Abstract
The human neurological disease known as ataxia with oculomotor apraxia 1 is caused by mutations in the APTX gene that encodes Aprataxin (APTX) protein. APTX is a member of the histidine triad superfamily of nucleotide hydrolases and transferases but is distinct from other family members in that it acts upon DNA. The target of APTX is 5′-adenylates at DNA nicks or breaks that result from abortive DNA ligation reactions. In this work, we show that APTX acts as a nick sensor, which provides a mechanism to assess the adenylation status of unsealed nicks. When an adenylated nick is encountered by APTX, base pairing at the 5′ terminus of the nick is disrupted as the adenylate is accepted into the active site of the enzyme. Adenylate removal occurs by a two-step process that proceeds through a transient AMP-APTX covalent intermediate. These results pinpoint APTX as the first protein to adopt canonical histidine triad-type reaction chemistry for the repair of DNA.
Aprataxin is a novel DNA repair protein whose dysfunction causes the neurodegenerative disease ataxia with oculomotor apraxia 1 (AOA1),2 which is characterized by cerebellar atrophy and sensorimotor neuropathy (1–3). Cells derived from AOA1 patients are sensitive to DNA single strand break (SSB)-inducing agents such as hydrogen peroxide and methyl methanesulfonate (4–6) and have been shown to accumulate SSBs after treatment with camptothecin and under conditions of oxidative stress (7, 8).
Human APTX is 342 amino acids in length and contains three functional domains: an N-terminal forkhead-associated (FHA) domain, a central histidine triad (HIT) domain, and a C-terminal zinc finger (ZF) domain (see Fig. 1A). The FHA domain mediates complex formation with XRCC1-DNA ligase IIIα by interaction with phosphorylated residues in XRCC1 (4–6, 9), supporting a role for APTX in DNA single strand break repair. The FHA domain of APTX also interacts with phosphorylated XRCC4, however, thereby mediating complex formation with XRCC4-DNA ligase IV, suggestive of an additional role in double strand break repair (4).
FIGURE 1.
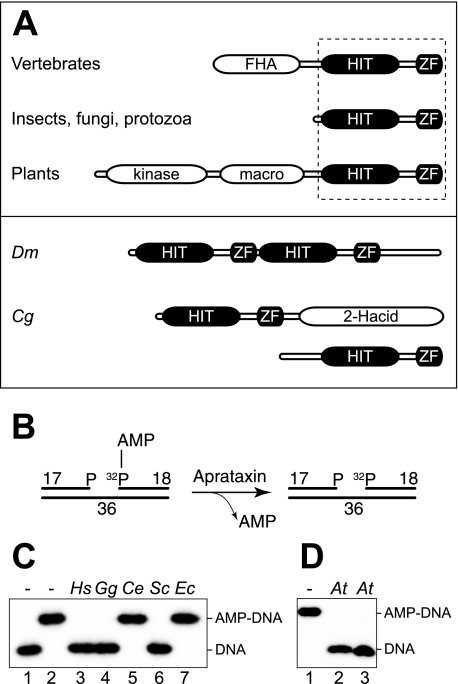
Aprataxins are evolutionary conserved HIT-ZF domain proteins. A, upper panel, Aprataxin orthologs are characterized by a common core consisting of a HIT-ZF domain combination (boxed) fused to additional functional domains in vertebrates and plants. Lower panel, atypical APTX orthologs include a dimeric head to tail fusion found in Drosophila melanogaster (Dm) and a protein pair found in C. globosum (Cg), one with the typical bipartite APTX structure found in other fungi and one with an additional d-isomer-specific 2-hydroxyacid dehydrogenase (2-Hacid) domain. B, synthetic DNA substrate that models an adenylated oxidative DNA single strand break. APTX releases the AMP group from the radiolabeled 18-mer. C, DNA deadenylation capacity of cell-free extracts (3 μg of total protein) from a variety of organisms (Hs, Homo sapiens; Gg, Gallus gallus; Ce, C. elegans; Ec, E. coli). Reaction products were analyzed by denaturing PAGE and detected by autoradiography. Lane 1, non-adenylated DNA; lane 2 adenylated DNA. D, a recombinant protein spanning the macro- and HIT-ZF domains of AtAPTX (At, A. thaliana) (10 nm) was tested for DNA deadenylation. Lane 1, adenylated DNA; lanes 2 and 3, reactions were carried out with adenylated DNA and AtAPTX in the presence of either 5 mm MgCl2 or 5 mm EDTA, respectively.
The actions of APTX are intimately linked to the mechanism by which DNA ligases promote break repair. Ligation occurs through a multistep process involving (i) the covalent binding of an adenylate (AMP) group to the active site lysine, (ii) transfer of the adenylate to the 5′-terminal phosphate group at the DNA break, and (iii) phosphodiester bond formation, which seals the DNA backbone and discharges the AMP moiety once again (10). Recently, however, it has become apparent that 5′-DNA adenylates are not always resolved in the course of a ligation reaction. For example, it has been shown that human DNA ligase IIIα attempts futile ligation at “dirty” oxidative SSBs, which often have phosphates at both 3′ and 5′ termini, and aborts the reaction after adenylation of the DNA (11). These dead-end adenylates appear to be the substrate for APTX as in vitro studies have shown that APTX can specifically remove 5′-adenylates, leading to the suggestion that APTX acts as a general proofreader for abortive DNA ligation reactions (3, 12, 13).
This discovery of the DNA deadenylation reaction catalyzed by APTX provides a molecular rationale for AOA1 because abortive ligation events are inevitable in vivo given that oxidative damage causes many dirty breaks per cell each day. Normally these dead-end DNA adenylates would be resolved by APTX, but in AOA1 patients they may accumulate as a consequence of ATPX dysfunction, leading to impaired gene transcription and eventual cell death. The specific sensitivity of neuronal cells seen in AOA1 may be due to high levels of oxidative stress coupled with their postmitotic status, which might not permit alternative (replication-dependent) options for the removal and repair of DNA adenylates (3, 14).
