Abstract
The Down syndrome candidate region 1 gene (DSCR1) can be expressed as four isoforms, one of which is the well-studied isoform 4 (DSCR1-4) that is induced by VEGF-A165 to provide a negative feedback loop in the VEGF-A165-induced angiogenesis. We reported previously that another DSCR1 isoform, DSCR1-1L, was also up-regulated by VEGF-A165 in cultured endothelial cells and in several in vivo models of pathological angiogenesis and that different from DSCR1-4, DSCR1-1L overexpression alone induced cultured endothelial cell proliferation and promoted angiogenesis in Matrigel assays. It was reported recently that tumor growth was greatly repressed in DSCR1 knock-out mice. Although DSCR1-4 transcription was primarily regulated by NFAT, the mechanism regulating DSCR1-1L expression was still unknown. We developed human DSCR1-1L promoter-driven luciferase system and found that deletion of a putative conserved M-CAT site located 1426-bp upstream of the translation start site blunted promoter activity. We further showed that knockdown of TEF3, not other members of TEF family inhibited VEGF-A165-induced DSCR1-1L expression. We also demonstrated that TEF3 directly interacted with the putative M-CAT site in the DSCR1-1L promoter in vitro and in vivo. Finally, overexpression of TEF3 isoform 1, not isoform 3, in HUVEC was sufficient to induce DSCR1-1L expression even in the absence of VEGF-A165 stimulation. Taken together, we elucidated a novel function of transcriptional factor TEF3. TEF3 was required for DSCR1-1L expression through binding to the M-CAT site in its promoter and could be an attractive target for anti-angiogenesis therapy.
Down syndrome candidate region 1 (DSCR1)2 is one of more than 50 genes present in the portion of trisomy 21, the chromosomal abnormality responsible for Down syndrome (1–3). DSCR1 is identical to MCIP1 (modulatory calcineurin-interacting protein) (4). It is homologous to Adapt78, an oxidant stress-inducible gene in hamsters (5–7) and also to Rcn1/nebula in Drosophila (8). Down syndrome is a major cause of mental retardation and is also associated with various cardiac and gastrointestinal anomalies, immune system defects, and Alzeimer disease. Down syndrome patients have increased risks of certain malignancies (leukemia, germ cell tumors) but an extremely low incidence of many solid tumors, particularly breast cancer (9, 10). On the other hand, clinically aggressive ovarian carcinomas often have trisomy of chromosome 21, whereas certain other solid tumors have deletions of this chromosome. Together these and other data suggest that genes such as DSCR1 that are triplicated in Down syndrome may play important roles in tumorigenesis (11, 12).
Interest in DSCR1 has recently been sparked by several reports indicating that it is up-regulated in cultured vascular endothelial cells by VEGF-A165 and further that it provides a negative feedback loop that inhibits VEGF-A165-induced angiogenesis (13–16). However, the conclusion was based on the studies of only one DSCR1 isoform, isoform 4 (DSCR1-4). The DSCR1 gene is comprised of seven exons, the first four of which can serve as start sites that then combine with exons 5–7 to produce four different mRNA transcripts (2, 17). Exon 1 was originally thought to encode a 29-amino acid peptide (DSCR1-1) (2, 17), but later studies by Genesca et al. (18) revealed a start site further upstream that encoded an 84-amino acid peptide (DSCR1-1L). Exon 2 is probably not translated into protein, because it lacks a methionine start site. Exon 3 encodes only 3 amino acids (2, 17). Exon 4, under the control of a different promoter from that regulating isoforms 1–3 (2, 17), encodes a 29-amino acid peptide that initiates a fourth DSCR1 isoform. The several DSCR1 isoforms have different expression patterns and likely different functions and regulatory mechanisms (2, 17). All four isoforms are expressed in heart and skeletal muscle (2, 17). Isoform 1 has also been detected in brain and isoform 4 in placenta and kidney (2, 17). DSCR1-1L has been found to play a protective role against cell stress (5–7). In addition to inhibiting angiogenesis (13–16), DSCR1-4 plays an inhibitory role in cardiac and skeletal muscle hypertrophy (19–21).
Recently, we reported that DSCR1-1L, like DSCR1-4, was up-regulated by VEGF-A165 in cultured endothelial cells and also in several types of pathological angiogenesis in vivo (22). DSCR1-1L was also expressed in the microvasculature of human ovarian cancers, but not in the tumor cells themselves nor in the microvessels of normal ovary (22). Of particular interest, the effects of DSCR1-1L in angiogenesis were antithetical to those of DSCR1-4 (22). Using a novel modification of the standard Matrigel assay, we found that overexpression of DSCR1-1L promoted endothelial cell proliferation in vitro and angiogenesis in vivo, whereas a DSCR1-1L-specific siRNA had opposite effects. Our data indicated that DSCR1-1 and -4 have opposing stimulatory and inhibitory effects on VEGF-A165-induced angiogenesis (22). Most recently, it was reported that tumor growth was greatly repressed in DSCR1 knock-out mice lacking all DSCR1 isoforms (23). Therefore, DSCR1-1L and the signaling pathway that regulates DSCR1-1L expression are excellent targets for cancer anti-angiogenic therapy.
It was shown that VEGF-A165-induced DSCR1-4 expression was mediated by the NFAT response element in the DSCR1-4 promoter that was located in its intron 3 (24). However, the regulatory element(s) that regulates DSCR1-1L has not been studied yet. To study the molecular mechanism that regulates DSCR1-1L expression in angiogenesis, we constructed a 1.4-kb human DSCR1-1L promoter-driven luciferase plasmid to study the molecular mechanism of DSCR1-1L expression.
EXPERIMENTAL PROCEDURES
Materials—Antibodies against TEF1 and TEF4 were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Rabbit antibody against human DSCR1-1L (hDSCR1) was purchased from Abcam, Inc. (Cambridge, MA). FLAG antibody was from Sigma-Aldrich. Mouse polyclonal antibody to TEF5 (TEAD3) was from Novus Biologicals. TEF3 antibody was developed previously (25). HUVEC (Clonetics, Biowhittaker, Inc. Walkersville, MD) were cultured and transduced with retroviruses carrying various constructs as previously described (26). EOMA cells and 293T cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and penicillin/streptomycin.
Immunohistochemistry—These experiments were carried out according to a protocol approved by the hospital's Committee on Clinical Investigation. Tumors were obtained at the time of surgery and were fixed in 4% paraformaldehyde, embedded in OCT compound, and prepared for immunohistochemistry. Frozen sections were then blocked with 5% goat serum, stained with mouse anti-hDSCR1-1L (Center for Biomedical Inventions, University of Texas Southwestern Medical School), washed three times with PBS, incubated for 1 h with biotinylated polyclonal anti-mouse IgG antibody (1:200 dilution, Vector Laboratories, Inc. Burlingame, CA), washed three times with PBS, reacted with the ABC peroxidase kit (Vector Laboratories, Inc. Burlingame, CA) at room temperature for 45 min, and washed with PBS prior to mounting for light microscopy and photography.
