptyl-β-d-thioglucoside and resuspended in the same buffer containing 150 mm NaCl to obtain 50% slurry. The immobilized enzyme was stable at -80 °C for at least six months.
Western Blot Analysis—The affinity-purified proteins were subjected to SDS-polyacrylamide gel electrophoresis (5-20% gradient gel), followed by Western blot analysis according to the manufacturer's instructions. The separated proteins were transferred onto polyvinylidene difluoride membrane and probed with an anti-FLAG peroxidase conjugate. Immune complexes were detected as positive bands using the SuperSignal detection system.
Preparation of 35S-Labeled Glycosaminoglycans and Digestion with Heparin Lyases—The reaction mixture (50 μl) containing 50 mm PIPES, pH 7.0, 10 mm MgCl2, 150 mm NaCl, 80 μg/ml of protamine chloride, 0.5% n-heptyl-β-d-thioglucoside, 1 mg/ml of glycosaminoglycan (heparan sulfate or heparin), 3 μm [35S]PAPS (about 1 × 106 dpm), and immobilized enzyme with the activity to yield a radiolabeled product of about 4 × 104 dpm, an amount that is sufficient for composition analysis, was incubated at 37 °C for 3 h. Then 35S-labeled glycosaminoglycan was precipitated with 2 volumes of ethanol containing 1.3% (w/v) potassium acetate. The precipitate was dissolved in 50 μl of water, filtered, and reprecipitated to remove [35S]PAPS and its degradation products. The dried precipitate was dissolved in 10 μl of 20 mm sodium acetate buffer, pH 7.0, containing 2 mm calcium acetate and 1 mg/ml of bovine serum albumin (digestion buffer). The 35S-labeled glycosaminoglycan was digested with a mixture of 30 milliunits of heparinase, 18 milliunits of heparitinase I, and 12 milliunits of heparitinase II in 6 μl of digestion buffer at 37 °C for 3 h. The reaction was stopped by heating at 100 °C for 1 min, and the reaction mixture was filtered. The digested products were then subjected to HPLC analysis. To evaluate incubation conditions, we employed a shorter incubation time (30 min). The same results were obtained with different isoforms.
HPLC on an Anion Exchange Column—Anion exchange (SAX) separation was performed on a CarboPac PA1 column (4 × 250 mm) as described elsewhere (31). A combination of five linear LiCl gradients was used: 0-5 min, 30-180 mm;5-8 min, 180-570 mm; 8-15 min, 0.57-1.14 m; 15-20 min, 1.14-2.1 m; 20-28 min, 2.1-2.28 m; and 28-35 min, 2.28 m. The flow rate was 0.8 ml/min, and the column temperature was 40 °C. Fractions of 0.2 ml were collected, and the radioactivity was determined by liquid scintillation counting.
Preparation of ΔDi-tetraS—The standard ΔDi-tetraS used in this study was prepared as follows. Three milligrams of heparin was incubated with 2.6 ml of reaction mixture, containing nonradioactive PAPS and 3OST-5, as described above. The 3-O-sulfated heparin was digested with a mixture of heparin lyases and separated by HPLC on a SAX column. The eluate was monitored at 232 nm, and a peak fraction of ΔDi-tetraS with a retention time of 28 min was collected. The obtained fraction was desalted and lyophilized.
Preparation of Heparan Sulfate from Rat Tissues—Heparan sulfates from various rat tissues were prepared as described (32) with some modifications. Briefly, excised tissues were frozen in liquid nitrogen immediately, crushed into small pieces, and homogenized with acetone using a Polytron homogenizer. The obtained powder was washed with acetone twice and dried under vacuum. Twenty milligrams of the defatted tissue powder was suspended in 200 μl of 0.5 m NaOH and incubated at 50 °C for 2 h to remove the glycosaminoglycan chains from its core protein by β-elimination. After neutralization with 1 m HCl, NaCl was added to a final concentration of 3 m. Insoluble materials were removed by centrifugation, and the pH of the supernatant was adjusted to 1.0 with 1 m HCl. Precipitated nucleotides were removed by centrifugation, and the supernatant was neutralized with 1 m NaOH. The crude glycosaminoglycans were precipitated by the addition of two volumes of ethanol containing 1.3% potassium acetate. After centrifugation, the precipitate was dissolved in 0.3 ml of distilled water, dialyzed against water at 4 °C for 12 h, and lyophilized. The obtained powder was dissolved in 10 μl of 0.2 m ammonium acetate and applied to 30 μl of DEAE-Sephacel column. The column was washed with 250 μl of 0.5 m ammonium acetate and eluted with 150 μl of 3 m ammonium acetate. Finally, the heparan sulfate fraction was lyophilized.
Analysis of the Heparan Sulfate by Post-column Fluorescent Labeling Method—The heparan sulfates derived from rat tissues were digested with a mixture of heparin lyases as described above. Unsaturated disaccharides produced by the digestion were separated by reverse-phase ion pair chromatography and detected by a post-column fluorescent labeling system as described by Toyoda et al. (33) with a minor modification of gradient formation. The eluents used were as follows: A, distilled water; B, 0.4 m NaCl; C, 10 mm tetra-n-butylammonium hydrogen sulfate; D, 50% acetonitrile. The gradient program was as follows: 0-10 min, 1-2% eluent B; 10-11 min, 2-8% eluent B; 11-20 min, 8-13% eluent B; 20-22 min, 13-27% eluent B; 22-29 min, 27% eluent B; 29-33 min, 27-70% eluent B; and 33-40 min, 70% eluent B. The proportions of eluents C and D were constant at 12 and 17%, respectively. Other reagents and system formations were similar to those of the previous report (33).
