Abstract
The postsynaptic N-methyl-d-aspartate (NMDA) receptor activates multiple kinases and changes the phosphorylation of many postsynaptic proteins organized in signaling networks. Because the NMDA receptor is known to regulate gene expression, it is important to examine whether networks of kinases control signaling to gene expression. We examined the requirement of multiple kinases and NMDA receptor-interacting proteins for gene expression in mouse hippocampal slices. Protocols that induce long-term depression (LTD) and long-term potentiation (LTP) activated common kinases and overlapping gene expression profiles. Combinations of kinases were required for induction of each gene. Distinct combinations of kinases were required to up-regulate Arc, Npas4, Egr2, and Egr4 following either LTP or LTD protocols. Consistent with the combinatorial data, a mouse mutant model of the human cognition disease gene SAP102, which couples ERK kinase to the NMDA receptor, showed deregulated expression of specific genes. These data support a network model of postsynaptic integration where kinase signaling networks are recruited by differential synaptic activity and control both local synaptic events and activity-dependent gene expression.
Molecular networks are now recognized as a basis for integration of signaling within cells. The potential for highly complex phosphorylation networks is suggested by phosphoproteomic studies that show thousands of phosphorylation sites in many cells (1, 2). Phosphoproteomics of the mammalian synapse have also revealed over 1000 phosphorylation sites occurring in vivo and over 60 postsynaptic kinases (3–6). Although the role of tyrosine, serine, and threonine kinases in synaptic plasticity, learning, and other forms of behavioral plasticity is well described (7, 8), little is known about their organization into networks and the properties of these networks.
The process of learning involves the conversion of information within patterns of neuronal electrical activity into biochemical changes within neurons. These biochemical changes are initiated at the synaptic level, where signal transduction pathways modulate local synaptic strength as well as signals to the nucleus to drive gene expression. In recent years it has become clear that learning is not a simple switch but a set of signaling events and cell biological processes that show diversity and complexity. For example, at the electrophysiological level, different patterns of activity either induce long-term depression (LTD)3 or long-term potentiation (LTP) of synaptic transmission by activating the N-methyl-d-aspartate (NMDA) receptor.
The potential exists for networks of kinases to regulate the gene expression associated with LTP and LTD. A kinase network model would be an attractive mechanism for orchestrating the differential expression of genes and would be expected to be revealed by addressing the following questions: First, to what extent do multiple kinases regulate any given gene? Second, do stimulation paradigms that initiate LTP and LTD regulate common sets of genes? Third, are genes that are both regulated by LTP and LTD share the same dependence on specific kinases? Fourth, do mutations in NMDA receptor complex (NRC) proteins (such as SAP102/dlg3) interfere with gene expression? To address these issues, we have studied NMDA receptor-activated gene expression in hippocampal slices from mice. The NMDA receptor was activated using chemical stimulation protocols that result in LTP or LTD. Using microarray profiling we identify a common set of genes induced by the LTP and LTD protocols. Inhibition of specific kinases shows that NMDA receptor activation of each gene is regulated by multiple kinases and that the set of kinases that regulates a gene may differ in LTP and LTD protocols. We also document that a mutation in NMDA receptor-associated proteins (SAP102/dlg3) impairs ERK kinase signaling and ERK-dependent gene expression. These data suggest a model where the synaptic kinase signaling networks coordinate gene expression through their differential activation during LTP and LTD.
EXPERIMENTAL PROCEDURES
Preparation of Hippocampal Slices—Experiments were performed on hippocampal slices obtained from 12–14-week old 129S5/SvEvBrd mice bred at the Wellcome Trust Sanger Institute essentially and were sacrificed by cervical dislocation in accordance with Schedule 1 to the UK Animals (Scientific Procedures) Act 1986. Hippocampal slices were prepared as described (30). Before the start of biochemical stimulations, slices rested under these conditions for at least 2 h after the last slice was cut.
Biochemical Stimulation—Experiments were usually performed on slices from four animals, simultaneously. We used two separate chambers for treated and untreated (solvent only) slices. One chamber had two wells, which were further divided into two halves by a thin silicone barrier to accommodate slices from four separate mice without mixing them up. In this way, 10–12 slices obtained from one mouse were evenly divided between two respective compartments in “treated” and “untreated” chambers yielding 5–6 slices per experimental condition. To stimulate biochemical pathways, we incubated slices in the “treated” chamber with one of the three drug-containing solutions: 1) 20 μm NMDA (3 min); 2) 50 μm forskolin + 100 nm rolipram in Mg2+-free ACSF (15 min). To facilitate solution exchange in the slice chamber well, the speed of solution flow was increased to 5–7 ml/min for 3 min during the start of incubation with drugs and upon their wash-out. In experiments using NMDA receptor or protein kinase inhibitors, perfusion of each inhibitor was used for 30 min prior to application of the stimulation drug solutions and also throughout drug stimulation and wash-out periods. Routinely, inhibitors were applied to one of the two wells of the “treated” chamber that contained slices from 2 animals. Slices from another pair of animals were stimulated with a drug solution that contained a concentration of DMSO equal to the one used to dissolve inhibitors in the other group. Slices in an untreated chamber also received the same solvent treatment, if necessary.