Construction of DSCR1-1L Promoter Reporter Plasmid and Its Mutants—To construct the DSCR1-1L promoter-luc plasmid, the human 1.4-kilobase promoter was cloned from a HUVEC genomic DNA by PCR with a primer set: forward primer with KpnI (5′-aaaaaaggtaccagaggggtgactttgaatggaatg) and reverse primer with HindIII (5′-aaaaaaaagcttcgttaaccccctcggaatc). The PCR products were digested with KpnI and HindIII and subcloned into the pGL3-basic vector (Promega, San Luis Obispo, CA) The identity of the promoter was confirmed by DNA sequencing, and this plasmid was named pD1L(-1450) (all numbering relative to the translation start site because there is no TATA box in the DSCR1-1L promoter). The primers used to create deletion mutants were listed below, all of which with KpnI and HindIII restriction sites in the 5′- and 3′-end, respectively. pD1L(-1344): 5′-aaaaaaggtaccacggttccaatttcttcacatcc (forward) and 5′-aaaaaaaagcttgccagccccgcccgtcac (reverse). pD1L(-1287): 5′-aaaaaaggtaccgatgggtattattattcctcttg (forward) and 5′-aaaaaaaagcttgccagccccgcccgtcac (reverse). pD1L(-1199): 5′-aaaaaaggtacccatggcaagttctgaataaatctc (forward) and 5′-aaaaaaaagcttgccacgccgtcctccatcc (reverse). pD1L(-1089): 5′-aaaaaaggtacctcacaacctccaaagaacc (forward) and 5′-aaaaaaaagcttgccacgccgtcctccatcc (reverse). The forward primers for the following deletion mutants were pD1L(-1426): 5′-aaaaaggtaccggaggctggttggctctaag; pD1L(-1421): 5′-aaaaaaggtaccctggttggctctaagcaa; pD1L(-1396): 5′-aaaaaaggtaccttagttttccttagtgattt; pD1L(-1374): 5′-aaaaaaggtacctgggggcccaagatttcc; pD1L(-1364): 5′-aaaaaaggtaccagatttccctttcacagt; pD1L(-1353): 5′-aaaaaaggtaccacagtatgacggttccaa; pD1L-ΔMCAT: 5′-aaaaaaggtaccagaggggtgacttggaggctggttggctctaagcaa. The reverse primer was 5′-aaaaaaaagcttcgttaaccccctcggaatc. The 3× M-CAT sequence was 3TEC1 (Kpn1/BglII)F, 5′-aaaaaaggtaccttgaatggaatggttgaatggaatggttgaatggaatggagatctggaaag (underline indicated the triple of M-CAT sequence), and 3TEC-1(Kpn1/BglII)R, 5′-ctttcgagatctccattccattcaaccattccattcaaccattccattcaaggtacctttttt. The mutated sequence was 3*2Mu-tec-1 (SacI/SmaI)F, 5′-ACACACGAGCTCTTGTACCGTACCTTGTACCGTACCTTGTACCGTACCCCCGGGATAAAC-3′ and 3*2Mu-tec-1 (SacI/SmaI)R, 5′-GTTTATCCCGGGGGTACGGTACAAGGTACGGTACAAGGTACGGTACAAGAGCTCGTGTGT.
Promoter Activity Assay—EOMA cells were seeded in 24-well plates at the density of 1–2 × 105 cells/well. Twenty hours later, cells were transfected with D1L promoter-luciferase plasmids and control luciferase plasmid (pRL-tk, Promega, Madison, WI) with Lipofectamine 2000 transfection reagent (Invitrogen, Carlsbad, CA). Two microliters of Lipofectamine 2000 were added directly to 50 μl of Dulbecco's modified Eagle's medium (DMEM) without serum and antibiotics and incubated at room temperature for 5 min. D1L promoter plasmid (0.4 μg) and pRL-tk (0.08 μg) were added to another 50 μl DMEM without serum and antibiotic. Lipofectamine 2000 dilution and plasmid were mixed and incubated for 20 min at room temperature. Then the mixture was added to EOMA cells and incubated in the absence of serum for 2 days. Cells were then washed with PBS and incubated with 100 μl of 1× passive lysis buffer of Dual-luciferase Reporter Assay System (Promega) at room temperature until cells were dissolved.
HUVEC (1 × 105 per well) were plated in 12-well plates. Twenty-four hours later, cells were transfected with pGL3-D1L plasmids and control luciferase plasmid (pRL-tk) using FuGENE-6 transfection reagent (Roche Diagnostics). Cells were washed twice with PBS. Two microliters of FuGENE was added directly to 50 μl of OPTI-MEM1 medium and incubated for 5 min. pD1L (0.5 μg) and pRL-tk (0.2 μg) were added to the mixture and incubated for 15 min, and then added to cells with 300 μl of medium. Twenty-four hours after transfection, cells were stimulated with VEGF-A165 for 6 h, then washed with PBS, and lysed with 100 μl of 1× passive lysis buffer until cells were dissolved. Luciferase activity was assayed and normalized to equal internal control luciferase activity according to the manufacturer's protocol.
Preparation of Cell Extracts—Cytoplasmic and nuclear extracts were prepared with the NE-PER Nuclear and Cytoplasmic Extraction Reagents (Pierce). Cell membranes were lysed in 200 μl of ice-cold Cytoplasmic Extraction Reagent I (CERI, 1× protease inhibitor mixture; Roche) for 10 min on ice. Then, ice-cold Cytoplasmic Extraction Reagent II (CERII) (5.5 μl) was added. Cells were incubated for 1 min and centrifuged (16000 × g at 4 °C for 10 min). The supernatants were collected as cytoplasmic fractions. The insoluble pellet that contains nuclei was washed two times with PBS to remove the remaining cytoplasmic proteins. The washed pellet was resuspended in 100 μl of ice-cold Nuclear Extraction Reagent (NER, 1× protease inhibitor mixture; Roche), incubated for 40 min on ice, and centrifuged (16000 × g at 4 °C for 10 min). The supernatant was used as nuclear extract. Extracts obtained by this procedure generally had less than 10% contamination between nuclear and cytoplasmic fractions. The protein concentration was determined with a commercial protein assay reagent (Bio-Rad Laboratories).
Electrophoresis Mobility Shift Assays (EMSA)—The probe with three copies of M-CAT sites and mutant probe were labeled with biotin 3′-end DNA labeling kit as described in the manufacturer's protocol (Pierce). EMSA was performed using a LightShift Chemiluminescent EMSA kit (Pierce) following the manufacturer's protocols with some modifications. Briefly, nuclear extracts (5 μg) and 20 fmol of biotin-labeled putative 3×M-CAT were incubated in a reaction mixture (10 mm Tris, 50 mm KCl, 10 mm dithiothreitol; pH 7.5, 50 ng/μl poly(dI·dC), 2.5% glycerol, 5 mm MgCl2, 0.05% Nonidet P-40, 0.05 mm dithiothreitol) at room temperature for 20 min. The mixtures were then separated by electrophoresis on a 6% polyacrylamide gel at 4 °C in 0.5× TBE at 100 V for 1–1.5 h. The samples were subsequently transferred to positively charged nylon membranes (Pierce) in 0.5× TBE at 380 mA for 1 h at 4 °C and exposed to UV light (120 mJ·cm-2, 1 min) to cross-link the DNA to the membrane. Detection of biotin-labeled DNA probe was performed in accordance with the manufacturer's protocol. For experiments with wild-type and mutant competitors, 0.2 pmol (10× excess) or 1 pmol (50× excess) unlabeled wild-type or mutant probe oligonucleotides were included in the nuclear protein-DNA reaction mixtures. In the experiment with antibody supershifting, antibodies against TEF1, TEF3, TEF4, and TEF5 (1 μg) were included in the reaction mixture.
RT-PCR Analysis—RNA was isolated from HUVEC. RT-PCR with specific primers for TEF1, TEF3, TEF4, and TEF5 was carried out as described (17). GAPDH served as a control for equal RNA loading. RT-PCR products were analyzed on 4% agarose gels. The primers used are listed below. TEF1-rt-F1120, 5′-TTGGTGGTAACAAACAGGGAT and TEF1-rt-R1209, 5′-ATGTTGTGCTCCGTGTTCA; TEF3-rt-F976, 5′-ATGATCATCACCTGCTCCAC and TEF3-rt-R1066, 5′-GTCCATTCTCATAGCGAGCA; TEF4-rt-F1135, 5′-ATGTGCGAGTACCTGGTGAA and TEF4-rt-R1231, 5′-CCTGGAGGATGGTGAAGTTT; TEF5-rt-F973, 5′-GATAGCATGACCATCAGCGT and TEF5-rt-R1065, 5′-GTTCTCCAGCCTGGCATACT. GAPDH F, 5′-TCCACCACCCTGTTGCTGTA and GAPDH R, 5′-ACCACAGTCCATGCCATCAC.
