Abstract
The balance between differentiation signals and signals maintaining the undifferentiated state of embryonic cells ensures proper formation of germ layers. The nodal/activin pathway represents one of the major signaling chains responsible for the differentiation of embryonic cells into mesodermal and endodermal germ layers, while Oct4 is one of the major players in the maintenance of an undifferentiated state. Here we show that Oct25, an Oct4 homologue in Xenopus, antagonizes the activity of nodal/activin signaling by inhibiting the transcription of its target genes, Gsc and Mix2. The inhibitory effect is achieved by forming repression complexes on the promoters of Gsc and Mix2 between Oct25 and the signal transducers of the nodal/activin pathway, WBSCR11, FAST1, and Smad2. We have analyzed the significance of the Oct binding site for its inhibitory effect within the Gsc promoter. Albeit VP16-Oct25 fusion protein demonstrated a stimulating effect and EVE-Oct25 revealed a repression effect on an artificial reporter that is composed of eight repeats of Oct binding motifs, both fusions, like wild-type Oct25, inhibited mesendoderm formation and the activity of Gsc and Mix2 promoters. These results suggest that the regulatory effect of Oct25 on the expression of Gsc and Mix2 is mediated by specific protein/protein interactions. Furthermore, we demonstrate that histone deacetylase activities are not required for the inhibitory effect of Oct25. Our results provide a novel view in that Oct25 controls the nodal/activin pathway and thus maintains the undifferentiated state of embryonic cells in preventing them from premature differentiation.
It has been well established that nodal/activin, two members of the transforming growth factor-β (TGF-β)2 superfamily of secreted proteins, are fundamental to the formation of mesoderm and endoderm in pre-gastrula and gastrula embryos of vertebrate animals. Incubation of animal explants from Xenopus laevis early blastula embryos with activin A protein leads to formation of a full spectrum of mesoderm- and endoderm-derived tissues in a dose-dependent fashion (1–3). Six different nodal-related proteins exist in Xenopus, Xnr1–6. Except for Xnr3 that is involved in neural induction, all other Xnrs display potent mesendoderm inducing activities in explants or in whole embryos (4–6). Likewise, mouse nodal, zebrafish Cyclops and Squint can also induce mesoderm formation (7–11). In contrast, blocking nodal activity results in failure in mesendoderm formation and, consequently, the failure in establishment of body axis not only in Xenopus, but also in other vertebrates such as mouse and zebrafish (7). Besides its function in mesendoderm formation, nodal is involved in the specification of left-right body axis (12).
Nodal/activin induces mesendoderm formation via triggering a distinct signal transduction process. The first step of the signal cascade is the binding of nodal or activin ligand to type I serine-threonine kinase activin receptor ActRIB (ALK4) together with the type II activin receptors ActRII (ActRIIA or ActRIIB) (11). Specific for the nodal pathway is that, unlike activin, nodal needs additional EGF-CFC co-receptors to transmit the signal (13). Upon ligand interaction, the type II receptor activates type I receptor, which consequently phosphorylates receptor-specific Smad transducers, Smad2 or Smad3. Activated Smad2 or Smad3 forms a complex with the co-Smad, Smad4, and is translocated to the nucleus. Because the binding affinity of Smads to DNA is relatively weak, it is generally thought that additional DNA binding cofactors, either tissue-specific or stage-specific, are required to assemble a high affinity Smad complex on the enhancer sequences of target genes to stimulate transcription (7, 10, 11). During mesendoderm formation, the winged-helix transcription factor FAST1 (FoxH1) is one of the best-understood interaction partners for Smad2 to induce the transcription of genes like Goosecoid (Gsc) or Mix2 (11, 14). Gsc encodes an organizer-specific homeobox protein that can instruct ventral tissue to form dorsal structures when ectopically expressed (15, 16). Mix2, similar to its close homologue Mix1, defines the domains of endoderm and mesoderm formation in Xenopus pre-gastrula and gastrula stage embryos (17).
The mechanisms governing how these mesendodermal genes respond to nodal/activin have been quite intensively investigated. The Gsc promoter contains a distal element (DE) that is responsive to activin/nodal (18). In addition, there is a Wnt-responsive proximal element (PE) in the promoter, which is probably responsible for the maintenance of Gsc transcription (14, 19). Mechanistic studies demonstrated that FAST1 binds to the PE via a FAST1 binding motif, AATATACA, and another transcription factor, WBSCR11, interacts with the DE. FAST1 associates with Smad2 as well as WBSCR11 to form a large complex thereby inducing Gsc transcription (14). In the Mix2 promoter, a small region from -215 to -166 was shown to be an activin-response element (ARE) and therefore is critical for Mix2 induction (20, 21). In fact, there also exists a FAST1 binding site, TGTGTATT, and an adjacent Smad binding site, GTCT, within the ARE. Therefore, FAST1 and Smad2 form a complex on the ARE to induce Mix2 transcription.
While promotion of nodal/activin activity is a prerequisite for mesendoderm formation, its negative regulation is equally important for normal embryogenesis. To guarantee correct specification of mesendoderm, the activity of the nodal/activin signaling pathway should be precisely tuned so that it can fulfill its function within correct locations at desired time, and at a correct level during early embryogenesis. Several studies have shown that regulation can occur at each level of signal transfer, for instance extracellular inhibition of nodal/activin ligands, cytoplasmic inhibition of Smad transducers or nuclear inhibition of FAST1 or Smad2 (8, 10). In this study we present a novel mechanism in which the POU factor Oct25 (22) regulates the nuclear response to nodal/activin. Oct25 is a Xenopus homologue to mammalian Oct4, a transcription factor of the POU family subclass V (POU-V) (23). Oct4 is well known for its crucial function in the maintenance of pluripotency and self-renewal of embryonic stem cells and for its importance in reprogramming of somatic cells (24–27). In Xenopus ectodermal explants or embryos, overexpressed Oct25 inhibited nodal/activin activity in the induction of mesendoderm formation (28, 29). Here we show that Oct25 inhibits transcription of nodal/activin signaling target genes, Gsc and Mix2. Mechanistic analyses demonstrated that Oct25 formed regulatory complexes with FAST1, WBSCR11, and Smad2 on the promoters of Gsc or Mix2. The results suggest that Oct25 inhibits mesendoderm formation via blocking nodal/activin target gene transcription and hence provide important insight into the mechanisms how embryonic cells are kept in an undifferentiated state by POU-V pluripotency factors.
EXPERIMENTAL PROCEDURES
Embryos and Explants—Methods for culturing embryos, excision, and culture of animal caps, activin A treatment of animal caps were as described (28, 30).