APTX is a member of the HIT superfamily of proteins, which takes its name from the active site HφHφHφφ histidine triad sequence motif (where φ is a hydrophobic residue) (15). HIT proteins catalyze nucleotide transfer from a number of specific substrates to water (hydrolase activity) or a second substrate (transferase activity) (16), and it is the HIT domain that gives APTX the capacity to deadenylate DNA (11).
In this study, we addressed the mechanism by which APTX interacts with DNA and resolves DNA adenylates. Significantly APTX is the only HIT superfamily member to act on DNA, and we provide evidence that the specificity for DNA adenylates is reflected in its HIT-ZF domain structure, which we propose is a defining and conserved feature of Aprataxin orthologs. We show that APTX possesses nick sensor activity, which is ideally suited to locate and repair 5′-DNA adenylates. The DNA deadenylation reaction involves two-step chemistry that proceeds through transient formation of a covalent AMP-enzyme intermediate.
EXPERIMENTAL PROCEDURES
Extracts and Proteins—Human APTX cDNA was PCR-amplified from a HeLa cDNA library (Invitrogen) and cloned into pET41 using the SpeI and BamHI restriction sites. This plasmid expressed recombinant APTX fused to an N-terminal glutathione S-transferase tag and a C-terminal His8 tag. The QuikChange II site-directed mutagenesis kit (Stratagene) was used to create mutant versions of APTX. Arabidopsis thaliana total RNA was kindly provided by Dr. Charles White. cDNA corresponding to a transcript starting at nucleotide 1146 to the end of APTX (AT5G01310) was cloned into pET41a using the NcoI and BamHI restriction sites. The resulting protein, containing glutathione S-transferase/His6 tags at the N terminus, was expressed in Escherichia coli. The tags used here have been shown previously not to interfere with APTX activity (11, 12) and were used to purify all recombinant APTXs to near homogeneity as described previously (12).
Extracts from human lymphoblastoid cells, chicken DT40 cells, and yeast were prepared as described previously (11), Caenorhabditis elegans extracts were a gift from Dr. Spencer Collis (17), and E. coli extracts were prepared by sonication of cells in a buffer containing 50 mm Tris-HCl, pH 8.0, 40 mm NaCl, 5 mm EDTA, 1 mm dithiothreitol, 100 μg/ml bovine serum albumin, and 5% glycerol followed by centrifugation to remove cell debris.
DNA Substrates—Synthetic DNA substrates were assembled from the following oligonucleotides: 1, 5′-ATTCCGATAGTGACTACA-3′; 2, 5′-CATATCCGTGTCGCCCT-3′P; 3, 5′-TGTAGTCACTATCGGAATGAGGGCGACACGGATATG-3′; 4, 5′-CATATCCGTGTCGCCCTCATTCCGATAGTGACTACA-3′; 1T, 5′-AGATTATCTTCGAGCTAC-3′; and 3T, 5′-GTAGCTCGAAGATAATCTGAGGGCGACACGGATATG-3′. All oligonucleotides were purchased from Sigma and purified by denaturing PAGE. Oligonucleotides were 5′-32P-labeled using T4 polynucleotide kinase (New England Biolabs) and [γ-32P]ATP (GE Healthcare), and 5′-phosphorylation was brought to completion by addition of excess cold ATP. SSB substrates (2 μm) were adenylated by abortive ligation with 400 units of T4 DNA ligase (New England Biolabs) in a buffer containing 50 mm Tris-HCl, pH 7.5, 10 mm MgCl2, 10 mm dithiothreitol, and 2 mm ATP for 12 h. The adenylated strand (oligo 1) was purified by 15% denaturing PAGE and reannealed with oligos 2 and 3 to form adenylated SSB substrates (11). The duplex substrate comprised oligo 3 with its complement (oligo 4). Neutral PAGE was used to purify all annealed DNA substrates before use. For the permanganate experiments, oligo 2 was annealed with oligos 1T and 3T.
Adenylated φX174 DNA was prepared as described previously (11). In brief, 95 nm φX174 replicative form I DNA (New England Biolabs) was treated with 10 μm H2O2, 0.1 mm FeCl3, 0.2 mm EDTA, 100 mm NaCl, and 1 mm NADH for 30 min at room temperature. Resulting oxidative DNA breaks were adenylated with [α-32P]ATP (GE Healthcare) by abortive ligation using T4 DNA ligase.
DNA Deadenylation Reactions—Synthetic SSB substrates (25 nm) and adenylated φX174 replication form I DNA (6 nm) were used in 10-μl reactions containing protein extract or recombinant APTX as indicated. Unless stated otherwise, incubation was for 1 min at 37 °C in 50 mm Tris-HCl, pH 8.0, 40 mm NaCl, 5 mm EDTA, 1 mm dithiothreitol, 100 μg/ml bovine serum albumin, and 5% glycerol. Reactions were scaled up for time course experiments. The deadenylation of synthetic SSB substrates was stopped by addition of formamide loading buffer and incubation at 95 °C for 3 min. Products were analyzed by 12% denaturing PAGE followed by autoradiography. Deadenylation reactions containing the φX174 DNA were stopped by addition of SDS (2% final concentration) and analyzed by 0.6% agarose gel electrophoresis or 10% SDS-PAGE followed by autoradiography. Data were quantified using a Storm 840 phosphorimaging system (GE Healthcare).
DNase I Footprinting—Reactions (5 μl) contained the DNA substrate (25 nm) in DNase I buffer (10 mm Tris-HCl, pH 7.6, 2.5 mm MgCl2, and 0.5 mm CaCl2). After 15 min of incubation at 25 °C, 0.1 unit of DNase I (New England Biolabs) was added. DNase I digestion was stopped after 1 min by addition of formamide loading buffer and incubation at 95 °C for 3 min. Reaction products were analyzed by 12% denaturing PAGE followed by autoradiography.
Chemical Cleavage of DNA—Reactions (10 μl) contained 25 nm DNA substrate, 50 mm Tris-HCl, pH 8.0, 30 mm NaCl, 5 mm EDTA, 1 mm dithiothreitol, 100 g/ml bovine serum albumin, 5% glycerol, and APTX as indicated. After preincubation for 20 min at 25 °C, KMnO4 was added to a final concentration of 5 mm. After 2 min β-mercaptoethanol was added to a final concentration of 1 m. After ethanol precipitation in the presence of 1 mg/ml glycogen, the DNA was resuspended in 1 m piperidine and incubated at 95 °C for 30 min. After ethanol precipitation, the DNA was resuspended in formamide loading buffer and analyzed by 12% denaturing PAGE followed by autoradiography.
