Abstract
Tryptases are serine proteases that are thought to be uniquely and proteolytically active as tetramers. Crystallographic studies reveal that the active tetramer is a flat ring structure composed of four monomers, with their active sites arranged around a narrow central pore. This model explains why many of the preferred substrates of tryptase are short peptides; however, it does not explain how tryptase cleaves large protein substrates such as fibronectin, although a number of studies have reported in vitro mechanisms for generating active monomers that could digest larger substrates. Here we suggest that alternate mRNA splicing of human tryptase genes generates active tryptase monomers (or dimers). We have identified a conserved pattern of alternate splicing in four tryptase alleles (αII, βI, βIII, and δI), representing three distinct tryptase gene loci. When compared with their full-length counterparts, the splice variants use an alternate acceptor site within exon 4. This results in the deletion of 27 nucleotides within the central coding sequence and 9 amino acids from the translated protein product. Although modeling suggests that the deletion can be easily accommodated by the enzymes structurally, it is predicted to alter the specificity by enlarging the S1′ or S2′ binding pocket and results in the complete loss of the “47 loop,” reported to be critical for the formation of tetramers. Although active monomers can be generated in vitro using a range of artificial conditions, we suggest that alternate splicing is the in vivo mechanism used to generate active tryptase that can cleave large protein substrates.
Tryptases belong to a family of serine proteases and are named based on their similarity to the pancreatic enzyme trypsin. The most intensively studied tryptases are the α/β tryptases (now known to be the products of two separate gene loci); they are reported to be selectively expressed in mast cells, where they are stored in granules in association with proteoglycans. Large quantities of these proteases are produced by mast cells, often representing around a quarter of the total cellular protein content (1).
Recently published reports by Caughey and co-workers (2, 3) (supported by data from the public and private genome data bases) indicate the presence of multiple gene loci on chromosome 16p13.3 that encode human tryptases. The first locus encodes a transmembrane tryptase called γ tryptase (3). The second is a locus whose allelic variants are βII and βIII tryptase (4). The third has allelic variants that include βI tryptase, αII tryptase, and probably αI tryptase (4). The fourth is a locus encoding δ tryptase (5), and the fifth is a more distantly related member named ε tryptase (6). With the recent report concerning the in vivo expression of δ tryptase (5), mRNA and protein products for all five loci have now been detected.
Native tryptase protein purified from tissue is biochemically heterogeneous (1, 7, 8), but the basis of this has not been fully determined. It may be due to the presence of multiple tryptase gene products, the presence of different post-translational modifications such as glycosylation, or more likely a combination of causes.
Tryptase has been implicated in the development of a number of clinical conditions, including asthma (9–11), inflammatory bowel disease (12), and inflammatory arthritis (13, 14). Furthermore, the detection of different forms of tryptase can be used as a diagnostic feature. For instance increased plasma levels of α tryptase, apparently constitutively expressed by all mast cells, are indicative of the increased mast cell burden of mastocytosis (15). Alternatively, the detection of β tryptase indicates the presence of activated mast cells of allergic conditions and anaphylaxis (16).
Interest in the biology of tryptases has increased because of their proposed role in inflammatory diseases such as asthma. Although the underlying cause of many of these diseases is not fully understood, multiple lines of evidence support a link between tryptases and an inflammatory phenotype. For example, inflamed tissue is often characterized by an increase in mast cell numbers (and tryptase levels) when compared with uninflamed control tissue (17), and both murine and human tryptases can act directly or indirectly to recruit inflammatory cells such as neutrophils and eosinophils (18, 19).
One of the most interesting findings that accompanied the publishing of the human genome was that the number of discrete genes was much lower than anticipated (20). Previously it was thought that the biochemical complexity of an organism was proportional to the size of its genome. Therefore, it was surprising that arguably the most complex organism, humans, possessed a genome not dramatically larger than that of an earthworm, and indeed significantly smaller than that of many plants (20). On closer inspection the relationship between the perceived complexity of an organism and the size of its genome holds true only for the so-called “lower organisms,” where there is roughly a 1:1 ratio between the number of individual genes and their resulting protein products. What becomes obvious is that in the “higher” organisms, complexity may be facilitated by increasing the number of distinct protein products that can be generated from a single gene. This may be accomplished by processes such as post-translational modifications or by generating multiple forms of mRNA from a single primary transcript, the process of alternate splicing. The human transcriptome is apparently distinguished from that of other organisms by the dramatically increased number of alternately spliced transcripts (20).
Here we report the cloning and initial characterization of alternately spliced forms of human αII, βI, βIII, and δI tryptase. The pattern of alternate splicing is identical in all four tryptases, and it results in the loss of 27 nucleotides from the mature transcript and 9 amino acids from the translated protein product, when compared with their full-length counterparts. The resulting proteases are predicted to have altered substrate specificities, at least partly due to the predicted inability of the splice variant tryptases to form tetramers. These splice variant tryptases were shown to be expressed in multiple human tissues. Alternate splicing is a mechanism that results in an increase in the number of functionally distinct human tryptases.
EXPERIMENTAL PROCEDURES
Sources of RNA—Total RNA from adult lung, heart, stomach, spleen, skin, and colon, fetal heart and fetal lung, and poly(A+) RNA isolated from human lung were obtained from commercial sources (Invitrogen). In addition, total RNA was isolated from human mast cell-1 (HMC-1)3 cells (5 × 106, a kind gift from Dr. J. H. Butterfield) using TRI Reagent™ (Sigma), according to the manufacturer's instructions, dissolved in distilled H2O, and stored at -80 °C until required.
Preparation of cDNA—First strand cDNAs were generated using the cDNA Cycle® kit (Invitrogen). Poly(A+) RNA (300 ng) or total RNA (1.5 μg) and 1 μl of oligo(dT) primer were heated to 65 °C for 10 min to remove secondary structure. Reverse transcription was performed for 1 h at 42 °C in a solution containing 1 μl of RNase inhibitor, 4 μl of 5× RT buffer, 1 μl of 100 mm dNTPs, 1 μl of 80 mm sodium pyrophosphate, and 0.5 μl of avian myeloblastosis virus reverse transcriptase. The reaction was terminated by incubating the mixture at 95 °C for 2 min and was then placed immediately on ice.
PCR Amplification and Cloning of αII and δI Tryptase cDNAs—PCR amplification of first strand cDNA was performed within 2 h of the reverse transcription reaction. Oligonucleotide primers used to amplify cDNAs were designed based on the published sequences of the αII and δI tryptase genes: αII, A2 forward, 5′-AGT GGC CAG GAT GCT GAG C-3′, and A2 reverse, 5′-GGA AGC AGT GGT GTT TTG GAC AG-3′; αII, nested NA2 forward, 5′-GCC TAC GCG GCC CCT GCC CCA GTC-3′, and NA2 reverse, 5′-CCC AGG TGG ACA CCC CAG GCC TGA-3′; δI, DF1, 5′-TGC AGC AAA CGG GCA TTG TTG-3′, and DR1 5′-AAA GCT GTG GCC CGT ATG GAG-3′.