Plasmid Construction—The coding region of Oct25 was PCR-amplified and ligated to the EcoRI/XbaI sites of pCS2+VP16-XWBSCR11 or pCS2+EVE-XWBSCR11 (14) after removal of the XWBSCR11 inserts to generate pCS2+VP16-Oct25 and pCS2+ EVE-Oct25. VP16 activation domain and EVE repression domain were generated from pCS2+VP16-XWBSCR11 or pCS2+EVE-XWBSCR11 separately and ligated to pCS2+ 4HAmcs vector3 after digestion with XbaI/EcoRI. The resulting plasmids were designated as pCS2+VP16mcs and pCS2+EVEmcs. GST-tagged constructs used in GST pull-down or EMSA assays, or HA-tagged constructs used in Co-IP assay were made by PCR-based strategy, the resulting plasmids were designated as pGEX-4T1-FAST1, pGEX-4T1-XWBSCR11, pGEX-4T1-Smad2, pCS2+XWBSCR11-HA, pCS2+Smad2-HA, or pCS2+FAST1-HA, respectively. For an analysis of the Oct binding site, eight catenated repeats of the canonical Oct binding site (in bold) (TGTTATGCAAATGGC)8 were ligated to the MluI/XhoI sites on pGL3-promoter vector (Promega) to generate the artificial luciferase reporter OctLuc8x. For Gsc and Mix2 promoter-luciferase reporter analysis, Gsc promoter -479/+3 (transcription start as +1) and -226/+3 were PCR-amplified from Gsc promoter reporter construct pOLUC(-1500) (17) and ligated to pGL3-basic vector (Promega) to generate GscLuc(-479) and GscLuc(-226). Mix2 promoter fragments -712/+13 (transcription start as +1), -457/+13 and -221/+13 were generated with PCR from Xenopus genomic DNA and ligated to pGL3-basic vector (Promega) to make reporter constructs Mix2Luc(-712), Mix2Luc(-457), and Mix2Luc(-221), respectively.
In Vitro RNA Transcription and Microinjection—After cutting Gsc plasmid with EcoRI and cutting pCS2+xMix2 with BamHI, antisense RNA probes for whole mount in situ hybridization to detect Gsc and Mix2 expression were transcribed with T7 RNA polymerase and then purified with RNeasy kit (Qiagen). Preparation of Xbra and Xsox17α antisense probes were described in Ref. 28. To prepare mRNAs used for overexpression, plasmids pCS2+Oct25, pCS2+FAST1, pCS2+Xnr1, pCS2+VP16mcs, and pCS2+EVEmcs were linearized with NotI and transcribed with Sp6 RNA polymerase; pCS2+VP16-Oct25, pCS2+EVE-Oct25, and pCS2+VP16-XWBSCR11 were linearized with SacII and transcribed with Sp6; pNRRX-Xnr5 was cut with XbaI and transcribed with T7, and pRNX-Smad2 was linearized with SfiI and transcribed with T3 polymerase. Transcription was performed using mMessage mMachine kits (Ambion), and transcripts were purified with RNeasy kit (Qiagen). The antisense morpholino oligo for Oct25 knockdown (Oct25MO) was as described (28). Xnr1 and Xnr5 mRNAs were injected at 100 pg each, Smad2, FAST1 and VP16-XWBSCR11 mRNAs were injected at 200 pg, and mRNAs for Oct25, VP16-Oct25, EVE-Oct25, VP16, and EVE were injected at 400 pg per embryo. Each reporter plasmid DNA was injected at 40 pg per embryo.
Whole Mount in Situ Hybridizations—Standard procedures were used (30).
Gene Expression Analysis using Real-time RT-PCR—The method for real-time RT-PCR and primers for Xbra, Xsox17α, Gsc, and H4 were as described (28, 31). The primers for Mix2 were forward: 5′-TCTTCCAAACAAACATGTACCCA-3′ and reverse: 5′-ACGGGACTCAGGGATGTAAATG-3′. Results are presented as histograms with relative units.
Luciferase Assays—Promoter reporter plasmid DNAs and mRNAs were injected into the animal pole, when caps were used, or injected into equatorial region, when whole embryos were used, of all blastomeres at the four-cell stage. Caps or whole embryos were collected at gastrula stage and the method for measuring luciferase activity was as described (31).
Cell Culture and Preparation of Cell Extracts—HEK293 cells (ATCC CRL 1573) were grown in Dulbecco's modified Eagle′s medium (Invitrogen) supplemented with 10% fetal calf serum. Cells were washed in phosphate buffered saline and suspended in 600 μl of ice-cold M-PER lysis reagent (Pierce) supplemented with protease inhibitors (Complete Mix, Roche Applied Science) and incubated on ice for 20 min. The lysates were cleared by centrifugation at 80,000 × g for 30 min. Extracts were used for Western blotting and immunoprecipitation experiments.
Coimmunoprecipitation (Co-IP) and Western Blotting— Co-IP was carried out using HEK-293 cell extracts 24 h after co-transfection with pCS2+Flag-Oct25 (31) alone or together with pCS2+Smad2-HA, pCS2+XWBSCR11-HA, or pCS2+ FAST1-HA, respectively. The HA-tagged proteins were precipitated using the Profound™ HA-Tag-IP/Co-IP-System (Pierce) according to the manufacturer's instructions. The eluates (each 20 μl) were resuspended in SDS-PAGE loading buffer and subjected to Western blotting, which was performed using standard procedures. The following antibodies were used: anti-Flag (M5, mouse monoclonal, Sigma), anti-HA (mouse monoclonal, MMS-101P, BaBCo). A peroxidase-conjugated sheep anti-mouse IgG (Amersham Biosciences) was used as secondary antibody. After washing, specific proteins were detected using an enhanced chemiluminescence system (GE Healthcare).
Chromatin Immunoprecipitation (ChIP)—ChIP was performed as described (31). One pair of primers was used for amplifying Gsc promoter region containing the Oct binding site, Gsc-A forward: 5′-CCTGTGGATTCCCTGTACCA-3′ (-1089/-1070) and Gsc-A reverse: 5′-GGCCTGGGATAATTTTTGGA-3′ (-837/-856); the other pair was used for amplifying the region covering the DE and PE, Gsc-B forward: 5′-TAATGTCCCATCACGCTCAA-3′ (-285/-266) and Gsc-B reverse: 5′-AAGCACAGCAGCTCCACTCT-3′ (-54/-73). Primers for detecting Mix2 promoter region that contains the ARE were: Mix2 forward 5′-GCCACAGTTCTGACAAAGCA-3′ (-318/-299) and Mix2 reverse 5′-CTCAGACCTGGCCTACAGGA-3′ (-74/-93).
GST Pull-down Assays—Preparation of GST fusion proteins and GST pull-down assays were performed in the same way as described previously (31, 32).
Electrophoresis Mobility Shift Assays (EMSA)—EMSAs were carried out essentially as described (31, 32).
TSA Treatment of Embryos—Uninjected or injected embryos were incubated in culture media containing 250 nm trichostatin A (TSA, Sigma) continuously until the desired stage when they were collected for analysis of gene expression.