Cloning and Expression of TEF3 Isoforms and siRNAs—TEF3 isoforms were cloned by RT-PCR using RNA isolated from HUVEC with primers in 5′-UTR (5′-GCGGACTCCTTGGAACTGGCTTAG) and 3′-UTR (5′-CATCTTGGGTTTATTTGGGGTTGG) of TEF3, respectively. The RT-PCR products were cloned to pTOPO vector (Invitrogen) and then subjected to DNA sequence analysis. TEF3-1 and TEF3-3 were subcloned to pMMP-FLAG vector with primers XholI-TEF3-1-F (5′-AACTCGAGTTGG AGGGCACGGCCGGCAC), XholI-TEF3-3-F (5′-AACTCGAGATGGCTGCCATGTCGTCTGC) and BamHI-TEF3-R (5′-AAGGATCCTCATTCTTTCACCAGCCTG, respectively, and overexpressed in HUVEC as described previously (26).
siRNAs were designed with the software from OligoEngine Co (Seattle, WA), cloned into pSUPER-retro vector (Oligo-Engine), and expressed in HUVEC. The siRNA sequences were siTEF1A, TACCGAATAAACCGCTCCC, siTEF1B, ATACCGAATAAACCGCTCC, siTEF1C, GATTCCACGCCCGACCTTC, siTEF3A, ACTACTCTTACCGCATCCA, siTEF3B, ACTCTTACCGCATCCACCG, siTEF4A, CCGTTCACCTTGTCACTGA, siTEF4B, GTTCACCTTGTCACTGACT, siTEF4C, CACCTTGTCACTGACTCCC siTEF5A, CCGTCTTCTCCACTTCCTC, siTEF5B, CGTCTTCTCCACTTCCTCG and siTEF5C, TCTTCTCCACTTCCTCGCG.
Chromatin Immunoprecipitation Assays (CHIP)—Chromatin immunoprecipitation assays were performed according to the protocol (CHIP assay, Upstate, Charlottesville, VA). Briefly, HUVEC pellet were sonicated and cross-linked with formaldehyde and then subjected to immunoprecipitation (IP) with an antibody against TEF1, TEF3, TEF4, TEF5, or IgG as a control overnight at 4 °C. The chromosomal DNA in the immunoprecipitated complexes was extracts with phenol-chloroform. After final ethanol precipitation, IP products were resuspended in TE buffer (10 mm Tris-HCl and 1 mm EDTA) and subjected to PCR assay with the primer set (forward: 5′-AGTACATTGGTTTGGTCTG and reverse: 5′-ACGGTTCTATGATTACTATTAT). The primers were designed according to the sequences of the putative M-CAT binding site on the proximal portion of the DSCR1-1 promoter. The supernatants before immunoprecipitation were served as total input chromatin control.
Statistics—The results are expressed based on triplet experiments as the means ± S.E. variance. The Tukey-Kramer multiple comparisons test was used to determine statistical significance. A p value of <0.05 was considered to be statistically significant.
RESULTS
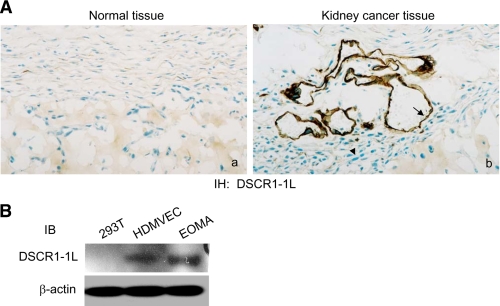
Expression of DSCR1-1L in Endothelial Cells—Previously, we reported that DSCR1-1L was expressed in the vascular structure of ovarian cancer tissues, not in tumor cells by immunohistochemical analysis (22). To find out whether the tumor vessel-specific expression of DCSR1-1L also occurs in other tumors, we analyzed the DSCR1-IL expression in human kidney cancer tissues by immunohistochemical staining. As expected, DSCR1-1L was selectively expressed in the tumor vascular endothelium but not in the tumor cells, nor in normal kidney tissues (Fig. 1A). We further confirmed these results by immunoblotting analysis with cellular extracts isolated from human kidney cancer cells. As shown in Fig. 1B, DSCR1-1L was also expressed in proliferating human microdermal vascular endothelial cells (HMDVEC), in addition to HUVEC (22), but not in proliferating human kidney carcinoma 293T cells (Fig. 1B). Because hemangioma EOMA cells, transformed mouse endothelial tumor cells, were shown to secrete high levels of VEGF that cause autocrine and VEGF-dependent growth (27), we also examined whether DSCR1-1L was expressed in the EOMA cells. Fig. 1B clearly indicated that DSCR1-1L was highly expressed in the hemangioma EOMA cells (Fig. 1B). Our data demonstrated that DSCR1 was highly expressed in endothelial cells, not in kidney cancer cells.
FIGURE 1.
Expression of DSCR1-1L in endothelial cells. A, immunohistochemical staining of a control human kidney tissues (a) and human kidney adenocarcinoma (b) with an antibody specific for human DSCR1-1L. DSCR1-1L expressed in vessels (arrow), not in tumor cells (arrowhead). Tissue from 1 of 5 different patients, all of which exhibited similar staining, is shown. B, cellular extracts from 293T cells, HMDVEC, and EOMA cells were immunoblotted with antibodies against DSCR1-1L (top panel) and β-actin (bottom panel) as a protein equal-loading control.
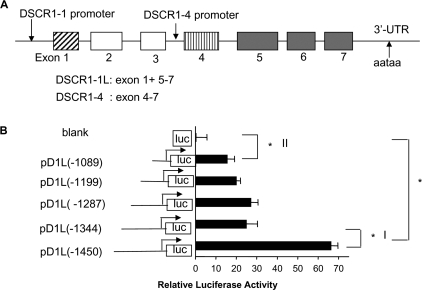
Identification of a Putative M-CAT Element in the DSCR1-1L Promoter in Hemangioma Cells—It was reported that DSCR1-1L and DSCR1-4 consisted of exons 1, 5, 6, 7 and exons 4, 5, 6, 7, respectively (Fig. 2A) and expression of DSCR1-4 was regulated by the calcineurin-NFAT signaling pathway (15, 28–32). The promoter regulating DSCR1-4 was found in the intron preceding exon 4 and contains several NFAT binding sites (Fig. 2A) (24). However, unlike DSCR1-4, DSCR1-1L is not likely to be induced through the Cn-NFAT pathway (13, 15, 16, 32). Therefore, we generated the 1.4-kb DSCR1-1L promoterluciferase reporter construct pD1L(-1450) as described under “Experimental Procedures.” The DSCR1-1L promoter activity was first analyzed in hemangioma cells (EOMA) because they were easier to be transfected than HUVEC and expressed high levels of DSCR1-1L protein as shown in Fig. 1B. EOMA cells were transfected with the pD1L(-1450) plasmid along with an internal control luciferase plasmid. The DSCR1-1L promoter activity in cell lysates was determined and normalized to control luciferase activity for equal transfection efficiency. As shown in Fig. 2B, the 1.4-kb DSCR1-1L promoter fragment induced high luciferase reporter activity (Fig. 2B, pD1L(-1450) versus blank, *, p < 0.001). We then created several deletion mutations of the D1L promoter as indicated in Fig. 2B. The data in Fig. 2B indicated that there was a major putative positive regulatory region ranging from -1450 bp to -1344 bp (p < 0.001).