Slices were incubated in the “treated” chamber with the following solutions: Wortmannin 200 nm (40 min), KN-62 10 μm (30 min), APV 50 μm (10 min), SB203580 10 μm (40 min), U0126 20 μm (40 min). All drugs were from Tocris Cooksoon Ltd (Bristol, UK). Standard techniques were used for Western blot analysis of the samples. For primary antibody information, see supplemental Table S3.
Nucleic Acid Isolation and Microarray Hybridization—Hippocampal slices were immediately frozen in dry ice or immersed into RNAlater reagent (Qiagen) on ice for RNA isolation. RNeasy Mini columns (Qiagen) were used to purify total RNA. On-column DNase treatment was carried out to remove traces of DNA. During all the purifications, the manufacturer's instructions were followed. Samples were concentrated with the addition of 1:10 volumes of 3 m NaOAc, pH 6 and 3 vol of 100% EtOH and precipitated at -20 °C. Once resuspended in 12 μl of RNase-free H2O, an aliquot was used to check the RNA integrity in 1% formaldehyde-agarose gels. For the transcriptome profiling, MG-430 2.0 arrays (Affymetrix) were used that contain 45,101 probe sets including controls, one array per sample. Four samples were analyzed, divided in two (stimulated and non-stimulated) for a total of eight hybridizations. Briefly, 1 μg of total RNA was reverse-transcribed, labeled, and hybridized using One Cycle Labeling kit instructions (Affymetrix). Fluidics Station 450 and GCS3000, both from Affymetrix, were used for the washing and scanning steps, respectively.
Microarray Statistical Analyses—Three procedures were used to obtain the changing genes after stimulation: (i) pairwise comparisons, (ii) GeneSpring analysis for comparison of multiple samples, and (iii) pairwise comparisons using a data extraction tool alternative to GCOS. Because of the high variability found in the response to stimulation, a balance was found by trial and error between permissibility and stringency in the results. To this aim, samples were analyzed in pairwise comparisons (stimulated versus non-stimulated slices from the same animal) to minimize the effect of different levels of transcript at the basal state. A Principal Component Analysis (PCA) showed a higher similarity between samples according to the animal processed rather than the treatment (data not shown). For the pairwise comparisons, two different tools were used that extracted the signal intensity values using different algorithms: Batch analysis (GCOS) and dChip analysis (31). In the Batch analysis, signal intensity values and detection flags were extracted using GCOS, using a Target value of 500 in the scaling step. Next, a Batch analysis (included in the GCOS package) was performed for each pair of samples. All the samples were found to be “Present” and no further filtering was required. In the dChip analysis, signal intensity values were extracted and analyzed using the PM-only model, considering each pair of samples (stimulated and non-stimulated) separately. In both analyses, we considered the probe sets that were retrieved in at least 3 of 4 comparisons in the same direction of change, with a fold change >1.2 (because of the low number of changing genes retrieved in NMDA experiments). These results were also compared with the results of a multiple comparison (all stimulated versus all non-stimulated samples) to study how many significant probe sets were sufficiently robust to overcome individual variability. GeneSpring v7 (Silicon Genetics) was used for the multiple comparison after the GCOS extraction procedure. First, the data were normalized by the median per gene and per array. Next, the resulting data were filtered using the following criteria: (i) analysis of variance test (p < 0.05) without assuming equal variances, (ii) signal intensity must be a Present or Marginal call in all untreated samples when down-regulated and treated samples when up-regulated, and (iii) applying the same fold change threshold as above. The final lists of positive probe sets consisted of those appearing in 2 of 3 types of analyses.