All PCR amplifications were performed with a GeneAmp PCR system 2400 (PerkinElmer Life Sciences). The PCRs were carried out with 2.5 units of AmpliTaq Gold™ (PerkinElmer Life Sciences) and 1 μl of the reverse transcriptase reaction mixture. The total reaction volume was 50 μl with a final concentration of 10 mm Tris/HCl, pH 8.3, 50 mm KCl, 2 mm MgCl2, 0.2 mm dNTPs, and 0.1 μm of the appropriate 5′ and 3′ primers. After an initial incubation for 5 min at 95 °C, samples were subjected to 35 cycles of PCR (45 s at 95 °C, 60 s at 58 °C, and 60s at 72 °C), followed by a final extension step of 72 °C for 10 min.
PCR products (10 μl) were visualized on a 1% agarose gel. Appropriately sized products were excised from the gel, purified with QIAquick gel extraction kit (Qiagen, Valencia, CA), and ligated into the plasmid vector pCR®2.1-TOPO (Invitrogen). The ligation mixture was then used to transform TOP10 Escherichia coli cells (Invitrogen). The transformation mixture was plated onto LB/agar plates containing ampicillin (50 μg/ml) and coated with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal) to enable blue/white color selection.
Screening of Plasmid Colonies—Plasmids containing the appropriate sized inserts were screened by PCR for the presence of αII and δI tryptase inserts using the appropriate primer sets listed above. Plasmid DNA was then purified using a commercial kit (Qiagen). Nucleotide sequencing was performed either in-house using an ABI Prism® BigDye terminator cycle sequencing ready reaction kit and an ABI377 PRISM DNA sequencer (PerkinElmer Life Sciences) or at a core facility (SUPAMAC, Sydney, Australia).
dbEST Data Base Screening—EST databases accessed through the National Institutes of Health information website (www.ncbi.org) were screened using the sequences of the clones identified above.
Real Time PCR Assays—A real time fluorescence-based RT-PCR assay was designed to detect all alternatively spliced (G234–G260) tryptase transcripts isolated in this study. To estimate the approximate ratio of alternatively spliced to full-length tryptase transcripts, an additional assay was designed to detect splice variant α/β/δ tryptase transcripts. Specificity was conferred by designing the forward primer spanning the junction of the deleted nucleotides (splice variant) or locating them in the deleted region (full length). The full-length primer was as follows: forward, 5′-CAC CCT CAG GGT GCA ACT G-3′, and reverse primer, 5′-ATG TAG AAC TGT GGG TGC ACG AT-3′; the splice variant was as follows: forward primer, 5′-TGG GAC CGG TGC AAC TG-3′, and reverse primer, 5′-TAG AAC TGT GGG TGC ACG AT-3′; Taqman probe was as follows: 5′-6-FAM-AGC AGC ACC TCT ACT ACC AGG ACC AGC TG-TAMRA-3′.
Reverse transcription of total human lung, heart, stomach, spleen, and skin RNA (1 μg) was performed using MultiScribe reverse transcriptase and random hexamers (PerkinElmer Life Sciences). The PCR was performed using the ABI Prism 7700 sequence detection system (PerkinElmer Life Sciences). The PCRs were carried out in a 25-μl mixture containing 2 μl of reverse transcript sample, 12.5 μl of 2× universal master mix buffer (PerkinElmer Life Sciences), 600 nm reverse primer, 600 nm forward primer, and 250 nm probe. After initial heating at 50 °C for 2 min, PCR consisted of 45 cycles, including a 15-s denaturation step at 95 °C and a 1-min annealing-extension at 59 °C. Each sample was tested in triplicate. Using the sequence detector system (version 1.7, PerkinElmer Life Sciences), fluorescence intensity was plotted against cycle number. Standard curves were constructed with serial dilutions of plasmid DNA containing a normally spliced (αII) or a splice variant (αIISV) tryptase insert, allowing the copy number to be calculated for each sample using appropriate software (Copy Calculator, version 1.1). Human tryptase transcripts resulting from the alternate splicing pattern described in this study will be annotated hereafter with superscript SV, e.g. αIISV tryptase, βISV tryptase, etc.
The specificity of the primers was tested by examining their ability to amplify cloned templates of full-length or splice variant tryptase cDNAs. Control amplifications were performed in the absence of template DNA.
Three-dimensional Protein Modeling—Three-dimensional structures of the novel tryptase sequences were modeled from the reported 3.0 Å x-ray structure of human βII tryptase (Protein Data Bank identification code 1AOL) (21). The nine-residue segment DVKDLATLR missing in the splice variants was deleted from a single monomer of the structure before energy minimization with AMBER 4.1 (22) to produce a new model. Modified monomers were superimposed on the wild-type tetramer structure for visualization.
Generation of Polyclonal Anti-peptide Antibodies to αIISV Tryptase—To determine whether splice variant tryptases are translated into protein products in vivo, we developed an antibody that recognized αIISV and βISV tryptase, but which may also cross-react with splice variant βII and δ tryptases. Anti-αIISV antibodies were raised in New Zealand White rabbits (Institute of Medical and Veterinary Science, Gilles Plains, SA, Australia) by immunizing with an αIISV-specific peptide, based on the “novel” juxtaposition of residues Pro48 and Val58. Therefore, an immunizing 10-mer peptide, His44-Cys45-Leu46-Gly47-Pro48-Val58-Gln59-Leu60-Arg61-Glu62, was designed consisting of the five amino acid residues on either side of the deletion, conjugated to diphtheria toxin (Mimotopes, Melbourne, Australia). Anti-αIISV antibodies were affinity-purified from antisera using the immunizing peptide conjugated to thiopropyl-Sepharose.
Western Blot Analysis—The specificity of the antibody was confirmed by Western blot. 1 μg of purified recombinant rβISV tryptase and rβI tryptase (Escherichia coli) and recombinant βII tryptase (Promega, Madison, WI) were separated on a 10% SDS-polyacrylamide gel and transferred to a nitrocellulose membrane. After blocking overnight at 4 °C with 5% skim milk powder, phosphate-buffered saline, 0.1% Tween 20, membranes were incubated with affinity-purified αIISV tryptase anti-peptide antibody (1 μg/ml in phosphate-buffered saline, 0.1% Tween 20, 1 h, room temperature). Bound primary antibody was detected using a goat anti-rabbit horseradish peroxidase-conjugated second antibody (Dako, Glostrup, Denmark) diluted 1:1000 for 30 min at room temperature, followed by exposure to a horseradish peroxidase chemiluminescence substrate for 30 s (ECL Western blotting substrate; Pierce). The resulting bands were visualized by exposure to Biomax ML photographic film (Eastman Kodak Co.). The blot was stripped (62.5 mm Tris/HCl, 2% SDS, 10 mm β-mercaptoethanol, pH 6.8) and re-probed with the mouse monoclonal anti-tryptase antibody AA1 diluted 1:1000 (Dako) followed by goat anti-mouse horseradish peroxidase conjugate.