Quantitative Analysis of 3OST Transcripts in Human Tissues using Real Time PCR—For quantification of 3OST transcripts in cDNAs of various human tissues, we employed the real time PCR method, as described in detail previously (34). To obtain the DNA of human glyceraldehyde-3-phosphate dehydrogenase (GAPDH), as an endogenous control, a DNA fragment was amplified by PCR using the Marathon Ready cDNA, as a template, and two primers, CTCCTCCTGTTCGACAGTCAG and GGTGATGGGATTTCCATTGATG. Standard curves for the 3OST isoforms and the GAPDH were generated by serial dilution of the pCR2.1-TOPO vector DNAs containing the cloned 3OST isoform, prepared as described above, and the control DNA of GAPDH. The primer sets and the probe with a minor groove binder were CGGCTGCTGCTCATCCT, TCTGCATGTGGTTGTAGAACACTT, and TCGGAGCGCGTGCTAT for 3OST-1; CGAGACACCAAGCTGATCGT, GGAGAGTGTCTGCGTGTAATCA, and TTGTGCGGAACCCTG for 3OST-2; GGGCAGATCACCATGGAGAAG, CCTTGGACATGGCCGAGATG, and ACGCCCAGTTACTTCG for 3OST-3; CAGGGCCATCTCTGACTACAC, AGGCCAGCACCTCAAAGG, and ACTGTCAAAGAAACCC for 3OST-4; and CAAAATGAACTCATCCATCAAGTTGTTGA, CCCCTCTAGCACCTGAGTATAATCA, and CAGGGAGCCAACCACA for 3OST-5, respectively (35). The primer set for 3OST-3 detects both 3OST-3A and -3B having the same sequence of the catalytic domain. For GAPDH, we used a TaqMan gene expression assay (assay ID Hs99999905_m1; Applied Biosystems). PCR products were continuously measured with an ABI PRISM 7300 sequence detection system (Applied Biosystems). The relative amounts of 3OST transcripts were normalized to the amount of GAPDH transcript in the same cDNA samples.
RESULTS
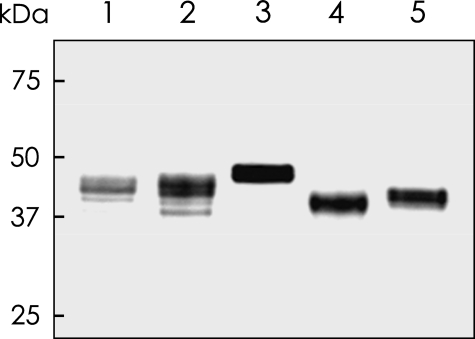
Molecular Cloning and Expression of 3OST Isoforms—cDNA fragments encoding the putative catalytic domains of five 3OST family enzymes, 3OST-1, 3OST-2, 3OST-3, 3OST-4, and 3OST-5, were obtained by PCR cloning using the human cDNA library as a template. FLAG-tagged recombinant proteins were produced by a baculovirus expression system and purified by anti-FLAG affinity gel as described under “Experimental Procedures.” Individual purified proteins were detected by Western blot analysis using an anti-FLAG peroxidase conjugate as shown in Fig. 2 and used for the subsequent enzyme reaction.
FIGURE 2.
Western blot analysis of recombinant 3OST isoforms. The FLAG-tagged 3OST proteins were expressed in insect cells, purified with an anti-FLAG agarose gel, and subjected to Western blot analysis as described under “Experimental Procedures.” Lanes 1-5 are 3OST-1, 3OST-2, 3OST-3, 3OST-4, and 3OST-5, respectively. The positions of molecular size standards (in kDa) are indicated on the left.
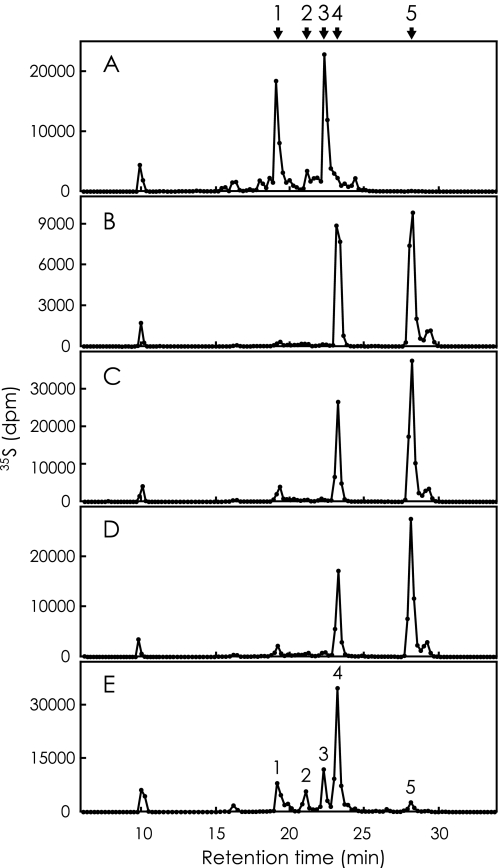
Analysis of 35S-Labeled Heparan Sulfate—35S-Labeled heparan sulfate prepared by incubating heparan sulfate with [35S]PAPS and one of the recombinant 3OST isoforms was digested with a mixture of heparin lyases and analyzed by HPLC on a SAX column as described under “Experimental Procedures.” Fig. 3 represents elution profiles of radiolabeled products derived from each reaction mixture. The elution profile is similar to that of our previous report in the case of 3OST-5 (Fig. 3E), and five peaks have been characterized as follows (27). Peaks 2, 4, and 5 are ΔDi-(N,3,6)S, ΔDi-(2,N,3)S, and ΔDi-tetraS, respectively. Peaks 1 and 3 are tetrasaccharides, named ΔTetra-1 and ΔTetra-2. The arrows in Fig. 3 indicate the positions of these products. The elution profile of the radiolabeled products derived from the 3OST-1 catalyzed reaction exhibits two major peaks at the positions of ΔTetra-1 and ΔTetra-2 (Fig. 3A). The molecular size of these products corresponds to tetrasaccharide by gel filtration chromatography (data not shown). Analysis of the radiolabeled products derived from the reaction of 3OST-2 (Fig. 3B), 3OST-3 (Fig. 3C), and 3OST-4 (Fig. 3D) shows similar elution patterns consisting of two peaks at the positions of ΔDi-(2,N,3)S and ΔDi-tetraS. The molecular sizes of all these products correspond to disaccharide by gel filtration chromatography (data not shown). A small peak following the ΔDi-tetraS is derived from ΔDi-tetraS itself because rechromatography of the parental peak resulted in the same profile. The peak with the retention time of 10 min, because it is not specific for the enzyme reaction, may be [35S]PAPS or its degradation product.