RESULTS
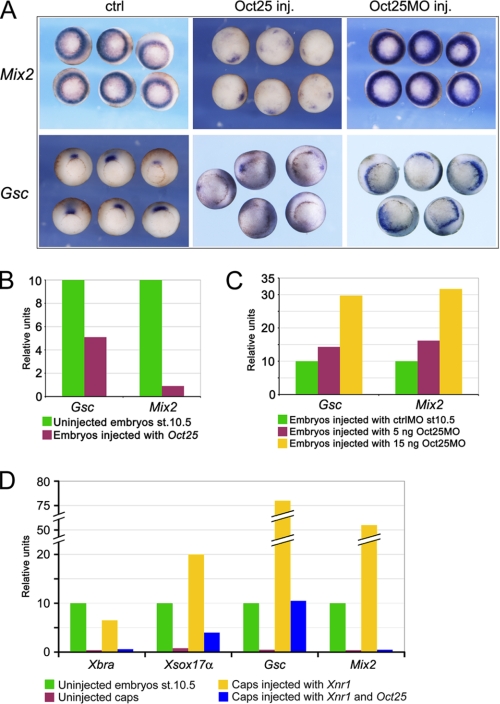
Oct25 Regulates Transcription of Gsc and Mix2—Oct25 was previously reported to inhibit the activity of nodal/activin signaling during Xenopus gastrulation (28). To substantiate these results and to explore the underlying mechanisms, we have investigated the expression of nodal/activin signaling direct target genes, Mix2 and Gsc, in response to overexpression or knockdown of Oct25. Whole mount in situ hybridization showed that both genes were severely repressed in embryos injected with Oct25 mRNA (Fig. 1A). In contrast, when endogenous Oct25 was knocked down by using an antisense morpholino oligonucleotide Oct25MO (27), Mix2 expression was significantly enhanced as revealed by higher signal intensity, and the Gsc expression domain extended toward the lateral marginal zone (Fig. 1A). Real-time RT-PCR also confirmed the inhibitory effect of Oct25 overexpression on Mix2 and Gsc expression (Fig. 1B), which, by contrast, was up-regulated upon Oct25 knockdown (Fig. 1C).
FIGURE 1.
Analysis of Gsc and Mix2 transcription in response to Oct25 overexpression or knockdown. A–C, whole mount in situ hybridization (A) or real-time RT-PCR in whole embryos (B and C) reveal an inhibition of Gsc and Mix2 by Oct25 gain of function and a dose-dependent increase in transcription of both genes by Oct25 loss of function. D, real-time RT-PCR shows that transcription of mesodermal and endodermal markers Xbra, Gsc, Xsox17a, and Mix2 is strongly enhanced in animal caps from embryos injected with Xnr1 mRNA, but is severely inhibited by co-injection with Oct25 mRNA.
Because Oct25 was shown to inhibit maternal VegT andβ-catenin activities being required for expression of nodal proteins (31) and β-catenin signaling is also involved in Gsc transcription (18, 19, 33), it might be argued that the observed inhibition of Gsc and Mix2 is indirect. We therefore performed nodal overexpression experiments in animal caps to exclude the possibility that the repression of Gsc and Mix2 expression by Oct25 overexpression was a secondary effect due to the inhibition of VegT and β-catenin activities in whole embryos. In animal caps, injection of Xnr1 mRNA resulted in a strong induction of mesendoderm formation, as revealed by the expression of Xbra and Xsox17α, and a strong induction of expression of direct target genes of nodal/activin signaling, Gsc and Mix2. However, co-injection of Oct25 with Xnr1 led to a dramatic inhibition of these genes (Fig. 1D). The result reflects that the interference of Oct25 with the activity of the nodal/activin signaling pathway is not a side effect of the inhibition of maternal VegT and/or β-catenin activities.
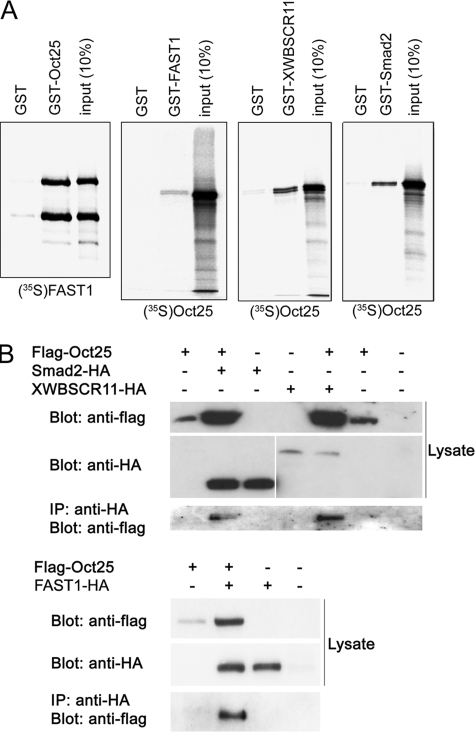
Oct25 Interacts with Components of the Nodal/Activin Signaling Pathway—To dissect the mechanism by which Oct25 regulates transcription of nodal/activin target genes, we first tested if Oct25 could interact with distinct components of the nodal/activin signaling pathway. GST pull-down assays showed a physical interaction between GST-tagged Oct25 and radiolabeled FAST1 (FoxH1), or vice versa, between GST-tagged FAST1 and radiolabeled Oct25 (Fig. 2A). A physical interaction between Oct25 and XWBSCR11 was also observed by using GST-tagged XWBSCR11 and radiolabeled Oct25. We could further demonstrate that Oct25 bound to the nodal/activin specific signal transducer, Smad2 (Fig. 2A). In these assays, the GST tag itself did not reveal any notable binding activity to the proteins of interest, indicating that the physical interactions between these proteins are specific (Fig. 2A). Co-immunoprecipitation assays revealed that the interactions also occur in vivo. In co-transfected cells, an anti-HA antibody could precipitate the immunocomplex comprising N-terminally Flag-tagged Oct25 and C-terminally HA-tagged Smad2, XWBSCR11 or FAST1 (Fig. 2B). In summary, both in vitro and in vivo experiments proved that Oct25 interacts with major signal transducers of the nodal/activin signaling pathway.
FIGURE 2.
Physical interactions between Oct25 and FAST1, Smad2, or XWBSCR11. A, GST pull-down assays reveal that Oct25 interacts with FAST1 (and vice versa), XWBSCR11, and Smad2. B, in transfected cells, Flag-tagged Oct25 formed complexes with HA-tagged Smad2, XWBSCR11, or FAST1 as displayed by precipitation with an anti-HA antibody.
