Abstract
Gene clusters encoding various type III secretion system (T3SS) injectisomes, frequently code downstream of the conserved atpase gene for small hydrophilic proteins whose amino acid sequences display a propensity for intrinsic disorder and coiled-coil formation. These properties were confirmed experimentally for a member of this class, the HrpO protein from the T3SS of Pseudomonas syringae pv phaseolicola: HrpO exhibits high α-helical content with coiled-coil characteristics, strikingly low melting temperature, structural properties that are typical for disordered proteins, and a pronounced self-association propensity, most likely via coiled-coil interactions, resulting in heterogeneous populations of quaternary complexes. HrpO interacts in vivo with HrpE, a T3SS protein for which coiled-coil formation is also strongly predicted. Evidence from HrpO analogues from all T3SS families and the flagellum suggests that the extreme flexibility and propensity for coiled-coil interactions of this diverse class of small, intrinsically disordered proteins, whose structures may alter as they bind to their cognate folded protein targets, might be important elements in the establishment of protein-protein interaction networks required for T3SS function.
Type III secretion systems (T3SS)4 are essential mediators of the interaction of many Gram-negative pathogenic proteobacteria (α, β, γ, and δ subdivisions) with their human, animal, or plant hosts and are evolutionarily related to bacterial flagella (1–3). They act as multiprotein nanomachines (injectisomes) that translocate a diverse repertoire of proteins (effectors) either to extracellular locations or directly into eukaryotic cells, in a Sec-independent manner (interkingdom protein transfer). The effectors modulate the function of crucial host regulatory molecules and trigger a range of highly dynamic cellular responses, which determine pathogen-host recognition, pathogen/symbiont accommodation and elicitation or suppression of defense responses by the eukaryotic hosts. T3SS have evolved into seven families distributed among Gram-negative proteobacteria by horizontal gene transfer (3).
The core of the T3SS apparatus is formed by a set of nine widely conserved proteins that share sequence and structural homologies that span the divide between flagellar and non-flagellar systems, as evidenced by the structural similarities between the homologous domains of HrcQB (4) and FliN (5), or the ATPases EscN (6) and FliI (7). On the other hand, despite considerable sequence diversity (2), many secreted T3SS components, exhibit sequence motifs strongly favoring coiled-coil formation, a property that they share with effectors and numerous other proteins involved in translocation and secretion regulation (8, 9).
The coiled-coil tertiary motif in proteins consists of 2–5 amphipathic α-helices winding around each other to form a supercoil structure and is associated with specific sequence patterns (heptad repeats) that reflect repeating variations of physicochemical properties (10, 11). The motif is highly versatile, serving a broad range of functions, and occurring both in monomeric form and a variety of oligomeric assemblies; its prevalence in T3SS suggests that it may contribute to fundamental requirements of their function in a manner that is not yet understood. In various biological systems, coiled-coils are frequently unfolded as monomers and fold only upon association and formation of quaternary complexes (12), while coiled-coil structures from early folding intermediates are frequently essential for molecular recognition (13, 14).
In this study the in vitro folding of HrpO, a soluble protein from the T3SS of the plant pathogen Pseudomonas syringae pv phaseolicola (15) will be presented, along with its interactions and relations to other T3SS proteins. Our interest in HrpO was provoked by recent insights into the function of soluble components of the export apparatus (16, 17), and in particular by its analogies to the flagellar export protein FliJ that has an essential function at the level of the export ATPase complex (18). HrpO and FliJ have similar size, are encoded by genes located immediately downstream of the atpase gene of their respective gene clusters and despite the absence of significant sequence homology, are predicted to contain coiled-coil segments. FliJ participates in an interaction network involving FliH (regulator of the FliI ATPase), the core proteins FlhA (19) and FliM (20) and the chaperones FlgN and FliT (18). Despite the progress already mentioned (16, 17), there are still many unresolved issues concerning in particular the structural basis of T3SS interaction networks associated with the export mechanism.
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification—N-terminally His6-tagged HrpO was expressed in Escherichia coli DH5α cells and purified using a Ni-NTA-agarose column and a Sephacryl S-100 gel filtration column. C-terminally His6-tagged HrpE was expressed in E. coli BL21(DE3) cells and purified using a Ni-NTA-agarose column (see supplemental data for details).
Gel Filtration Analysis—Gel filtration was performed at 20 °C using anÄKTA purifier system (Amersham Biosciences) with a calibrated XK16/100 column packed with Sephacryl S100. The flow rate was 0.8 ml/min, and elution was monitored at 280 nm. Sample solutions were prepared to a concentration of 5–30 mg/ml and loaded using a 2-ml loop.