Quantitative RT-PCR Analysis—For the retrotranscription, 1 μg of total RNA was used for the retrotranscription, following the recommendations of Invitrogen (SuperScript II Reverse Transcriptase). PCR reactions were set up per sample using 0.4 μl of cDNA, 0.3 μm each primer, 10 μl of 2× SYBR Green PCR mix (either Quantitect SYBR Green PCR Master Mix, Qiagen, or Power SYBR Green PCR Master Mix, Applied Biosystems) to a final volume of 20 μl. The sequences of the primers are reported in supplemental Table S4. Reactions were run out in the 7500 Real Time PCR System (Applied Biosystems) with the following conditions: 1 cycle of 95 °C for 15 min, 35 cycles of 94 °C for 30 s, 55 °C for 30 s, 72 °C for 50 s (when using Qiagen SYBR Green mix), 1 cycle of 95 °C for 10 min, 40 cycles of 95 °C for 15 s, 60 °C for 1 min (when using SYBR Green mix). The resulting amplicons were checked for specificity and size in agarose gels. The data generated were analyzed in 7500 System SDS v1.2.2 software that calculated the cycle threshold (CT) and the fold change (2-ΔΔCT). GAPDH was used as endogenous control. Results were retrieved from at least two different days of stimulation (eight animals, four treated with the agonist and four treated with the agonist plus the signaling pathway inhibitor). After GAPDH normalization, fold changes were compared between pairs of stimulated/non-stimulated slices from the same animal. To calculate the effect of the inhibitors, % of change was calculated related to full stimulation (without inhibitor). However, those cases where the fold change was negative or very mild (i.e. <1.3) were considered as non-induced, and the % of the basal (1) was calculated. To analyze the significance of the changes observed in individual genes, Student's t test was used. To cluster the qRT-PCR results for the tested genes (FSK treatment, see Fig. 4 and text), smooth correlations of the % changes were used to build a hierarchical tree (GeneSpring).
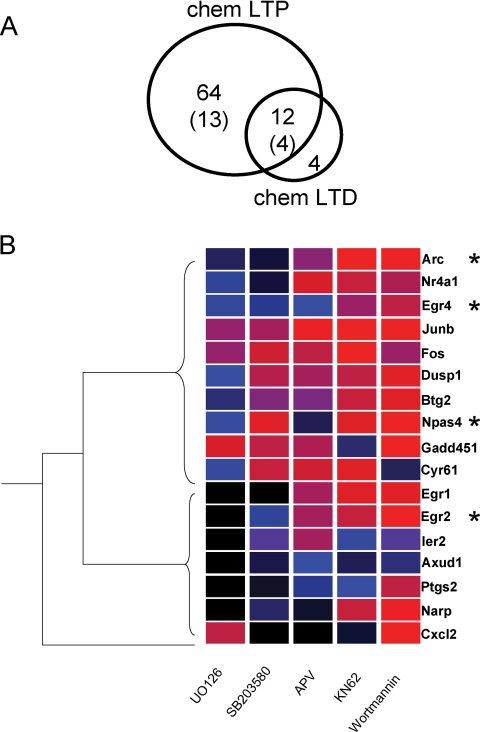
FIGURE 4.
A, number of genes induced after 1 h of stimulation by either Chem-LTD or Chem-LTP. Venn diagrams show the number of significantly changing genes in each treatment as retrieved in microarray analyses. See supplemental Tables S1 and S2 for further details. B, hierarchical clustering of the effects of signaling pathway inhibitors on Chem-LTP-induced gene expression. qRT-PCR assays were performed on 17 genes in Chem-LTP-stimulated and non-stimulated hippocampal slices. Changes in induced gene expression after kinase inhibition were normalized to samples without kinase inhibitors and clustered by the inhibitory effect on each pathway. Each gene shows a unique profile of regulation (no two genes have an identical profile of colored boxes) and the branched tree (left) shows these genes can be grouped into two main sets. This indicates the combinatorial nature of the regulatory network. Asterisks (*) indicate genes also modulated by Chem-LTD stimulation. Bright red: 100% (no effect); bright blue: 50% inhibition; black, 0% (total abolition of induction).
RESULTS
NMDA Receptor Activation of Multiple Kinases and Genes—Hippocampal slices were treated with bath application of the glutamate receptor agonist NMDA (20 μm), which is known to induce long-term depression (Chem-LTD) (9, 10). To characterize a set of kinases regulated by this protocol, proteins were extracted at different time points (3′, 20′, and 40′) and immunoblotted with phosphospecific antibodies and quantified (Fig. 1, A and B). We chose to study 7 kinases from 4 different groups; (i) AGC kinases, Akt, (ii) CMGC kinases, Erk2, p38, JNK1, (iii) STE kinases PAK1/3, MEK1/2, and (iv) PTK kinases, Src. Consistent with previous studies, all kinases were modulated by NMDA stimulation: the 6 serine/threonine kinases showed robust increased phosphorylation with a peak at 3 min, and Src tyrosine kinase showed a decrease in phosphorylation. All the changes in phosphorylation returned to basal levels after 40-min post-stimulation. To identify a candidate set of immediate early genes (IEGs) that might be dependent on these kinases, we isolated total RNA from slices at 60 min after NMDA receptor stimulation and performed microarray profiling. The profiling time of 60 min was selected after performing quantitative RT-PCR assays of well known IEGs at different time points (data not shown). As shown in supplemental Table S1, we identified 16 genes that were activated by NMDA using MG-430 2.0 arrays and qRT-PCR assays.