RESULTS
Molecular Architecture and Evolutionary Conservation of Aprataxin—Human APTX features a tripartite FHA-HIT-ZF domain structure (11, 12). To determine whether the co-occurrence of a ZF with a HIT domain might indicate authentic Aprataxins, we performed data base searches to identify potential orthologs in virtually all sequenced eukaryotic genomes (Fig. 1A). One exception was found in C. elegans where, despite the presence of several HIT superfamily proteins, we were unable to detect the presence of a ZF in any of these proteins. HIT-ZF domain proteins were also absent from prokaryotes. When cell-free extracts prepared from a variety of organisms were incubated with a synthetic 5′-adenylated nicked DNA substrate with a one-nucleotide gap and phosphates at both the 3′ and the 5′ termini (Fig. 1B; this substrate mimics the product of an abortive ligation event that would occur at an SSB induced by oxidative DNA damage), we observed a perfect correlation between our HIT-ZF predictions and DNA deadenylation activity (Fig. 1C).
The HIT-ZF domain proteins identified in plants are characterized by a more elaborate domain structure including a number of additional modules (Fig. 1A). To determine whether these are true Aprataxins, we cloned recombinant A. thaliana basic helix-loop-helix family protein AT5G01310.1 and tested its ability to catalyze DNA deadenylation. The recombinant protein spanned the macro-, HIT, and ZF domains of the protein and was proficient for DNA deadenylation in the presence or absence of Mg2+ ions, indicative of true APTX-like metal-independent DNA deadenylation activity (Fig. 1D, lanes 2 and 3) (11). Taken together, these data show that APTX is functionally conserved in animals, fungi, and plants but not in prokaryotes and establish the combination of a HIT domain and a ZF domain as the structural hallmark of the Aprataxins.
Nick Sensor Activity of Aprataxin—Previous studies have shown that APTX binds linear double-stranded DNA and nicked DNA substrates with similar affinity and that adenylated DNA is bound more avidly than non-adenylated substrates (12, 18). To define these interactions in more detail, we used DNase I protection assays to analyze complexes formed between APTX and DNA substrates containing 5′-adenylated nicks. To inhibit adenylate removal, it was necessary to use human APTX carrying the inactivating mutation H260A. Previous studies have shown that recombinant APTXH260A, with an N-terminal glutathione S-transferase tag and a C-terminal His8 tag, exhibits wild-type substrate binding properties (12).
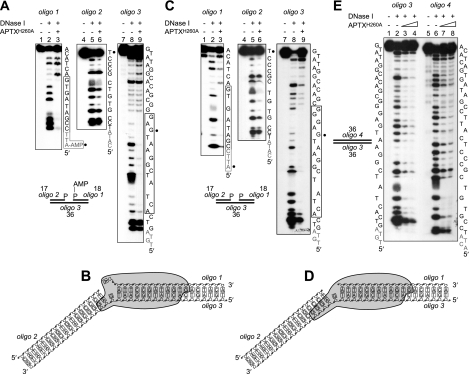
APTXH260A was incubated with the adenylated SSB substrate (Fig. 2A; this contains a one-nucleotide gap and is composed of three oligonucleotides: an 18-mer with a 5′-adenylate (oligo 1), a 17-mer with a 3′-phosphate terminus at the 5′-side of the nick (oligo 2), and a continuous 36-mer (oligo 3)), and the resulting complexes were probed using DNase I. In the presence of APTX, regions of protection were observed over ∼12 nucleotides of oligo 1 and 14 nucleotides of oligo 3. In contrast, no significant changes were observed with oligo 2, indicating that Aprataxin binds asymmetrically to the DNA almost exclusively at the 3′-side of the adenylate (Fig. 2A and indicated schematically in Fig. 2B).
FIGURE 2.
DNase I footprinting of APTX-DNA complexes. A, adenylated SSB DNA 5′-32P-end-labeled on strand 1, 2 or 3 as indicated was incubated with DNase I in the presence or absence of APTXH260A (50 nm) as described under “Experimental Procedures.” Reaction products were resolved by 12% denaturing PAGE and visualized by autoradiography. Nucleotides protected from DNase I are indicated with a box. Gray font denotes 5′-terminal nucleotides not resolved by PAGE; black circles mark the position of the nick. B, graphic representation of the binding of APTX (indicated as a gray shape) to the adenylated nicked DNA. C and D, same as A and B except the DNA was non-adenylated. E, same as A except non-adenylated linear duplex DNA was used. APTX was present at 50 and 100 nm.
When similar experiments were carried out with non-adenylated nicked DNA, we were surprised to find very similar protection patterns (Fig. 2, C and D) as again strong protection patterns were observed at the 3′-side of the nick. To ensure it was the nick that determined the specific positioning of APTX, we next analyzed complexes formed between APTXH260A and linear duplex DNA of the same length. In this case, we failed to see a clear protection pattern and instead observed weak protection over the entire DNA duplex (Fig. 2E; note that the apparent protection at the 5′ terminus of oligos 3 and 4 was due to the inhibition of multiple cleavage events in the presence of APTX). The nick sensor activity of APTX was disrupted when the ZF domain was inactivated by mutation of the critical zinc-coordinating residues Cys-319 and Cys-322 as the APTXH260A/C319A/C322A mutant protein failed to protect either adenylated or non-adenylated SSB substrates from DNase I digestion (data not shown).