FIGURE 2.
Functional analysis of human DSCR1 promoter in hemangioma EOMA cells. A, schematic representation of the different promoter for DSCR1 isoform 1 and isoform 4. B, serial deletions of the DSCR1-1L promoter reporters and their relative luciferase activity in EOMA cells. A major positive regulatory region from -1450 bp to -1344 bp was identified in the DSCR1-1L promoter. The data are expressed as the means ± S.E. of three separate experiments (n = 6, *, denotes p < 0.001).
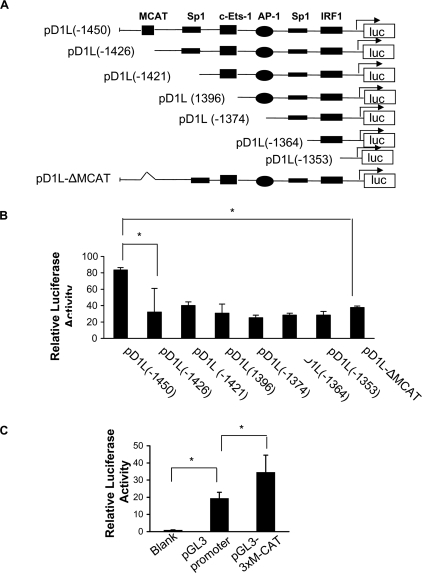
We further analyzed the region (-1450 bp ∼-1344 bp) and found 6 putative binding sites (Fig. 3A). Deletion constructs within the region of the D1L promoter were created (Fig. 3A) and their luciferase activity was measured in EOMA cells. Our results showed that the deletion of a 25-bp fragment containing a putative M-CAT site from -1450 to -1426 caused significant reduction of the DSCR1-1L promoter activity (Fig. 3B, left two bars, p < 0.001). To further identify which element within this 25-bp region was involved, we made another deletion construct lacking just the M-CAT site (pD1L-ΔM-CAT). Compared with pD1L(-1450), transfection of pD1L-ΔM-CAT construct into EOMA cells resulted in comparable reduction of DSCR1-1L promoter activity (Fig. 3B, *, p < 0.001) suggesting that the M-CAT site mediates the effect. To further confirm the functionality of the M-CAT site, we cloned the three copies of the M-CAT site upstream of a promoter luciferase vector (pGL3-promoter) that contained the SV40 basal promoter. Promoter activity analysis showed that the M-CAT site (pGL3–3xM-CAT) significantly increased the SV40 basal promoter activity (Fig. 3C, *, p < 0.001).
FIGURE 3.
Identification of an M-CAT-positive regulation site. A, diagram of serial shortening of the positive region I in the D1L promoter and the M-CAT deletion construct (ΔM-CAT). B, relative promoter activity of these deletion construct mutants in EOMA cells (n = 6, *, denotes p < 0.001). C, activity of the M-CAT reporter construct in EOMA cells (n = 6, *, denotes p < 0.001). The data are representative of three independent experiments.
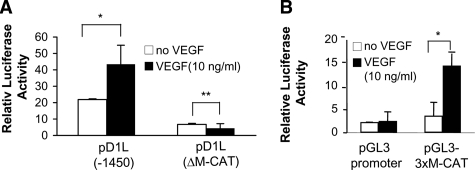
Requirement of the M-CAT Site in the DSCR1-1L Promoter for VEGF-A165-induced DSCR1-1L Expression in HUVEC—We then tested whether the M-CAT site was required for VEGF-A165-stimulated DSCR1-1L promoter activity in HUVEC. Serum-starved HUVEC were transfected with the full-length DSCR1-1L promoter pD1L(-1450), M-CAT deletion mutant (pD1L-ΔM-CAT), 3xM-CAT in promoter vector (pGL3–3xM-CAT), and control promoter vector (pGL3-Promoter) and then stimulated with VEGF-A165 for 6 h. As shown in Fig. 4A, VEGF stimulated about 2-fold of DSCR1-1L promoter activity (Fig. 4A, *, p < 0.001). However, this VEGF-A165-induced promoter activity was completely lost in the ΔM-CAT construct (Fig. 4A, **, p > 0.05). The reporter activity driven by the 3xM-CAT site was significantly increased in response to VEGF-A165 stimulation (Fig. 4B, *, p < 0.001).
FIGURE 4.
Requirement of the M-CAT element for VEGF-A165-induced DSCR1-1L promoter activity. A and B, relative promoter activity as indicated was measured in serum-starved HUVEC with or without VEGF-A165-stimulation for 6 h (n = 6, * denotes p < 0.001, ** denotes p > 0.05). The data represent three independent experiments.
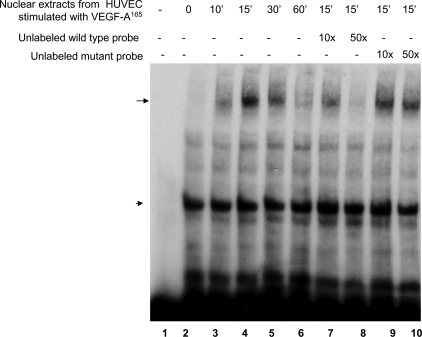
VEGF-A165 Induces the Formation of a Nuclear Complex with the M-CAT Probe—We further tested whether M-CAT associated with nuclear proteins in HUVEC with or without VEGF-A165 stimulation. Nuclear extracts were isolated from serum-starved HUVEC stimulated with VEGF-A165 for 0, 10′, 15′, 30′, and 60′ and subjected to DNA electrolytic mobility gel shift assay (EMSA) with biotin-labeled 3xM-CAT oligonucleotide probe. As shown in Fig. 5, there was one DNA-associated complex in the absence of VEGF-A165 stimulation (Fig. 5, lane 2, arrowhead). After VEGF-A165 stimulation, a new DNA protein band was detected (indicated by arrow) at 10′, 15′, and 30′ after stimulation (Fig. 5, lanes 3, 4, and 5). To further confirm the specificity of this protein-DNA interaction, we included 10-fold and 50-fold excess of unlabeled wild-type probes and mutant probes in the binding reaction. The data showed that excess of unlabeled wild-type probe competed off the shift of the binding complex (Fig. 5, lanes 7 and 8), but the mutant unlabeled probe did not (Fig. 5, lanes 9 and 10), indicating that the binding was specific. Similar specific nuclear-DNA interaction complex was detected in EOMA cells (data not shown).
FIGURE 5.
VEGF-A165 induced the formation of a transcriptional complex with M-CAT cis-element in HUVEC. HUVEC were stimulated with VEGF-A165 for 0, 10′, 15′, 30′, and 60′. Nuclear extracts were used for EMSA assay using a biotin-labeled DNA oligo probe corresponding to the M-CAT site of the DSCR1-1L promoter. Specific DNA-protein complex induced by VEGF-A165 was indicated by a long arrow and nonspecific shifted band by a short arrow. 10-fold and 50-fold excess of unlabeled wild-type (lanes 7 and 8) or mutant probes (lanes 9 and 10) were included in the reaction mixtures with nuclear extracts from HUVEC stimulated by VEGF-A165 for 15 min. The VEGF-A165-induced shifted band was titrated off by an excess of unlabeled wild-type probes, but not unlabeled mutant probes. The data represent three independent experiments.