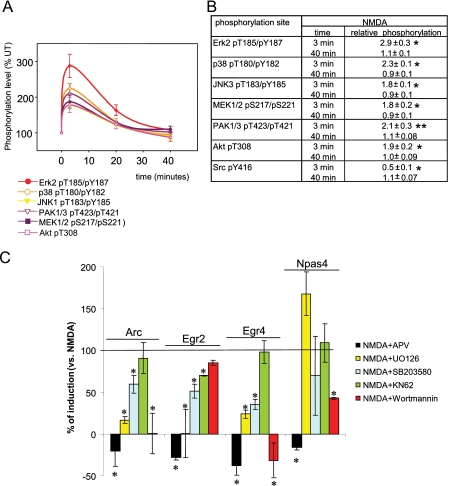
FIGURE 1.
A, time course of phosphorylation levels for different kinases after NMDA stimulation of hippocampal slices. B, summary of the results. *, significant changes (p < 0.001, Student's t test); **, p < 0.05, Student's t test. C, effect of signaling pathway inhibitors on induced gene expression of Arc, Egr2, Egr4, and Npas4 in qRT-PCRs assays from stimulated and non-stimulated hippocampal slices. Resulting fold changes from the co-application of NMDA and the corresponding inhibitor were normalized by the fold changes obtained with the stimulatory drug only, shown as % (see “Experimental Procedures”). *, significant changes (p < 0.05, Student's t test) to 100% of induction.
To map the requirement of specific kinases in the induction of gene expression, we pretreated the hippocampal slices with different kinase inhibitors or the NMDA antagonist APV prior to NMDA stimulation and then measured the amount of gene induction. We focused on 4 specific kinase antagonists that interfere with signaling pathways downstream of the NMDA receptor (Fig. 1, A and B): MAPK pathways; MEK/ERK (inhibited by UO126) and p38 (inhibited by SB203580); Ca2+-calmodulin-dependent kinase pathway (inhibited by KN-62) and phoshoinositide-3-kinase (PI3K)-dependent pathway (inhibited by wortmannin). As expected, these inhibitors were effective in reducing or preventing the phosphorylation of their respective kinase substrates (supplemental Fig. S1). We selected 4 NMDA-regulated genes (Arc, Egr2, Egr4, and Npas4) as reporters and monitored their expression using qRT-PCR and, as expected, APV prevented the activation of all 4 genes (Fig. 1C).
We next addressed the role of individual kinases in the activation in the Chem-LTD protocol. We observed that each gene was regulated by multiple kinase inhibitors with differential effects (Fig. 1C). This indicates that each of the NMDA-dependent kinases tested contributed to gene expression. However, no single kinase could account for the modulation of the total set of genes. For example Arc, Egr2, and Egr4 were inhibited by MEK and p38 inhibitors but only Egr2 was insensitive to PI3K inhibition. In contrast to these three genes, Npas4 was insensitive to MEK and p38 inhibitors and only required PI3K pathway for activation. Fig. 1C summarizes the differential kinase requirements of the 4 genes in NMDA receptor activation and illustrates the network of kinase-gene interactions.
The fact that multiple genes were regulated by multiple kinases suggests that integration of synaptic kinase pathways provide subtlety and graded responses to the output of synaptic signaling in the gene expression programming. In this model, shifting the activity of a kinase might alter the output and relative levels of gene expression. To address this issue, we selected a different protocol, known to induce LTP (Chem-LTP) (11). This protocol combines activation of NMDAR and PKA with a combination of forskolin (PKA activator), rolipram (phosphodiesterase inhibitor that enhances cAMP levels), and low Mg2+ (facilitates NMDA receptor activation).