Immunohistochemistry—Immunohistochemistry was performed on 4-μm serial sections cut from formalin-fixed and paraffin-embedded samples of human aorta adventitia, lung, colon, spleen, lymph node, and breast cancer. Sections were deparaffinized, dehydrated, and rinsed in tap water. Antigen retrieval was performed by microwaving the sections in citrate buffer (10 mm trisodium citrate, 0.05% Tween 20, pH 6.0) for 10 min on high and then allowed to cool for 10 min. Sections were then rinsed with TBS and blocked with 20% normal goat serum/TBS at room temperature for 20 min. Sections were incubated with primary antibody diluted in TBS, 2% bovine serum albumin (αIISV tryptase = 4 μg/ml overnight at 4 °C, normal rabbit IgG = 4 μg/ml overnight at 4 °C, and AA1 anti-tryptase antibody = 1:1000-dilution for 1 h at room temperature). Sections were then washed four times for 5 min in TBS and then incubated at room temperature for 30 min with the appropriate biotinylated secondary antibody diluted 1:200 in TBS, 2% bovine serum albumin/goat anti-rabbit for αIISV tryptase and normal rabbit IgG, and goat anti-mouse for AA1 tryptase antibody. Sections were washed four times for 5 min in TBS, incubated with avidin-conjugated alkaline phosphatase (Vector Laboratories, Burlingame, CA) for 30 min at room temperature, and then washed four times for 5 min in TBS. The sections were incubated in the dark for ∼15 min with alkaline phosphatase substrate (Vector Red, Vector Laboratories), which gives a red reaction product in the presence of alkaline phosphatase. All incubations were performed in a humidified chamber. Sections were rinsed in tap water, counterstained with hematoxylin for 30 s, rinsed in tap water and coverslipped with CrystalMount (Biomeda, Foster City, CA). Stained sections were examined using an Olympus BX-60 microscope and images captured using a SPOT digital camera (Diagnostic Instruments, Sterling Heights, MI).
Generation of Recombinant βI Tryptase and βISV Tryptase—When compared with their full-length counterparts, the splice variant tryptases identified in this study have a 9-amino acid deletion, Asp49–Arg57. To determine whether a splice variant tryptase may be proteolytically active, recombinant βI tryptase and its splice variant βISV were expressed in bacterial cells to test splice variant antibody specificity and in yeast to test for the ability to cleave a trypsin-sensitive substrate.
The recombinant fusion protein included an N-terminal His6 tag (to enable purification), an enterokinase (EK) recognition site (to allow activation of the pro-enzyme), and the mature βIor βISV tryptase sequence. The construct was made in bacteria (E. coli) using the pET30 EK/ligation-independent cloning vector (Merck), and the following primers: forward, 5′-GAC GAC GAC AAG AAT ATC GTC GGG GGT CAG-3′, and reverse; 5′-GAG GAG AAG CCC GGT TCA CGG CTT TTT GGG-3′, using ligation independent cloning, and the construct was used to transform BL21 DE3 cells. Following the addition of isopropyl β-thiogalactopyranoside (0.5 mm final concentration), the bacterial cells were incubated for 6 h at 37 °C while being agitated vigorously. The cells were pelleted by centrifugation and resuspended in lysis buffer (Cellytic B, Sigma). The lysate was centrifuged to remove cellular debris, and the His-6 tagged recombinant protein was purified from the supernatant using a nickel-nitrilotriacetic acid column (Qiagen).
The construct was made in yeast (Pichia pastoris) by insertion of the mature βIor βISV tryptase sequence into the pPIC9κ vector (Invitrogen), similar to a method described previously (23), using the following primers: EKBI forward, 5′-GAT GAC GAC GAC AAG ATC GTT GGG GGT-3′, and BII reverse, 5′-ACG ACG GCG GCC GCT TCA CGG CTT TTT GGG GAC ATA GTG-3′; HISEKF forward, 5′-CAT CAT CAT CAT CAT CAC GAT GAC GAC GAC AAG-3′; KEXHIS forward, 5′-GGG CCC CTC GAG AAA AGA CAT CAT CAT CAT CAT CAT-3′. These primers were used to amplify βII tryptase sequence and to add a His6 and EK cleavage site upstream of the mature tryptase sequence. The KEXHIS forward primer encodes the KEXI cleavage site, as well as an XhoI restriction site. The reverse primer encodes a NotI restriction site and the mature β tryptase sequence. The vectors were linearized with SalI, and GS115 cells were transformed using electroporation (XCell II, Bio-Rad). These cells were grown on minimal dextrose/agar plates, followed by minimal methanol/agar plates to screen for colonies expressing βISV tryptase, as described previously (24). Positive colonies were grown in BMMY media, containing 1% methanol.
Recombinant βI and βISV tryptase purified as above were activated for 16 h at room temperature in cleavage buffer (10 mm MES, 150 mm NaCl, 200 mm CaCl2, pH 6.1) using 1 unit of EK. After enzyme activation, EK was removed using EKapture-agarose beads (EMD Biosciences-Novagen) as per the manufacturer's instructions.
The enzymatic activity of the recombinant βISV tryptase was evaluated by testing its ability to cleave peptide substrates, and Nα-benzoyl-dl-arginine p-nitroanilide (BAPNA) (Sigma) or N-(p-tosyl)-Gly-Pro-Arg p-nitroanilide (tosyl-GPR-pNA) (Sigma), or the physiological substrate human fibronectin. The cleavage pattern was compared with that of βI tryptase.
For the tosyl-GPR-pNA assay, βI tryptase was purified using a nickel-nitrilotriacetic acid column (Qiagen), whereas βISV tryptase was purified using Affi-Gel Hz (Bio-Rad) conjugated to the AA1 tryptase antibody (Dako), as per the manufacturers' protocol. EK activated and unactivated (as described above) βI and βISV tryptase (25 μl), and an EK control was incubated with tosyl-GPR-pNA (0.5 μl, 20 mg/ml) in 200 μl of Dulbecco's phosphate-buffered saline, pH 7.4, for 2 h at room temperature. The plate was read at 405 nm. The EK control consisted of 1 unit of EK, which was then removed using Ekapture-agarose beads; this was done to control for any residual EK in the tryptase preparations activated with EK.
For the BAPNA assay, yeast supernatants were concentrated five times using a Microcon centrifugal filter (Millipore, Billerica, MA), and 40 μl of concentrated supernatant was added to 110 μl of reaction buffer (2 mm BAPNA, 100 mm Tris, HCl, 1 m glycine, pH 8.0) in a 96-well plate and incubated for 1 h at 37 °C. The plate was read at 405 nm.