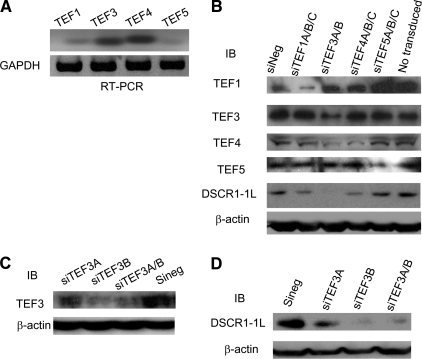
Requirement of TEF3, not Other TEF Family Members, for VEGF-A165-induced DSCR1-1L Expression—M-CAT (muscle-specific CAT element, 5′-CATTCCT) is a highly conserved sequence bound by TEF family proteins (33). There are 4 members of this family in human, which share a nearly identical N-terminal TEA/ATTS DNA binding domain (34, 35). The requirement for TEA/ATTS family transcription factors is conserved throughout eukaryotic development. However, it was not known whether TEF family members played any role in VEGF-A165 signaling. To identify which TEF family members played a role in VEGF-A165-stimulated DSCR1-1L promoter activity, we first examined, which TEF family members were expressed in HUVEC by RT-PCR with RNA isolated from HUVEC. As shown in Fig. 6A, all of four TEF family members were expressed in HUVEC (Fig. 6A).
FIGURE 6.
Requirement of TEF3, not other TEF1 family members, for VEGF-A165-induced DSCR1-1L expression. A, expression of TEF1, TEF3, TEF4, and TEF5 mRNAs in HUVEC was detected by RT-PCR with the specific primers for TEF1, TEF3, TEF4, and TEF5, respectively. B, HUVEC were transduced with siTEF1A/B/C (siTEF1A, siTEF1B, and siTEF1C), siTEF3A/B (siTEF3A and siTEF3B), siTEF4A/B/C (siTEF4A, siTEF4B, and siTEF4C), siTEF5A/B/C (siTEF5A, siTEF5B, and siTEF5C) or siNeg as a control. Cellular extracts were analyzed by immunoblotting with antibodies against TEF1, TEF3, TEF4, TEG5, DSCR1-1L, and β-actin as the protein equal-loading control. C and D, HUVEC were transduced with siTEF3A siTEF3B individually or in combination. Cellular extracts were analyzed by immunoblotting with antibodies against TEF3 (C) and DSCR1-1L (D). Bottom panels are protein equal-loading controls. The data represent three independent experiments.
Because all of the TEF family members were expressed in HUVEC, we developed retroviral vector expressing at least 2 siRNAs for each TEF family members: TEF1 (siTEF1A, siTEF1B, siTEF1C), TEF3 (siTEF3A, siTEF3B), TEF4 (siTEF4A, siTEF4B, siTEF4C), TEF5 (siTEF5A, siTEF5B, and siTEF5C). We first verified the specificity of these siRNAs for their respective TEF family members and then determined their effect on DSCR1-1L expression. Cellular extracts from HUVEC transduced without or with the siRNA in combination, siTEF1(A/B/C), siTEF3(A/B), siTEF4(A/B/C), siTEF5(A/B/C), and control siRNA (siNeg) were subjected to immunoblotting analysis with antibodies against TEF1, TEF3, TEF4, and TEF5, respectively. As shown in Fig. 6B, the siRNAs in combination specifically inhibited the expression of their corresponding TEF members, respectively, except that siTEF4(A/B/C) slightly inhibited the expression of TEF3 (Fig. 6B, top 4 panels). The same cellular extracts were subjected to immunoblotting analysis with an antibody against DSCR1-1L. We found that it was siTEF3(A/B), not others, that significantly inhibited DSCR1-1L expression (Fig. 6B panel 5). siTEF4(A/B/C) slightly inhibited the expression of DSCR1-1L. The bottom panel showed the protein equal-loading control with an antibody against β-actin (Fig. 6B, bottom panel).
We then tested the efficiency of each siTEF3s on DSCR1-1L expression. HUVEC were transduced with siTEF3A, siTEF3B, siTEF3A/B, and negative siRNA (siNeg), respectively. Cellular extracts were subjected to immunoblotting analysis with an antibody against TEF3. As shown in Fig. 6C, expression of siTEF3A partially inhibited the expression of TEF3, while expression of siTEF3B alone or in combination with siTEF3A almost completely inhibited the expression of TEF3 (Fig. 6C, top panel). The same cellular extracts were immunoblotted with an antibody against DSCR1-1L. As expected, expression of siTEF3A partially inhibited the expression of DSCR1-1L, while expression of siTERF3B alone or in combination with siTERF3A almost completely inhibited the expression of DSCR1-1L (Fig. 6D, top panel).
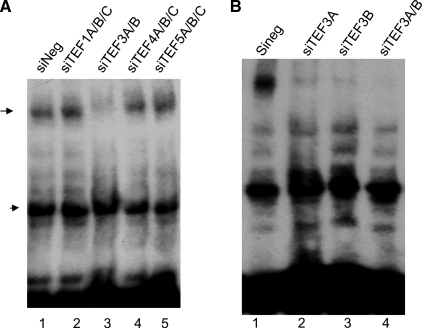
We then went onto to determine whether TEF3 was required for VEGF-A165-induced the formation of the M-CAT transcriptional complex. Nuclear extracts from VEGF-A165-stimulated HUVEC that were transduced without or with TEF siRNAs as indicated were subjected to EMSA. As shown in Fig. 7A, the DNA-protein complex induced by VEGF-A165 was almost completely inhibited by expression of siTEF3(A/B), not significantly by TEF1, TEF4, and TEF5 siRNAs (Fig. 7A, band indicated by arrow). Fig. 7B also indicated that expression of siTEF3A and siTEF3B individually greatly inhibited the VEGF-A165-induced nuclear protein interaction with M-CAT element (Fig. 7B).
FIGURE 7.
Requirement of TEF3, not other TEF1 family members, for VEGF-A165-induced M-CAT nuclear complex. A and B, HUVEC were transduced with siTEF1A/B/C (siTEF1A, siTEF1B, and siTEF1C), siTEF3A/B (siTEF3A and siTEF3B), siTEF4A/B/C (siTEF4A, siTEF4B, and siTEF4C), siTEF5A/B/C (siTEF5A, siTEF5B, and siTEF5C) or siNeg as control (A), with siTEF3A siTEF3B individually or in combination (B). After serum starvation, cells were stimulated with VEGF-A165 for 15 min. Nuclear extracts were subjected to DNA mobility gel shift assays. The data represent three independent experiments.
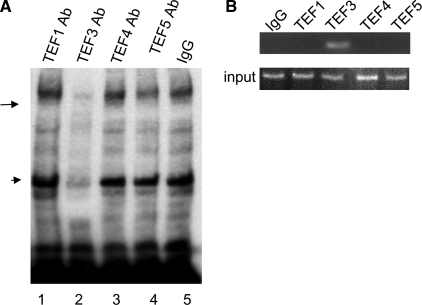
TEF3 Directly Interacted with M-CAT Cis-element in Vitro and in Vivo—To further confirm that TEF3 may be the major nuclear protein in the M-CAT-nuclear protein complex induced by VEGF-A165, we did antibody supershift assay by addition of TEF3 antibody in the DNA binding reaction. We found that TEF3 antibody, not the control IgG, nor antibodies against any other TEF family members supershifted the VEGF-A165-induced M-CAT protein complex (Fig. 8A, arrow). Surprisingly, the TEF3 antibody also supershifted the un-stimulated protein-DNA complex, suggesting an unknown closely related protein other than those known TEF family members in HUVEC cross-reacted with TEF3 antibody (Fig. 8A, band indicated by arrowhead).
FIGURE 8.
TEF3 associated with M-CAT cis element in vitro and in vivo. A, EMSA was performed as above with antibodies against TEF1, TEF3, TEF4, and TEF5, respectively. TEF3 antibody, not other antibodies, supershifted the M-CAT nuclear complex (lane 3). B, CHIP assay. HUVEC extracts were immunoprecipitated with antibodies against TEF1, TEF3, TEF4, TEF5, and IgG as a control. The immunoprecipitates were subjected to PCR to detect the associated DSCR1-1L promoter element (top panel). The bottom is the input DNA without immunoprecipitation. The data represent three independent experiments.