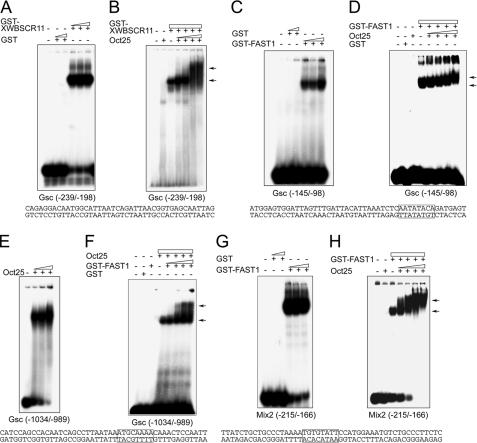
Oct25 Binds to Gsc and Mix2 Promoters—We next analyzed whether Oct25 would interact with the promoter regions of Gsc and Mix2 genes by EMSA. As reported previously (14), GST-tagged XWBSCR11 bound to the activin-responsive distal element (DE) (Fig. 3A) and a GST-tagged FAST1 bound to the Wnt-responsive proximal element (PE) (Fig. 3C) in the Gsc promoter, while the GST peptide itself did not (Fig. 3, A and C). Because Oct25 interacts with XWBSCR11 or FAST1, we tested whether Oct25 and XWBSCR11 or FAST1 would form complexes on the Gsc promoter. Supershift assays clearly revealed that Oct25 actually formed a complex with XWBSCR11 on the DE and with FAST1 on the PE, because the electrophoretic mobilities of XWBSCR11-DNA (Fig. 3B) or FAST1-DNA complexes were retarded when Oct25 was added (Fig. 3D). These data suggest that Oct25 may regulate Gsc transcription via interaction with XWBSCR11 or FAST1 that are bound to the promoter. We have also asked whether Oct25 could interact directly with the Gsc promoter. Analysis of the Gsc promoter sequence revealed two elements resembling the canonical octamer ATGCAAAT motif. One ATGCAAAA motif is present at position -1007 to -1000, the other TTTAGCAT motif is located at -486 to -479. EMSAs demonstrated strong binding between Oct25 and the first octamer motif (Fig. 3E), but no specific interaction was observed for Oct25 and the second one (data not shown). By means of supershift assays, we further found that FAST1 formed a complex with Oct25 on the Oct binding site at -1007 (Fig. 3F).
FIGURE 3.
In vitro binding of Oct25 to Gsc and Mix2 promoters. A, increasing amounts of a GST-tagged XWBSCR11 fusion protein bind to an oligonucleotide containing the DE of the Gsc promoter (position -239 to -198), while GST itself does not show any binding. B, increasing amounts of Oct25 and GST-XWBSCR11 form a complex on the DE as displayed by a supershift. C, FAST1 specifically binds to an oligonucleotide containing the PE of the Gsc promoter (position -145 to -98) with a FAST1 binding motif. D, Oct25 forms a complex with FAST1 on the PE as revealed by supershifts. E, Oct25 binds to an oligonucleotide (position -1034 to -989) of the Gsc promoter containing an octamer-like motif. F, incubation of this region of the Gsc promoter with Oct25 and increasing amounts of GST-FAST1 fusion protein leads to a supershift. G, FAST1 interacts with the ARE of the Mix2 promoter (position -215 to -166) that contains a FAST1 binding motif. H, supershift generated with GST-FAST1 and increasing amounts of Oct25. The sequences corresponding to the regions of Gsc and Mix2 promoters used for EMSA and supershift assays are listed below each panel and protein binding motifs for FAST1 or Oct25 are boxed. The arrows denote supershifting.
We then investigated the binding of Oct25 on the Mix2 promoter. The promoter contains an ARE (21) comprising a FAST1 binding motif TGTGTATT from -196 to -189 and a Smad binding motif GTCT from -178 to -175. EMSA experiments demonstrated an interaction between a GST-tagged FAST1 protein and an oligonucleotide containing the ARE (Fig. 3G). Importantly, when Oct25 was added to the FAST1-DNA complex, we observed a severe retardation of the electrophoretic mobility, showing that Oct25 and FAST1 formed a complex on the ARE (Fig. 3H). On the -712 promoter region, there exist also octamer-like sequences, for instance ATGTATTA from -623 to -616 and AATAACAT from -457 to -450. However, we did not observe specific interactions between Oct25 and either of these sequences (data not shown).
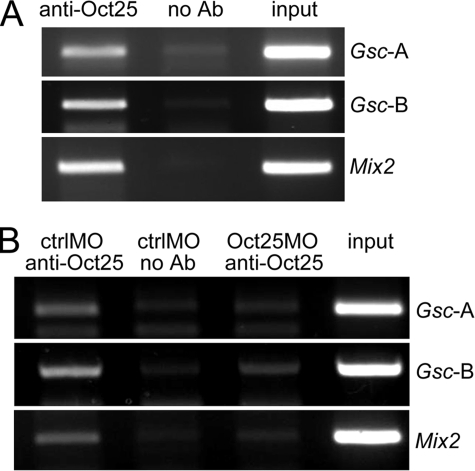
The in vitro analyses may not necessarily mean that Oct25 interacts with these promoters in wild-type embryos. Therefore, ChIP were carried out to confirm the binding between Oct25 and Gsc or Mix2 promoters in vivo. We could show that an anti-Oct25 peptide antibody (31) precipitated two regions of the Gsc promoter, one region Gsc-A containing the Oct binding site and the other region Gsc-B spanning the DE and PE. Moreover, the antibody also precipitated the Mix2 promoter region that contains the ARE (Fig. 4A). When Oct25MO was injected into embryos to knock down endogenous Oct25, the quantities of precipitated Gsc or Mix2 promoter fragments were much less than those precipitated from ctrlMO injected embryos (Fig. 4B), validating the specificity of protein-promoter interactions. Hence, experimental results obtained from in vitro and in vivo experiments suggest that Oct25 binds to the promoters of Gsc and Mix2 genes. This binding may occur indirectly via interaction with XWBSCR11 and FAST1 or it may occur directly as in the case of Gsc promoter at position -1007.
FIGURE 4.
In vivo interaction of Oct25 to Gsc and Mix2 promoters. A, ChIP assays demonstrate that Gsc promoter regions containing either the Oct25 binding site (Gsc-A) or containing the DE and PE (Gsc-B) as well as the Mix2 promoter region spanning the ARE were amplified from chromatin precipitated by an Oct25 antibody. This amplification failed in the absence of antibody (no Ab). B, when Oct25 translation was blocked by Oct25MO, PCR products for Gsc and Mix2 promoter regions were reduced as compared with ctrlMO injections.
Oct25 Represses Gsc and Mix2 Promoter Activities—To further characterize the effect of Oct25 on Gsc transcription, we made a series of Gsc promoter/luciferase reporter constructs (Fig. 5A). First we tested how the Gsc promoter responded to Oct25 overexpression. A -1500 promoter fragment was strongly stimulated by injection of Xnr1 mRNA or by incubation with activin A protein. Moreover, it was also stimulated by injection of mRNA coding for Smad2. In any case, when Oct25 mRNA was co-injected, luciferase activity was severely inhibited (Fig. 5B), reflecting that Oct25 represses Gsc promoter activity. We then analyzed whether different regions of Gsc promoter would have different response to Oct25 activity. Two deletion mutants, -479 and -226 were fused to luciferase reporter. Similar to the -1500 promoter region, both promoter mutants were stimulated by injection of Xnr1 or Smad2 mRNA or by incubation with activin A. Again, overexpression of Oct25 inhibited the activity of both promoter deletion constructs (Fig. 5, C and D). Because XWBSCR11 is required for the induction of Gsc and interacts with Oct25, we subsequently examined the effect of Oct25 on XWB-SCR11-induced promoter activity. The DE of Gsc promoter is responsive to XWBSCR11 (14). As reported, an artificial promoter reporter composed of 6 catenated repeats of DE in front of the -104 minimal promoter (6×DE) (14) was significantly stimulated by injection of VP16-XWB-SCR11 mRNA encoding an active form of XWBSCR11. An even higher level of stimulation was observed when VP16-XWBSCR11 and FAST1 were applied together (Fig. 5E). We found that Oct25 repressed not only the basal activity of 6xDE but also the activity stimulated by VP16-XWB-SCR11 alone or by VP16-XWB-SCR11 and FAST1 together (Fig. 5E). All these data above provide solid evidence that Oct25 represses Gsc promoter activity.
