Abstract
Human T-cell leukemia virus type 1 (HTLV-1) encodes an antisense viral gene product termed HTLV-1 basic leucine-zipper factor (HBZ). HBZ forms heterodimers with c-Jun, a member of the AP-1 family, and promotes its proteasomal degradation. Although most proteasomal substrates are targeted for degradation via conjugation of polyubiquitin chains, we show that ubiquitination is not required for HBZ-mediated proteasomal degradation of c-Jun. We demonstrate that HBZ directly interacts with both the 26 S proteasome and c-Jun and facilitates the delivery of c-Jun to the proteasome without ubiquitination. HBZ acts as a tethering factor between the 26 S proteasome and its substrate, thereby bypassing the targeting function of ubiquitination. These findings disclose a novel viral strategy to utilize the cellular proteolytic system for viral propagation.
Protein degradation plays a variety of roles in fundamental cellular processes, including the cell cycle, apoptosis, immune response, and disposal of misfolded or oxidized proteins (1-3). The ubiquitin-proteasome system has evolved as a key machinery in the selective degradation of intracellular short-lived regulatory or abnormal proteins. Most proteasomal substrates are tagged with polyubiquitin, which serves as a recognition signal for the 26 S proteasome. The cellular machinery that adds ubiquitin to substrates consists of three main enzyme classes. The E1 ubiquitin-activating enzyme transfers activated ubiquitin to the E2 ubiquitin-conjugating enzyme, which, in combination with E3 ubiquitin ligase, transfers ubiquitin to the substrate. The E3 ubiquitin ligase associates with the substrate and confers substrate specificity (3).
The proteasome is a major nonlysosomal proteolytic apparatus. The catalytic core of this multisubunit proteolytic complex is the 20 S proteasome. The addition of a 19 S regulatory complex to either or both ends of the 20 S proteasome forms the 26 S proteasome. The 19 S regulatory complex recognizes polyubiquitin chains on substrates and catalyzes deubiquitination, denaturation, and translocation of the unfolded substrate into the 20 S catalytic core for degradation (4). Thus, the substrates for the actual proteolysis are unfolded and nonubiquitinated. Therefore, if a protein can be delivered to the proteasome in a denatured or partially unfolded state, ubiquitination should not be required for its degradation (5, 6). Recently, there have been emerging reports of proteasome-dependent, ubiquitin-independent degradation of eukaryotic proteins (7), including ornithine decarboxylase (ODC)2 (8), p53 (9), p21waf1/cip1 (10, 11), and retinoblastoma (Rb) protein (12, 13), suggesting the significance of this alternative pathway for various cellular events. However, the mechanism by which proteasomes recognize nonubiquitinated substrates is not well understood.
A variety of viruses are known to utilize the host ubiquitin-proteasome system to dysregulate cellular functions for their benefit. Notably, human papillomavirus type-16 E6 protein acts as a part of the E3 ubiquitin ligase complex to promote ubiquitination and subsequent degradation of the tumor suppressor, p53 (14). There are an increasing number of viral proteins that utilize cellular ubiquitination machinery (15-17), suggesting that the cellular proteolytic system is an important tool for viral propagation.
Human T-cell leukemia virus type-1 (HTLV-1) causes adult T-cell leukemia in 2-5% of carriers after a long latent period (18). HTLV-1-induced disruption of cellular transcription is associated with the development of adult T-cell leukemia. Tax, one of the HTLV-1-encoded proteins, is postulated to play a pivotal role in the development of adult T-cell leukemia (19). Although Tax promotes proliferation and inhibits apoptosis of infected cells, it is a major target of cytotoxic T lymphocytes (20), thus the overexpression of viral protein is disadvantageous for the survival of infected cells. There are significant involvements of other viral factors that control viral expression (21, 22). Recently, a novel viral protein, HTLV-1 basic leucine-zipper factor (HBZ), which is encoded in the complementary strand of the HTLV-1 genome, was identified (23). HBZ is a nuclear protein that contains a transactivation domain and a basic leucine-zipper (bZIP) domain in its N and C termini, respectively. HBZ interacts with cellular bZIP proteins, particularly the AP-1 family of transcription factors, and regulates their transcriptional activities, allowing the virus to control viral gene transcription from the HTLV-1 promoter (23-25). We previously reported that HBZ interacts with c-Jun, an AP-1 family member, and suppresses its transcriptional activity by two distinct mechanisms. HBZ not only impairs the DNA-binding activity of c-Jun but also promotes its proteasomal degradation (26).