Small Angle X-ray Scattering (SAXS)—Data were collected at 4 °C for three different HrpO concentrations (1.8, 3.6, and 7.3 mg/ml) at beamline X33 at EMBL/DESY using a MAR345 detector (21) at a sample-detector distance of ∼3 m (covering a momentum transfer range of 0.08 < s < 0.45 Å-1), mica cells with 1-mm path length, and a collection time of 2 min per frame (21). Using PRIMUS (22) the data were analyzed for radiation damage, averaged after normalization to the intensity of the incident beam, and corrected for the detector response. PRIMUS was also used for the buffer subtraction, data scaling, calculation of the radius of gyration Rg from the slope of the Guinier plot (lnI(s) versus s2) (23) and the radius of gyration of the cross section (Rg,CS) (24), based on the Guinier approximation for a rod particle (scattering intensity: I; momentum transfer: s = 4πsin(θ)/λ;2θ: scattering angle; λ = 1.5 Å: wavelength of x-rays). The forward scattering intensity I(0) (proportional to the number of electrons of the particle) was estimated through extrapolation of the Guinier plot at 2θ = 0° and used to deduce molecular mass of HrpO in solution. MW calibration was based on scattering data from a 5 mg/ml bovine serum albumin solution in 20 mm HEPES, pH 7.5. The GNOM package (25) was used to calculate the pair distribution function P(r) and to estimate the maximum particle dimension (Lmax). Kratky plots were obtained by plotting I(s) × s2 versus s and used to judge the folding of the protein.
EOM (26), was used to quantitatively characterize the disorder of HrpO by allowing for the coexistence of different conformations contributing to the experimental scattering pattern. These conformers were selected using a genetic algorithm (GAJOE) from a pool containing 10,000 models randomly generated by RanCh (26) to cover the conformational space.
Circular Dichroism—CD spectra were acquired using a J-815 CD spectropolarimeter (Jasco Inc., Easton, MD) with a 1-mm path length quartz cuvette, at HrpO concentrations of 0.02–0.21 mg/ml, 20 mm phosphate buffer, pH 8.5, 50 mm KF. Far-UV spectra were measured with 50 nm/min scanning speed, 1 min response time, and 3 accumulations. Far-UV CD spectra were also collected for the HrpE protein at concentrations 0.16 and 0.33 mg/ml in 20 mm phosphate buffer, pH 8.5, 50 mm KF. The Spectra Manager program (Jasco Corp.) was used for buffer subtraction and unit conversions to molar residual ellipticities. Thermal denaturation data were collected monitoring the CD signal at 222 nm in the range of 0–90 °C with a temperature increase of 80 °C/h and a waiting time of 3 s for stabilization. Full far-UV CD spectra (260–195 nm) were also recorded in the range of 4–64 °C during the above procedure in steps of 4 °C. Singular Value Decomposition (SVD) (27) of the thermal denaturation spectra using the program SVD1 was performed to determine significant independent states in the unfolding transition. The significant species needed to model the CD data as a function of temperature were determined on the basis of the characteristics of the SVD basis vectors ui[θ] and the temperature dependence of their associated coefficients vi. The thermal unfolding curve was fitted using SigmaPlot, assuming a two-state model (N → U) (28). Far-UV CD spectra were also recorded at 4 °C after the sample was heated at 90 °C to verify the reversibility of the thermal unfolding transition.
Yeast Two-hybrid System—Yeast two-hybrid assay was performed using MATCHMAKER Two-hybrid System 2 (Clontech). The hrpO gene was inserted into the pAS2-1 plasmid to create an in-frame fusion with the GAL4 DNA-binding domain. The hrpE gene was cloned in the pACT2 plasmid and fused in frame with the transcription activation domain of GAL4. The plasmids were transformed into Saccharomyces cerevisiae strain PJ69–4A carrying three different GAL4-responsive reporter genes (ade, his, and lacZ), each driven by a different GAL4-responsive promoter, to reduce the incidence of false positives. The interaction of hybrid proteins was examined by growing the transformed cells on complete minimal medium lacking Trp, Leu, Ade, and His for 3 days at 30 °C. Plasmids pVA3-1 and pTD1-1 are DNA-binding domain and activation domain fusion plasmids that provide a positive control for interacting proteins.
Co-purification of HrpE and HrpO—Co-expressed His10-HrpE and HrpO were co-purified by metal affinity chromatography from E. coli BL21(DE3) cells co-expressing His10-HrpE and HrpO. Details are given in supplemental data.
Sequence Analysis—T3SS gene clusters were retrieved from GenBank™. For all T3SS families products of genes located immediately downstream the T3SS atpase gene, and having a comparable size to HrpO were analyzed for the detection of intrinsic unfolding (program FoldIndex, Ref. 29), coiled-coil propensities (COILS (11) and MATCHER (30)) and secondary structure (PSIPRED (31)). Three-dimensional structural profile recognition from sequences was performed with PHYRE (32). Homologues of HrpE were found using PSI-BLAST. Protein physicochemical parameters were calculated with ProtParam (33).
RESULTS
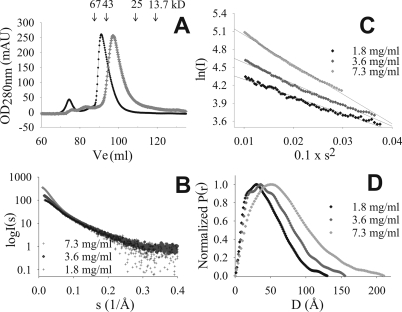
HrpO Forms Heterogeneous Populations of Oligomers in Vitro— When subjected to size-exclusion chromatography at high protein concentrations, the 147-residue HrpO protein elutes with an asymmetric peak profile at an apparent molecular weight (MW) of ∼51 kDa (Fig. 1A), corresponding either to a dimer or trimer (assuming globularity) or to a highly extended non-globular conformation. The broadness of the peak indicates polydispersity. At lower concentrations the elution profiles shifts toward smaller apparent MW.