FIGURE 5.
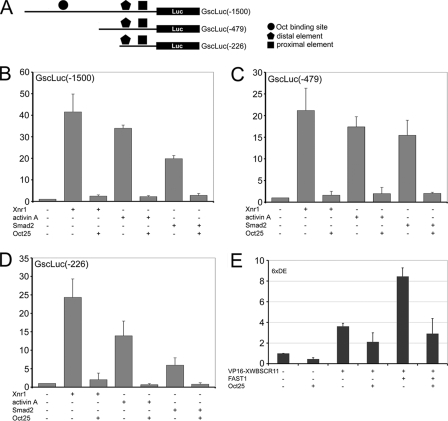
Gsc promoter assays. A, diagram depicting the generation of Gsc promoter-luciferase reporter constructs. B, Oct25 represses Gsc -1500/+3 promoter activities stimulated by overexpression of Xnr1, Smad2, or by incubation with activin A protein. C and D, the same effect is observed for truncated promoters -479/+3(C) and -226/+3(D). E, VP16-XWBSCR11 alone or together with FAST1 stimulates an artificial reporter for DE, which is then repressed by Oct25.
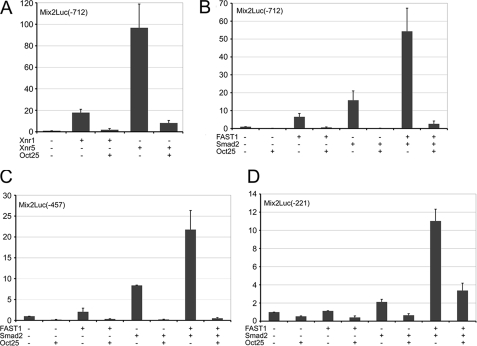
Next we have analyzed the influence of Oct25 on Mix2 promoter activity. A -712 promoter fragment of Mix2 was isolated from genomic DNA and fused to the luciferase reporter vector. Promoter activity was strongly stimulated by injection of Xnr1 or Xnr5 mRNA, albeit to different levels. In contrast, co-injection of Oct25 mRNA resulted in a dramatic decrease of promoter activity (Fig. 6A). Because Oct25 interacts with FAST1 and Smad2, we examined how Oct25 would affect Mix2 promoter activity induced by FAST1 and/or Smad2. Overexpression of FAST1 or Smad2 alone led to a significant stimulation of Mix2 promoter activity, and a synergistic inducing effect was found when FAST1 and Smad2 were overexpressed together. However, co-injection of Oct25 mRNA caused always a severe inhibition of Mix2 promoter activity induced by FAST1 or Smad2 alone or by the two factors together (Fig. 6B), indicating that Oct25 indeed represses Mix2 promoter activity. We also examined whether different regions within the Mix2 promoter exhibit any difference in the inhibitory response to Oct25. Two truncated promoter constructs, -457 and -221, both containing the ARE, were strongly stimulated by FAST1 or Smad2 individually or in combination. As a matter of fact, overexpression of Oct25 showed the same inhibitory effects on these two reporter constructs (Fig. 6, C and D). Therefore, we conclude that Oct25 represses Mix2 promoter activity by interference with the Fast1/Smad2 complex.
FIGURE 6.
Mix2 promoter assays. A and B, Oct25 inhibits Mix2 -712/+13 promoter activity stimulated not only by overexpression of the ligand of nodal signaling, Xnr1 or Xnr5 (A), but also by overexpression of the transducers, Smad2 and/or FAST1 (B). C and D, Oct25 shows a repressive effect on the truncated Mix2 promoters -457/+13 (C) and -221/+13 (D), which are stimulated by Smad2 and/or FAST1.
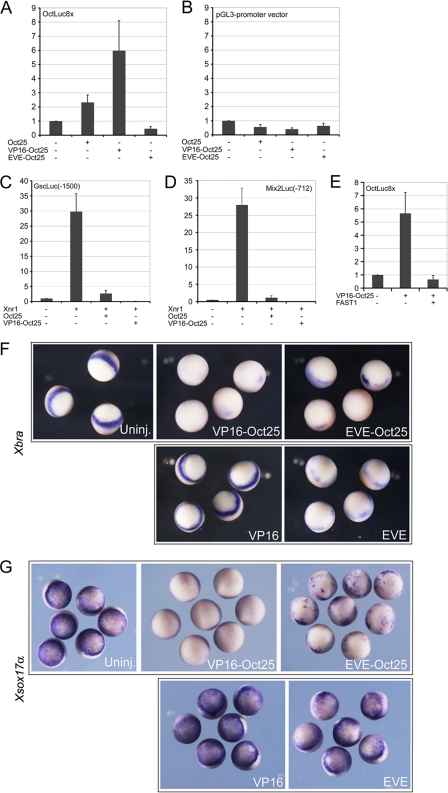
The Oct Binding Site Is Dispensable for the Inhibitory Effect of Oct25—In the promoter of Gsc, there is an Oct binding site that is responsible for Oct25 binding. Since we have shown that the Gsc promoter reporter deletion mutants lacking this site were still repressed by Oct25 (Fig. 5, C and D), it was interesting to investigate the functional role of this Oct binding site in the regulation of Gsc. Therefore, we made use of an artificial promoter reporter, OctLuc8x, which was generated by ligating 8 tandem repeats of a canonical Oct binding site, ATGCAAAT, to the pGL3-promoter vector. In addition, we synthesized a VP16-fusion and an EVE-fusion with Oct25 and tested the response of OctLuc8x to these fusion proteins. VP16 is the transcriptional activator domain of herpes simplex virus. Fusion to VP16 resulted in a constitutively active version of Oct25, while fusion to EVE, the Drosophila even-skipped repression domain, generated a repressive form of Oct25. We observed that OctLuc8x was mildly stimulated by wild-type Oct25 but strongly stimulated by VP16-Oct25. In contrast, EVE-Oct25 exhibited a repressive effect on the promoter activity (Fig. 7A). In a control test, we observed a somewhat repressive effect by Oct25, VP16-Oct25 or EVE-Oct25 on the pGL3-promoter vector (Fig. 7B), reflecting that the response of OctLuc8x to different versions of Oct25 was specific. This result suggested that Oct25 acted as a transcriptional activator with regard to its direct binding to promoter via the octamer sequence. Although putative binding sites for other Oct factors were reported for the pGL3 vector (see technical notes, Promega), this experiment suggests that no Oct25 binding site is present in this vector that confers activation of reporter expression. We then examined the response of the wild-type Gsc (-1500) promoter to VP16-Oct25. As already shown, the dramatic stimulation of Gsc promoter by Xnr1 mRNA was strongly repressed by co-injection of Oct25 mRNA. However, instead of stimulating Gsc promoter activity, VP16-Oct25 revealed an even stronger repressive effect than Oct25, because luciferase activity was barely detectable when VP16-Oct25 was expressed (Fig. 7C). In case of the Mix2 promoter, where no specific Oct binding sites are present, VP16-Oct25 also exerted a more severe repressive effect than Oct25 on Xnr1 induced promoter activity (Fig. 7D). These results suggest that the presence of an Oct binding motif does not necessarily mean an activation of gene expression via Oct25 binding or that it is a pre-requisite for gene repression. Considering the fact that Oct25 interacts with FAST1, XWBSCR11, and Smad2, we then investigated whether these proteins would affect stimulation of OctLuc8x by VP16-Oct25. Reporter assay demonstrated that luciferase activity induced by VP16-Oct25 was significantly repressed by co-injection of FAST1 mRNA (Fig. 7E) but not by co-injection of XWBSCR11 or Smad2 mRNA (data not shown). This result implies that, at least, FAST1 may convey a repressive effect on Oct25 induced gene activation.