Human fibronectin (1 μg; Invitrogen) was added to the activated tryptases and incubated for 18 h at room temperature. To ensure tetramer formation, heparin (1 mg, Sigma) was added to the activated enzymes. Digests were then separated on a non-reduced 7.5% Tris-Tricine SDS-polyacrylamide gel, fixed, and visualized by silver staining.
RESULTS
Identification and Cloning of Alternatively Spliced Human Tryptase cDNAs—RT-PCR was performed using αII tryptase-specific primers on total RNA derived from human lung and using δ tryptase-specific primers on total RNA derived from the HMC-1 cell line. When the resulting PCR products were electrophoresed and visualized on an agarose gel, the amplimers from each reaction appeared to run as a “doublet.” One amplimer in each lane was of the expected size for a human tryptase, and the second, or doublet, was slightly smaller (data not shown). All PCR products were isolated from the gel, cloned into the pCR2.1 vector, and sequenced.
Sequencing revealed that the larger cDNA generated using αII tryptase primers was 884 nucleotides in length and included an ATG translation start codon and TGA stop codon at position 826. The sequence of this fragment (Fig. 1A, submitted as GenBank™ accession number AF206665) matched the predicted exonic sequence of the αII tryptase gene (4). Sequencing of the smaller cDNA generated using these same primers revealed an 805-bp cDNA fragment that matched the αII-tryptase cDNA exactly, except for a 27-nucleotide deletion from nucleotides 234 to 260 (Fig. 1A, submitted as GenBank™ accession number AF206666).
FIGURE 1.
A, cDNA and putative amino acid sequence of αII and αIISV tryptase. The top line represents the translated amino acid sequence; the middle represents the nucleotide sequence ofαII tryptase (uppercase); and the bottom line represents the nucleotide sequence of αIISV tryptase (lowercase). The nucleotides deleted in the splice variant are denoted by a hyphen. Nucleotide numbering begins from the translation initiation codon (Met). B, partial cDNA and amino acid sequences of δI and δISV tryptase. C, partial cDNA and amino acid sequences of βI and βISV tryptase. D, partial cDNA and amino acid sequences of βIII and βIIISV tryptase.
Analysis of cloned PCR products generated using the δ tryptase-specific primers revealed similar results. The sequence of the larger PCR product matched that of δI tryptase, as described by us previously (5). The sequence of the smaller cDNA also matched that of δI tryptase, except for the same 27-nucleotide deletion present in the truncated αII cDNA described above (Fig. 1B, submitted as GenBank™ accession number AF421357).
Using the truncated tryptase sequences as a query, a BLAST search of dbEST data base identified two similarly truncated human transcripts. The sequence of the first (GenBank™ accession number T50832) was isolated from human fetal spleen (25) and matched that of βI tryptase, but possessed the 27-nucleotide deletion as described above. The original clone was obtained (ID code 72786, Genome Systems Inc., St. Louis, MO) and sequenced (partial sequence shown in Fig. 1C, full sequence submitted as GenBank™ accession number AF206667). The second transcript (GenBank™ accession number BQ720404) matched the sequence of βIII tryptase (again with the 27-nucleotide deletion) and was isolated from sympathetic trunk (Fig. 1D).
Using oligonucleotide primers specific for β tryptases, additional amplicons were obtained by RT-PCR of human lung mRNA (data not shown). These PCR products were cloned into the pCR2.1 vector and sequenced. Clones were identified with sequences matching that of full-length βI, βII, and βIII tryptase cDNAs (data not shown), as well as a truncated βI tryptase cDNA containing the 27-nucleotide deletion. This confirms that truncated transcripts of both αII and βI tryptase are present in human lung.
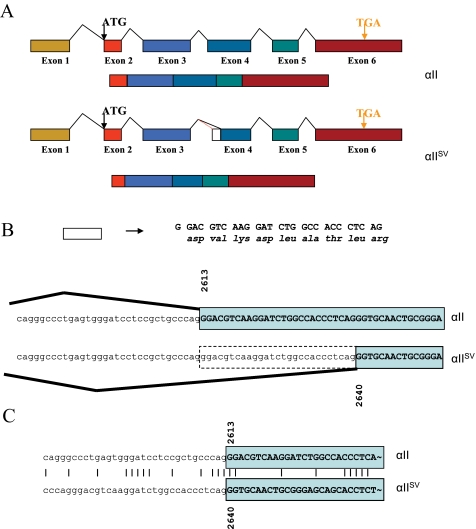
mRNA Splicing Pattern—Comparison of the sequence of the truncated transcripts of αII, βI, βIII, and δI tryptase transcripts with their full-length counterparts and the corresponding genes (located on chromosome 16p13.3) (αII, AF098328; βI, M33491; βI, M33492; βIII, M33493; δI, AY055427) reveals that the truncated transcripts are the result of alternate mRNA splicing. A comparison of the splicing pattern for the splice variant and truncated transcripts is shown diagrammatically for the αII tryptase gene only (Fig. 2). In the conventionally spliced transcript, intron 3 extends to the 3′ splice site at AG2612 (nucleotide numbering taken from GenBank™ entry AF098328), with exon 4 beginning at G2613 (Fig. 2, A and B). In mRNA splicing that leads to the production of the truncated transcript, there is recognition of an alternate 3′ splice site at AG2639, with the new exon 4 now beginning at G2640. The effect of this alternate splicing pattern is that exon 4 is shortened by 27 nucleotides and consequently 9 amino acids, with no shift in the reading frame.
FIGURE 2.
mRNA splicing pattern. A, in αIISV tryptase, recognition of a novel 3′ splice site within exon 4 results in shortening of exon 4. B, detail of intron 3/exon 4 splice site junctions for αII and αIISV tryptase. C, nucleotide sequence alignment of intron 3/exon 4 splice site junctions for αII and αIISV tryptase.
Some recent reports suggest that the nucleotide sequence consensus of alternate splice sites is weak when compared with that of normal splice sites (26). To investigate this for human tryptases, we aligned the nucleotide sequence of the normal αII tryptase intron three 3′ splice site with that of the alternate 3′ splice site (Fig. 2C). When the sequence region surrounding the novel intron 3/exon 4 boundary was compared with the normal intron 3/exon 4 boundary, a high degree of similarity was observed, with four of the last five nucleotides of the respective introns being identical, as well as the first two nucleotides of the exon. Additionally, data base searches revealed that no SNPs have been detected in this region (www.ncbi.nlm.nih).
Comparison of Deduced Amino Acid Sequences—The deduced amino acid sequences of αIISV, βISV, βIIISV, and δISV tryptase were aligned with the published sequences of their full-length counterparts and βII tryptase and annotated with the location of the catalytic triad and the 47 loop (Fig. 3). The six surface loops (21) are named the 22 (37), 47 (60), 62–69 (70–80), 86 (97), 136 (147), and 163 (173) loops, respectively; chymotrypsin numbering is given in parentheses.