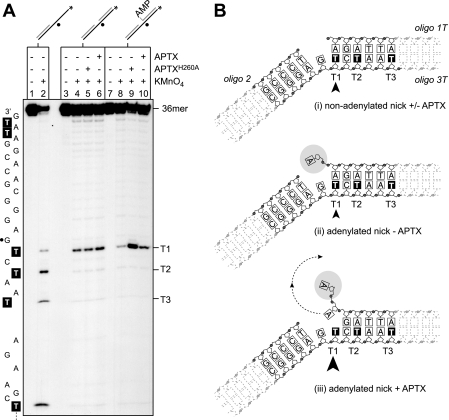
APTX Disrupts Watson-Crick Base Pairing at DNA Adenylates—Reasoning that to sample for and/or bind a 5′-AMP moiety, APTX might need to separate the Watson and Crick strands, we next determined whether the interaction of Aprataxin with 5′-adenylates results in base pair disruption. To do this, the formation of unpaired thymines close to the adenylate was monitored using potassium permanganate (KMnO4) as a specific probe. To enable these analyses, we incorporated thymine residues directly adjacent to the nick (T1) and at positions three (T2) and six (T3) nucleotides downstream of the nick on the continuous strand of the SSB substrate (oligo 3T), which was annealed with oligonucleotides 1T and 2 (Fig. 3B).
FIGURE 3.
APTX disrupts Watson-Crick base pairing at DNA adenylates. A, the indicated non-adenylated and adenylated substrates with a 5′-32P label (asterisk) on the 36-mer oligo 3T were incubated with potassium permanganate in the presence or absence of APTX or APTXH260A (50 nm) followed by treatment with piperidine. Reaction products were resolved by 12% denaturing PAGE and visualized by autoradiography. Lanes 1 and 2, partially double-stranded control; lanes 3–6, non-adenylated nicked SSB substrate; lanes 7–10, adenylated nicked SSB substrate. Black circles mark the position of the nick in the SSB substrate. The DNA sequence of oligo 3T is indicated with thymine residues highlighted. Thymine residues T1, T2, and T3 at the 3′-side of the nick are indicated. B, schematic representation of the results shown in A. The arrowhead indicates thymine T1 that is hypersensitive to permanganate attack in the adenylated SSB substrate. Arrow size relates to the level of hypersensitivity.
Preliminary experiments using oligo 3T annealed with oligo 2 only (single-stranded/tailed duplex DNA) confirmed that thymine residues T1–T3 were readily accessible to the chemical probe when they were unpaired (Fig. 3A, lane 2). When oligo 3T was present in a non-adenylated SSB substrate, we observed that T1 remained sensitive to chemical cleavage in the absence of APTX, whereas all other thymine residues were protected (lane 5). These results indicate that the chemical probe retains access to the base adjacent to the nick possibly as a result of DNA breathing. In the presence of APTXH260A (lane 5) or wild-type APTX (lane 6), the degree of sensitivity at T1 was unchanged, and we did not find induced sensitivity at other thymine residues. These results show that although APTX binds the 3′-side of the nick (Fig. 2) binding per se does not lead to separation of the Watson and Crick strands (Fig. 3B, panel i).
Parallel experiments were carried out with adenylated SSB substrates. Without APTX, we found that T1 was relatively insensitive to chemical cleavage (Fig. 3A, lane 8, and indicated schematically in Fig. 3B, panel ii), but the presence of APTXH260A resulted in a significant hypersensitivity of residue T1 to chemical attack (lane 9). In contrast, wild-type APTX failed to induce such a level of hypersensitivity as expected because the active protein will remove the adenylate under these reaction conditions (lane 10).
These results show that the stable binding of APTX to its target lesion results in measurable base disruption to the nucleotide pair directly flanking the adenylated 5′-terminus of the nick (Fig. 3B, panel iii). No sensitivity was induced at T2 or T3, demonstrating that strand separation is limited to the immediate proximity of the adenylated base pair. These results demonstrate that the interaction of APTX with the DNA adenylate induces structural changes in the DNA that are a likely prerequisite for DNA deadenylation.
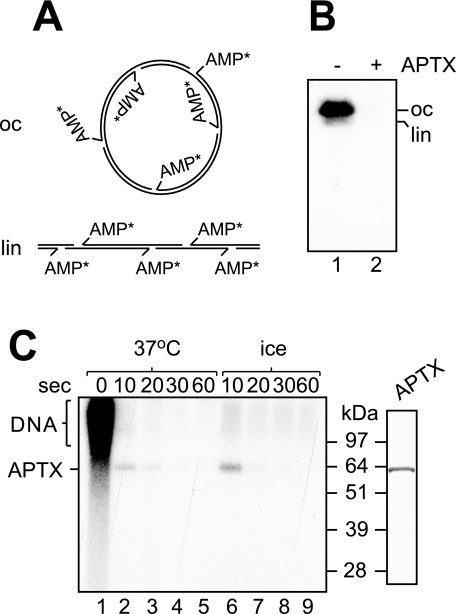
A Covalent AMP-Enzyme Intermediate—Previously it was shown that DNA deadenylation by APTX results in the release of free AMP (11), but the mechanism of adenylate hydrolysis remains unknown. The HIT hydrolases generally catalyze a two-step reaction, which includes (i) nucleotide transfer to the active site of the enzyme and (ii) hydrolysis of the resulting nucleotidylated enzyme intermediate (19). To probe for a covalent AMP-APTX intermediate, we incubated APTX with plasmid DNA that contained 32P-labeled adenylates (Fig. 4A). This substrate was produced by treating plasmid DNA with hydrogen peroxide to produce dirty breaks and subsequent treatment of the damaged plasmid with DNA ligase in the presence of [α-32P]ATP (Fig. 4A) (11). As expected, incubation with APTX resulted in rapid deadenylation of the DNA such that all radioactive label was lost (Fig. 4B, compare lanes 1 and 2). However, when aliquots of the deadenylation reaction were taken 0, 10, 20, 30, and 60 s after initiation of the reaction, and the products were resolved by SDS-PAGE, we observed the formation of a discrete 32P-labeled product that comigrated with recombinant APTX (Fig. 4C, lane 2). The formation of 32P-labeled APTX was even more apparent when reactions were carried out on ice (lane 6). These results demonstrate that DNA deadenylation proceeds through a short lived covalent AMP-APTX intermediate.
FIGURE 4.