In this study, we show that ubiquitination is not required for HBZ-mediated proteasomal degradation of c-Jun. We demonstrate that HBZ directly interacts with both the 26 S proteasome and c-Jun and facilitates the delivery of c-Jun to the proteasome without ubiquitination. HBZ acts as a tethering factor between the 26 S proteasome and its substrate, thereby bypassing the targeting function of ubiquitination.
EXPERIMENTAL PROCEDURES
Cell Culture, Transfection, and Reagents—HEK-293T cells were cultured in Dulbecco's modified Eagle's medium (Nissui) supplemented with 10% fetal bovine serum (Invitrogen), 4 mm l-glutamine, and 0.1 mg/ml kanamycin sulfate at 37 °C in a 5% CO2 atmosphere. Mouse ts20 cells (kindly provided by Dr. Harvey Ozer) were cultured similarly, but maintained at 35 °C (the permissive temperature). C8166 cells were maintained in RPMI 1640 medium (Nissui) supplemented with 10% fetal bovine serum and 4 mm l-glutamine. Transfections were performed using FuGENE6 (Roche Applied Sciences) for HEK-293T cells, or Lipofectamine 2000 (Invitrogen) for ts20 cells following the manufacturer's protocol. However, in the experiment in Fig. 1B, HEK-293T cells were transfected with Lipofectamine 2000. Cells were treated with the proteasome inhibitor MG132 (20 μm, Peptide Institute) for 12 h prior to collection.
FIGURE 1.
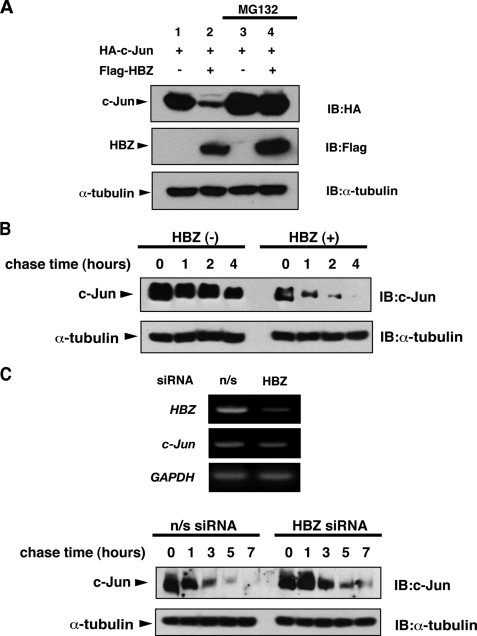
HBZ promotes proteasomal degradation of c-Jun. A, HEK-293T cells were transfected with 0.5 μg of pcDNA3-HA-c-Jun and 3 μg of either pcDNA3 or pcDNA3-FLAG-HBZ. After 12 h, the cells were treated with or without MG132 for 12 h. Cell lysates were immunoblotted with the antibodies indicated. B, HEK-293T cells were transfected with 5 μg of pCAG-FLAG-HBZ or control vector. After 36 h, the cells were treated with cycloheximide (50 μg/ml) and collected at the indicated times. Cell lysates were analyzed by immunoblot analysis. C, upper panel: C8166 cells were transfected using lentiviruses transcribing short hairpin RNAs against HBZ or a nonspecific (n/s) sequence. The relative mRNA expression of HBZ, c-Jun or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was evaluated by semiquantitative reverse transcription-PCR 7 days after transfection. Efficiencies of lentivirus vector transfection, which were determined by green fluorescent protein expression, were >95%. Lower panel: C8166 cells were transfected as indicated in the upper panel. After 7 days, the cells were treated with cycloheximide (50 μg/ml) and collected at the indicated times. Cell lysates were analyzed by immunoblot analysis.