FIGURE 3.
Amino acid sequences of αIISV, βISV, and δISV tryptase compared with their full-length counterparts and to βII tryptase. A dash indicates the presence of an identical amino acid. Numbering begins at the first residue of the mature enzyme, which is indicated by a down arrow. The location of the 47 loop is indicated by a thick bar. His, Asp, and Ser of the catalytic triad are marked with a number sign.
Importantly for the potential proteolytic activity of the alternately spliced tryptases, no residues of the catalytic triad (Ser194, His44, and Asp91) are located in the deleted nine amino acid segment. However the deletion is located only five amino acids C-terminal to the His44 residue.
The major structural consequence of the deletion is the loss of all but the initial proline residue of the 47 loop, PDVKD. This loop has been identified as a crucial component of one of the monomer-monomer interaction sites that mediate the formation of tetramers (21).
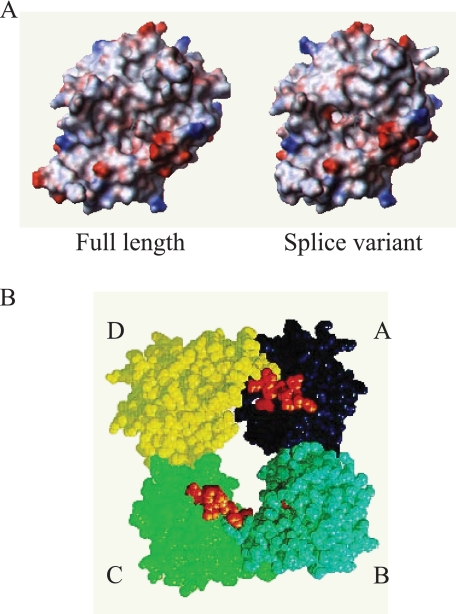
Modeling of Splice Variant Tryptases—Fig. 4A includes surface electrostatic models of αII tryptase and αIISV tryptase monomers. Deletion of the nine-residue segment absent from the 47 loop in the splice variants can be accommodated relatively easily by the structure but causes significant changes in the vicinity of the active site. The major result is the formation of an enlarged pocket behind His44 that could provide a new S1′ or S2′ binding pocket. In addition to this local structural alteration at the active site, loop truncation also affects the A–D interface of the tryptase tetramer, where the 47 loop of monomer D contacts the 163 loop of monomer A (Fig. 4B). Alteration of this contact may profoundly affect the ability of splice variant tryptases to form tetramers.
FIGURE 4.
A, three-dimensional surface electrostatic models of wild-type (or full-length) αII tryptase (left panel) and αIISV tryptase (right panel). B, surface representation of βII tryptase as reported by Pereira et al. (21) showing the tryptase tetramer ring structure. The 9 amino acids that are deleted in the splice variants are colored orange, and due to the orientation of the individual monomers they are visible in the A and C monomers only. Monomers A–D are depicted as dark blue, light blue, green and yellow, respectively.
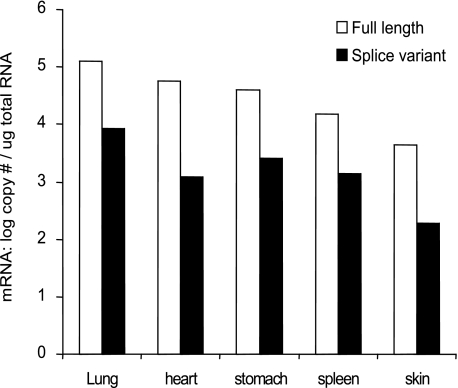
In Vivo Splice Variant Tryptase mRNA Expression—The abundance of splice variants and full-length tryptase transcripts was estimated in various human tissues (lung, heart, stomach, spleen, and skin) using a real time PCR assay. The expression of splice variant tryptase transcripts was highest in the lung (8.7 × 103 copies/μg RNA) but was also detected (in order of abundance) in the stomach, spleen, heart, and skin (Fig. 5). The highest proportion of full-length to splice variant transcripts was detected in the spleen (∼10%), and the lowest proportion was detected in the heart (∼2%).
FIGURE 5.
Splice variant tryptases are transcribed in various tissues. The relative abundance of splice variant tryptase transcripts in a range of human tissues was determined using RT-quantitative PCR. The data represents the mean (± S.D.) from a single experiment. All samples were tested in triplicate and have been tested in at least two independent experiments.
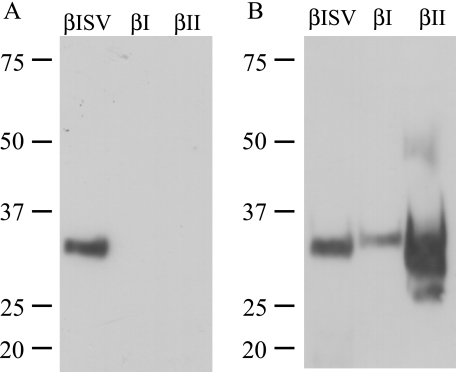
Specificity of Anti-splice Variant Tryptase Antibody—A Western blot analysis revealed that the anti-αIISV anti-peptide antibody specifically recognizes recombinant αIISV (data not shown) and βISV (Fig. 6) tryptase but not full-length αII tryptase, βI tryptase, or a commercial preparation of recombinant βII tryptase. The commercially available AA1 antibody recognized recombinant αII, αIISV, βI, βISV, and as expected recombinant βII tryptase.
FIGURE 6.
A, affinity-purified anti-αIISV tryptase antibody recognizes rβISV tryptase but not rβIorrβII tryptase. B, anti-tryptase antibody AA1 recognizes rβISV, rβI, and rβII tryptases. Sizes are as indicated in kDa. rβISV and rβI tryptases were expressed in E. coli.
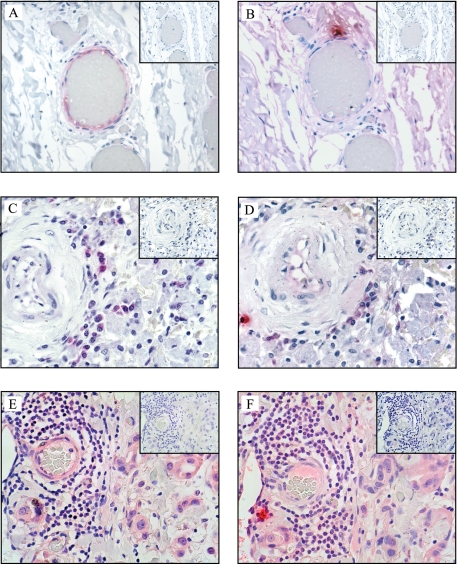
Immunohistochemistry—Immunohistochemical analysis of a limited range of formalin-fixed paraffin-embedded human tissue specimens revealed expression of splice variant tryptase protein in sections of aorta, spleen, and breast tumor (Fig. 7). Expression was most prominent in the endothelial cells of some blood vessels surrounding the aorta, as well as blood vessels surrounding the tumor. These cells stained only weakly with the AA1 anti-tryptase antibody, suggesting they may be expressed at lower levels than full-length tryptases. This was consistent with the finding that AA1 can recognize splice variant tryptase protein by Western blotting (Fig. 6). A number of mast cells in the section stained strongly with AA1 antibody but were negative for the splice variant tryptases, indicating that at least in these sections mast cells were not expressing splice variant tryptases. No positive-staining cells were observed in any tissue when the primary antibody was omitted or when normal nonimmune rabbit Ig was used as the primary antibody.