FIGURE 7.
Analysis of Oct25 VP16 and EVE fusions on promoter activity and mesendoderm formation. A, wild-type Oct25 and VP16-Oct25 stimulate in contrast to EVE-Oct25 an artificial octamer reporter, suggesting that Oct25 is a transcriptional activator with regard to its octamer binding property. B, the controls demonstrate that stimulation is not due to the pGL3 vector, to which the octamer motifs were ligated. C and D, both Oct25 and VP16-Oct25 reveal repressive effect on Gsc (C) and Mix2 (D) promoters. E, stimulation of octamer reporter by VP16-Oct25 is inhibited by FAST1. F and G, either or revealed inhibition of Xbra (F) and Xsox17α (G), while in controls, the VP16 alone had no effect and EVE alone had weak effect on the expression of the two genes.
We next tested the effect of VP16-Oct25 or EVE-Oct25 overexpression on early embryogenesis. It was previously shown that vegetal overexpression of Oct25 led to an inhibition of mesendoderm formation (28). Here we show that vegetal injection of either VP16-Oct25 or EVE-Oct25 mRNA also resulted in severe inhibition of mesendoderm formation, because nearly no Xbra and Xsox17α expression could be detected (Fig. 7F). In control injections, VP16 alone did not reveal any significant effect at all on expression of Xbra and Xsox17α. Although the EVE domain exhibited a slight inhibition of Xbra and Xsox17α expression, it could not account for the strong effect displayed by EVE-Oct25 (Fig. 7F). In summary, the modification of Oct25 by fusion with either an extra activation domain or a repression domain does surprisingly not alter its inhibitory function on mesendoderm formation.
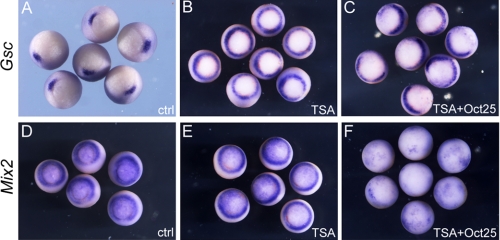
The Repressive Effect of Oct25 on Gsc and Mix2 Transcription Is Independent of HDAC Activity—In a previous report, it was shown that Oct4 interacts with HDAC2 (34), a class I histone deacetylase (35), suggesting that Oct4 or its Xenopus orthologue Oct25 may regulate gene expression via modification of the histone acetylation status. This prompted us to examine whether repression of nodal/activin target genes by Oct25 was indeed dependent on the activity of HDACs. Hence, we tested the repressive effect of Oct25 on the expression of Gsc and Mix2 when HDAC activity was blocked by trichostatin A (TSA), a specific inhibitor of HDACs (36). Interestingly, when uninjected embryos were treated with TSA, Gsc transcription was ectopically activated and showed a circular pattern around the blastopore (Fig. 8B). This clearly demonstrates that repression of Gsc transcription at the ventral side of wild-type embryos requires HDAC activity. However, dorsal injection of Oct25 mRNA in TSA treated embryos eliminated Gsc expression at the dorsal side, in that circular expression was reduced to a semicircle at the ventral side (Fig. 8C). Therefore, TSA mediated inhibition of HDAC activity does not influence the repression of Gsc by Oct25. In contrast to Gsc, TSA-treated embryos did not display a significant change in Mix2 expression as compared with untreated control embryos (Fig. 8E). Again, Oct25 overexpression still exerted efficiently the inhibitory effect on Mix2 expression when embryos were treated with TSA (Fig. 8F). Therefore, these experiments imply that the repressive effect of Oct25 on Gsc or Mix2 expression is not dependent on HDAC activity.
FIGURE 8.
Inhibition of HDAC activity did not alter the inhibitory effect of Oct25 on Gsc and Mix2. A, Gsc expression in wild-type gastrulae. B, circular expression of Gsc in TSA-treated embryos. C, dorsal injection of Oct25 mRNA leads to the disappearance of Gsc expression at the dorsal side in TSA-treated embryos. D and E, Mix2 showed similar expression in control (D) or TSA (E)-treated embryos. F, Oct25 overexpression leads to an inhibition of Mix2 expression in TSA-treated embryos.
DISCUSSION
One of the key issues to understand early embryogenesis is the mechanism that maintains the balance between the undifferentiated and differentiated state of embryonic cells. Accordingly, there exist two types of signals during embryogenesis, one is responsible for inducing early embryonic cells to differentiate to form germ layers and subsequently different types of tissues, and the other type is for preventing embryonic cells from premature differentiation. The nodal/activin pathway is a major signal to induce mesoderm and endoderm formation in all vertebrates. In Xenopus, the nodal activity, which is zygotic, is induced by the upstream maternal factors VegT and β-catenin. On the other hand, Oct4 or its homologues in other species represent the key players of the regulatory circuitry that maintains the undifferentiated state of embryonic cells (37, 38). We have recently demonstrated that Xenopus Oct4 homologous proteins disable maternal VegT and β-catenin activity thereby inhibiting the formation of mesendoderm (31). Moreover, it was previously shown that Oct25 blocks mesendoderm inducing activity mediated by nodal/activin signaling (28, 29). These results are extended and further substantiated by the analyses shown in the present study. By means of both, overexpression and knockdown assays in whole embryos or animal caps, we could show that Oct25 represses transcription of nodal/activin target genes, Gsc and Mix2, implying that Oct25 regulates the nuclear transcriptional response to nodal/activin. Biochemical analyses revealed that Oct25 forms complexes with major nuclear components of the nodal/activin pathway, Smad2, FAST1, or WBSCR11. These complexes are directed to the promoters of Gsc or Mix2 via the FAST1 binding site or WBSCR11 binding site and, as further confirmed by a series of promoter luciferase reporter assays, inhibit transcription of these genes. This inhibition is not due to a decrease of transcripts encoding these signal transducers by overexpression of Oct25 (data not shown). Therefore, joining of Oct25 turns the activating complexes of Smad2, FAST1, or WBSCR11 for nodal/activin target gene transcription into inhibitory ones. This type of regulatory mechanism for Oct25 is not unique for the nodal/activin pathway, since a similar one has also been observed for the regulation of target genes of VegT and maternal β-catenin signaling (31). Therefore, this principle might also be applicable to other signaling pathways that promote embryonic cell or tissue differentiation.