FIGURE 3.
HPLC analysis of 35S-labeled products derived from heparan sulfate. Heparan sulfate was incubated with [35S]PAPS and one of the recombinant 3OST isoforms. The 35S-labeled products were digested with a mixture of heparin lyases, and then the digests were separated by HPLC on a SAX column as described under “Experimental Procedures.” The fractions were collected, and the radioactivity was determined by liquid scintillation counting. The five panels (A-E), show the elution profiles of radiolabeled products derived from the reactions of 3OST-1, 3OST-2, 3OST-3, 3OST-4, and 3OST-5, respectively. The five peaks of panel E have been identified as ΔTetra-1 (peak 1), ΔDi-(N,3,6)S (peak 2), ΔTetra-2 (peak 3), ΔDi-(2,N,3)S (peak 4), and ΔDi-tetraS (peak 5) in a previous study (27). These positions are also indicated above the figure. For the abbreviations, see Fig. 1.
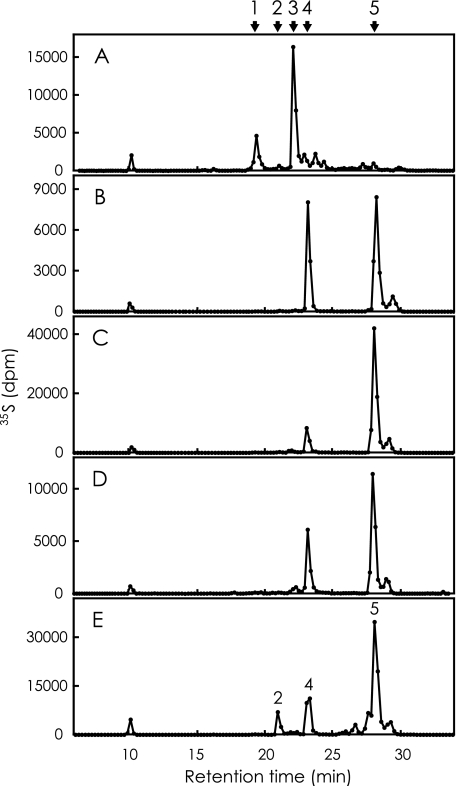
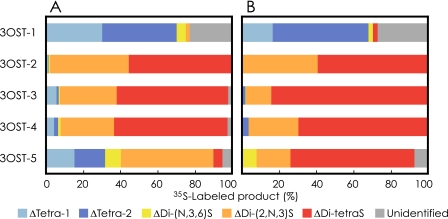
Analysis of 35S-Labeled Heparin—35S-Labeled heparin was prepared and analyzed in the same way as described above. Fig. 4 represents the elution profiles of radiolabeled products derived from each reaction mixture. The elution profile is also similar to that of our previous report for the case of 3OST-5 (Fig. 4E). The arrows in Fig. 4 indicate the positions of the products described above. Analysis of the radiolabeled products derived from the reaction of four isoforms resulted in chromatograms similar to Fig. 3. The products of 3OST-1 indicate two major peaks at the positions of ΔTetra-1 and ΔTetra-2 (Fig. 4A). The products of 3OST-2 (Fig. 4B), 3OST-3 (Fig. 4C), and 3OST-4 (Fig. 4D) demonstrate two peaks at the positions of ΔDi-(2,N,3)S and ΔDi-tetraS. 3OST-5 is the only enzyme that shows different specificity to two glycosaminoglycans. ΔDi-tetraS is a prominent peak, and tetrasaccharides are not detected when heparin is the acceptor substrate. Fig. 5 was calculated from Figs. 3 and 4 to review the composition of the reaction product from heparan sulfate and heparin, (panels A and B), respectively. Three isoforms, 3OST-2, 3OST-3, and 3OST-4, produce Di-tetraS units as a major product from both glycosaminoglycans. 3OST-5 also produces this unit as a major product from heparin, but the production from heparan sulfate is very low. 3OST-1 produces a negligible amount of Di-tetraS units.
FIGURE 4.
HPLC analysis of 35S-labeled products derived from heparin. Heparin was incubated with [35S]PAPS and one of the recombinant 3OST isoforms. The 35S-labeled products were digested with a mixture of heparin lyases and analyzed by HPLC on a SAX column as described under “Experimental Procedures.” The five panels(A-E) show the elution profile of radio labeled products derived from the reactions of 3OST-1, 3OST-2, 3OST-3, 3OST-4, and 3OST-5, respectively. The arrows indicate the elution positions of ΔTetra-1 (peak 1), ΔDi-(N,3,6)S (peak 2), ΔTetra-2 (peak 3), ΔDi-(2,N,3)S (peak 4), and ΔDi-tetraS (peak 5). For the abbreviations, see Fig. 1.
FIGURE 5.