To determine whether TEF3, not other TEF1 family members, actually binds to the M-CAT site in the DSCR1-1L promoter in vivo, we performed CHIP assays. HUVEC were treated with VEGF-A165 as above, cross-linked with formaldehyde and then subjected to immunoprecipitation (IP) with antibodies against TEF1, TEF3 TEF4, TEF5, or IgG as a control. The chromosomal DNA in the immunoprecipitated complexes was extracts and subjected to PCR assay. The two primers were designed according to the sequences to the putative M-CAT binding site on the DSCR1-1 promoter, respectively. As shown in Fig. 8B, a PCR-amplified band was detected in the immunoprecipitated complex with an antibody against TEF3, not with other antibodies, nor control IgG (Fig. 8B). Our data clearly indicated that TEF3 directly interacted with the M-CAT cis-element in the DSCR1-1L promoter to mediate VEGF-A165-induced DSCR1-1L expression.
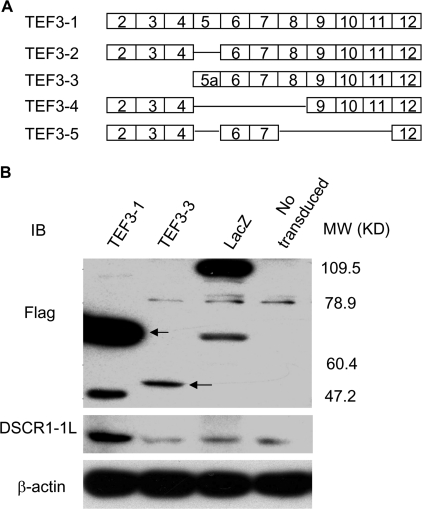
TEF3 Isoform 1 Induces DSCR1-1L Expression—So far, five TEF3 isoforms have been reported in different cells (Fig. 9A) (36–39). However, it was not known which isoform(s) are expressed in HUVEC. We cloned the TEF3 isoforms from the RNA isolated from HUVEC by RT-PCR using the primers in the 5′-untranslated region and 3′-untranslated region of TEF3 cDNA, respectively. By DNA sequencing analysis, we obtained the TEF3 isoform 1 (TEF3-1) and TEF3-isoform 3 (TEF3-3) cDNAs. The cDNA fragments were in-frame fused to the FLAG tag in the N terminus and cloned into a retroviral expression vector, which gave out almost 100% infection yield (26). The retroviruses expressing these two genes were used to infect HUVEC. As shown in Fig. 9B, both FLAG-TEF3-1 and FLAG-TEF3-3 were overexpressed in HUVEC (Fig. 9B, top panel). The same samples were analyzed by immunoblotting with the antibody against DSCR1-1L. Our data indicated that TEF3-1, not TEF3-3, up-regulated DSCR1-1L expression (Fig. 9B, middle panel).
FIGURE 9.
Induction of DSCR1-1L expression by overexpression of TEF3 isoform 1, not isoform 3. A, schematic representation of TEF3 isoforms. B, cellular extracts from HUVEC that were transduced without or with LacZ, TEF3-3, and TEF3-1 were analyzed by immunoblotting with antibodies against FLAG (top panel), DSCR1-1L (middle panel), and β-actin as a protein equal-loading control (bottom panel). The data represent three independent experiments.
DISCUSSION
Extensive studies have shown that angiogenesis and its associated microvessel permeability play critical roles in tumor growth. VEGF-A165 is likely the most important cytokine involved in tumor angiogenesis. Avastin, an antibody that blocks VEGF-A165, has shown therapeutic effect in several types of human cancers. However, Avastin has a number of side effects. Therefore, it is desirable to study the downstream molecules that play a specific role in tumor angiogenesis, but not in normal vascular structure. Our previous studies indicated that Down syndrome candidate region 1 isoform 1L was up-regulated by VEGF-A165 in cultured endothelial cells and in several types of mouse angiogenesis model in vivo (22). VEGF-A165-induced endothelial cell proliferation and angiogenesis was greatly inhibited by knocking down the expression of DSCR1-1L with DSCR1-1L-specific siRNA (22). More recently, Ryeom et al. (23) reported that tumor growth was greatly repressed in DSCR1 knock-out mice lacking all DSCR1 isoforms, indicating the complexity of this family of protein in tumorigenesis. Excitingly, we found recently that DSCR1-1L was specifically expressed in tumor vascular structure, not in normal vascular structure of human ovarian cancer tissues (22). Our present work further showed that DSCR1-1L was expressed in typical enlarged tumor microvessels of human kidney carcinoma not in tumor epithelial cells. Therefore, DSCR1-1L may represent a promising and specific target for tumor anti-angiogenic therapy.
Because of the unique feature of DSCR1-1L in tumor angiogenesis, elucidation of the molecular mechanism responsible for DSCR1-1L up-regulation in tumor microvessels became very important. Because the study of the unique DSCR1 isoform DCSR1-1L is very limited, not to mention its gene regulation, our present work became the first one to characterize how this gene is regulated in any conditions. Unlike DSCR1-4, DSCR1-1L is not likely to be induced through the Cn-NFAT pathway (13–16) as the DNA sequence upstream of exon 1 lacks any putative NFAT binding sites (13–16). Cloning and analysis of the 1.4-kb upstream region of DSCR1-1L gene showed that this DSCR1-1L promoter drove strong luciferase reporter activity in endothelial cells. Further analysis of this region led to identification of a unique family of transcriptional factors TEF, which is responsible for VEGF-A165-induced DSCR1-1L gene transcription based on a number of evidence. Deletion of the putative TEF binding M-CAT site 1425-bp upstream of the DSCR1-1L translation start site abolishes most of DSCR1-1L promoter activity induced by VEGF-A165. Further, a reporter construct containing three copies of this M-CAT site can be functionally activated by VEGF-A165. Therefore this M-CAT reporter plasmid can be used to measure transcriptional activity of the TEF family of transcriptional factors.
M-CAT (muscle-specific CAT element) sequence (5′-CATTCCT) is a highly conserved sequence bound by transcription enhancer factor-1 (TEF1) family proteins (33). The human transcriptional enhancer factor (TEF) family includes TEF1 (NTEF-1/Tead1) (40), TEF3 (RTEF-1/ETFR-2/FR-19/Tead4) (38, 39, 41–43), TEF4 (ETF/Tead2) (38, 42), and TEF5 (DTEF-1/ETFR-1/Tead3) (42, 44, 45). The TEFs share a highly conserved 68-amino acid TEA/ATTS DNA binding domain in N-terminal, which binds to SV40 GT-IIC (GGAATG), SphI (AGTATG), SphII (AGCATG), and muscle-specific M-CAT (GGTATG) enhancers. The tissue distribution of TEF1 family members has been examined by a number of studies (39, 40, 42, 43, 45, 46). Indeed, TEF1, TEF3, and TEF5 are widely expressed in multiple tissues including the skeletal muscle, pancreas, placenta, lung, and heart. In contrast to these three factors, TEF4 is selectively expressed in a subset of embryonic tissues including the cerebellum, testis, and distal portion of the forelimb and hindlimb buds as well as the tail bud, but it is essentially absent from the adult tissues. Our data clearly indicated that all of the four TEF1 family members were expressed in HUVEC. TEF1 was originally identified to transactivate SV40 large T antigen (47). Later, it was found to regulate the development of cardiac, skeletal, and smooth muscles (37, 41, 48). Consistent with this, TEF1 knock-out mice have defective heart development and embryonic lethal phenotypes in mice (49). TEF3 (RTEF-1, TEAD-4) was cloned from human heart cDNA library and had similar tissue distribution to TEF1 (TEAD-1) with highest abundance in skeletal muscle and lower levels in pancreas, placenta, and heart (39). Most recently, TEF3 was also found to be up-regulated in hypoxic endothelial cells to regulate VEGF promoter (25). TEF3 is specifically expressed in skeletal muscle precursors and implicated in muscle-specific gene expression (38). TEF4 (TEAD-2) appears to be involved in central nervous system development (38). TEF5 (ETFR-1, DTEF-1) is expressed in placenta and regulates human somatomammotropin gene enhancer and influences fetal development (38, 45). However, nothing was known about their roles in VEGF-A165 signaling. In our studies, we found for the first time that it was TEF3, not other TEF1 family members that is mainly required for the formation of M-CAT nuclear protein complex induced by VEGF-A165 and for VEGF-A165-induced DSCR1-1L expression. Further studies indicated that TEF3 directly interacted with M-CAT in vitro and in vivo. The function of other TEF1 family members in HUVEC remained to be elucidated. Our data that TEF4 siRNAs slightly inhibited the expression of TEF3 and DSCR1-1L may be due to two possibilities. TEF4 regulated the expression of TEF3 and hence regulated the expression of DSCR1-1L because we did not detect TEF4 interacted with DSCR1-1L in antibody supershift assay, not in CHIP assay. These data indicated that, even if TEF4 regulated DSCR1-1L expression, it was regulated indirectly. The other possibility is the nonspecific effect of the TEF4 siRNAs.