Oct4 was initially characterized as a transcriptional activator in embryonic stem cells via interaction with the octamer motif ATGCAAAT on the promoters of its target genes (39), including FGF4, nanog, Sox2, and the Oct4 gene itself. We here found in the Gsc promoter a similar sequence ATGCAAAA, which can be also occupied by Oct25. However, Oct25 did not activate Gsc promoter activity, but instead displayed a strong inhibitory effect, even when this sequence was removed from the promoter. We therefore examined the effect of Oct25 and its constitutively active version VP16-Oct25 or repressive version EVE-Oct25 specifically on the activity of an artificial promoter luciferase reporter that is composed of eight repeats of the octamer motif. In agreement with the findings on Oct4 as transcriptional activator, both Oct25 and VP16-Oct25 stimulated the reporter activity, but the latter showed an apparently stronger stimulating effect. In contrast, EVE-Oct25 exerted an inhibitory influence. The result implies that Oct25 alone indeed activates gene transcription via direct binding to the octamer motif. Nevertheless, just like the case of wild-type protein, both Oct25 fusion proteins repress mesendoderm formation and Gsc and Mix2 promoter activities. Therefore, the presence of an octamer motif per se does not necessarily mean activation by Oct25. Instead, the regulatory effect of Oct25 on the expression of Gsc and Mix2 is dependent on its interaction partners. This notion is also strengthened by the observation that, when FAST1 associated with Oct25 on the octamer motif, a repressive effect was observed on the octamer reporter activity stimulated by VP16-Oct25. However, it has to be emphasized that both injection and reporter studies were assessed under conditions of overexpression. Whether our findings do also persist under physiological levels of expression and chromatin environment, remains to be ascertained.
It should be noted that the octamer consensus motif is also utilized for DNA binding by other POU-family transcription factors (40). However, except for Oct4 and its homologues in other species, other POU factors, for example Oct1, seem not to be involved in the maintenance of pluripotency and self-renewal of embryonic stem cells. Thereby, POU factors of different subclasses should regulate distinct sets of target genes, which cannot be explained solely by interaction to target gene promoters via the octamer motif. Therefore, it is reasonable to assume that the functions of different POU factors are not entirely dependent on their DNA binding capability. On the other hand, since detailled information about other, non-canonical Oct25 binding sites is missing, we cannot definitely rule out the possibility that specific DNA binding of Oct25 might also contribute to the observed transcriptional inhibition of Gsc and Mix2. However, it is an interesting concept that specific interaction partners for Oct factors determine which target genes are selected together with the mode of the regulatory effect.
Many transcription factors regulate target gene expression through interaction with chromatin modifiers. A physical interaction between Oct4 and HDAC2 in embryonic stem cells was previously reported (34), suggesting that Oct4 might repress gene expression by the way of reducing the acetylation level of histones on chromatin. However, we here show that repression of Gsc or Mix2 by Oct25 is not dependent on HDAC activity in gastrulating Xenopus embryos, because Oct25 still repressed expression of these genes when HDAC activity was blocked. This discrepancy may be due to differences between gene regulatory mechanisms in mesendodermal cells and in embryonic stem cells. Furthermore, the HDAC inhibitor TSA used in the present study is effective on HDACs of class I and II (41), we therefore cannot exclude the possibility that the repression effect of Oct25 is related to HDACs of class III. However, besides histone acetylation other ways of chromatin modification exist, for instance histone methylation. In embryonic stem cells, most transcriptionally silent genes for developmental regulators are targeted by both Oct4 and the Polycomb group (PcG) proteins (42–44). PcG proteins are epigenetic regulators that modify chromatin structure via H3-lysine-27 methylation (H3K27), thereby repressing target gene transcription (45). Based on these observations, it is reasonable to assume that Oct25 works in concert with PcG proteins to inhibit nodal/activin target gene transcription. This possibility is also applicable to the former observation that Oct25 represses target genes of maternal β-catenin/Tcf3 signaling and VegT (31). Oct4 and its homologues in other species can both activate and repress gene transcription. The regulatory effect and the specificity of target genes regulated by Oct proteins might be determined coordinately by the spatial-temporal distributions of Oct proteins, their interaction partners and PcG proteins. Whether this holds true not only in embryonic stem cells but also during embryogenesis remains an intriguing topic to investigate in future studies.
Acknowledgments
We thank Cornelia Donow, Nicole Heymann, and Sabine Schirmer for skillful technical assistance. We are indebted to Drs. K. Cho, M. Whitman, and M. Taira for their generous gifts of plasmids.
This work was supported by Grants (SFB497/A1) (to W. K.) and (SFB497/B9) (to F. O.) from the Deutsche Forschungsgemeinschaft. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: TGF-β, transforming growth factor-β; FAST, forkhead activin signal transducer; WBSCR11, Williams-Beuren syndrome critical region 11; Smad, derived from Caenorhabditis elegans small and Drosophila MAD genes; Gsc, goosecoid; GST, glutathione S-transferase; RT, reverse transcriptase; ALK4, activin receptor like kinase 4; ChIP, chromatin immunoprecipitation assay; EMSA, electrophoretic mobility shift assay; HA, hemagglutinin; TSA, trichostatin A.