A comparison of 35S-labeled products derived from heparan sulfate and heparin by the reaction of 3OST isoforms. Percentages of radiolabeled products were calculated from Figs. 3 and 4. A, compositions of radiolabeled products derived from heparan sulfate by the reactions of 3OST isoforms, as indicated on the left. B, compositions of radiolabeled products derived from heparin by the reactions of 3OST isoforms.
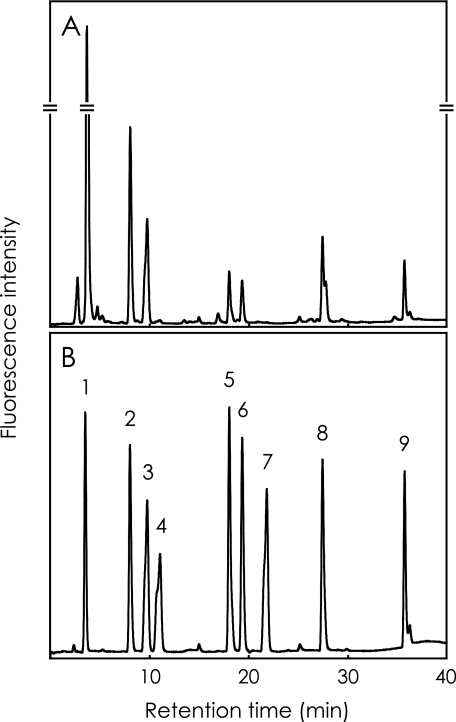
Analysis of Heparan Sulfate from Various Rat Tissues—Heparan sulfate was purified from various rat tissues and digested with a mixture of heparin lyases as described under “Experimental Procedures.” The digestion products were analyzed by reverse-phase ion pair chromatography with a post-column fluorescence labeling system. The elution condition was optimized to elute ΔDi-tetraS, the most negatively charged disaccharide unit. Fig. 6 shows a typical chromatogram of the digestion products of heparan sulfate from rat liver (panel A), and of standard disaccharides (panel B). ΔDi-tetraS is successfully detected with this analytical system. Table 1 shows the disaccharide composition of heparan sulfate from various rat tissues. ΔDi-tetraS was detected in all of the tissues analyzed here. Heparan sulfate from liver and spleen contain relatively high levels of Di-tetraS units. On the other hand, the content of this unit in heart heparan sulfate is very low. Other 3-O-sulfated components were not determined because we focused on the Di-tetraS unit in the present study. Some very small peaks around the ΔDi-(2,N,6)S may contain such rare components.
FIGURE 6.
Typical chromatograms of unsaturated disaccharides prepared from rat tissue heparan sulfate. A, heparan sulfate isolated from rat liver was digested with a mixture of heparin lyases, and the resultant disaccharides were subjected to reverse-phase ion pair chromatography with post-column fluorescence labeling as described under “Experimental Procedures.” B, nine disaccharide standards are ΔDi-0S (peak 1), ΔDi-NS (peak 2), ΔDi-6S (peak 3), ΔDi-2S (peak 4), ΔDi-(N,6)S (peak 5), ΔDi-(2,N)S (peak 6), ΔDi-(2,6)S (peak 7), ΔDi-(2,N,6)S (peak 8), and ΔDi-tetraS (peak 9). For the abbreviations, see Fig. 1.
TABLE 1.
The disaccharide composition of heparan sulfate from various rat tissues (molar percent)
| Di-0Sa | Di-NS | Di-6S | Di-2S | Di-(N,6)S | Di-(2,N)S | Di-(2,6)S | Di-(2,N,6)S | Di-tetraS | |
|---|---|---|---|---|---|---|---|---|---|
| Cerebrum | 52b | 19 | 7.4 | 0.85 | 5.9 | 8.3 | NDc | 6.0 | 0.26 |
| Cerebellum | 53 | 18 | 6.6 | 0.62 | 5.9 | 7.4 | ND | 8.0 | 0.20 |
| Lung | 52 | 21 | 8.4 | 1.6 | 4.5 | 7.4 | ND | 4.1 | 0.16 |
| Heart | 62 | 19 | 6.4 | 0.35 | 3.6 | 4.4 | ND | 3.9 | 0.026 |
| Kidney | 44 | 19 | 15 | 0.63 | 7.8 | 5.7 | ND | 7.4 | 0.21 |
| Liver | 47 | 19 | 8.2 | 0.46 | 4.2 | 6.0 | ND | 13 | 1.6 |
| Spleen | 46 | 13 | 17 | 0.14 | 15 | 2.1 | ND | 5.7 | 0.95 |
| Pancreas | 58 | 21 | 7.6 | 0.45 | 4.5 | 4.4 | ND | 4.4 | 0.20 |
| Stomach | 59 | 21 | 6.2 | 0.56 | 3.0 | 5.9 | ND | 4.6 | 0.22 |
| Ileum | 60 | 22 | 5.7 | 1.0 | 2.6 | 4.9 | ND | 4.2 | 0.13 |
| Large intestine | 60 | 21 | 5.7 | 0.61 | 2.6 | 5.8 | ND | 4.2 | 0.12 |
| Testis | 60 | 16 | 7.7 | 0.35 | 3.6 | 7.3 | ND | 4.3 | 0.29 |
For the abbreviations, see Fig. 1.
The values represent the averages of two independent experiments.
ND, not detected.