FIGURE 1.
In vitro polydispersity of HrpO. A, typical asymmetric gel filtration elution profiles at high (black circles: 30 mg/ml) and low protein concentration (open circles: 5 mg/ml). The heights of the two elution peaks are normalized to the same A280 value. Arrows indicate the elution position of MW markers. The first peak (at 75 ml) corresponds to aggregates (void volume). B, SAXS data (corrected for buffer effects) for three protein concentrations. The shape of the curves in the small angle range indicates the presence of particles of different size/mass for the 3 concentrations (self-association phenomena). C, Guinier plots show deviations from linearity with increasing concentration, an indication for polydispersity. D, normalized pair distribution functions (P(r)) showing that the maximum diameter of the protein particle in solution is concentration-dependent.
SAXS was used to analyze the HrpO oligomerization. From the forward scattering intensity I(0) a MW of ∼23 kDa was determined at low concentrations (<2 mg/ml) suggesting a prevalence of the monomeric form (supplemental Table S1). The I(0) values are concentration-dependent (Fig. 1B) suggesting self-association effects. At 7 mg/ml the average MW is ∼77 kDa, corresponding to tetramers or higher order oligomers. Guinier plots of the scattering curves (Fig. 1C) show increasing deviations from linearity at higher concentrations, an indication of polydisperse oligomer populations and of self-association toward complexes of increasingly higher average size (Fig. 1C).
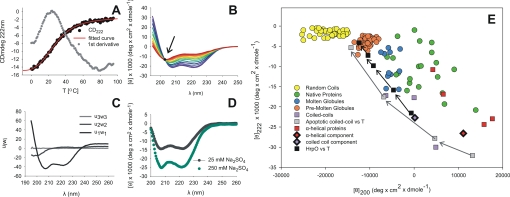
HrpO Is Highly α-Helical and Undergoes a Two-state Thermal Unfolding with Very Low Melting Temperature—Far-UV CD spectra of HrpO show the characteristic minima of α-helical proteins at 208 and 222 nm (Fig. 2). From the mean residual ellipticity at 222 nm, an α-helical content of ≈60% is estimated (34) at 4 °C, which decreases to ≈50% at 20 °C and to ≈20% at 60 °C (HrpO concentration, 0.1 mg/ml). The α-helical content is concentration dependent and increases by ∼50% when concentration increases from 0.02 to 0.1 mg/ml (supplemental Fig. S1). An increase of the ionic strength of the solution from 25 to 250 mm Na2SO4 also increases α-helical content by ∼50% (Fig. 2D).
FIGURE 2.
Thermal unfolding of HrpO monitored by CD. A, variation of the CD signal at 222 nm with the temperature (black dots). The fit (red line) to the experimental sigmoid curve under assumption of a two-state transition yields a Tm value of 21 °C that agrees with the value determined by the first derivative (gray dots) of the experimental curve. B, smoothed far-UV CD spectra at different temperatures. The isodichroic point (arrow) at 203 nm indicates an α-helix to random coil transition in the absence of intermediate states. C, SVD analysis of the thermal unfolding farUV-CD spectra. The first three basis vectors ui, weighted by the singular values wi, are shown. The first two states suffice to model the CD spectra, while all other states are negligible. D, effects of the ionic strength effect of the solution on the CD spectra. An increase in salt concentration increases the α-helical content. CD data were collected at a protein concentration of 0.1 mg/ml in 10 mm CAPS, pH 11.0. E, double-wavelength plot of [θ]200 versus [θ]222 including: (i) colored circles for random coils (yellow), pre-molten globules (orange), molten globules (blue), and native proteins (green) from previous reports (39). (ii) red squares for α-helical, non coiled-coil proteins (myoglobin, hemoglobin, human serum albumin, cytochrome c (53). (iii) violet squares for coiled-coils (ROP (54), Jun/Fos dimer (55), GCN4-like leucine zipper (56)). (iv) black/red and black/violet diamonds for spectra of globin-like (CATH ID: 1.10.490) and coiled-coil proteins (CATH ID: 1.20.120) respectively, from the reference data base for CD spectroscopy (37). (v) Thermal unfolding data of: the apoptotic coiled-coil protein (gray squares, from right to left: at 0, 10, 20, and 45 °C) (57) and HrpO (black squares, from right to left: at 4, 20, 40, 64, 90 °C).
The thermal unfolding of HrpO was monitored by the temperature dependence of the CD signal at 222 nm (Fig. 2A). The 1st derivative of the thermal unfolding data reveals an unusually low transition temperature of 21 °C. Fitting the thermal unfolding data by a curve under the assumption of a two-state transition (Fig. 2A, red line), yields a melting temperature of 21 ± 4 °C, in good agreement with the value obtained by the 1st derivative of the thermal unfolding data. The 203-nm isodichroic point of the CD spectra collected at different temperatures (Fig. 2B) is also consistent with a two-state unfolding via an α-helix to random coil transition (35). The SVD analysis (36) of the far-UV CD spectra confirms the above transition, as it produces only two significant species of linearly independent CD spectra, all other SVD basis vectors being negligible (Fig. 2C). The first significant species corresponds to coiled-coil (37), the second one to random coil (38). The temperature dependence of the coefficients of the basis vectors is shown in supplemental Fig. S2. The double wavelength plot (39) [θ]200 versus [θ]222 of molar residual ellipticities at 200 nm and 222 nm (Fig. 2E) reflects a propensity for structural disorder (see “Discussion”).