Plasmids—pcDNA3-FLAG-HBZ, pcDNA3-HA-c-Jun, and pcDNA3-c-Jun-Myc-His have been described previously (26). Deletions and point mutants of HBZ and c-Jun were generated by PCR amplification of pcDNA3-FLAG-HBZ and pcDNA3-HA-c-Jun, respectively. These PCR fragments were then subcloned into expression vectors such as pcDNA3-FLAG, pcDNA3-HA, pcDNA3-His (Invitrogen), pGEX-6P-1 (Amersham Biosciences), or pGBT9 (Clontech). In addition, pcDNA3-FLAG-HBZ-SI was generated by PCR using pcDNA3-FLAG-HBZ as a template. The cDNAs were amplified by reverse transcription-PCR using total RNA from MT-2 cells (for c-Jun, JunB, JunD, c-Fos, ATF1, ATF2, ATF4, CREB, p53, and GADD34), 293T cells (for ubiquitin, Rpn5, MCM5, MCM7, and DET1), and Saos-2 cells (for COP1). They were subcloned into pcDNA3-FLAG, pcDNA3-Myc, pcDNA3-HA, pcDNA3-His, or pCAG vectors. Site-directed mutagenesis was used to generate the dominant-negative ubiquitin mutant (Ub-K48R), which contains an Arg substitution at Lys-48. To generate pcDNA3-FLAG-c-JunLZ/p53, the fragment encoding the c-Jun leucine-zipper region (273-310 amino acids) was PCR-amplified from pcDNA3-HA-c-Jun, digested with BamHI, and subcloned in-frame into BamHI-linearized pcDNA3-FLAG-p53.
Immunoblot Analysis—Proteins were fractionated by SDS-PAGE, transferred to a polyvinylidene difluoride membrane, and hybridized with the appropriate primary and horseradish peroxidase-conjugated secondary antibodies for subsequent detection by enhanced chemiluminescence (ECL, PerkinElmer Life Sciences). The antibodies used in this study were specific for FLAG (M2, Sigma), Myc (9E10, Santa Cruz Biotechnology), HA (3F10, Roche Applied Science), α-tubulin (Ab-1, Calbiochem), c-Jun (H-79, Santa Cruz Biotechnology), c-Fos (sc-52, Santa Cruz Biotechnology), α3 (MCP257, Affiniti Research Products), α7 (MCP72, Affiniti), Rpt6 (p45-110, Affiniti), and Rpn10 (S5a-18, Affiniti). Horseradish peroxidase-conjugated goat antibodies to mouse, rabbit. or rat IgG were obtained from Amersham Biosciences.
Immunoprecipitations—Cells were lysed in 1 ml of lysis buffer, which contained 50 mm Tris-HCl (pH 8.0), 100 mm NaCl, 1 mm EDTA, 0.5% Nonidet P-40, 1 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, 50 mm sodium fluoride, 2 mm sodium orthovanadate, and a protease inhibitor mixture (Complete EDTA-free, Roche Applied Science) at 4 °C for 30 min. After centrifugation, the supernatant was incubated with 10 μl of agarose-immobilized anti-FLAG (M2, Sigma) or anti-HA (Sigma) antibodies at 4 °C for 3 h. Resins were washed four times with 700 μl of lysis buffer. Bound proteins were eluted by boiling for 10 min in 1× sample buffer (62.5 mm Tris-HCl (pH 6.8), 2% SDS, 10% glycerol, and 5% 2-mercaptoethanol) and then subjected to SDS-PAGE, followed by immunoblot analysis.
Pulse-Chase Analysis—HEK-293T cells were seeded on 6-well plates (3 × 105 cells/well) and transfected with the appropriate expression plasmids. After 24 h, cells were labeled metabolically for 1 h with 100 μCi of [35S]methionine/cysteine (ICN) in methionine/cysteine-free Dulbecco's modified Eagle's medium (ICN) supplemented with 10% dialyzed fetal bovine serum and 4 mm l-glutamine. After washing with phosphate-buffered saline, cells were chased for the indicated intervals in complete medium. Harvested cells were then solubilized with lysis buffer that also contained 0.1% SDS and 0.5% sodium deoxycholate. The soluble fractions were incubated for 1 h with 2 μg of anti-c-Myc antibody and adsorbed to protein G-Sepharose (Amersham Biosciences). Immunoprecipitates were resolved by SDS-PAGE and detected by autoradiography. Band intensities were measured with scanning densitometry (Fluor-S MultiImager, Bio-Rad).