M. Taira, unpublished data.
References
- 1.Ariizumi, T., Sawamura, K., Uchiyama, H., and Asashima, M. (1991) Int. J. Dev. Biol. 35 407-414 [PubMed] [Google Scholar]
- 2.Green, J. B., and Smith, J. C. (1990) Nature 347 391-394 [DOI] [PubMed] [Google Scholar]
- 3.Gurdon, J. B., Harger, P., Mitchell, A., and Lemaire, P. (1994) Nature 371 487-492 [DOI] [PubMed] [Google Scholar]
- 4.Agius, E., Oelgeschlager, M., Wessely, O., Kemp, C., and De Robertis, E. M. (2000) Development 127 1173-1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones, C. M., Kuehn, M. R., Hogan, B. L., Smith, J. C., and Wright, C. V. (1995) Development 121 3651-3662 [DOI] [PubMed] [Google Scholar]
- 6.Joseph, E. M., and Melton, D. A. (1997) Dev. Biol. 184 367-372 [DOI] [PubMed] [Google Scholar]
- 7.Schier, A. F., and Shen, M. M. (2000) Nature 403 385-389 [DOI] [PubMed] [Google Scholar]
- 8.Schier, A. F. (2003) Annu. Rev. Cell Dev. Bio.l 19 589-621 [DOI] [PubMed] [Google Scholar]
- 9.Schier, A. F., and Talbot, W. S. (2005) Annu. Rev. Genet. 39 561-613 [DOI] [PubMed] [Google Scholar]
- 10.Shen, M. M. (2007) Development 134 1023-1034 [DOI] [PubMed] [Google Scholar]
- 11.Whitman, M. (2001) Dev. Cell 1 605-617 [DOI] [PubMed] [Google Scholar]
- 12.Ramsdell, A. F., and Yost, H. J. (1998) Trends Genet. 14 459-465 [DOI] [PubMed] [Google Scholar]
- 13.Shen, M. M., and Schier, A. F. (2000) Trends Genet. 16 303-309 [DOI] [PubMed] [Google Scholar]
- 14.Ring, C., Ogata, S., Meek, L., Song, J., Ohta, T., Miyazono, K., and Cho, K. W. (2002) Genes Dev. 16 820-835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cho, K. W. Y., Blumberg, B., Steinbeisser, H., and DeRobertis, E. M. (1991) Cell 67 1111-1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Niehrs, C., Keller, R., Cho, K. W. Y., and DeRobertis, E. M. (1993) Cell 72 491-503 [DOI] [PubMed] [Google Scholar]
- 17.Lemaire, P., Darras, S., Caillol, D., and Kodjabachian, L. (1998) Development 125 2371-2380 [DOI] [PubMed] [Google Scholar]
- 18.Watabe, T., Kim, S., Candia, A., Rothbacher, U., Hashimoto, C., Inoue, K., and Cho, K. W. (1995) Genes Dev. 9 3038-3050 [DOI] [PubMed] [Google Scholar]
- 19.Laurent, M., Hashimoto, C., Blitz, I. L., Rothbacher, U., and Cho, K. W. Y. (1997) Development 124 4905-4916 [DOI] [PubMed] [Google Scholar]
- 20.Huang, H.-C., Murtaugh, L. C., Vize, P. D., and Whitman, M. (1995) EMBO J. 14 5965-5973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vize, P. D. (1996) Dev. Biol. 177 226-231 [DOI] [PubMed] [Google Scholar]
- 22.Hinkley, C. S., Martin, J. F., Leibham, D., and Perry, M. (1992) Mol. Cell. Biol. 12 638-649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nichols, J., Zevnik, B., Anastassiadis, K., Niwa, H., Klewe-Nebenius, D., Chambers, I., Scholer, H., and Smith, A. (1998) Cell 95 379-391 [DOI] [PubMed] [Google Scholar]
- 24.Boyer, L. A, Mathur, D., and Jaenisch, R. (2006) Curr. Opin. Genet. Dev. 16 455-462 [DOI] [PubMed] [Google Scholar]
- 25.Chambers, I. (2004) Cloning Stem Cells 6 386-391 [DOI] [PubMed] [Google Scholar]
- 26.Yu, J., Vodyanik, M. A., Smuga-Otto, K., Antosiewicz-Bourget, J., Frane, J. L., Tian, S., Nie, J., Jonsdottir, G. A., Ruotti, V., Stewart, R., Slukvin, I. I., and Thomson J. A. (2007) Science 318 1917-1920 [DOI] [PubMed] [Google Scholar]
- 27.Takahashi, K., Tanabe, K., Ohnuki, M., Narita, M., Ichisaka, T., Tomoda, K., and Yamanaka, S. (2007) Cell 131 861-872 [DOI] [PubMed] [Google Scholar]
- 28.Cao, Y., Siegel, D., and Knöchel, W. (2006) Mech. Dev. 123 614-625 [DOI] [PubMed] [Google Scholar]
- 29.Morrison, G. M., and Brickman, J. M. (2006) Development 133 2011-2022 [DOI] [PubMed] [Google Scholar]
- 30.Harland, R. M. (1991) Methods Cell Biol. 36 685-695 [DOI] [PubMed] [Google Scholar]
- 31.Cao, Y., Siegel, D., Donow, C., Knöchel, S., Yuan, L., and Knöchel, W. (2007) EMBO J. 26 2942-2954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cao, Y., Knöchel, S., Donow, C., Miethe, J., Kaufmann, E., and Knöchel, W. (2004) J. Biol. Chem. 279 43735-43743 [DOI] [PubMed] [Google Scholar]
- 33.Steinbeisser, H., De Robertis, E. M., Ku, M., Kessler, D. S., and Melton, D. A. (1993) Development 118 499-507 [DOI] [PubMed] [Google Scholar]
- 34.Wang, J., Rao, S., Chu, J., Shen, X., Levasseur, D. N., Theunissen, T. W., and Orkin, S. H. (2006) Nature 444 364-368 [DOI] [PubMed] [Google Scholar]
- 35.Khochbin, S., Verdel, A., Lemercier, C., and Seigneurin-Berny, D. (2001) Curr. Opin. Genet. Dev. 11 162-166 [DOI] [PubMed] [Google Scholar]
- 36.Yoshida, M., Kijima, M., Akita, M., and Beppu, T. (1990) J. Biol. Chem. 265 17174-17179 [PubMed] [Google Scholar]
- 37.Zhou, Q., Chipperfield, H., Melton, D. A., and Wong, W. H. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 16438-16443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim, J., Chu, J., Shen, X., Wang, J., and Orkin, S. H. (2008) Cell 132 1049-1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pesce, M., and Schöler, H. R. (2000) Mol. Reprod. Dev. 55 452-457 [DOI] [PubMed] [Google Scholar]
- 40.Ryan, A. K., and Rosenfeld, M. G. (1997) Genes Dev. 11 1207-1225 [DOI] [PubMed] [Google Scholar]
- 41.Mariadason, J. M. (2008) Epigenetics 3 28-37 [DOI] [PubMed] [Google Scholar]
- 42.Bernstein, B. E., Mikkelsen, T. S., Xie, X., Kamal, M., Huebert, D. J., Cuff, J., Fry, B., Meissner, A., Wernig, M., Plath, K., Jaenisch, R., Wagschal, A., Feil, R., Schreiber, S. L., and Lander, E. S. (2006) Cell 125 315-326 [DOI] [PubMed] [Google Scholar]
- 43.Boyer, L. A., Plath, K., Zeitlinger, J., Brambrink, T., Medeiros, L. A., Lee, T. I., Levine, S. S., Wernig, M., Tajonar, A., Ray, M. K., Bell, G. W., Otte, A. P., Vidal, M., Gifford, D. K., Young, R. A., and Jaenisch, R. (2006) Nature 441 349-353 [DOI] [PubMed] [Google Scholar]
- 44.Lee, T. I., Jenner, R. G., Boyer, L. A., Guenther, M. G., Levine, S. S., Kumar, R. M., Chevalier, B., Johnstone, S. E., Cole, M. F., Isono, K., Koseki, H., Fuchikami, T., Abe, K., Murray, H. L., Zucker, J. P., Yuan, B., Bell, G. W., Herbolsheimer, E., Hannett, N. M., Sun, K., Odom, D. T., Otte, A. P., Volkert, T. L., Bartel, D. P., Melton, D. A., Gifford, D. K., Jaenisch, R., and Young, R. A. (2006) Cell 125 301-313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ringrose, L., and Paro, R. (2004) Annu. Rev. Genet. 38 413-443 [DOI] [PubMed] [Google Scholar]