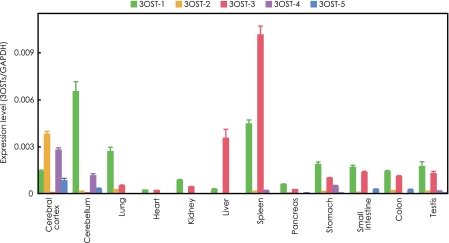
Quantitative Analysis of 3OST Isoforms in Human Tissues—The tissue distribution and expression levels of 3OST transcripts were determined by a real time PCR method. The expression levels of 3OST transcripts in various tissues are shown in relative amounts compared with the GAPDH transcripts (Fig. 7). The 3OST-1 transcripts (green bar) are widely distributed in various tissues. The expression of 3OST-2 (orange bar) is very specific for cerebral cortex. 3OST-3 (red bar) transcripts are also widely distributed in many tissues, but prominent in spleen and liver. 3OST-4 (violet bar) and 3OST-5 (blue bar) transcripts are mainly expressed in cerebral cortex and cerebellum, but the expression levels of 3OST-5 are relatively low in all tissues.
FIGURE 7.
Quantitative analysis of 3OST isoform transcripts in human tissues using real time PCR. Standard curves for 3OST isoforms and GAPDH were generated by serial dilution of plasmid DNA or control DNA as described under “Experimental Procedures.” The expression levels of 3OST isoforms were normalized to that of the GAPDH transcript, which was measured in the same cDNAs. The data were obtained from triplicate experiments and given as the means ± S.D.
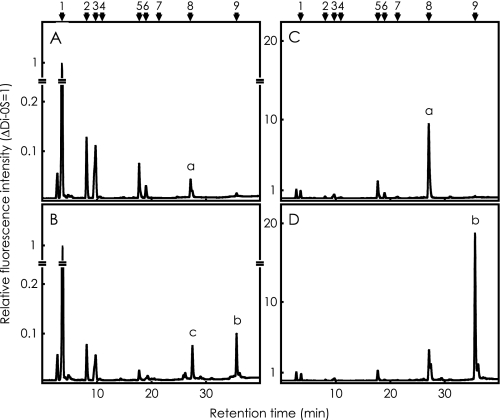
Analysis of Heparan Sulfate and Heparin Sulfated by 3OST-3—Heparan sulfate or heparin was incubated with nonradioactive PAPS and excess amounts of 3OST-3 in the standard reaction mixture, digested with a mixture of heparin lyases, and analyzed by reverse-phase ion pair chromatography with a post-column fluorescence labeling system as described under “Experimental Procedures.” Panels A and B in Fig. 8 show the disaccharide analysis of heparan sulfate, before and after the sulfation reaction with 3OST-3, respectively. Panels C and D show that of heparin. Most ΔDi-(2,N,6)S in both acceptor substrates, peak a in panels A and C, was converted to ΔDi-tetraS, peak b in panels B and D, respectively. A peak c in panel B with a retention time later than ΔDi-(2,N,6)S is an unidentified 3-O-sulfated product that may be ΔDi-(2,N,3)S.
FIGURE 8.
Analysis of heparan sulfate and heparin sulfated by excess amounts of 3OST-3. Ten micrograms of heparan sulfate or heparin were incubated with nonradioactive PAPS and an excess amount of 3OST-3 in the standard reaction mixture and digested with a mixture of heparin lyases, as described under “Experimental Procedures.” The resultant disaccharides were subjected to reverse-phase ion pair chromatography with post-column fluorescence labeling. A and C show the analysis of heparan sulfate and heparin, without sulfation reaction, respectively. B and D show the analysis of reaction products from heparan sulfate and heparin, respectively. The elution positions of standard disaccharides are indicated by numbers; see legend to Fig. 6.
DISCUSSION
We previously cloned human 3OST-5, expressed it in insect cells, and characterized the reaction products of the expressed enzyme (27). Three disaccharide structures, Di-(N,3,6)S, Di-(2,N,3)S, and Di-tetraS, were determined as the reaction products by 3OST-5 using heparan sulfate and heparin as acceptor substrate. We then analyzed bovine intestinal heparan sulfate and successfully detected Di-tetraS units as a minor component (0.15% of total disaccharides). This was the first report to demonstrate the natural presence of Di-tetraS units. However, there has been no report showing the tissue distribution of this unique structure or the content of Di-tetraS units in natural glycosaminoglycans except intestinal heparan sulfate. The present study was conducted to obtain a better understanding of the physiological function of this unique structure as an ordinary component of heparan sulfate.
First, we investigated the potential of other 3OST isoforms to produce Di-tetraS and analyzed the reaction products of five human 3OST family enzymes in the same way as our previous study using heparan sulfate and heparin as acceptor substrates. The result obtained with 3OST-5 is consistent with our previous report (27), indicating reproducibility. A summary of the composition analysis of the reaction products (Fig. 5) suggests that the reaction products of each of the 3OST isoforms reflected the physiological activities of the products previously reported. For example, 3OST-1 is a specific enzyme for the production of AT-binding heparan sulfate (36), and the product is resistant to the enzyme digestion, giving tetrasaccharides of ΔTetra-1 and -2. It has been reported that the glucosaminidic linkage adjacent to GlcA-GlcNS3S±6S units, which is a critical component of the AT-binding site, is resistant to heparin lyases (37, 38). Therefore, ΔTetra-1 and -2 may contain these disaccharide units at the reducing terminal. 3OST-5 has also been reported to produce AT-binding heparan sulfate (21) and also produces ΔTetra-1 and -2. Four 3OST isoforms except 3OST-1 have been reported to make cells susceptible to HSV-1 entry (21, 30) and produce two disaccharides of ΔDi-(2,N,3)S and ΔDi-tetraS as common products. On the other hand, N-unsubstituted and 3-O-sulfated glucosamine residue, which has been identified as the product of 3OST-3, has been reported as a specific component for heparan sulfate binding to gD (26). Therefore, it may be necessary to re-examine the components essential for HSV-1 entry. 3OST-5 is a unique enzyme having broad specificity to produce both AT-binding and gD-binding heparan sulfate, and the product is partially resistant to enzyme digestion, giving both tetrasaccharides and disaccharides as described above.