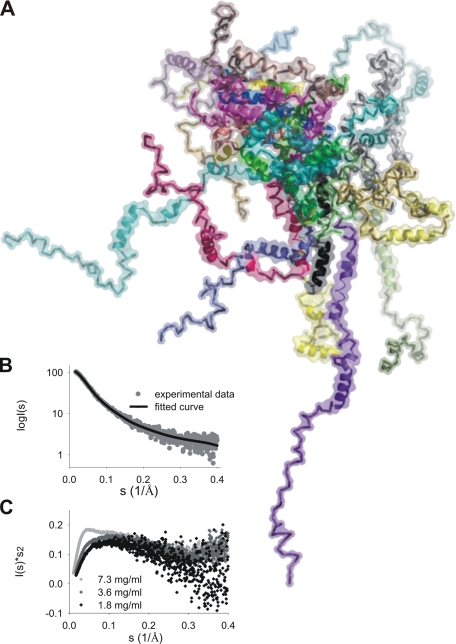
HrpO Does Not Possess a Well Folded Structure in Vitro— Kratky plots (40) from SAXS data were used to judge the folding of HrpO (Fig. 3C). The absence of a bell-shaped curve with a pronounced peak indicates the absence of globular structure, although the presence of some residual structure is suggested by the tail of the plot that reaches a plateau at higher s values. At 1.8 mg/ml an average radius of gyration (Rg) of 35 Å was calculated from the Guinier plot, which is considerably larger than the value expected (17 Å) for a monomeric, globular protein of 147 residues. On the other hand, a value of 41 Å is expected for HrpO in a completely unfolded state (27, 41). We interpret these results as being consistent with an unfolded molecule that retains some secondary structure. The radius of gyration of the cross section (Rg,CS), an estimate of the average transverse dimensions of the molecule, is 12.9 ± 0.1 Å (calculated for the angular range 0.5 > sRg,CS > 1.01), and is consistent with the presence of α-helices. Taking into account that high α-helical content was determined by CD even at lower HrpO concentrations compared with SAXS experiments, we assume that the Kratky plots reflect a highly disordered structure containing to a significant extent α-helical segments. To test this hypothesis, the Ensemble Optimization Method (EOM) (26) was used to characterize the HrpO disorder, under the constraint that all HrpO models include four α-helical segments (∼50% of the sequence) strongly predicted by the PHYRE (32) fold recognition techniques (see “Experimental Procedures”), the rest of the protein being random chain. An ensemble of 20 models shown in Fig. 3A was selected on the basis of the best fit to the experimental SAXS data (Fig. 3B). The quality of the fit (χ = 1.19) supports the model of a highly α-helical, highly disordered structure.
FIGURE 3.
SAXS analysis of HrpO. A, optimized ensemble of HrpO monomers from the EOM software (26). B, fit of the ensemble to the experimental data. C, Kratky plots for three different concentrations.
The HrpO Sequence Exhibits Characteristics of an Intrinsically Disordered Coil-coil and Has Extensive Analogies to Proteins from Other T3SS and the Flagellum—Despite the absence of a significant homology (the residue identity is less than 18%), the HrpO and FliJ sequences share specific characteristics, which suggest similar properties: Based on their average hydrophobicity and net charge, both proteins are predicted (29) to be unfolded and highly α-helical with a pronounced coiled-coil propensity. For HrpO the predicted properties agree with the experimental observations. A set of 27 additional proteins (supplemental Table S2) also encoded by genes immediately downstream of the atpase gene in various T3SS gene clusters, possess similar sequence characteristics: They are small (145 ± 34 residues) and most probably extensively disordered due to a high content of disorder-promoting and a low content of order-promoting residues (42). High α-helical content and coiled-coil propensity is also strongly predicted for all. Despite these analogies, their sequences are diverse, lacking overall similarities. Some homologies can be detected if they are compared at the level of individual T3SS families (3): Members of the Ysc and Hrc1 families show the highest similarities, although even in these cases the level residue identities does not exceed 12–18%. The conserved residues are overwhelmingly either disorder-promoting (Arg, Gln, Glu, Lys) or Leu and Ala, i.e. amino acids strongly associated with coiled-coil proteins (10, 11).
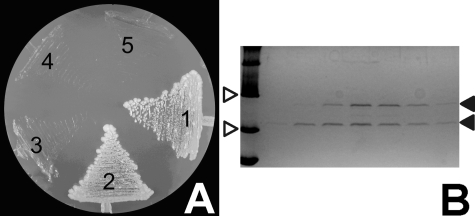
HrpO Interacts with the HrpE Protein in Vivo—HrpO was found to interact in vivo with the HrpE protein using yeast 2-hybrid system and co-purification assays (Fig. 4). Both proteins are coded by terminal ORFs of separate operons in the respective pathogenicity islands of P. syringae pathovars and are not translocated through the T3SS (43), being considered instead injectisome core components (44). HrpE belongs to the FliH/YscL-like proteins that are homologous with the second-stalk components of the FoF1 and v ATPases (44). FliH regulates the activity of the FliI ATPase (45). In addition, FliH interacts with FliJ (19), which is the HrpO counterpart in the flagellum; this suggests analogous roles for the FliJ/FliH and HrpO/HrpE interactions in different systems.