Glutathione S-Transferase Pulldown Assay—GST and GST fusion proteins were expressed in Escherichia coli strain BL21 and affinity-purified using glutathione-Sepharose beads (Amersham Biosciences). For in vitro translation, [35S]methionine (Amersham Biosciences)-labeled proteins were produced using the TnT quick-coupled transcription/translation system (Promega). In vitro translated proteins were incubated at 4 °C for 3 h with GST or GST fusion protein in 700 μl of GST-binding buffer containing 20 mm Tris-HCl (pH 8.0), 0.5% Nonidet P-40, 150 mm NaCl, 1 mm EDTA, 1 mm dithiothreitol, 10% glycerol, 1 mm phenylmethylsulfonyl fluoride, and a protease inhibitor mixture. Beads were washed three times with GST-binding buffer, and then bound proteins were resolved by SDS-PAGE and detected by autoradiography.
In Vivo Ubiquitination Assay—HEK-293T cells were transfected with pcDNA3-His-c-Jun or pcDNA3-His-c-Fos, pcDNA3-HA-HBZ, and pcDNA3-FLAG-ubiquitin (Ub) in different combinations. After treatment with MG132 (20 μm) for 12 h, cells were lysed in buffer A (8 m urea, 10 mm imidazole, 0.5% Triton X-100, 100 mm Hepes (pH 7.5)) and incubated with 20 μl of Ni-NTA beads (Qiagen) for 2 h at room temperature. The beads were washed with buffer B (8 m urea, 50 mm imidazole, 0.5% Triton X-100, 100 mm Hepes (pH 7.5), 0.5 m NaCl). The bound proteins were eluted with buffer C (8 m urea, 500 mm imidazole, 100 mm Hepes (pH 7.5)) and then subjected to SDS-PAGE, followed by immunoblot analysis.
Degradation Analysis in Temperature-sensitive ts20 Cells—ts20 cells in 10-cm plates were transfected with the appropriate expression plasmids and incubated for 12 h at 35 °C (permissive temperature). Cells were split into two dishes and incubated for an additional 12 h at 35 °C. Then, appropriate plates were moved to the restrictive temperature (39 °C). Cells were harvested after 24 h and subjected to SDS-PAGE, followed by immunoblot analysis.
Yeast Two-hybrid Screen—A human spleen cDNA library fused to the GAL4 activation domain of pACT2 (Clontech) was screened against the bait protein, the GAL4 DNA-binding domain fused to the HBZ N-terminal 120 amino acids (pGBT9-HBZ-N). pGBT9-HBZ-N and the cDNA library were co-introduced into the yeast cell line Y190 using the lithium acetate transformation method (27). The transformants were plated onto selective medium lacking histidine, tryptophan, and leucine, and containing 150 mm 3-aminotriazole to isolate clones with histidine prototrophy. Selected clones were assayed for another marker, β-galactosidase activity, as described in the Clontech protocol. Plasmid DNA was extracted from positive clones and analyzed by partial DNA sequencing.
Glycerol Density Gradient Analysis—HEK-293T cells transiently expressing HA-tagged HBZ were lysed in 1 ml of lysis buffer containing 25 mm Tris-HCl (pH 7.5), 1 mm dithiothreitol, 2 mm ATP, and 0.2% Nonidet P-40. After centrifugation, the supernatant was subjected to 10-40% (v/v) glycerol density gradient centrifugation in a Hitachi RPS40T rotor (22 h, 83,000 × g). The gradient was separated into 30 fractions from the bottom.
Lentiviral Vector Construction and Transfection—The lentivirus-based transfection system was a generous gift from Dr. H. Miyoshi. To express short hairpin RNAs against HBZ, complementary DNA oligonucleotides were inserted into the lentiviral vector (pCS-RfA-EG) under control of the H1 promoter. The target sequence against HBZ was 5′-GCAGATTGCTGAGTATTTG-3′. We cotransfected 4.5 μg of the above lentiviral vector (pCS-RfA-EG-shHBZ) with 3.5 μg of pMDLg/pRRE and 1.5 μg of pCMV-VSV-G-RSV-Rev into packaging cells (293FT) using Lipofectamine 2000. After a 48-h incubation, the culture medium was harvested and concentrated 100- to 200-fold by ultrafiltration. The titer of concentrated virus was measured on 293T cells based on their green fluorescent protein expression. Cells were transfected at a multiplicity of infection of 30 in the presence of Polybrene (4 μg/ml, Sigma). Cells were collected 7 days later, and green fluorescent protein expression was analyzed by FACSCalibur instrument (BD Biosciences).