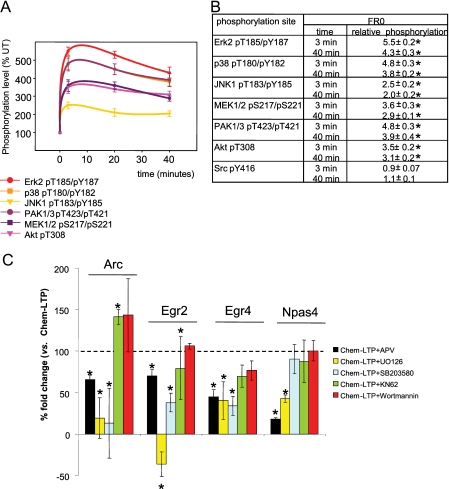
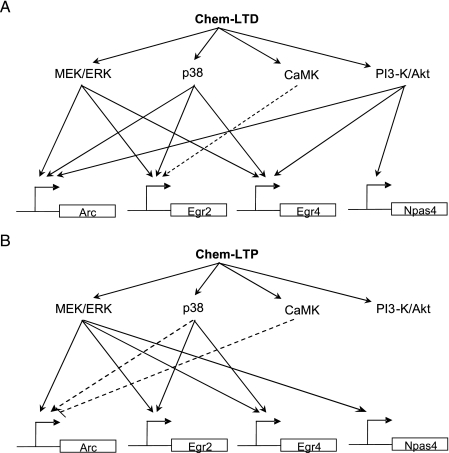
Similar to the experiments with the Chem-LTD protocol we next examined the activation of the same set of kinases using phosphorylation assays using the Chem-LTP protocol (Fig. 2A). The same set of kinases was activated, although higher and more persistent levels of kinase phosphorylation were observed (lasting at least 40-min poststimulation). Next, we examined the requirement for NMDA receptor using the reporter genes and found a partial blockade of Arc, Egr2, and Egr4, and the most effective blockade in Npas4 (Figs. 1C and 2C). Differential kinase requirements were observed in each gene, and importantly, a different pattern to that seen with Chem-LTD. For example, Arc, Egr4, and Npas4 were sensitive to PI3K inhibition in the Chem-LTD protocol but insensitive in the Chem-LTP protocol. Despite this shift in the PI3K sensitivity of these 3 genes between the LTP and LTD protocols, their sensitivity to MEK and p38 was maintained. The comparison of both protocols indicates that kinases that are not required for the expression of particular genes in particular protocols become essential under other conditions. This switching of kinase dependence was further illustrated in the NMDAR-dependent activation of Npas4: PI3K was required to obtain a 100% induction of Npas4 in the Chem-LTD protocol; however, in the Chem-LTP protocol the PI3K pathway was no longer required; and Npas4 expression was largely contributed to by a MEK-dependent pathway. As summarized in Fig. 3A, these differential kinase requirements and differential gene expression profiles indicate that considerable plasticity in gene expression profiles can be achieved by differentially recruiting a combination of kinases.
FIGURE 2.
A, time course of phosphorylation of kinases after Chem-LTP stimulation of hippocampal slices. B, summary of the results. *, significant changes (p < 0.001, Student's t test). C, effect of signaling pathway inhibitors on induced gene expression of Arc, Egr2, Egr4, and Npas4 in qRT-PCRs assays from stimulated and non-stimulated hippocampal slices. Resulting fold changes from the co-application of NMDA and the corresponding inhibitor were normalized by the fold changes obtained with the stimulatory drug only, shown as % (see “Experimental Procedures”). *, significant changes (p < 0.05, Student's t test) to 100% of induction.
FIGURE 3.
A, model of the signaling pathways that were activated by Chem-LTD and B Chem-LTP that influenced gene expression, based upon the data in Fig. 1C and Fig. 2C. Solid arrow indicates a strong and significant effect (>2-fold, p < 0.05). Dashed arrow indicates either a slight but significant effect (<2-fold, p < 0.05, Student's t test) or large but not significant effect of the inhibitor (>2-fold, p > 0.05, Student's t test). B, differences in the signaling pathways for gene induction were not due to differences in the magnitude of change because only Egr2 was significantly different in both treatments (p < 0.05, Student's t test). Data are expressed as mean ± S.E.
As the above studies show, two distinct protocols associated with two opposing forms of synaptic plasticity result in the induction of the same 4 genes, and moreover, the magnitude of gene induction by each protocol (Chem-LTD, Chem-LTP) was not significantly different for Npas4 (5.34 ± 0.89 versus 5.0 ± 0.40, p = 0.36), Arc (2.70 ± 0.23 versus 3.53 ± 0.45, p = 0.06), or Egr4 (3.09 ± 0.31 versus 4.23 ± 0.65, p = 0.06). Only Egr2 presented a significant higher fold induction with the Chem-LTP protocol: 2.92 + 0.30 in NMDA versus 4.87 ± 0.41, p = 0.0002). This suggests that these genes are important for the biological outcome (perhaps memory storage) associated with both LTP and LTD, and that the signaling networks are arranged so as to activate these genes using different kinases recruited under the different LTP and LTD paradigms. This raises the question: does LTP and LTD drive the same set of genes or are there other sets that are activated in either protocol? To investigate this, we used microarrays profiling for the Chem-LTP protocol and compared the overlapping and distinct genes in a Venn diagram (Fig. 4A and supplemental Table S1). Twelve genes were found to be common to both chemical protocols, representing 75% of the Chem-LTD genes and 16% of Chem-LTP. These data show that there is a significant overlap in the two gene sets induced by these protocols, as well as distinct differences.