DNA deadenylation proceeds through a covalent AMP-APTX intermediate. A, schematic representation of the open circle (oc)/linear (lin) plasmid DNA substrate adenylated with [32P]AMP (indicated by the asterisks). B, incubation of the adenylated DNA with APTX (50 nm) results in loss of [32P]AMP from the DNA as visualized by agarose gel electrophoresis followed by autoradiography. C, incubation of APTX (150 nm) with the 32P-labeled adenylated DNA at 37 °C or on ice results in the formation of radiolabeled APTX as detected by SDS-PAGE. Marker lane on the right, glutathione S-transferase (GST)/His-tagged APTX (0.3 μg) was resolved by SDS-PAGE and stained with Coomassie Blue.
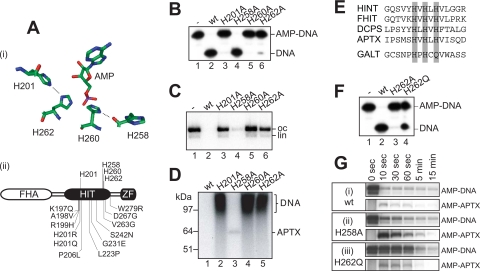
Catalytic Role of Four Conserved Histidine Residues—Next we assessed the significance of a conserved network of active site hydrogen-bonding interactions for the DNA deadenylation reaction. Structural analysis of the HIT hydrolases HINT and fragile histidine triad protein has pinpointed two histidine residues within the histidine triad that make direct contacts with the α-phosphate of the nucleotide ligand (19–22). These residues are stabilized by two further histidine residues, one situated within the histidine triad and one positioned upstream in the primary sequence. By analogy, we predict that APTX His-260 contacts the α-phosphate of the adenylate group with its Nε atom and is stabilized by a hydrogen bond between the Nδ atom and the carbonyl oxygen atom of His-258, whereas His-262 contacts the α-phosphate and is stabilized by a hydrogen bond between its Nδ atom and the Nε atom of the upstream residue His-201 (Fig. 5A).
FIGURE 5.
The four conserved active site histidine residues are critical for DNA deadenylation by APTX. A, panel i, arrangement of the histidine triad residues His-258, His-260, His-262, and His-201 in the active site of APTX based on crystallographic analysis of human HINT1, a related HIT hydrolase, with bound AMP (19). Hydrogen bonds between His-201 and His-262 and between His-258 and His-260 are indicated. His-260 is the nucleophilic residue that forms the phosphohistidine bond with the substrate. Panel ii, primary structure of APTX indicating the four histidines and the amino acid changes found in AOA1 patients. B, DNA deadenylation by wild-type (wt) and active site mutants of APTX (10 nm) determined using the oligonucleotide-based SSB substrate shown in Fig. 1B. Deadenylation was determined by denaturing PAGE followed by autoradiography. C, DNA deadenylation by wild-type and active site mutants of APTX (50 nm) determined using the adenylated DNA substrate of Fig. 4A. After 5 min at 37 °C, remaining 32P-labeled DNA adenylates were visualized by agarose gel electrophoresis followed by autoradiography. oc, open circle; lin, linear. D, aliquots from reactions shown in C were analyzed by SDS-PAGE/autoradiography to detect 32P-labeled adenylated DNA and 32P-labeled APTX. E, sequence alignment of the indicated HIT superfamily proteins from humans. The hydrolases feature histidines at all three position of the HIT motif (gray shading), whereas the transferase galactose-1-phosphate uridylyltransferase (GALT) is characterized by a glutamine in the third position of the motif. FHIT, fragile histidine triad protein; DCPS, decapping enzyme, scavenger. F, a transferase-like version of APTX with glutamine at His-262 (APTXH262Q; 10 nm) was analyzed for deadenylation activity in comparison with APTX and APTXH262A as described in B. G, time course of the deadenylation activity and covalent AMP-APTX intermediate formation of wild-type APTX (panel i) and the APTXH258A (panel ii) and APTXH262Q (panel iii) mutants as determined in C and D.
To test this model, we generated recombinant APTXH201A, APTXH258A, and APTXH262A mutant proteins and compared their activities with APTXH260A and the wild-type protein. First the activity of each protein was determined using the oligonucleotide-based adenylated SSB substrate (Fig. 5B). We observed substantial residual activity for APTXH258A (lane 4), weak activity for APTXH201A and APTXH262A (lanes 3 and 6), and no detectable activity with APTXH260A (lane 5). Similar catalytic activities were observed when we measured the loss of radiolabel from plasmid DNA that was adenylated with [32P]AMP (Fig. 5C). Interestingly when the products of the [32P]AMP plasmid reactions were analyzed by SDS-PAGE after 5 min, we observed a long lived covalent AMP-enzyme intermediate with APTXH258A indicative of impaired enzyme-adenylate hydrolysis (Fig. 5D, lane 3).
The last position of the histidine triad is not occupied by a histidine but by a glutamine residue in the HIT nucleotide transferases (e.g. galactose-1-phosphate uridylyltransferase) (Fig. 5E). They also differ from the HIT nucleotide hydrolases by reacting with a second substrate rather than with water to resolve the nucleotidylated enzyme intermediate. To determine the effect of a glutamine residue at position 262 in APTX, we generated mutant APTXH262Q. In contrast to APTXH262A, this mutant showed significant residual DNA deadenylation activity without the need for a second substrate (Fig. 5F).
Given that mutation of the four conserved histidine residues, His-201, His-258, His-260, and His-262, differentially affect the DNA deadenylation activity of APTX and that the H258A mutant exhibits a specific defect in the second reaction step, those mutants that retained substantial residual activity were examined more closely. We found that wild-type APTX deadenylated over 90% of the DNA substrate within the first 10 s of the reaction (Fig. 5G). In contrast, mutant APTXH262Q was defective in step 1 (90% DNA deadenylation was achieved only after 5 min of incubation), and yet the rate of release of the AMP from the AMP-APTXH262Q was not significantly affected. With APTXH258A, DNA deadenylation was only slightly slowed with 86% deadenylation taking place within 10 s. At the same time point, 30% of the 32P-labeled AMP was covalently linked to the protein compared with just 3% with the wild-type protein.
Taken together, the results presented in Fig. 5 underline the importance of the hydrogen-bonding network that the four conserved histidines form among each other and with the substrate in HIT hydrolases. Consistent with the biochemical results, AOA1-related point mutations are found almost exclusively in the vicinity of the active site and particularly around the histidine triad and histidine residue 201 (Fig. 5A, panel ii) (3, 23).