FIGURE 4.
HrpO-HrpE interactions. A, yeast cells expressing the hybrid proteins DBD-HrpO and activation domain HrpE (area 2) grown in the absence of Trp, Leu, Ade, and His formed white colonies, as did the positive control cells, indicating that the hybrid proteins interact with each other. Plasmids pVA3-1 and pTD1-1 were used as positive controls, (area 1) and the vectors pAS2-1 and pACT2 without inserts served as negative controls (area 5). Proteins HrpO and HrpE were also tested for autoactivation (areas 3 and 4, respectively). B, SDS-PAGE of step-elution of His10-HrpE and HrpO co-purification experiment stained with Coomassie Brilliant Blue. White arrows indicate two MW markers (MWM) of 20.1 and 30 kDa. Black arrows indicate the His10-HrpE protein at ∼28 kDa (upper band) and the HrpO protein at ∼21 kDa (lower band). Beginning from left, the first lane is loaded with MWM while the rest with various elution samples from the Ni-NTA-agarose column with increasing imidazole concentration (80, 100, 150, 200, 250, 300 mm, respectively).
The sequences of HrpE, FliH, and YscL possess extensive heptad repeat patterns, comparable to those found for their respective HrpO-like interaction partners (supplemental Table S2). It is thus possible that association via intermolecular coiled-coil formation underlies the HrpO/HrpE and FliJ/FliH interactions.
Using an approach that has been used for the study of flagellar protein interactions (46), far-UV CD spectra of HrpO, HrpE and the HrpO-HrpE complex were recorded and compared. They reveal possible conformational changes accompanying complex formation. Both HrpO and HrpE exhibit CD spectra that are characteristic for proteins with significant α-helical content, in accordance to secondary structure predictions and the presence of heptad patterns. Comparison of the sum of the HrpO and HrpE spectra with the spectrum of the HrpO-HrpE complex shows a decrease of the CD signal at 222 nm, indicating α-helical ordering as a result of interaction (supplemental Fig. S3). Similar α-helical ordering effects were observed for HrpO with increasing protein concentrations (supplemental Fig. S4). As these concentrations fall within the range that favors the monomeric form of HrpO, these effects are probably due to monomer-monomer interactions.
DISCUSSION
This study shows that the HrpO protein forms low to high order oligomers at different concentrations. It is highly α-helical with coiled-coil characteristics, very low melting temperature, and does not posses a well folded structure. HrpO interacts with HrpE, another α-helical T3SS protein that is predicted to possess coiled-coils. This interaction was identified through yeast two-hybrid data and confirmed by co-purification. Sequence analysis suggests that HrpO has characteristics of an intrinsically disordered coiled-coil with extensive analogies to FliJ and to several other proteins across all T3SS families, despite low levels of sequence similarities.
Proteins span a continuum from totally disordered to well folded structures. The extreme flexibility of intrinsically disordered proteins, whose structures may alter dramatically as they bind to their cognate folded protein target, has been suggested to represent a strategy for optimizing the search and interaction with their targets (47). This strategy may also apply to the binding of HrpO to its protein targets, as suggested by its pronounced flexibility and the observed α-helical ordering effects associated with the HrpO/HrpE interaction. The observed properties of HrpO and the predicted properties of its analogues from other T3SS, suggest that HrpO represents a class of intrinsically disordered proteins present in all T3SS families and the flagellum that bind target molecules (e.g. HrpE) probably via coiled-coil interactions. The pronounced self-association propensity of HrpO that produces quaternary complexes in which coiled-coil interactions play a role, as suggested by CD spectra, has probably analogies to the interactions of HrpO with its coiled-coil target proteins. Intrinsic disorder is reflected in the double wavelength plot (Fig. 2E) of [θ]200 versus [θ]222 (39), which reveals that in the temperature range 4–20 °C HrpO exists at the borderline between a folded (coiled-coil) and a molten globule-like state, with more features of intrinsic disorder (molten globule and pre-molten globule states) being gradually established beyond 20 °C. Thermal unfolding studies reveal a two-state transition, with a strikingly low transition temperature of ∼21 °C. SAXS data suggest that at low concentrations HrpO exists as an ensemble of disordered, highly α-helical conformers characterized by lack of globularity, although it is not completely unfolded. We believe that at concentrations favoring the monomeric form, HrpO contains a core formed by some secondary structure elements, with prevailing coiled-coil interactions, as evidenced by the characteristics of CD spectra. The remainder of the polypeptide chain is disordered. The lack of globularity i.e. of a tightly packed core, deduced from the Kratky plot at low concentrations, indicates that this core is probably small and loosely packed. More globular features are induced at higher concentrations, probably due to association effects producing higher order multimers, as evidenced by the Kratky plot and I(0). Presumably, either the association of monomers occurs first leading to the induction of additional structure, or under the specific conditions, the population of more structured intermediates increases, which then associate. It is likely that the association of disordered intermediates occurs via coiled-coil interactions, thereby stabilizing partial secondary structural elements and leading to the formation of additional helices, as observed by CD. Some analogies exist with the process via which α-helical peptides stabilize their helical structures upon association at high concentrations (48), or with the association-induced folding of partially folded intermediates of staphylococcal nuclease (49). In contrast to the above cases however, only limited additional globular characteristics are induced upon increase of the HrpO α-helical content due to oligomerization.