We confirmed that most of the Di-(2,N,6)S units in heparan sulfate and heparin are available as substrates for the Di-tetraS formation. It was reported that most of the Di-(2,N,6)S units in heparan sulfate contain iduronic acid (8), i.e. IdoA2S-GlcNS6S unit. Therefore the Di-tetraS units should contain the same hexuronic acid isomer, i.e. iduronic acid. However, Di-tetraS unit having glucuronic acid residue is still possible because the GlcA2S-GlcNS6S unit has been reported to be a minor component of heparin (39).
We examined the disaccharide compositions of the heparan sulfate preparations from various rat tissues to clarify the tissue distribution of Di-tetraS units. Interestingly, all tissues analyzed here contain this unique disaccharide unit. Heparan sulfate from liver and spleen in particular contained relatively high levels of this unit. This is the first report demonstrating the content of Di-tetraS in heparan sulfate from various tissues, although several reports show the disaccharide compositions of heparan sulfate from various sources (8, 33, 40-42). Some liver-specific functions of heparan sulfate have been reported. For example, exogenous and endogenous triglycerides enter the circulation as chylomicron and very low density lipoprotein, respectively. After the release of fatty acids for the peripheral tissues by lipoprotein lipase, the remnant particles are captured by hepatocytes. Hepatic clearance of these triglyceride-rich lipoproteins is mediated by heparan sulfate (43). In malaria infection, Plasmodium sporozoites injected into the skin by infected mosquitoes are carried to the liver in the bloodstream, where the sporozoites invade hepatocytes and develop into exoerythrocytic forms. It has been shown that the liver-specific heparan sulfate activates sporozoites to begin the invasion process (44). There is evidence that infections caused by some viruses in liver, such as hepatitis B virus, hepatitis C virus, and dengue virus, are also mediated by liver-specific heparan sulfate (45-47). Di-tetraS-rich heparan sulfate in liver may be a target of infection by these pathogens.
To discover how the Di-tetraS units are derived, we examined the expression levels of 3OST isoforms in various human tissues by quantitative real time PCR. Northern blot analysis of the 3OST isoforms, from 3OST-1 to -4, has already been reported by others (20). Our present results are consistent with the reported expression pattern, such as brain-specific expression of 3OST-2 and -4. Further, our quantitative analyses give more detailed information. For example, high expression levels of 3OST-3 in spleen and liver account for the high levels of Di-tetraS units in these tissues. Therefore, some physiological roles of 3OST-3 in liver-specific phenomena such as those described above can be expected. The expression profiles of isoforms in cerebral cortex and cerebellum are clearly different from each other. 3OST-2 is very specific for the cerebral cortex, and 3OST-1 is highly expressed in the cerebellum. Some cerebellum-specific function of 3OST-1 can also be expected.
As shown in Fig. 7, some 3OST isoforms are coexpressed in the same tissues. Synergistic effects have been reported between heparan sulfate synthesizing enzymes or isoforms. For example, complex formation of EXT1 and 2 elicits heparan sulfate polymerase activities (48, 49). Uronosyl 5-epimerase needs interaction with 2-O-sulfotransferase for its activity and localization to Golgi (50). However, there is no evidence for a synergistic effect of 3OSTs. We mixed some 3OST isoforms and examined the sulfotransferase activities, but no synergistic effect was observed (data not shown). The observed activities were the total sum of each isoform, suggesting that a team of isoforms coexpressed in the same tissue produced the functional domains. More detailed studies on coexpression in mammalian cells are required to clarify these questions.
The biological significance of 3-O-sulfate other than the regulation of blood coagulation and HSV-1 entry remains unknown at the present time. However, there are a few important reports that suggest the physiological importance of this modification. Hs3st1-/- knock-out mice showed postnatal lethality and intrauterine growth retardation (51). Reduction of 3OST-B, a Drosophila homolog of 3OST family enzyme, by transgenic RNA interference technique revealed phenotypes of Notch signaling deficiency (52). Kuberan et al. (53) analyzed heparan sulfate from rat pineal gland using mass spectrometry and detected Di-tetraS units only from daytime pineal gland. In addition, we have investigated the expressions of 3OSTs in various tissues and found that, corresponding to the tissue-specific expression of 3OST isoforms, Di-tetraS units of heparan sulfate increased, suggesting the existence of a regulation mechanism of 3-O-sulfation by the enzyme. Overall, the present study has demonstrated that some 3OST isoforms are Di-tetraS-producing enzymes and that heparan sulfate in most tissues possess this rare and unique component, suggesting that it may have some significant physiological function.