DISCUSSION
When APTX was discovered to be the causative gene of the neurodegenerative disease AOA1, a connection to DNA break repair was immediately obvious given the similarity between the FHA domain of APTX and that of 5′-polynucleotide kinase 3′-phosphatase (2). Subsequently it was established that APTX utilizes this FHA domain to associate with XRCC1-DNA ligase IIIα and with XRCC4-DNA ligase IV, the end-joining complexes involved in single strand break repair and double strand break repair, respectively (4–6, 9). The specific role of APTX in DNA repair became apparent when the DNA deadenylation activity was detected, and it has been proposed that APTX acts as a proofreader for abortive ligation events that occur at sites of oxidative DNA damage (3, 11).
In the present work, we have shown that a HIT domain coupled to a DNA binding ZF domain effectively provides a structural definition of the Aprataxins. Based on this structural definition and the demonstration of APTX activity in a variety of cell-free extracts from organisms containing HIT-ZF proteins, it appears that most eukaryotes encode Aprataxins. We also found that APTX is functionally conserved in plants where the protein has a more elaborate domain structure. The presence of a macro- and a kinase domain in plant APTX is intriguing: macrodomains are high affinity ADP-ribose/poly(ADP-ribose) binding modules (24), and ADP-ribose is an important signaling molecule in DNA repair (25). Thus, it is tempting to speculate that the macrodomain could be used to recruit plant APTX to sites of DNA damage flagged with poly(ADP-ribose) synthesized by poly(ADP-ribose) polymerase. Such a mechanism would be analogous to the recruitment of vertebrate APTX through the interaction of its FHA domain with phosphorylated XRCC1, which itself is recruited to poly(ADP-ribosyl)ated poly(ADP-ribose) polymerase at sites of DNA damage (26). The presence of a potential kinase domain implicates plant APTX as a multifunctional DNA break repair protein. It is also noteworthy that one of the two APTX orthologs found in the fungus Chaetomium globosum contains a d-isomer-specific 2-hydroxyacid dehydrogenase domain with an embedded NAD binding site (27). Reminiscent of the situation in plants, this domain may allow cross-talk between APTX and poly(ADP-ribose) polymerase, which consumes cellular NAD when synthesizing poly(ADP-ribose).
Taken together, these results suggest that the HIT-ZF domain core of APTX is uniquely tailored for DNA deadenylation. However, although the two domains in combination are required and sufficient for efficient DNA deadenylation, additional modules such as the FHA domain and possibly ADP-ribose/poly(ADP-ribose) binding modules coordinate the function of APTX in DNA repair with other repair factors.
Our analyses of the complexes formed between APTX and adenylated and non-adenylated SSB substrate revealed almost identical DNase I footprints, leading us to suggest that APTX alone can act as a nick sensor without the need for specific targeting by XRCC1-DNA ligase IIIα. This result is supported by the fact that mutations in the FHA domain of APTX have not been found in patients with the neurological disease AOA1 (23). Remarkably the APTX footprints are quite asymmetric with regard to the nick as they are found almost exclusively at the 3′-side of the strand break. The location of APTX to one side of the break site may be important for the interplay between APTX and the XRCC1-DNA ligase IIIα complex, and in this regard it is possible that the primary role of the FHA domain of APTX relates to the tight assembly of the APTX-XRCC1-DNA ligase IIIα complex at the strand break rather than specific targeting.
Consistent with our proposal that APTX acts as a nick sensor and thereby can continually sample SSBs for the presence of dead-end adenylates, it is important to note that the APTXs of insects, fungi, and protozoa lack an FHA domain and therefore may function as a stand-alone enzymatic activity. The nick sensor activity of APTX may also be important to allow it to compete with DNA ligase IIIα, which has nick sensor activity of its own (28, 29). The recently proposed “jackknife model” for DNA ligase III action supposes that the ligase initially binds SSBs in an open conformation to allow access to the nick by other repair enzymes before closing over the SSB like the blade of a jackknife to fully encircle the DNA (30). If abortive ligation occurs, the DNA ligase must relax its tight grip and allow APTX access to the DNA adenylate. One possibility is that the ability of APTX to specifically recognize nicks allows competitive binding interactions that help reset the ligase into the open DNA binding conformation. This may also facilitate access of other repair enzymes, which is likely to be required given that dead-end adenylates arise at SSBs with unconventional termini.
Although the DNase I protection patterns observed with adenylated and non-adenylated nicked DNA were almost identical, one distinct difference was observed when similar complexes were probed with permanganate ions that specifically attack unpaired thymines. We found that the binding of APTX to adenylated nicks resulted in the disruption of base pairing at the nucleotide pair directly flanking the adenylated 5′ terminus. Base pair disruption was dependent on the presence of the adenylate and appears to be a consequence of the AMP moiety being accepted into the HIT domain active site. In support of this conclusion, base pair disruption was abrogated, but not completely lost, when similar experiments were carried out using the ZF finger mutant APTXH260A/C319A/C322A (data not shown). The opening of the adenylated base pair may be necessary to avoid steric hindrance between the protein and the DNA and to expose the scissile pyrophosphate bond to the specific amino acid groups involved in its coordination and cleavage. Strand separation was not observed to extend away from the nick, indicating that sufficient flexibility at the 5′ terminus is achieved by breaking only the base pair that carries the adenylate group.
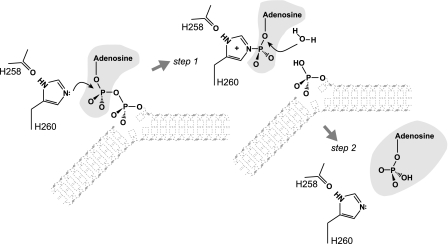
We found that DNA adenylate hydrolysis by APTX involves two-step catalysis that takes place via the transient formation of an AMP-APTX covalent intermediate (Fig. 6). First APTX attacks the DNA adenylate to break the phosphodiester bond between the AMP and the DNA leaving group (step 1). During this step, APTX restores the DNA 5′-phosphate terminus by transferring the AMP group onto itself. Second the resulting AMP-APTX intermediate is resolved by hydrolysis of the phosphohistidine bond to release AMP and restore catalytically active enzyme. Thus, APTX catalyzes AMP removal using canonical HIT superfamily reaction chemistry (19) and is the first example of a DNA repair protein making use of two-step catalysis involving a covalent intermediate with the excised lesion. The mode of action is somewhat reminiscent of the way in which alkylated DNA is repaired by suicide enzymes such as E. coli O6-methylguanine transferase, which transfers a methyl group to an active site cysteine (31). However, O6-methylguanine transferase gets inactivated in the process, whereas APTX is regenerated by hydrolysis of the AMP-APTX intermediate.