Transcripts of the TEF3 gene were first identified in chicken tissue and demonstrated to be enriched in cardiac and skeletal muscle (37). Full-length human homolog of TEF3 was identified from a heart cDNA library (39, 50). Northern blot analysis of human tissues indicated the highest levels of expression in skeletal muscle and pancreas, with lower levels in the heart, kidney, and placenta, whereas the message was not detected in the liver, lung, or brain (39). Northern blot analysis of the mouse homolog of TEF3 indicated a different tissue expression pattern when compared with that in humans. Adult mouse lung tissue expresses the highest level, with very low levels in kidney, heart, and skeletal muscle and undetectable amounts in liver, thymus, spleen, and brain, whereas TEF3 message is abundant in mouse embryonic skeletal muscle (43). TEF3 binds to the myocyte-specific CAT (M-CAT) cis DNA elements to regulate the expression of muscle-specific genes, and it requires muscle-specific cofactors for full transcriptional activation (51). Analysis of TEF3 indicates that it contains 11 exons. So far five isoforms were identified to be expressed in muscle cells or human ocular vascular endothelial cells and murine retina (36). TEF3-1 (1305 bp) and TEF3-4 (936 bp) are expressed under normoxic conditions, while TEF3-5 (447 bp) is induced by hypoxic condition in human retina vascular endothelial cells (RVECs) (36). TEF3-1, TEF3-4, and TEF3-5 induced VEGF promoter activity with 4-, 3-, and 12-fold, respectively. Analysis with deletion promoter constructs showed that all isoforms required the presence of Sp1 elements for efficient activation and that the hypoxia response element (HRE) was not essential for enhancement (25). However, prior to our studies, no evidence suggested that TEF3 was involved in the angiogenic effect of VEGF-A165. We cloned two TEF3 isoforms (TEF3 isoform 1 and TEF3 isoform 3) from HUVEC RNA and overexpressed them in HUVEC. Our results indicated that full-length TEF3 isoform 1, not truncated TEF3 isoform 3, regulated the expression of DSCR1-1L.
In summary, we have identified a novel transcriptional mechanism that mediates VEGF-A165-induced DSCR1-1L expression, in which VEGF-A165 induced interaction of TEF3 with the M-CAT cis element of DSCR1-1L promoter. Based on our previous report showing that DSCR1-1L may act as a proangiogenic factor, we assume that TEF3 may also function to promote angiogenic response. Future study will aim to elucidate the signaling pathway by which VEGF-A165 induces the activation of TEF3 and also to investigate the role of TEF3-1 and other TEF3 isoforms in tumor angiogenesis.
This work was supported by ACS Grant RSG-05-118-01-CSM. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement”in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: DSCR, Down syndrome candidate region; VEGF-A165, vascular endothelial growth factor-A165; M-CAT, muscle-specific CAT element; TEF, transcription enhancer factor; siTEF, TEF-specific siRNA; siNeg, control siRNA; FTEF, FLAG-fused TEF; HUVEC, human umbilical vein endothelial cells; HMVEC, human microdermal vascular endothelial cells; EOMA, hemangioma cells; EMSA, electrophoresis mobility shift assays; PBS, phosphate-buffered saline; UTR, untranslated region; ChiP, chromatin immunoprecipitation assay; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
References
- 1.Hassold, T. J., and Jacobs, P. A. (1984) Annu. Rev. Genet. 18 69-97 [DOI] [PubMed] [Google Scholar]
- 2.Fuentes, J. J., Pritchard, M. A., and Estivill, X. (1997) Genomics 44 358-361 [DOI] [PubMed] [Google Scholar]
- 3.Fuentes, J. J., Pritchard, M. A., Planas, A. M., Bosch, A., Ferrer, I., and Estivill, X. (1995) Hum. Mol. Genet. 4 1935-1944 [DOI] [PubMed] [Google Scholar]
- 4.Rothermel, B., Vega, R. B., Yang, J., Wu, H., Bassel-Duby, R., and Williams, R. S. (2000) J. Biol. Chem. 275 8719-8725 [DOI] [PubMed] [Google Scholar]
- 5.Crawford, D. R., Leahy, K. P., Abramova, N., Lan, L., Wang, Y., and Davies, K. J. (1997) Arch. Biochem. Biophys. 342 6-12 [DOI] [PubMed] [Google Scholar]
- 6.Leahy, K. P., Davies, K. J., Dull, M., Kort, J. J., Lawrence, K. W., and Crawford, D. R. (1999) Arch. Biochem. Biophys. 368 67-74 [DOI] [PubMed] [Google Scholar]
- 7.Ermak, G., Morgan, T. E., and Davies, K. J. (2001) J. Biol. Chem. 276 38787-38794 [DOI] [PubMed] [Google Scholar]
- 8.Gorlach, J., Fox, D. S., Cutler, N. S., Cox, G. M., Perfect, J. R., and Heitman, J. (2000) EMBO J. 19 3618-3629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hasle, H. (2001) Lancet Oncol. 2 429-436 [DOI] [PubMed] [Google Scholar]
- 10.Smucker, J. D., Roth, L. M., Sutton, G. P., and Hurteau, J. A. (1999) Gynecol. Oncol. 74 512-514 [DOI] [PubMed] [Google Scholar]
- 11.Faruqi, S. A., Noumoff, M. J., Deger, R. B., Jalal, S. M., and Antoniades, K. (2002) Cancer Genet. Cytogenet. 138 165-168 [DOI] [PubMed] [Google Scholar]
- 12.Verhest, A., Nedoszytko, B., Noel, J., Simon, P., and Limon, J. (1991) Fourth International Workshop on Chromosomes in solid Tumors, Abstract B24
- 13.Liu, D., Jia, H., Holmes, D. I., Stannard, A., and Zachary, I. (2003) Arterioscler. Thromb. Vasc. Biol. 23 2002-2007 [DOI] [PubMed] [Google Scholar]
- 14.Minami, T., Horiuchi, K., Miura, M., Abid, R., Takabe, W., Kohro, T., Ge, X., Aburatani, H., Hamakubo, T., Kodama, T., and Aird, W. C. (2004) J. Biol. Chem. 279 50537-50554 [DOI] [PubMed] [Google Scholar]