RNA Isolation and Reverse Transcription-PCR—After 7 days of lentiviral infection, total RNA was extracted with Sepasol RNA I super reagent (Nacalai Tesque). We evaluated the relative expression of each mRNA by semiquantitative reverse transcription-PCR using a One-step RNA PCR kit (Takara). The following primers were used. HBZ: 5′-GAGAAGAAGGCCGCTGAC-3′ and 5′-TTATTGCAACCACATCGC-3′, c-Jun: 5′-GAACTGCACAGCCAGAAC-3′ and 5′-GGCGATTCTCTCCAGCTT-3′, and glyceraldehyde-3-phosphate dehydrogenase: 5′-ATGGGGAAGGTGAAGGTCGG-3′ and 5′-TGGAGGGATCTCGCTCCTGG-3′.
RESULTS
HBZ Promotes Proteasomal Degradation of c-Jun—We previously showed that HBZ heterodimerized with c-Jun via respective leucine-zipper domains and targeted c-Jun for proteasomal degradation (26). When c-Jun and HBZ were ectopically coexpressed in cells, c-Jun expression was significantly reduced. The reduction in c-Jun protein levels by HBZ was prevented by treating with the proteasome inhibitor, MG132, as confirmed in Fig. 1A. In addition, ectopically expressed HBZ reduced the half-life of endogenous c-Jun in HEK-293T cells (Fig. 1B). We next evaluated the effect of HBZ on c-Jun stability in HTLV-1-infected cells. We suppressed the HBZ gene in HTLV-1-infected C8166 cells by using lentivirus vector transcribing short hairpin RNAs against HBZ (Fig. 1C, upper panel). As shown in Fig. 1C (lower panel), HBZ knockdown prolonged the half-life of endogenous c-Jun in C8166 cells. These results indicate that HBZ regulates c-Jun stability by promoting its proteasomal degradation.
HBZ Does Not Promote the Ubiquitination of c-Jun—Most proteasomal substrates are targeted for degradation by the conjugation of polyubiquitin chains (28). The conjugation reaction is catalyzed by a variety of E3 ubiquitin ligases. Because c-Jun is also degraded by a ubiquitin-proteasome pathway that is mediated by several E3 ubiquitin ligases (29, 30), we speculated that HBZ acts as a component of E3 ubiquitin ligase. Therefore, we examined whether HBZ promotes polyubiquitination of c-Jun. Histidine-tagged c-Jun and epitope-tagged HBZ and ubiquitin were co-expressed in HEK-293T cells in different combinations. After purification with Ni-NTA beads, polyubiquitinated c-Jun species were specifically detected by immunoblotting. Interestingly, not only did we not detect an increase, but we also were unable to detect any ubiquitinated species of c-Jun in the presence of HBZ (Fig. 2A). This is not likely due to decreased c-Jun protein levels, because we also failed to detect any ubiquitinated c-Jun species in the presence of HBZ even after the treatment with MG132 (Fig. 2B). However, this abrogation of ubiquitination did not seem to be the result of a global inhibition of ubiquitination machinery, because HBZ did not alter the ubiquitination level of c-Fos (Fig. 2C), another AP-1 family member, which does not interact with HBZ (26). Conceivably, HBZ promotes c-Jun degradation without promoting its ubiquitination.
FIGURE 2.
HBZ does not promote the ubiquitination of c-Jun. A, HEK-293T cells were transfected with 2 μg of pcDNA3-His-c-Jun, pcDNA3-FLAG-Ub, and pcDNA3-HA-HBZ as indicated. Following purification with Ni-NTA beads, bound proteins were detected by immunoblot analysis. B, HEK-293T cells were transfected as indicated in A. After 12 h, the cells were treated with MG132 for 12 h. Following Ni-NTA purification, bound proteins were detected by immunoblot analysis. C, HEK-293T cells were transfected with 2 μg of pcDNA3-His-c-Fos, pcDNA3-FLAG-Ub, and pcDNA3-HA-HBZ as indicated. After 12 h, the cells were treated with MG132 for 12 h. Following Ni-NTA purification, bound proteins were detected by immunoblot analysis.