To further characterize the combinatorial action of kinases on sets of genes described above, we extended these studies to a larger set of genes in the Chem-LTP protocol. We selected 17 representing ∼27% of the total and 4 were in the overlapping Chem-LTD set (Fig. 4B). Changes in expression levels were quantitated by qRT-PCR, and the resulting patterns of inhibitor sensitivity (% of inhibition related to full gene stimulation) were correlated using a hierarchical clustering method (see “Experimental Procedures”). For each gene, the signaling pathway profile or induction code was represented in a row and the inhibitor profile for many genes is shown as a column (Fig. 4B). Several points became clear: first, a wide range of unique profiles were observed, each gene had a unique profile. Second, the patterns were non-random and general patterns were observed, with genes grouped into two principal sets. Third, inhibitors with similar profiles were clustered in adjacent positions in the following order: MEK, p38, NMDA receptor, CamK, PI3K. The MAPK pathways (MEK and p38) generated more closely related profiles (i.e. inhibiting a similar subset of genes) and thereby reflecting the cooperativity between both MAPK pathways. As shown in Fig. 4B, we indicate those genes that are regulated by Chem-LTP and Chem-LTD highlighting the overlapping and distinct sets of genes induced. Together these data show that the combinations of kinases regulate many genes, and differential recruitment of kinases can read out distinct profiles comprised of overlapping and unique genes.
Altered Gene Expression in SAP102/dlg Mutant Mice— The NMDA receptor-dependent activation of synaptic plasticity and behavior has been shown to require the function of membrane-associated guanylate kinases (MAGUK) family proteins. Three vertebrate MAGUK paralogues (SAP102, PSD-95, PSD-93) bind directly to the C terminus of NMDA 2 subunits (NR2) (12). Knockouts in SAP102 (13), PSD-95 (14–16);4 and PSD-934 result in altered synaptic plasticity. Our previous characterization of SAP102 knock-out mice (13), showed that this mutation caused a specific impairment of the NMDAR-dependent Erk2 activation. We therefore tested if the reduced activation of Erk2 by NMDAR in SAP102 mutant mice affected the expression of genes associated with Erk2 modulation in hippocampal slices. Consistent with the predictions presented above, the induction of Arc and Egr2 was significantly reduced compared with wild-type animals in 38 ± 2% (p = 0.003, Student's t test) and 27 ± 6% (p = 0.023, Student's t test), respectively. Moreover, consistent with SAP102 not being involved in the modulation of the PI3K-Akt pathway the NMDAR-dependent induction of Npas4 was not affected (p = 0.136, Student's t test). These data indicate that specific mutations in NMDA receptor-associated proteins can uncouple the ERK-dependent pathway to ERK-dependent gene expression.
DISCUSSION
We examined signaling from NMDA receptor to gene expression focusing on the differential roles played by multiple kinases in regulating downstream genes. We asked whether multiple kinases regulate a given gene and whether stimulation paradigms that initiate LTP and LTD regulate common and distinct sets of genes. We observed that multiple kinases contributed to the transcriptional activation of a given gene and using LTP or LTD induction paradigms found these protocols shifted the relative contribution made by a kinase in the induction of a target gene. A common set of genes was regulated by the LTP and LTD protocols as well as subsets specific to each protocol. We also addressed the role of NMDA receptor-associated proteins coupling to gene expression and found that specific kinases and genes could be functionally uncoupled from NMDA receptor signaling by mutations of SAP102.
Our data are consistent with postsynaptic kinase networks with the following features: (i) a stimulus activates multiple kinases, and different stimuli trigger different sets of kinases, (ii) each kinase phosphorylates multiple types of postsynaptic proteins including receptors, adaptors, enzymes, structural proteins, trafficking, and translational regulators, (iii) each kinase regulates multiple transcription factors (TF), and (iv) each TF regulates multiple genes. Together these steps allow differential receptor activation or multiple receptors, to drive overlapping sets of genes.
An interesting characteristic of the recruitment of a combination of kinases that regulate a set of genes, is that the same gene could be induced by two different protocols (e.g. Chem-LTD and Chem-LTP) yet have completely different kinase dependencies. The most dramatic effect was observed in the activation of Npas4: Npas4 only required the modulation of the PI3K pathway to induce its expression with Chem-LTD, while with Chem-LTP the MEK/ERK pathway was essential for the expression of Npas4, with no requirement for the PI3K cascade. Interestingly, this lack of a requirement for PI3K in Chem-LTP induction of Npas4 occurs even though PI3K was activated (Fig. 3A). In other words, PI3K was being activated but not used for gene expression. Thus, there is a switching between the utilization of different kinases depending on upstream activation protocols. Switching in the dependence of ERK and PI3K has been reported for the induction of LTP: high-frequency (100 Hz)-induced LTP is not blocked by MEK inhibitors but is inhibited by PI3K inhibitors. Importantly, this contrasts to theta-pulse (5Hz) induced LTP that required both MEK and PI3K (16). Moreover the dependence on MEK in TPS-LTP was abolished in mice carrying a mutation in the NMDA receptor-interacting protein PSD-95 (16). Understanding the details of the switching mechanisms may require the mapping of phosphorylation sites on substrates and their interactions.
How the combinatorial activation of signaling pathways is translated into patterns of gene expression is likely explained by the phosphorylation of TFs and their protein interactors. Recently an integrative model of transcription factors, activated by cannabinoid receptor 1 (CB1R), described a signaling network connecting CB1R to 23 activated transcription factors. In this model, the use of pharmacological inhibitors for different protein kinases revealed a kinase-transcription factor network organization of CB1R-induced neurite outgrowth (17).