FIGURE 7.
Splice variant tryptase protein is expressed in human aorta (adventitia), spleen, and breast tumor tissues. Sections were stained immunohistochemically using anti-αIISV tryptase antibody (A, C, and E), the monoclonal anti-tryptase antibody AA1 (B, D, and F), and isotype control sections are shown in the inset. A and B, C and D, and E and F are serial sections.
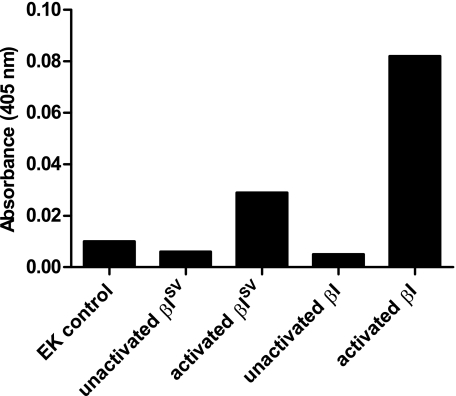
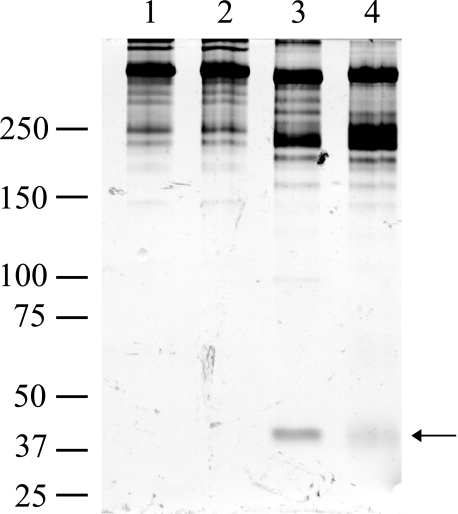
Expression and Activity of Recombinant βISV Tryptase—rβI and rβISV tryptase were expressed in yeast cells (P. pastoris), and their ability to cleave several substrates was investigated. Purified and EK-activated rβI and rβISV tryptase were able to cleave the chromogenic substrates tosyl-GPR-pNA (Fig. 8) and BAPNA (data not shown). The EK control (as described under “Experimental Procedures”) had activity similar to unactivated tryptase. Although rβISV tryptase appeared to have less proteolytic activity for the small substrate BAPNA than rβI tryptase (Fig. 8), cleavage of the large substrate human fibronectin was more effective by rβISV tryptase (Fig. 9), and the pattern of fibronectin cleavage products was different for rβISV tryptase compared with rβI tryptase (compare lanes 3 and 4, Fig. 9).
FIGURE 8.
βISV tryptase expressed in yeast and activated by EK cleavage of the propeptide cleaves the substrate tosyl-GPR-pNA, whereas unactivated βISV tryptase does not. EK activated, but not unactivated, βI tryptase also cleaves tosyl-GPR-pNA. The EK control had a similar level of activity as the unactivated proteins.
FIGURE 9.
βISV tryptase cleaves human fibronectin. Purified and activated βISV tryptase (lane 3) and βI tryptase (lane 4) were incubated with human fibronectin (1 mg) in the presence of heparin for 18 h at room temperature. Samples were separated on a nonreduced 7.5% Tris-Tricine SDS-polyacrylamide gel and silver-stained. Arrow indicates a fibronectin digestion band. Controls are human fibronectin alone (lane 1) and fibronectin with EK (lane 2). Sizes are as indicated in kDa.
DISCUSSION
Recent reports indicate that the human genome contains more tryptase or tryptase-like genes than originally anticipated. Here we report that the diversity of functionally distinct human tryptases may be further increased through the process of alternate splicing of pre-mRNA transcribed from multiple tryptase genes. We isolated or identified alternately spliced transcripts of αII tryptase, βI tryptase, βIII tryptase, and δI tryptase. As αII and βI tryptase are likely allelic variants of the one gene locus (4), this pattern of alternate splicing is present in at least three separate tryptase gene loci.
The consequence of the alternate splicing is a deletion of 27 nucleotides from the internal region of the mRNA (when compared with their full-length counterparts) and a deletion of nine amino acids from the translated protein product. The alternate splicing pattern arises from suppression of the normal 3′ splice site of intron 3 and the activation of an alternate or cryptic 3′ splice site within what is normally exon 4. This form of alternate splicing is often termed exon shortening. In a review of protein isoform diversity and alternate splicing patterns, Nakao et al. (26) reported that the pattern of alternate splicing leading to an in-frame insertion or deletion into the central part of a protein is one of the most important and conserved splicing mechanisms in the human genome, accounting for ∼20% of splicing events.
Up to 60% of human genes show alternate splicing. However a large number of alternate splicing events cause frameshift and the introduction of premature termination codons. This often results in the rapid degradation of the transcript preventing translation, as we have demonstrated for the mouse tryptase gene, mmcp-7, in the C57BL/6 mouse strain (27). Under these circumstances, the alternately spliced mMCP-7 transcript was barely detectable, and no protein product could be demonstrated. Alternately spliced transcripts that do not contain premature termination codons are not often targeted for such rapid degradation (27). The alternately spliced products described in this study do not contain premature stop codons; the transcripts were easily detected, and their protein products were detected in vivo.