Ubiquitination Is Not Required for HBZ-mediated Proteasomal Degradation of c-Jun—We next determined the role of ubiquitination in HBZ-mediated c-Jun degradation. In the first approach, we constructed a c-Jun mutant (c-JunK/R) in which all lysine residues (potential sites of ubiquitin conjugation) were mutated to arginine. We confirmed that the c-JunK/R mutant was not ubiquitinated in cells (Fig. 3A, left panel). Wild-type c-Jun and the mutant were confirmed to associate with HBZ comparably (data not shown). As shown in Fig. 3A (right panel), the c-JunK/R mutant was degraded by HBZ as efficiently as wild-type c-Jun. This result led us to hypothesize that HBZ targets c-Jun for proteasomal degradation in a ubiquitin-independent manner.
FIGURE 3.
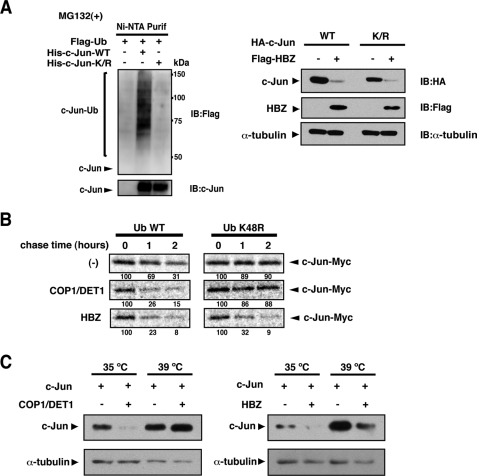
HBZ promotes c-Jun degradation in a ubiquitin-independent manner. A, HBZ promotes the degradation of lysine-free c-Jun. Left panel: HEK-293T cells were transfected with 2 μg of either pcDNA3-His-c-Jun or pcDNA3-His-c-JunK/R in the presence of pcDNA3-FLAG-Ub as indicated. After 12 h, the cells were treated with MG132 for 12 h. Following Ni-NTA purification, bound proteins were detected by immunoblot analysis. Right panel: HEK-293T cells were transfected with 0.5 μg of either pcDNA3-HA-c-Jun or pcDNA3-HA-c-JunK/R and 3 μg of either pcDNA3 or pcDNA3-FLAG-HBZ. Cell lysates were immunoblotted using the antibodies indicated. B, HBZ-mediated degradation of c-Jun is not inhibited by a dominant-negative ubiquitin. HEK-293T cells were transfected with 0.3 μg of pcDNA3-c-Jun-Myc together with 0.5 μg of pcDNA3-HA-COP1 and 1.5 μg of pcDNA3-HA-DET1, or 2 μg of pcDNA3-HA-HBZ in the presence of 3 μg of pcDNA3-FLAG-Ub or -Ub K48R. After 24 h, the cells were pulse-labeled with [35S]methionine/cysteine for 1 h and then chased for the indicated intervals. Labeled c-Jun was immunoprecipitated and resolved by SDS-PAGE. c-Jun levels were detected by autoradiography. The intensity of each band was quantified and listed below each panel. C, HBZ promotes c-Jun degradation in the absence of a functional ubiquitination system. ts-20 cells were transfected with 1 μg of pcDNA3-FLAG-c-Jun in the presence of 2 μg of pcDNA3-HA-COP1 and pcDNA3-HA-DET1 or 4 μg of pcDNA3-HA-HBZ, as indicated. The cells were then split and incubated at 35 or 39 °C for an additional 24 h. Cell lysates were immunoblotted with anti-FLAG or anti-α-tubulin antibodies.