Acknowledgments
We are grateful to Kennichi Maeyama and Kazuhiro Kojima for skillful technical support, to Hiroshi Maeda and Takehiko Nakamura for helpful suggestions, to Akira Tawada and Masaki Kosemura for fruitful discussion, and to Cecilia Hamagami for correcting the manuscript in English.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: 3OST, heparan sulfate d-glucosaminyl 3-O-sulfotransferase; PAPS, 3′-phosphoadenosine 5′-phosphosulfate; PIPES, piperazine-1,4-bis(2-ethanesulfonic acid); HexA, hexuronic acid; GlcA, d-glucuronic acid; IdoA, l-iduronic acid; GlcN, d-glucosamine; 2S, 2-O-sufate; NS, 2-N-sulfate; 3S, 3-O-sulfate; 6S, 6-O-sulfate; AT, antithrombin; HSV-1, herpes simplex virus type-1; gD, glycoprotein D; SAX, strong anion exchange; HPLC, high performance liquid chromatography; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
References
- 1.Yanagishita, M., and Hascall, V. C. (1992) J. Biol. Chem. 2679451 -9454 [PubMed] [Google Scholar]
- 2.Bernfield, M., Götte, M., Park, P. W., Reizes, O., Fitzgerald, M. L., Lincecum, J., and Zako, M. (1999) Annu. Rev. Biochem. 68729 -777 [DOI] [PubMed] [Google Scholar]
- 3.Esko, J. D., and Selleck, S. B. (2002) Annu. Rev. Biochem. 71435 -471 [DOI] [PubMed] [Google Scholar]
- 4.Lin, X. (2004) Development 1316009 -6021 [DOI] [PubMed] [Google Scholar]
- 5.Bishop, J. R., Schuksz, M., and Esko, J. D. (2007) Nature 4461030 -1037 [DOI] [PubMed] [Google Scholar]
- 6.Turnbull, J. E., and Gallagher, J. T. (1991) Biochem. J. 273553 -559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lyon, M., Deakin, J. A., and Gallagher, J. T. (1994) J. Biol. Chem. 26911208 -11215 [PubMed] [Google Scholar]
- 8.Maccarana, M., Sakura, Y., Tawada, A., Yoshida, K., and Lindahl, U. (1996) J. Biol. Chem. 27117804 -17810 [DOI] [PubMed] [Google Scholar]
- 9.Murphy, K. J., Merry, C. L. R., Lyon, M., Thompson, J. E., Roberts, I. S., and Gallagher, J. T. (2004) J. Biol. Chem. 27927239 -27245 [DOI] [PubMed] [Google Scholar]
- 10.Brickman, Y. G., Ford, M. D., Gallagher, J. T., Nurcombe, V., Bartlett, P. F., and Turnbull, J. E. (1998) J. Biol. Chem. 2734350 -4359 [DOI] [PubMed] [Google Scholar]
- 11.Lindahl, U., Kusche-Gullberg, M., and Kjellen, L. (1998) J. Biol. Chem. 27324979 -24982 [DOI] [PubMed] [Google Scholar]
- 12.Gallagher, J. T. (2001) J. Clin. Investig. 108357 -361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakato, H., and Kimata, K. (2002) Biochim. Biophys. Acta 1573312 -318 [DOI] [PubMed] [Google Scholar]
- 14.Esko, J. D., and Lindahl, U. (2001) J. Clin. Investig. 108169 -173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Norgard-Sumnicht, K., and Varki, A. (1995) J. Biol. Chem. 27012012 -12024 [DOI] [PubMed] [Google Scholar]
- 16.van den Born, J., Gunnarsson, K., Bakker, M. A., Kjellen, L., Kusche-Gullberg, M., Maccarana, M., Berden, J. H. M., and Lindahl, U. (1995) J. Biol. Chem. 27031303 -31309 [DOI] [PubMed] [Google Scholar]
- 17.Habuchi, O. (2000) Biochim. Biophys. Acta 1474115 -127 [DOI] [PubMed] [Google Scholar]
- 18.Sugahara, K., and Kitagawa, H. (2002) IUBMB Life. 54163 -175 [DOI] [PubMed] [Google Scholar]
- 19.Shworak, N. W., Liu, J., Fritze, L. M. S., Schwartz, J. J., Zhang, L., Logeart, D., and Rosenberg, R. D. (1997) J. Biol. Chem. 27228008 -28019 [DOI] [PubMed] [Google Scholar]
- 20.Shworak, N. W., Liu, J., Petros, L. M., Zhang, L., Kobayashi, M., Copeland, N. G., Jenkins, N. A., and Rosenberg, R. D. (1999) J. Biol. Chem. 2745170 -5184 [DOI] [PubMed] [Google Scholar]
- 21.Xia, G., Chen, J., Tiwari, V., Ju, W., Li, J.-P., Malmström, A., Shukla, D., and Liu, J. (2002) J. Biol. Chem. 27737912 -37919 [DOI] [PubMed] [Google Scholar]
- 22.Lindahl, U., Bäckström, G., Thunberg, L., and Leder, I. G. (1980) Proc. Natl. Acad. Sci. U. S. A. 776551 -6555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu, J., Shworak, N. W., Sinaÿ, P., Schwartz, J. J., Zhang, L., Fritze, L. M. S., and Rosenberg, R. D. (1999) J. Biol. Chem. 2745185 -5192 [DOI] [PubMed] [Google Scholar]
- 24.Liu, J., Shriver, Z., Blaiklock, P., Yoshida, K., Sasisekharan, R., and Rosenberg, R. D. (1999) J. Biol. Chem. 27438155 -38162 [DOI] [PubMed] [Google Scholar]
- 25.Shukla, D., Liu, J., Blaiklock, P., Shworak, N. W., Bai, X., Esko, J. D., Cohen, G. H., Eisenberg, R. J., Rosenberg, R. D., and Spear, P. G. (1999) Cell 99 13-22 [DOI] [PubMed] [Google Scholar]
- 26.Liu, J., Shriver, Z., Pope, R. M., Thorp, S. C., Duncan, M. B., Copeland, R. J., Raska, C. S., Yoshida, K., Eisenberg, R. J., Cohen, G., Linhardt, R. J., and Sasisekharan, R. (2002) J. Biol. Chem. 27733456 -33467 [DOI] [PubMed] [Google Scholar]
- 27.Mochizuki, H., Yoshida, K., Gotoh, M., Sugioka, S., Kikuchi, N., Kwon, Y.-D., Tawada, A., Maeyama, K., Inaba, N., Hiruma, T., Kimata, K., and Narimatsu, H. (2003) J. Biol. Chem. 27826780 -26787 [DOI] [PubMed] [Google Scholar]