It is now evident that many alternate splicing events involve the deletion or insertion of whole protein domains without disrupting the rest of the polypeptide (28). The consequence of the alternate splicing mechanism described here is the deletion of 9 amino acids from the center of the molecule, and located within this 9-amino acid segment is the 47 loop, a domain reported to be crucial in the formation of tryptase tetramers. Pereira et al. (21), in a landmark study, reported the crystal structure of human βII tryptase and concluded that all monomer-monomer contacts are made through the six loops that form ridges around the substrate binding cleft as follows: the 22, 47, 62–69, 86, 136, and the 163 loops, respectively. If the four monomers that comprise the tetramer are named A, B, C, and D in a clockwise manner (see Fig. 4B), three of the loops are involved in the binding of monomer A to B (and C to D), the 147 loop, the 62–69 loop, and the 22 loop. The other three loops are involved in the binding of monomer A to monomer D (and B to C). In this interaction the 86 loops of each monomer bind to each other, whereas the respective 163 flaps from each monomer interact with the 47 loop of the opposing monomer. The phenolic side chain of Tyr167 inserts into a hydrophobic cleft made by the 47 loop of one monomer and the 86 loop of the other. Therefore, there are two different monomer-monomer interactions as follows: the monomer A to monomer D (and B to C) interaction, which has a surface area of ∼1100 Å, and the monomer A to monomer B (and C to D) interaction, with a surface area of ∼500 Å. Although intrinsically a weaker bond, it is thought that the A to B (and B to C) interaction is strengthened by the linking of positively charged patches on each monomer with heparin. Pereira et al. (21) postulated that this AB (or CD) bond may be a “natural shear point,” where the tetramer could first break into AD (BC) dimers and finally into monomers. Because of the crystallographic evidence concerning the importance of the 47 loop to the formation of tryptase tetramers, we postulate that the absence of this loop in the splice variant tryptases will prevent them from forming tetramers. However, it is not clear whether they will be able to form homodimers, interacting through the AB interface only and stabilized with heparin, or will exist only as monomers.
Restriction of the splice variant tryptases to either the monomer or dimer configuration would have a significant effect on their biological activity. The biological characteristics of the normal tetrameric tryptases, i.e. a preference for short peptide substrates rather than larger proteins, and the lack of effect of any of the known endogenous protease inhibitors, is dictated by the tetrameric formation and the location of the active sites around the central pore. Presumably, large protein substrates and inhibitors would be excluded from the small (50 × 30 Å) pore. According to Pereira et al. (21), it is not the surface loops that restrict access of large potential substrates to the active site but the presence of the neighboring monomers. Based on this we would predict two new biological characteristics of the splice variant tryptases (in either the monomer or dimer configuration); their activity will be inhibited by endogenous protease inhibitors that are normally ineffective against tetrameric tryptase, and they will be more efficient in cleaving large protein substrates.
There is evidence to support this hypothesis from studies of the mouse tryptase mMCP-6. Hallgren et al. (29) reported that they could produce active monomers and tetramers of mMCP-6 in vitro, by incubating mMCP-6 with heparin-oligosaccharide of 10 and 20 unit lengths, respectively. Monomeric mMCP-6 cleaved a large protein, fibronectin, and its proteolytic activity against a tryptase sensitive substrate (H-d-Ile-Pro-Arg-pNa) was inhibited by bovine pancreatic trypsin inhibitor, whereas tetrameric mMCP-6 was unable to cleave fibronectin and was resistant to inhibition by bovine pancreatic trypsin inhibitor. On the basis of homotetrameric structure, several human tryptase inhibitors have been designed (30, 31) and used in therapeutic trials for asthma (11) and ulcerative colitis (12). The alternatively spliced tryptases described in this study may provide new models for the design of tryptase inhibitors.
Our results indicate that alternately spliced tryptase transcripts are detected in a wide range of tissues, but the protein may have a more restricted pattern of expression. Our preliminary immunohistochemical findings suggest that splice variant tryptases (at least αIISV and βISV recognized by our anti-peptide antibody) are expressed in vivo but are rarely expressed in the same cells as their full-length counterparts. This would be consistent with the literature regarding the expression of alternately spliced transcripts, as the full-length and splice variant transcripts would require the expression and activity of different splicing factors. However, it may be that our antibody is not detecting splice variants of other tryptases such as βIII and δI tryptases, which may be expressed in mast cells. Future studies are needed to determine exactly which cell types (e.g. the type of blood vessels), and under what conditions spliced variant tryptases are expressed.
We chose to initially examine the recombinant form of a single splice variant tryptase as a “proof of principle” for splice variant tryptases in general. We expressed βISV tryptase because the activity of αII tryptase remains controversial. It has variously been reported that recombinant forms of αII are proteolytically active (32), have reduced activity (33), or have no proteolytic activity (34). The results of our study indicate that recombinant forms of βI and βISV tryptase both cleave two tryptase-sensitive substrates, tosyl-GPR-pNA and BAPNA, although there is no indication that this is a preferred substrate of either. In addition, βISV tryptase was able to cleave the physiological substrate fibronectin, and this cleavage was more efficient than that of βI tryptase. The ability of βI tryptase to cleave fibronectin was most likely due to incomplete tetramer formation, with residual monomers or dimers that were able to cleave fibronectin. However, it does demonstrate that βISV tryptase is able to cleave a large physiological substrate, namely fibronectin.
Although the crystal structure elegantly answered many of the questions concerning the unique biochemical and biological characteristics of tetrameric human tryptase, a conundrum remained; tryptase was known to cleave large protein substrates that could not possibly be accommodated within the central pore. A number of reports have suggested in vitro mechanisms of producing proteolytically active monomers, but none are realistic propositions for an in vivo mechanism. Here we report that alternate splicing results in the expression of two forms of each tryptase gene as follows: a full-length version and a splice variant with an internal 9-amino acid deletion. We predict that the full-length version will form tetramers, be restricted to cleaving short peptide substrates, and be resistant to endogenous serine protease inhibitors, although the splice variant will not form tetramers, will be able to cleave large protein substrates, and will be sensitive to endogenous protease inhibitors. This model would explain all the observed biological characteristics of tryptases.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBank™/EBI Data Bank with accession number(s) AF206665, AF206666, AF206667 and AF421357.
This work was supported by the National Health and Medical Research Council of Australia and Australian Research Council Linkage Grant LP0455637. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: HMC-1, human mast cell-1; BAPNA, Nα-benzoyl-dl-arginine p-nitroanilide; EK, enterokinase; SV, splice variant; tosyl-GPR-pNA, N-(p-tosyl)-Gly-Pro-Arg-p-nitroanilide; RT, reverse transcription; TBS, Tris-buffered saline; MES, 4-morpholineethanesulfonic acid; Tricine, N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine.