FIGURE 6.
Proposed mechanism of two-step catalysis. Step 1, APTX breaks the phosphodiester bond between the AMP and the DNA 5′-phosphate. The AMP group becomes covalently linked to His-260 at the central position of the histidine triad that acts as a nucleophile. His-260 is stabilized by hydrogen bonding with His-258. Step 2, APTX activates water to hydrolyze the phosphohistidine bond to release AMP and regenerate the enzyme. A schematic contour of the DNA is indicated, whereas the α- and β-phosphates of the adenylate are depicted fully.
Our studies show that histidine residues His-258, His-260, His-262, and His-201 play important roles in the reaction chemistry similar to that of other HIT superfamily members (19, 22, 32, 33). By analogy with other HIT hydrolases, His-201 of APTX contacts His-262, which in turn makes contact with the adenylated DNA substrate, and His-258 contacts His-260, which initially attacks the α-phosphate of the adenylate group. We found that DNA deadenylation by APTXH201A and APTXH262A is strongly impaired, APTXH260A is inactive, and APTXH258A is proficient in forming the enzyme-adenylate reaction intermediate but is impaired for hydrolysis of the phosphohistidine bond to release the AMP group. Disruption of the hydrogen bond between His-201 and His-262 in APTXH201A may change the charge and position of His-262, resulting in the dramatic decrease in DNA deadenylation activity. Interestingly more than half of all amino acid substitutions linked to the AOA1 disease map to residue His-201 or its immediate vicinity (see Fig. 5A, panel ii), indicating the crucial catalytic role played by this residue. His-262 occupies the last position in the histidine triad, and in other HIT members, this residue has been considered to be the possible acid/base catalyst for the protonation of the leaving group in reaction step 1 and the activation of water in reaction step 2 (16, 20). Our finding that APTXH262A is strongly impaired for DNA deadenylation concurs with this notion, but the fact that activity is substantially restored by a glutamine at position 262 argues against it. Indeed APTXH262Q proved to be impaired in step 1 of the DNA deadenylation reaction, whereas hydrolysis of the phosphohistidine bond was not affected. This indicates that His-262 is important for substrate recognition and suggests that a glutamine may partially substitute for histidine at this position by enabling a bifurcated interaction between its side-chain NH2 and the adenylate similar to the interaction between Gln-168 of galactose-1-phosphate uridylyltransferase and its substrate (34).
Complete loss of activity for APTXH260A confirms His-260 as the nucleophile in reaction step 1. By analogy to HINT and fragile histidine triad protein, His-260 is properly orientated and activated for nucleophilic attack by its interaction with His-258 (20). However, His-258 interacts with His-260 at its carbonyl oxygen atom, and it is therefore not surprising that APTXH258A retains substantial DNA deadenylation activity. Indeed the first position of the histidine triad, which is occupied by His-258, is not strictly conserved in APTX orthologs. For example, plant APTX contains a QφHφHφφ triad motif, whereas insects feature an RφHφHφφ sequence, and because we have shown that Arabidopsis APTX is competent for DNA deadenylation, these do not appear to be inactivating variations.
In summary, our results on the mechanism of DNA deadenylation by APTX confirm the biochemical uniformity of the HIT superfamily. Despite the similarities, however, APTX is unique in its specificity for DNA substrates, which is afforded by a unique combination of the HIT domain with a DNA binding ZF domain. The ZF domain endows APTX with nick sensor activity, which allows the enzyme to detect SSBs to sample for DNA adenylates. The use of HIT-type reaction chemistry seems ideally suited to resolve DNA adenylates as it allows the protein to act as an acceptor for the offending AMP group and repair the DNA lesion by direct reversal.
Acknowledgments
We thank our laboratory colleagues for comments and suggestions throughout the course of this work.
This work was supported by Cancer Research UK, the Louis-Jeantet Foundation, and the European Union DNA Repair Consortium. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement”in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: AOA1, ataxia with oculomotor apraxia 1; HIT, histidine triad; SSB, single strand break; APTX, Aprataxin; FHA, forkhead-associated; ZF, zinc finger; HINT, histidine triad nucleotide-binding protein.