There are several well known TFs including CREB and other members of the family, the ternary complex Elk-1/SRF, Egr members, MEF2, AP-1, and NF-kB that have a demonstrated role in synaptic plasticity processes (18–21). Focusing on the NMDA receptor-activated genes Arc, Egr2, Egr4, and Npas induction, it is reported that Arc expression can be activated by CREB or Egr and Egr2 activated by CREB and SRF (22–25, 27). We examined the computationally predicted TF binding sites in the 5′-region (1-kb upstream and 200-bp downstream of the transcription start site) of Arc, Egr2, Egr4, and Npas4 and found they share similar TF sites for NF-κB, SRF, and CREB among others) albeit in different locations and combinations (supplemental Fig. S2). The differences in the promoter architecture may account for differential strength (or affinity) of the TF, the recruitment of other TFs and the direct binding of specific TFs. Therefore, differential phosphorylation of these TFs and their interacting proteins may explain induction of either different genes or common genes under different stimulation protocols.
A similar model of cooperation of multiple signaling pathways in regulating gene expression has been reported for CD40 receptor activation in B lymphocytes (28). Following receptor activation and gene expression profiling in the presence of specific kinase inhibitors (including those used in the present study), it was reported that overlapping and distinct sets of genes were regulated by PI3K, p38, and NF-κB pathways. Three mechanisms were proposed to account for the regulation by multiple signaling pathways downstream of single receptor of gene expression: independent, collective, and redundant control. The independent control refers to situations where each pathway regulates a distinct set of genes; the collective control is when a set of pathways regulate a single or common large set of genes that constitute the entire expression profile; in redundant control, different pathways can substitute for one another. Both independent and collective regulation was observed in the response of B cells to CD40 activation.
These studies on the organization of postsynaptic signaling networks and the postsynaptic proteome are revealing a signaling system with features of complexity, combinatorial functions, and redundancy. In addition to their basic roles in information processing (both electrophysiological and biochemical) studies on mice carrying mutations in these proteins reveal changes in cognitive function. Many of these proteins have also been implicated in the etiology of multiple brain diseases (29) and it is likely that some aspects of the phenotype of these diseases results from changes in the signaling networks. Understanding the organization of the networks may help identify drug targets for modulating the disease processes.
Supplementary Material
Acknowledgments
We thank V. J. Robinson for mouse colony maintenance, D. G. Fricker, E. C. Sotheran, and N. H. Komiyama for genotyping, and J. V. Turner for editorial assistance.
The arrays reported in this paper has been submitted to ArrayExpress Database under accession number E-MEXP-1184.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2 and Tables S1–S3.
Footnotes
The abbreviations used are: LTD, long-term depression; LTP, long-term potentiation; NMDA, N-methyl-d-aspartate; PI3K, phoshoinositide-3-kinase; TF, transcription factors; IEG, immediate early genes; MEK, mitogen-activated protein kinase/extracellular signal-regulated kinase kinase; ERK, extracellular signal-regulated kinase; ER, endoplasmic reticulum.
Carlisle, H. J., Fink, A. E., Grant, S. G. N., and O' Dell, T. J. (2008) J. Physiol. 586, in press.
References
- 1.Olsen, J. V., Blagoev, B., Gnad, F., Macek, B., Kumar, C., Mortensen, P., and Mann, M. (2006) Cell 127 635-648 [DOI] [PubMed] [Google Scholar]
- 2.Villen, J., Beausoleil, S. A., Gerber, S. A., and Gygi, S. P. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 1488-1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Munton, R. P., Tweedie-Cullen, R., Livingstone-Zatchej, M., Weinandy, F., Waidelich, M., Longo, D., Gehrig, P., Potthast, F., Rutishauser, D., Gerrits, B., Panse, C., Schlapbach, R., and Mansuy, I. M. (2007) Mol. Cell Proteomics 6 283-293 [DOI] [PubMed] [Google Scholar]