The properties of HrpO at even relatively low temperatures resemble those of a molten globule state although it is not clear how relevant they are physiologically. Intrinsic disorder and the coiled-coil interactions may play an important role in the recognition of HrpO binding partners in T3SS, where coiled-coils containing proteins (e.g. HrpE) are abundant. A more structured state of HrpO with specific contacts may be established upon association to its partner, similarly to what we observe in HrpO oligomerization and in the HrpO-HrpE interaction. This process could be also comparable to the interaction of the intrinsically disordered pKID domain of gene transcription factor CREB with the KIX domain of the CREB-binding protein in the cell nucleus (47). Conformational flexibility and coiled-coil interactions were recently confirmed as necessary requirements for complex formation in the case of tropomyosin-actin binding (13).
Generally, intrinsic disorder is a distinctive and common characteristic of functional interactions (50), and more specifically of hub proteins (proteins that interact with more than 10 partners) in eukaryotic interactomes (51). Disorder could be thus a determinant of functional interactivity of HrpO and its analogues from other T3SS gene clusters (supplemental Table S2) and provide a basis for a widespread coiled-coil interaction mode between proteins of the HrpO family and their coiled-coil targets. This possibility is supported by the properties of the HrpO target HrpE that is α-helical and is predicted to form extensive coiled-coil regions. These regions could be involved at the stage of the initial encounter complex between HrpO and HrpE and facilitate the transition toward more structured states/α-helical ordering, in agreement with the finding that α-helical molecular recognition elements (interaction-prone short segments of disorder that become ordered upon specific binding) are abundant in protein-protein interaction networks (52). HrpE homologues with coiled-coil propensity are found for nearly every T3SS in which HrpO-analogues have been identified (supplemental Table S3).
The structures of the interaction partners of FliJ, the flagellum counterpart of HrpO, also support the concept of a coiled-coil interaction mode: FliJ interacts with FliH, a HrpE homologue, for which high α-helical and coiled-coil segments are predicted. In addition, FliJ interacts directly with the flagellar export chaperone FlgN (18), a 4-α-helical bundle (PDB ID: 2fup). A 20-residue coiled-coil region of FlgN has been mapped to be the common binding site for FliJ and for the export substrate FlgK (PDB ID: 2d4y) the structure of which has as its most prominent feature a coiled-coil domain comprising ∼50% of all residues. The association of FliJ or FlgK to their common binding site is thus probably mediated through interactions between coiled-coil segments.
If the properties of HrpO can be projected to FliJ (an assumption that is supported by the characteristics of the FliJ protein (19)), then the properties of intrinsic disorder and coiled-coil propensity may be also understood under the aspect of the regulation of the flagellar T3SS export apparatus: At the level of the export ATPase complex, two activities have been reported for FliJ, i.e. a T3SS chaperone escort activity and a stimulation of the FliI ATPase activity (18). The escort mechanism for export chaperones (FlgN, FliT) clears the membrane FliI ATPase complex of unloaded chaperones, enhances the cycling of chaperones and increases the efficiency of chaperone-export substrate formation and its docking to the ATPase complex. This activity is established via interactions between FliJ and chaperones that are coiled-coil proteins. The stimulation of the FliI ATPase activity by FliJ occurs either via a transient FliJ-FliI interaction or via interactions of FliJ with other flagellar components, e.g. the FliH protein (the flagellar counterpart of HrpE) with a predicted coiled-coil structure. FliH is a known regulator of FliI activity that binds to FliI and suppresses its oligomerization and ATPase activity. ATP hydrolysis, which is increased in the presence of FliJ, induces the essential dissociation step of the FliH-FliI complex prior to unfolding and translocation of the export substrate that is driven proton motive force (17).
FliJ is thus probably involved in T3S regulation through two activities at the ATPase complex, and these require interactions with several coiled-coil proteins of different structures. Coiled-coiled propensity and a considerable structural plasticity, reflected in intrinsic structural disorder, are thus essential prerequisites for FliJ function. It should be noted that FliJ also interacts with further T3SS components, e.g. the C-ring protein FliM (20), which also exhibits coiled-coil regions, so that this pronounced interactivity is probably enabled by a considerable structural disorder (19).
In summary, the observed properties of HrpO, the properties of the FliJ interactions network, a large body of sequence and structural data and the prevalence of the coiled-coil motif, support the hypothesis of the existence of a widespread coiled-coil interaction mode in all T3SS families and the flagellum mediated by intrinsically disordered proteins. The functional implications of this interaction mode are not clear yet; however they may include a role in the regulation of the T3SS export apparatus.
Supplementary Material
Acknowledgments
We thank Dr. D. Svergun and his group for assistance with SAXS data collection.
This work was supported by PENED, PYTHAGORAS, and PEP (KP-15) Grants from the Greek Ministry of Education, GSRT, and the EPEAEK-Plant Molecular Biology and Biotechnology graduate program. This work was also supported by the EMBL/Hamburg Outstation and the EU through the EU-I3 access grant from the EU Research Infrastructure Action under the FP6 “Structuring the European Research Area Programme,” Contract Number RII3/CT/2004/5060008. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4 and Tables S1–S3.