In the second approach, we used a dominant-negative ubiquitin mutant (Ub-K48R) that bears a substitution of lysine 48 with arginine. Ub-K48R exerts a chain-terminating effect and blocks ubiquitin-dependent proteasomal degradation (31). In a pulse-chase analysis, the expression of Ub-K48R stabilized c-Jun (Fig. 3B, upper panel), because c-Jun is basally targeted for ubiquitin-dependent degradation (32). COP1 and DET1, a ubiquitin ligase complex for c-Jun (29), promoted c-Jun degradation in a ubiquitin-dependent manner. Therefore, the expression of Ub-K48R blocked its degradation (Fig. 3B, middle panel). In contrast, HBZ-mediated degradation of c-Jun was not blocked by Ub-K48R expression (Fig. 3B, lower panel).
Finally, we performed degradation experiments using the mouse ts20 cell line. These cells have a temperature-sensitive E1 ubiquitin-activating enzyme that is inactivated at the restrictive temperature of 39 °C (33). As expected, ubiquitin-dependent degradation of c-Jun mediated by COP1/DET1 was blocked at 39 °C (Fig. 3C, left panel), whereas HBZ still promoted c-Jun degradation at the restrictive temperature (Fig. 3C, right panel). Taken together, these results suggest that HBZ promotes proteasomal degradation of c-Jun through a ubiquitin-independent mechanism.
HBZ Promotes Degradation of Proteins That Bind Its bZIP Domain but Not Its N Terminus—We next examined the specificity of HBZ-mediated proteasomal degradation. In the first approach, we investigated whether HBZ promotes degradation of other bZIP proteins. HBZ has been shown to associate with JunB, JunD, and ATF4 via their bZIP domains (23-25). We found that HBZ also associates with ATF2 through its bZIP domain in vivo (Fig. 4A, lower panel). As shown in Fig. 4A (upper panel), HBZ promoted degradation of these bZIP-binding proteins, and degradation of these proteins were prevented by MG132 treatment (data not shown). On the other hand, HBZ failed to promote the degradation of CREB and ATF1. Recently, HBZ has been shown to interact with CREB and ATF1 via their bZIP domains in vivo and in vitro (34). However, we failed to detect these interactions under the experimental conditions in which we readily detected interaction between c-Jun and HBZ (data not shown), likely because these interactions (HBZ-CREB or -ATF1) were very labile or very weak. We previously reported that HBZ does not promote the degradation of c-Fos, a protein that does not interact with HBZ (26). From these results, we predicted a relationship between HBZ association and degradation.
FIGURE 4.
HBZ promotes degradation of proteins that bind its bZIP domain but not its N terminus. A, HBZ promotes the degradation of proteins that bind its bZIP domain. Upper panel: HEK-293T cells were transfected with pcDNA3-Myc-JunB, pcDNA3-HA-JunD, pCAG-FLAG-ATF1, pCAG-FLAG-ATF2, pCAG-FLAG-ATF4, or pcDNA3-HA-CREB, with or without HBZ. Cell lysates were immunoblotted with the appropriate epitope-tag antibodies. Lower panel: HEK-293T cells were transfected with 2.5 μg of pcDNA3-FLAG-ATF2 together with 2.5 μg of pcDNA3-HA-HBZ or pcDNA3-HA-HBZ-ΔLZ, followed by treatment with 20 μm MG132 for 12 h. Cell lysates were immunoprecipitated with an anti-HA antibody, followed by immunoblotting with an anti-FLAG antibody. Immunoblotting of whole cell lysates was performed using the antibodies indicated. B, an interaction with HBZ is responsible for target protein degradation. Upper panel: schematic diagram of c-Jun and c-JunΔLZ used in this study. Middle panel: HEK-293T cells were transfected as indicated and then treated with MG132 for 12 h. Cell lysates were immunoprecipitated with an anti-FLAG antibody and then immunoblotted with an anti-HA antibody. LC indicates the immunoglobulin light chain. Lower panel: HEK-293T cells were transfected with pcDNA3-HA-c-JunΔLZ, with or without HBZ. Cell lysates were immunoblotted with an anti-HA antibody. C, upper panel: schematic diagram of p53 and c-JunLZ/p53. Middle panel: HEK-293T cells were transfected as indicated and then treated with MG132 for 12 h. Cell lysates were immunoprecipitated with an anti-FLAG antibody and then immunoblotted with an anti-HA antibody. Lower panel: HEK-293T cells were transfected with pcDNA3-FLAG-p53 or pcDNA3-FLAG-c-JunLZ/p53, with or without HBZ. Cell lysates were immunoblotted with an anti-FLAG antibody. D, HBZ does not promote degradation of proteins that bind its N terminus. Upper panel: HEK-293T cells were transfected with pcDNA3-HA-GADD34, pcDNA3-HA-MCM5, or pcDNA3-HA-MCM7, with or without HBZ. Cell lysates were immunoblotted with an anti-HA antibody. Lower panel: HEK-293T cells were transfected with 2.5 μg of pcDNA3-HA-GADD34, pcDNA3-HA-MCM5, or pcDNA3-HA-MCM7, together with 2.5 μg of pcDNA3-FLAG-HBZ, pcDNA3-FLAG-HBZ-N, or pcDNA3-FLAG-HBZ-ΔN60. Cell lysates were immunoprecipitated with an anti-FLAG antibody and then immunoblotted with an anti-HA antibody.