References
- 1.Date, H., Onodera, O., Tanaka, H., Iwabuchi, K., Uekawa, K., Igarashi, S., Koike, R., Hiroi, T., Yuasa, T., Awaya, Y., Sakai, T., Takahashi, T., Nagatomo, H., Sekijima, Y., Kawachi, I., Takiyama, Y., Nishizawa, M., Fukuhara, N., Saito, K., Sugano, S., and Tsuji, S. (2001) Nat. Genet. 29 184–188 [DOI] [PubMed] [Google Scholar]
- 2.Moreira, M. C., Barbot, C., Tachi, N., Kozuka, N., Uchida, E., Gibson, T., Mendonca, P., Costa, M., Barros, J., Yanagisawa, T., Watanabe, M., Ikeda, Y., Aoki, M., Nagata, T., Coutinho, P., Sequeiros, J., and Koenig, M. (2001) Nat. Genet. 29 189–193 [DOI] [PubMed] [Google Scholar]
- 3.Rass, U., Ahel, I., and West, S. C. (2007) Cell 130 991–1004 [DOI] [PubMed] [Google Scholar]
- 4.Clements, P. M., Breslin, C., Deeks, E. D., Byrd, P. J., Ju, L., Bieganowski, P., Brenner, C., Moreira, M. C., Taylor, A. M., and Caldecott, K. W. (2004) DNA Repair 3 1493–1502 [DOI] [PubMed] [Google Scholar]
- 5.Gueven, N., Becherel, O. J., Kijas, A. W., Chen, P., Howe, O., Rudolph, J. H., Gatti, R., Date, H., Onodera, O., Taucher-Scholz, G., and Lavin, M. F. (2004) Hum. Mol. Genet. 13 1081–1093 [DOI] [PubMed] [Google Scholar]
- 6.Luo, H., Chan, D. W., Yang, T., Rodriguez, M., Chen, B. P., Leng, M., Mu, J. J., Chen, D., Songyang, Z., Wang, Y., and Qin, J. (2004) Mol. Cell. Biol. 24 8356–8365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mosesso, P., Piane, M., Palitti, F., Pepe, G., Penna, S., and Chessa, L. (2005) CMLS Cell. Mol. Life Sci. 62 485–491 [DOI] [PubMed] [Google Scholar]
- 8.Hirano, M., Yamamoto, A., Mori, T., Lan, L., Iwamoto, T. A., Aoki, M., Shimada, K., Furiya, Y., Kariya, S., Asai, H., Yasui, A., Nishiwaki, T., Imoto, K., Kobayashi, N., Kiriyama, T., Nagata, T., Konishi, N., Itoyama, Y., and Ueno, S. (2007) Ann. Neurol. 61 162–174 [DOI] [PubMed] [Google Scholar]
- 9.Sano, Y., Date, H., Igarashi, S., Onodera, O., Oyake, M., Takahashi, T., Hayashi, S., Morimatsu, M., Takahashi, H., Makifuchi, T., Fukuhara, N., and Tsuji, S. (2004) Ann. Neurol. 55 241–249 [DOI] [PubMed] [Google Scholar]
- 10.Ellenberger, T., and Tomkinson, A. E. (2008) Annu. Rev. Biochem. 77 313–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ahel, I., Rass, U., El-Khamisy, S. F., Katyal, S., Clements, P. M., McKinnon, P. J., Caldecott, K. W., and West, S. C. (2006) Nature 443 713–716 [DOI] [PubMed] [Google Scholar]
- 12.Rass, U., Ahel, I., and West, S. C. (2007) J. Biol. Chem. 282 9469–9474 [DOI] [PubMed] [Google Scholar]
- 13.Lavin, M. F., Gueven, N., and Grattan-Smith, P. (2008) DNA Repair 7 1061–1076 [DOI] [PubMed] [Google Scholar]
- 14.McKinnon, P. J., and Caldecott, K. W. (2007) Annu. Rev. Genomics Hum. Genet. 8 37–55 [DOI] [PubMed] [Google Scholar]
- 15.Seraphin, B. (1992) DNA Seq. 3 177–179 [DOI] [PubMed] [Google Scholar]
- 16.Brenner, C. (2002) Biochemistry 41 9003–9014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Polanowska, J., Martin, J. S., Garcia-Muse, T., Petalcorin, M. I., and Boulton, S. J. (2006) EMBO J. 25 2178–2188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kijas, A. W., Harris, J. L., Harris, J. M., and Lavin, M. F. (2006) J. Biol. Chem. 281 13939–13948 [DOI] [PubMed] [Google Scholar]
- 19.Lima, C. D., Klein, M. G., and Hendrickson, W. A. (1997) Science 278 286–290 [DOI] [PubMed] [Google Scholar]
- 20.Lima, C. D., D'Amico, K. L., Naday, I., Rosenbaum, G., Westbrook, E. M., and Hendrickson, W. A. (1997) Structure (Lond.) 5 763–774 [DOI] [PubMed] [Google Scholar]
- 21.Lima, C. D., Klein, M. G., Weinstein, I. B., and Hendrickson, W. A. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 5357–5362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brenner, C., Garrison, P., Gilmour, J., Peisach, D., Ringe, D., Petsko, G. A., and Lowenstein, J. M. (1997) Nat. Struct. Biol. 4 231–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caldecott, K. W. (2008) Nat. Rev. Genet. 9 619–631 [DOI] [PubMed] [Google Scholar]
- 24.Karras, G. I., Kustatscher, G., Buhecha, H. R., Allen, M. D., Pugieux, C., Sait, F., Bycroft, M., and Ladurner, A. G. (2005) EMBO J. 24 1911–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malanga, M., and Althaus, F. R. (2005) Biochem. Cell Biol. 83 354–364 [DOI] [PubMed] [Google Scholar]
- 26.Masson, M., Niedergang, C., Schreiber, V., Muller, S., Menissier-de Murcia, J., and de Murcia, G. (1998) Mol. Cell. Biol. 18 3563–3571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bellamacina, C. R. (1996) FASEB J. 10 1257–1269 [DOI] [PubMed] [Google Scholar]
- 28.Caldecott, K. W., Aoufouchi, S., Johnson, P., and Shall, S. (1996) Nucleic Acids Res. 24 4387–4394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mackey, Z. B., Niedergang, C., Murcia, J. M., Leppard, J., Au, K., Chen, J., de Murcia, G., and Tomkinson, A. E. (1999) J. Biol. Chem. 274 21679–21687 [DOI] [PubMed] [Google Scholar]
- 30.Cotner-Gohara, E., Kim, I. K., Tomkinson, A. E., and Ellenberger, T. (2008) J. Biol. Chem. 283 10764–10772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Olsson, M., and Lindahl, T. (1980) J. Biol. Chem. 255 10569–10571 [PubMed] [Google Scholar]
- 32.Barnes, L. D., Garrison, P. N., Siprashvili, Z., Guranowski, A., Robinson, A. K., Ingram, S. W., Croce, C. M., Ohta, M., and Huebner, K. (1996) Biochemistry 35 11529–11535 [DOI] [PubMed] [Google Scholar]
- 33.Gu, M., Fabrega, C., Liu, S. W., Liu, H., Kiledjian, M., and Lima, C. D. (2004) Mol. Cell 14 67–80 [DOI] [PubMed] [Google Scholar]
- 34.Wedekind, J. E., Frey, P. A., and Rayment, I. (1995) Biochemistry 34 11049–11061 [DOI] [PubMed] [Google Scholar]