- 15.Yao, Y. G., and Duh, E. J. (2004) Biochem. Biophys. Res. Commun. 321 648-656 [DOI] [PubMed] [Google Scholar]
- 16.Hesser, B. A., Liang, X. H., Camenisch, G., Yang, S., Lewin, D., Scheller, R., Ferrara, N., and Gerber, H. P. (2004) Blood 104 149-158 [DOI] [PubMed] [Google Scholar]
- 17.Ermak, G., Harris, C. D., and Davies, K. J. (2002) Faseb. J. 16 814-824 [DOI] [PubMed] [Google Scholar]
- 18.Genesca, L., Aubareda, A., Fuentes, J. J., Estivill, X., De La Luna, S., and Perez-Riba, M. (2003) Biochem. J. 374 567-575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Molkentin, J. D., Lu, J. R., Antos, C. L., Markham, B., Richardson, J., Robbins, J., Grant, S. R., and Olson, E. N. (1998) Cell 93 215-228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Musaro, A., McCullagh, K. J., Naya, F. J., Olson, E. N., and Rosenthal, N. (1999) Nature 400 581-585 [DOI] [PubMed] [Google Scholar]
- 21.Semsarian, C., Wu, M. J., Ju, Y. K., Marciniec, T., Yeoh, T., Allen, D. G., Harvey, R. P., and Graham, R. M. (1999) Nature 400 576-581 [DOI] [PubMed] [Google Scholar]
- 22.Qin, L., Zhao, D., Liu, X., Nagy, J. A., Hoang, M. V., Brown, L. F., Dvorak, H. F., and Zeng, H. (2006) Mol. Cancer Res. 4 811-820 [DOI] [PubMed] [Google Scholar]
- 23.Ryeom, S., Baek, K. H., Rioth, M. J., Lynch, R. C., Zaslavsky, A., Birsner, A., Yoon, S. S., and McKeon, F. (2008) Cancer Cell 13 420-431 [DOI] [PubMed] [Google Scholar]
- 24.Fuentes, J. J., Genesca, L., Kingsbury, T. J., Cunningham, K. W., Perez-Riba, M., Estivill, X., and de la Luna, S. (2000) Hum. Mol. Genet 9 1681-1690 [DOI] [PubMed] [Google Scholar]
- 25.Shie, J. L., Wu, G., Wu, J., Liu, F. F., Laham, R. J., Oettgen, P., and Li, J. (2004) J. Biol. Chem. 279 25010-25016 [DOI] [PubMed] [Google Scholar]
- 26.Zeng, H., Dvorak, H. F., and Mukhopadhyay, D. (2001) J. Biol. Chem. 276 26969-26979 [DOI] [PubMed] [Google Scholar]
- 27.Ilan, N., Tucker, A., and Madri, J. A. (2003) Lab. Investig. 83 1105-1115 [DOI] [PubMed] [Google Scholar]
- 28.Chan, B., Greenan, G., McKeon, F., and Ellenberger, T. (2005) Proc. Natl. Acad. Sci. U. S. A 102 13075-13080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cho, Y. J., Abe, M., Kim, S. Y., and Sato, Y. (2005) Arch. Biochem. Biophys. 439 121-128 [DOI] [PubMed] [Google Scholar]
- 30.Iizuka, M., Abe, M., Shiiba, K., Sasaki, I., and Sato, Y. (2004) J. Vasc. Res. 41 334-344 [DOI] [PubMed] [Google Scholar]
- 31.Mann, K. M., Ray, J. L., Moon, E. S., Sass, K. M., and Benson, M. R. (2004) J. Cell Biol. 165 483-491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Minami, T., Horiuchi, K., Miura, M., Abid, M. R., Takabe, W., Noguchi, N., Kohro, T., Ge, X., Aburatani, H., Hamakubo, T., Kodama, T., and Aird, W. C. (2004) J. Biol. Chem. 279 50537-50554 [DOI] [PubMed] [Google Scholar]
- 33.Kaneko, K. J., and DePamphilis, M. L. (1998) Dev. Genet. 22 43-55 [DOI] [PubMed] [Google Scholar]
- 34.Andrianopoulos, A., and Timberlake, W. E. (1991) Plant Cell 3 747-748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burglin, T. R. (1991) Cell 66 11-12 [DOI] [PubMed] [Google Scholar]
- 36.Appukuttan, B., McFarland, T. J., Davies, M. H., Atchaneeyasakul, L. O., Zhang, Y., Babra, B., Pan, Y., Rosenbaum, J. T., Acott, T., Powers, M. R., and Stout, J. T. (2007) Investig. Ophthalmol. Vis. Sci. 48 3775-3782 [DOI] [PubMed] [Google Scholar]
- 37.Farrance, I. K., and Ordahl, C. P. (1996) J. Biol. Chem. 271 8266-8274 [DOI] [PubMed] [Google Scholar]
- 38.Jacquemin, P., Hwang, J. J., Martial, J. A., Dolle, P., and Davidson, I. (1996) J. Biol. Chem. 271 21775-21785 [DOI] [PubMed] [Google Scholar]
- 39.Stewart, A. F., Richard, C. W., 3rd, Suzow, J., Stephan, D., Weremowicz, S., Morton, C. C., and Adra, C. N. (1996) Genomics 37 68-76 [DOI] [PubMed] [Google Scholar]
- 40.Xiao, J. H., Davidson, I., Matthes, H., Garnier, J. M., and Chambon, P. (1991) Cell 65 551-568 [DOI] [PubMed] [Google Scholar]
- 41.Hsu, D. K., Guo, Y., Alberts, G. F., Copeland, N. G., Gilbert, D. J., Jenkins, N. A., Peifley, K. A., and Winkles, J. A. (1996) J. Biol. Chem. 271 13786-13795 [DOI] [PubMed] [Google Scholar]
- 42.Yasunami, M., Suzuki, K., and Ohkubo, H. (1996) Biochem. Biophys. Res. Commun. 228 365-370 [DOI] [PubMed] [Google Scholar]
- 43.Yockey, C. E., Smith, G., Izumo, S., and Shimizu, N. (1996) J. Biol. Chem. 271 3727-3736 [DOI] [PubMed] [Google Scholar]
- 44.Azakie, A., Larkin, S. B., Farrance, I. K., Grenningloh, G., and Ordahl, C. P. (1996) J. Biol. Chem. 271 8260-8265 [DOI] [PubMed] [Google Scholar]
- 45.Jiang, S. W., Wu, K., and Eberhardt, N. L. (1999) Mol. Endocrinol. 13 879-889 [DOI] [PubMed] [Google Scholar]
- 46.Azakie, A., Lamont, L., Fineman, J. R., and He, Y. (2005) Am. J. Physiol. Cell Physiol. 289 C1522-C1534 [DOI] [PubMed] [Google Scholar]
- 47.Davidson, I., Xiao, J. H., Rosales, R., Staub, A., and Chambon, P. (1988) Cell 54 931-942 [DOI] [PubMed] [Google Scholar]
- 48.Mar, J. H., and Ordahl, C. P. (1990) Mol. Cell. Biol. 10 4271-4283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen, J. X., Lawrence, M. L., Cunningham, G., Christman, B. W., and Meyrick, B. (2004) J. Appl. Physiol. 96 612-620 [DOI] [PubMed] [Google Scholar]
- 50.Frigerio, J. M., Berthezene, P., Garrido, P., Ortiz, E., Barthellemy, S., Vasseur, S., Sastre, B., Seleznieff, I., Dagorn, J. C., and Iovanna, J. L. (1995) Hum. Mol. Genet. 4 37-43 [DOI] [PubMed] [Google Scholar]
- 51.Stewart, A. F., Suzow, J., Kubota, T., Ueyama, T., and Chen, H. H. (1998) Circ. Res. 83 43-49 [DOI] [PubMed] [Google Scholar]