- 4.Collins, M. O., Yu, L., Coba, M. P., Husi, H., Campuzano, I., Blackstock, W. P., Choudhary, J. S., and Grant, S. G. (2005) J. Biol. Chem. 280 5972-5982 [DOI] [PubMed] [Google Scholar]
- 5.Trinidad, J. C., Specht, C. G., Thalhammer, A., Schoepfer, R., and Burlingame, A. L. (2006) Mol. Cell Proteomics 5 914-922 [DOI] [PubMed] [Google Scholar]
- 6.Trinidad, J. C., Thalhammer, A., Specht, C. G., Lynn, A. J., Baker, P. R., Schoepfer, R., and Burlingame, A. L. (2007) Mol. Cell Proteomics 7 684-696 [DOI] [PubMed] [Google Scholar]
- 7.Greengard, P. (2001) Science 294 1024-1030 [DOI] [PubMed] [Google Scholar]
- 8.Kandel, E. R. (2001) Science 294 1030-1038 [DOI] [PubMed] [Google Scholar]
- 9.Lee, H. K., Kameyama, K., Huganir, R. L., and Bear, M. F. (1998) Neuron 21 1151-1162 [DOI] [PubMed] [Google Scholar]
- 10.Malenka, R. C., and Bear, M. F. (2004) Neuron 44 5-21 [DOI] [PubMed] [Google Scholar]
- 11.Otmakhov, N., Khibnik, L., Otmakhova, N., Carpenter, S., Riahi, S., Asrican, B., and Lisman, J. (2004) J. Neurophysiol. 91 1955-1962 [DOI] [PubMed] [Google Scholar]
- 12.Kornau, H. C., Schenker, L. T., Kennedy, M. B., and Seeburg, P. H. (1995) Science 269 1737-1740 [DOI] [PubMed] [Google Scholar]
- 13.Cuthbert, P. C., Stanford, L. E., Coba, M. P., Ainge, J. A., Fink, A. E., Opazo, P., Delgado, J. Y., Komiyama, N. H., O'Dell, T. J., and Grant, S. G. (2007) J. Neurosci. 27 2673-2682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Migaud, M., Charlesworth, P., Dempster, M., Webster, L. C., Watabe, A. M., Makhinson, M., He, Y., Ramsay, M. F., Morris, R. G., Morrison, J. H., O'Dell, T. J., and Grant, S. G. (1998) Nature 396 433-439 [DOI] [PubMed] [Google Scholar]
- 15.Komiyama, N. H., Watabe, A. M., Carlisle, H. J., Porter, K., Charlesworth, P., Monti, J., Strathdee, D. J., O'Carroll, C. M., Martin, S. J., Morris, R. G., O'Dell, T. J., and Grant, S. G. (2002) J. Neurosci. 22 9721-9732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Opazo, P., Watabe, A. M., Grant, S. G., and O'Dell, T. J. (2003) J. Neurosci. 23 3679-3688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bromberg, K. D., Ma'ayan, A., Neves, S. R., and Iyengar, R. (2008) Science 320 903-909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deisseroth, K., Mermelstein, P. G., Xia, H., and Tsien, R. W. (2003) Curr. Opin. Neurobiol. 13 354-365 [DOI] [PubMed] [Google Scholar]
- 19.Winstanley, C. A., LaPlant, Q., Theobald, D. E., Green, T. A., Bachtell, R. K., Perrotti, L. I., DiLeone, R. J., Russo, S. J., Garth, W. J., Self, D. W., and Nestler, E. J. (2007) J. Neurosci. 27 10497-10507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McClung, C. A., and Nestler, E. J. (2008) Neuropsychopharmacology 33 3-17 [DOI] [PubMed] [Google Scholar]
- 21.Bloomer, W. A., VanDongen, H. M., and VanDongen, A. M. (2008) J. Biol. Chem. 283 582-592 [DOI] [PubMed] [Google Scholar]
- 22.Han, J. H., Kushner, S. A., Yiu, A. P., Cole, C. J., Matynia, A., Brown, R. A., Neve, R. L., Guzowski, J. F., Silva, A. J., and Josselyn, S. A. (2007) Science 316 457-460 [DOI] [PubMed] [Google Scholar]
- 23.Li, L., Carter, J., Gao, X., Whitehead, J., and Tourtellotte, W. G. (2005) Mol. Cell. Biol. 25 10286-10300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramanan, N., Shen, Y., Sarsfield, S., Lemberger, T., Schutz, G., Linden, D. J., and Ginty, D. D. (2005) Nat. Neurosci. 8 759-767 [DOI] [PubMed] [Google Scholar]
- 25.Ying, S. W., Futter, M., Rosenblum, K., Webber, M. J., Hunt, S. P., Bliss, T. V., and Bramham, C. R. (2002) J. Neurosci. 22 1532-1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deleted in proof.
- 27.Watanabe, T., Hongo, I., Kidokoro, Y., and Okamoto, H. (2005) Dev. Biol. 277 508-521 [DOI] [PubMed] [Google Scholar]
- 28.Dadgostar, H., Zarnegar, B., Hoffmann, A., Qin, X. F., Truong, U., Rao, G., Baltimore, D., and Cheng, G. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 1497-1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grant, S. G., Marshall, M. C., Page, K. L., Cumiskey, M. A., and Armstrong, J. D. (2005) Hum Mol Genet 14 R225-R234 [DOI] [PubMed] [Google Scholar]
- 30.Kopanitsa, M. V., Afinowi, N. O., and Grant, S. G. (2006) BMC Neurosci. 7 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li, C., and Wong, W. H. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 31-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.