References
- 1.Schwartz, L., Lewis, R., and Austen, K. (1981) J. Biol. Chem. 256 11939-11943 [PubMed] [Google Scholar]
- 2.Vanderslice, P., Ballinger, S., Tam, E., Goldstein, S., Craik, C., and Caughey, G. (1990) Proc. Natl. Acad. Sci. U. S. A. 87 3811-3815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caughey, G. H., Raymond, W. W., Blount, J. L., Hau, L. W.-T., Pallaoro, M., Wolters, P. J., and Verghese, G. M. (2000) J. Immunol. 164 6566-6575 [DOI] [PubMed] [Google Scholar]
- 4.Pallaoro, M., Fejzo, M. S., Shayesteh, L., Blount, J. L., and Caughey, G. H. (1999) J. Biol. Chem. 274 3355-3362 [DOI] [PubMed] [Google Scholar]
- 5.Wang, H.-W., McNeil, H. P., Husain, A., Liu, K., Tedla, N., Thomas, P. S., Raftery, M., King, G. C., Cai, Z. Y., and Hunt, J. E. (2002) J. Immunol. 169 5145-5152 [DOI] [PubMed] [Google Scholar]
- 6.Wong, G. W., Yasuda, S., Madhusudhan, M. S., Li, L., Yang, Y., Krilis, S. A., Sali, A., and Stevens, R. L. (2001) J. Biol. Chem. 276 49169-49182 [DOI] [PubMed] [Google Scholar]
- 7.Harvima, R. J., Harvima, I. T., Dull, D., Dunder, U. K., and Schwartz, L. B. (1999) Arch. Dermatol. Res. 291 73-80 [DOI] [PubMed] [Google Scholar]
- 8.Peng, Q., McEuen, A. R., Benyon, R. C., and Walls, A. F. (2003) Eur. J. Biochem. 270 270-283 [DOI] [PubMed] [Google Scholar]
- 9.Clark, J. M., Abraham, W. M., Fishman, C. E., Forteza, R., Ahmed, A., Cortes, A., Warne, R. L., Moore, W. R., and Tanaka, R. D. (1995) Am. J. Respir. Crit. Care Med. 152 2076-2083 [DOI] [PubMed] [Google Scholar]
- 10.Sekizawa, K., Caughey, G., Lazarus, S., Gold, W., and Nadel, J. (1989) J. Clin. Investig. 83 175-179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krishna, M. T., Chauhan, A., Little, L., Sampson, K., Hawksworth, R., Mant, T., Djukanovic, R., Lee, T., and Holgate, S. (2001) J. Allergy Clin. Immunol. 107 1039-1045 [DOI] [PubMed] [Google Scholar]
- 12.Tremaine, W. J., Brzezinski, A., Katz, J. A., Wolf, D. C., Fleming, T. J., Mordenti, J., Strenkoski-Nix, L. C., and Kurth, M. C. (2002) Aliment. Pharmacol. Ther. 16 407-413 [DOI] [PubMed] [Google Scholar]
- 13.He, S., Gaca, M. D. A., and Walls, A. F. (2001) Eur. J. Pharmacol. 412 223-229 [DOI] [PubMed] [Google Scholar]
- 14.Gotis-Graham, I., and McNeil, H. P. (1997) Arthritis Rheum. 40 479-489 [DOI] [PubMed] [Google Scholar]
- 15.Schwartz, L. B., Sakai, K., Bradford, T. R., Ren, S., Zweiman, B., Worobec, A. S., and Metcalfe, D. D. (1995) J. Clin. Investig. 96 2702-2710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buckley, M. G., Variend, S., and Walls, A. F. (2001) Clin. Exp. Allergy 31 1696-1704 [DOI] [PubMed] [Google Scholar]
- 17.Holgate, S. T., Hardy, C., Robinson, C., Agius, R. M., and Howarth, P. H. (1986) J. Allergy Clin. Immunol. 77 274-282 [DOI] [PubMed] [Google Scholar]
- 18.Huang, C., Friend, D. S., Qiu, W.-T., Wong, G. W., Morales, G., Hunt, J., and Stevens, R. L. (1998) J. Immunol. 160 1910-1919 [PubMed] [Google Scholar]
- 19.He, S., Peng, Q., and Walls, A. (1997) J. Immunol. 159 6216-6225 [PubMed] [Google Scholar]
- 20.Gamba, G. (2001) Am. J. Physiol. 281 F781-F794 [DOI] [PubMed] [Google Scholar]
- 21.Pereira, P. J. B., Bergner, A., Macedo-Ribeiro, S., Huber, R., Matschiner, G., Fritz, H., Sommerhoff, C. P., and Bode, W. (1998) Nature 392 306-311 [DOI] [PubMed] [Google Scholar]
- 22.Pearlman, D., Case, D., Caldwell, J., Ross, W., Cheatham, T., Ferguson, D., Seibel, G., Singh, U., Weiner, P., and Kollman, P. (1995) AMBER, Version 4.1, University of California, San Francisco, CA
- 23.Niles, A. L., Maffitt, M., Haak-Frendscho, M., Wheeless, C. J., and Johnson, D. A. (1998) Biotechnol. Appl. Biochem. 28 125-131 [PubMed] [Google Scholar]
- 24.Wung, J. L., and Gascoigne, N. R. (1996) BioTechniques 21 808, 810, 812 [DOI] [PubMed] [Google Scholar]
- 25.Hillier, L. D., Lennon, G., Becker, M., Bonaldo, M. F., Chiapelli, B., Chissoe, S., Dietrich, N., DuBuque, T., Favello, A., Gish, W., Hawkins, M., Hultman, M., Kucaba, T., Lacy, M., Le, M., Le, N., Mardis, E., Moore, B., Morris, M., Parsons, J., Prange, C., Rifkin, L., Rohlfing, T., Schellenberg, K., and Marra, M. (1996) Genome Res. 6 807-828 [DOI] [PubMed] [Google Scholar]
- 26.Nakao, M., Barrero, R. A., Mukai, Y., Motono, C., Suwa, M., and Nakai, K. (2005) Nucleic Acids Res. 33 2355-2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hunt, J. E., Stevens, R. L., Austen, K. F., Zhang, J., Xia, Z., and Ghildyal, N. (1996) J. Biol. Chem. 271 2851-2855 [DOI] [PubMed] [Google Scholar]
- 28.Cline, M., Shigeta, R., Wheeler, R., Siani-Rose, M., Kulp, D., and Loraine, A. (2004) Pac. Symp. Biocomput. 17-28 [DOI] [PubMed]
- 29.Hallgren, J., Spillmann, D., and Pejler, G. (2001) J. Biol. Chem. 276 42774-42781 [DOI] [PubMed] [Google Scholar]
- 30.Burgess, L. E., Newhouse, B. J., Ibrahim, P., Rizzi, J., Kashem, M. A., Hartman, A., Brandhuber, B. J., Wright, C. D., Thomson, D. S., Vigers, G. P. A., and Koch, K. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 8348-8352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rice, K., Tanaka, R., Katz, B., Numerof, R., and Moore, W. (1998) Curr. Pharm. Des. 4 381-396 [PubMed] [Google Scholar]
- 32.Mirza, H., Schmidt, V. A., Derian, C. K., Jesty, J., and Bahou, W. F. (1997) Blood 90 3914-3922 [PubMed] [Google Scholar]
- 33.Huang, C., Li, L., Krilis, S. A., Chanasyk, K., Tang, Y., Li, Z., Hunt, J. E., and Stevens, R. L. (1999) J. Biol. Chem. 274 19670-19676 [DOI] [PubMed] [Google Scholar]
- 34.Selwood, T., Wang, Z.-M., McCaslin, D. R., and Schechter, N. M. (2002) Biochemistry 41 3329-3340 [DOI] [PubMed] [Google Scholar]