Footnotes
The abbreviations used are: T3SS, type III secretion system; SAXS, small angle X-ray scattering; CAPS, 3-(cyclohexylamino)propanesulfonic acid; NTA, nitrilotriacetic acid; PDB, Protein Data Bank.
References
- 1.Dale, C., and Moran, N. A. (2006) Cell 126 453-465 [DOI] [PubMed] [Google Scholar]
- 2.Tampakaki, A. P., Fadouloglou, V. E., Gazi, A. D., Panopoulos, N. J., and Kokkinidis, M. (2004) Cell. Microbiol. 6 805-816 [DOI] [PubMed] [Google Scholar]
- 3.Troisfontaines, P., and Cornelis, G. R. (2005) Physiology 20 326-339 [DOI] [PubMed] [Google Scholar]
- 4.Fadouloglou, V. E., Tampakaki, A. P., Glykos, N. M., Bastaki, M. N., Hadden, J. M., Phillips, S. E., Panopoulos, N. J., and Kokkinidis, M. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 70-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown, P. N., Mathews, M. A. A., Joss, L. A., Hill, C. P., and Blair, D. F. (2005) J. Bacteriol. 187 2890-2902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zarivach, R., Vuckovic, M., Deng, W., Finlay, B. B., and Strynadka, N. C. J. (2007) Nat. Struct. Mol. Biol. 14 131-137 [DOI] [PubMed] [Google Scholar]
- 7.Imada, K., Minamino, T., Tahara, A., and Namba, K. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 485-490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delahay, R. M., and Frankel, G. (2002) Mol. Microbiol. 45 905-916 [DOI] [PubMed] [Google Scholar]
- 9.Pallen, M. J., Beatson, S. A., and Bailey, C. M. (2005) FEMS Microbiol. Rev. 29 201-229 [DOI] [PubMed] [Google Scholar]
- 10.Paliakasis, C. D., and Kokkinidis, M. (1992) Prot. Engineering 5 739-749 [DOI] [PubMed] [Google Scholar]
- 11.Lupas, A., Van Dyke, M., and Stock, J. (1991) Science 252 1162-1164 [DOI] [PubMed] [Google Scholar]
- 12.Uversky, V. N., Gillespie, J. R., and Fink, A. L. (2000) Proteins Struct. Func. Genet. 41 415-427 [DOI] [PubMed] [Google Scholar]
- 13.Singh, A., and Hitchcock-DeGregori, S. E. (2006) Structure 14 43-50 [DOI] [PubMed] [Google Scholar]
- 14.Steinmetz, M. O., Jelesarov, I., Matousek, W. M., Honnappa, S., Jahnke, W., Missimer, J. H., Frank, S., Alexandrescu, A. T., and Kammerer, R. A. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 7062-7067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lindgren, P. B., Peet, R. C., and Panopoulos, N. J. (1986) J. Bacteriol. 168 512-522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paul, K., Erhardt, M., Hirano, T., Blair, D., and Hughes, K. T. (2008) Nature 451 489-493 [DOI] [PubMed] [Google Scholar]
- 17.Minamino, T., and Namba, K. (2008) Nature 451 485-489 [DOI] [PubMed] [Google Scholar]
- 18.Evans, L. D. B., Stafford, G. P., Ahmed, S., Fraser, G. M., and Hughes, C. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 17474-17479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fraser, G. M., Gonzalez-Pedrajo, B., Tame, J. R. H., and Macnab, R. M. (2003) J. Bacteriol. 185 5546-5554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gonzalez-Pedrajo, B., Minamino, T., Kihara, M., and Namba, K. (2006) Mol. Microbiol. 60 984-998 [DOI] [PubMed] [Google Scholar]
- 21.Roessle, M. W., Klaering, R., Ristau, U., Robrahn, B., Jahn, D., Gehrmann, T., Konarev, P., Round, A., Fiedler, S., Hermes, C., and Svergun, D. (2007) J. Appl. Crystallogr. 40 s190-s194 [Google Scholar]
- 22.Konarev, P. V., Volkov, V. V., Sokolova, A. V., Koch, M. H. J., and Svergun, D. I. (2003) J. Appl. Crystallogr. 36 1277-1282 [Google Scholar]
- 23.Guinier, A., and Fournet, G. (1955) Small-Angle X-ray Scattering of X-rays, Wiley, New York
- 24.Ishimuro, Y., Hamada, F., and Nakajima, A. (1979) J. Polym. Sci. 17 1811-1819 [Google Scholar]
- 25.Svergun, D. I. (1993) J. Appl. Crystallogr. 26 258-267 [Google Scholar]
- 26.Bernadó, P., Mylonas, E., Petoukhov, M. V., Blackledge, M., and Svergun, D. I. (2007) J. Am. Chem. Soc. 129 5656-5664 [DOI] [PubMed] [Google Scholar]
- 27.Pérez, J., Vachette, P., Russo, D., Desmadril, M., and Durand, D. (2001) J. Mol. Biol. 308 721-743 [DOI] [PubMed] [Google Scholar]