- 28.Wu, Z. L., Lech, M., Beeler, D. L., and Rosenberg, R. D. (2004) J. Biol. Chem. 2791861 -1866 [DOI] [PubMed] [Google Scholar]
- 29.Lawrence, R., Kuberan, B., Lech, M., Beeler, D. L., and Rosenberg, R. D. (2004) Glycobiology 14 467-479 [DOI] [PubMed] [Google Scholar]
- 30.Lawrence, R., Yabe, T., HajMohammadi, S., Rhodes, J., McNeely, M., Liu, J., Lamperti, E. D., Toselli, P. A., Lech, M., Spear, P. G., Rosenberg, R. D., and Shworak, N. W. (2007) Matrix Biol. 26442 -455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kariya, Y., Yoshida, K., Morikawa, K., Tawada, A., Miyazono, H., Kikuchi, H., and Tokuyasu, K. (1992) Comp. Biochem. Physiol. 103B473 -479 [DOI] [PubMed] [Google Scholar]
- 32.Heinegård, D., and Sommarin, Y. (1987) Methods Enzymol. 144319 -372 [DOI] [PubMed] [Google Scholar]
- 33.Toyoda, H., Kinoshita-Toyoda, A., and Selleck, S. B. (2000) J. Biol. Chem. 2752269 -2275 [DOI] [PubMed] [Google Scholar]
- 34.Iwai, T., Inaba, N., Naundorf, A., Zhang, Y., Gotoh, M., Iwasaki, H., Kudo, T., Togayachi, A., Ishizuka, Y., Nakanishi, H., and Narimatsu, H. (2002) J. Biol. Chem. 27712802 -12809 [DOI] [PubMed] [Google Scholar]
- 35.Kutyavin, I. V., Afonina, I. A., Mills, A., Gorn, V. V., Lukhtanov, E. A., Belousov, E. S., Singer, M. J., Walburger, D. K., Lokhov, S. G., Gall, A. A., Dempcy, R., Reed, M. W., Meyer, R. B., and Hedgpeth, J. (2000) Nucleic Acids Res. 28 655-661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu, J., Shworak, N. W., Fritze, L. M. S., Edelberg, J. M., and Rosenberg, R. D. (1996) J. Biol. Chem. 27127072 -27082 [DOI] [PubMed] [Google Scholar]
- 37.Yamada, S., Yoshida, K., Sugiura, M., Sugahara, K., Khoo, K.-H., Morris, H. R., and Dell, A. (1993) J. Biol. Chem. 2684780 -4787 [PubMed] [Google Scholar]
- 38.Desai, U. R., Wang, H.-M., and Linhardt, R. J. (1993) Arch. Biochem. Biophys. 306461 -468 [DOI] [PubMed] [Google Scholar]
- 39.Yamada, S., Murakami, T., Tsuda, H., Yoshida, K., and Sugahara, K. (1995) J. Biol. Chem. 2708696 -8705 [PubMed] [Google Scholar]
- 40.Toida, T., Yoshida, H., Toyoda, H., Koshiishi, I., Imanari, T., Hileman, R. E., Fromm, J. R., and Linhardt, R. J. (1997) Biochem. J. 322499 -506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamada, S., Van Die, I., Van den Eijnden, D. H., Yokota, A., Kitagawa, H., and Sugahara, K. (1999) FEBS Lett. 459327 -331 [DOI] [PubMed] [Google Scholar]
- 42.Warda, M., Toida, T., Zhang, F., Sun, P., Munoz, E., Xie, J., and Linhardt, R. J. (2006) Glycoconj. J. 23 555-563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.MacArthur, J. M., Bishop, J. R., Stanford, K. I., Wang, L., Bensadoun, A., Witztum, J. L., and Esko, J. D. (2007) J. Clin. Investig. 117153 -164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Coppi, A., Tewari, R., Bishop, J. R., Bennett, B. L., Lawrence, R., Esko, J. D., Billker, O., and Sinnis, P. (2007) Cell Host. Microbe 15316 -327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schulze, A., Gripon, P., and Urban, S. (2007) Hepatology 461759 -1768 [DOI] [PubMed] [Google Scholar]
- 46.Barth, H., Schäfer, C., Adah, M. I., Zhang, F., Linhardt, R. J., Toyoda, H., Kinoshita-Toyoda, A., Toida, T., van Kuppevelt, T. H., Depla, E., von Weizsäcker, F., Blum, H. E., and Baumert, T. F. (2003) J. Biol. Chem. 27841003 -41012 [DOI] [PubMed] [Google Scholar]
- 47.Chen, Y., Maguire, T., Hileman, R. E., Fromm, J. R., Esko, J. D., Linhardt, R. J., and Marks, R. M. (1997) Nat. Med. 3866 -871 [DOI] [PubMed] [Google Scholar]
- 48.Senay, C., Lind, T., Muguruma, K., Tone, Y., Kitagawa, H., Sugahara, K., Lidholt, K., Lindahl, U., and Kusche-Gullberg, M. (2000) EMBO Rep. 1 282-286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McCormick, C., Duncan, G., Goutsos, K. T., and Tufaro, F. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 668-673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pinhal, M. A. S., Smith, B., Olson, S., Aikawa, J., Kimata, K., and Esko, J. D. (2001) Proc. Natl. Acad. Sci. U. S. A. 9812984 -12989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.HajMohammadi, S., Enjyoji, K., Princivalle, M., Christi, P., Lech, M., Beeler, D., Rayburn, H., Schwartz, J. J., Barzegar, S., de Agostini, A. I., Post, M. J., Rosenberg, R. D., and Shworak, N. W. (2003) J. Clin. Investig. 111989 -999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kamimura, K., Rhodes, J. M., Ueda, R., McNeely, M., Shukla, D., Kimata, K., Spear, P. G., Shworak, N. W., and Nakato, H. (2004) J. Cell Biol. 1661069 -1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kuberan, B., Lech, M., Borjigin, J., and Rosenberg, R. D. (2004) J. Biol. Chem. 2795053 -5054 [DOI] [PubMed] [Google Scholar]