- 28.Greenfield, N. J. (2006) Nat. Prot. 1 2527-2535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prilusky, J., Felder, C. E., Zeev-Ben Mordehai, T., Rydberg, E. H., Man, O., Beckmann, J. S., Silman, I., and J. L., S. (2005) Bioinformatics 21 3435-3438 [DOI] [PubMed] [Google Scholar]
- 30.Fischetti, V. A., Landau, G. M., Schmidt, J. P., and Sellers, P. (1993) Inform. Process Lett. 45 11-18 [Google Scholar]
- 31.Jones, D. T. (1999) J. Mol. Biol. 292 195-202 [DOI] [PubMed] [Google Scholar]
- 32.Kelley, L. A., MacCallum, R. M., and Sternberg, M. J. E. (2000) J. Mol. Biol. 299 499-500 [DOI] [PubMed] [Google Scholar]
- 33.Gasteiger, E., Hoogland, C., Gattiker, A., Duvaud, S., Wilkins, M. R., Appel, R. D., and Bairoch, A. (2005) The Proteomics Protocols Handbook, Humana Press
- 34.Chen, Y.-H., Yang, J. T., and Chau, K. H. (1974) Biochemistry 13 3350-3359 [DOI] [PubMed] [Google Scholar]
- 35.Holtzer, M. E., and Holtzer, A. (1995) Biopolymers 36 365-379 [DOI] [PubMed] [Google Scholar]
- 36.Wall, M. E., Rechtsteiner, A., and Rocha, L. M. (2003) Singular Value Decomposition and Principal Component Analysis, Kluwer, Norwell
- 37.Lees, J. G., Miles, A. J., Wien, F., and Wallace, B. A. (2006) Bioinformatics 22 1955-1962 [DOI] [PubMed] [Google Scholar]
- 38.Uversky, V. N. (2002) FEBS J. 269 2-12 [Google Scholar]
- 39.Uversky, V. N. (2003) Cell Mol. Life Sci. 60 1852-1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hagihara, Y., Hoshino, M., Hamada, D., Kataoka, M., and Goto, Y. (1998) Folding Des. 3 195-201 [DOI] [PubMed] [Google Scholar]
- 41.Jha, A. K., Colubri, A., Freed, K. F., and Sosnick, T. R. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 13099-13104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dunker, A. K., Brown, C. J., Lawson, J. D., Iakoucheva, L. M., and Obradovic, Z. (2002) Prot. Funct. Biochem. 41 6573-6582 [DOI] [PubMed] [Google Scholar]
- 43.Ramos, A. R., Morello, J. E., Ravindran, S., Deng, W.-L., Huang, H.-C., and Collmer, A. (2007) J. Bacteriol. 189 5773-5778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pallen, M. J., Bailey, C. M., and Beatson, S. A. (2006) Prot. Sci. 15 935-941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lane, M. C., O'Toole, P. W., and Moore, S. A. (2006) J. Biol. Chem. 281 508-517 [DOI] [PubMed] [Google Scholar]
- 46.Muskotal, A., Kiraly, R., Sebestyen, A., Gugolya, Z., Vegh, B. M., and Vonderviszt, F. (2006) FEBS Lett. 580 3916-3920 [DOI] [PubMed] [Google Scholar]
- 47.Sugase, K., Dyson, H. J., and Wright, P. E. (2007) Nature 447 1021-1025 [DOI] [PubMed] [Google Scholar]
- 48.Yoshida, K., Shibata, T., Masai, J., Sato, K., Noguti, T., Go, M., and Yanagawa, H. (1993) Biochemistry 32 2162-2166 [DOI] [PubMed] [Google Scholar]
- 49.Uversky, V. N., Karnoup, A. S., Khurana, R., Segel, D. J., Doniach, S., and Fink, A. L. (1999) Protein Sci. 8 161-173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dyson, H. J., and Wright, P. E. (2005) Nat. Rev. Mol. Cell Biol. 6 197-208 [DOI] [PubMed] [Google Scholar]
- 51.Haynes, C., Oldfield, C. J., Ji, F., Klitgord, N., Cusick, M. E., Radivojac, P., Uversky, V. N., Vidal, M., and Iakoucheva, L. M. (2006) PLoS Comp. Biol. 2 890-901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dunker, A. K., Cortese, M. S., Romero, P., Iakoucheva, L. M., and Uversky, V. N. (2005) FEBS J. 272 5129-5148 [DOI] [PubMed] [Google Scholar]
- 53.Matsuo, K., Yonehara, R., and Gekko, K. (2004) J. Biochem. 135 405-411 [DOI] [PubMed] [Google Scholar]
- 54.Vlassi, M., Steif, C., Weber, P., Tsernoglou, D., Wilson, K. S., Hinz, H.-J., and Kokkinidis, M. (1994) Nat. Struct. Biol. 1 706-716 [DOI] [PubMed] [Google Scholar]
- 55.Mason, J. M., Schmitz, M. A., Müller, K. M., and Arndt, K. M. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 8989-8994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Holtzer, M. E., Braswell, E., Angeletti, R. H., Mints, L., Zhu, D., and Holtzer, A. (2000) Biophys. J. 78 2037-2048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dutta, K., Alexandrov, A., Huang, H., and Pascal, S. M. (2001) Protein Sci. 10 2531-2540 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.