To further investigate this relationship, we constructed a c-Jun mutant lacking the leucine-zipper region (c-JunΔLZ, Fig. 4B, upper panel) and found that it was unable to associate with HBZ (Fig. 4B, middle panel). Although HBZ degraded intact c-Jun (Fig. 1), it did not promote degradation of the c-JunΔLZ mutant (Fig. 4B, lower panel). Furthermore, HBZ did not promote the degradation of p53 (Fig. 4C, lower panel), which does not associate with HBZ (Fig. 4C, middle panel, lane 2). However, an artificial fusion protein that contained the c-Jun leucine-zipper region and p53 (c-JunLZ/p53) could interact with HBZ (Fig. 4C, middle panel, lane 4) and was degraded by HBZ (Fig. 4C, lower panel). These results suggest that the association with HBZ is essential for target protein degradation.
Does HBZ promote the degradation of all its binding partners? To further investigate this question, we examined whether HBZ promotes degradation of proteins that bind its N terminus. We identified growth arrest and DNA damage-inducible transcript 34 (GADD34), minichromosome maintenance protein 5 (MCM5) and MCM7 as cellular proteins that bind the HBZ N terminus using a yeast two-hybrid screen with the HBZ N-terminal 120 amino acids as bait. These interactions were confirmed in vivo (Fig. 4D, lower panel) and in vitro (data not shown). Interestingly, HBZ did not promote degradation of these N-terminal-binding proteins (Fig. 4D, upper panel).
The N-terminal Region of HBZ (Especially Some Hydrophobic Residues) Is Essential for c-Jun Destabilization—In our previous report, we identified the regions of HBZ responsible for the destabilization of c-Jun (26). We have shown that not only the leucine-zipper region, as an interaction domain with c-Jun, but also the N-terminal 60 amino acids of HBZ are responsible for the destabilization of c-Jun. Notably, several hydrophobic residues (especially Leu, Ile, and Val) are concentrated in the N-terminal 60 amino acids of HBZ (Fig. 5A). We performed alanine-scanning mutagenesis on this region to identify point mutants that were unable to destabilize c-Jun. Alanine substitutions were focused primarily on Leu, Ile, and Val residues (Fig. 5B). These mutants associated with c-Jun comparably to wild-type HBZ (data not shown). As shown in Fig. 5C, some of these alanine mutants promoted c-Jun degradation less effectively than wild-type. In particular, the triple mutant (HBZ-3.8.10) had little ability to degrade c-Jun. These results suggest that the N-terminal 60 amino acids of HBZ (particularly certain hydrophobic residues) are essential for the destabilization of c-Jun.
FIGURE 5.
The N-terminal region of HBZ (especially some hydrophobic residues) is responsible for destabilization of c-Jun. A, the N-terminal region of HBZ is rich in hydrophobic residues. The percentage of hydrophobic residues (Leu/Ile/Val) in the respective regions is indicated below. B, alanine-scanning substitutions in the N-terminal region of HBZ. The gray letters indicate hydrophobic residues (Leu/Ile/Val). C, some alanine mutants promoted c-Jun degradation less effectively than wild-type. HEK-293T cells were transfected with c-Jun and either pcDNA3-FLAG-HBZ or its mutants. Cell lysates were analyzed by immunoblotting.
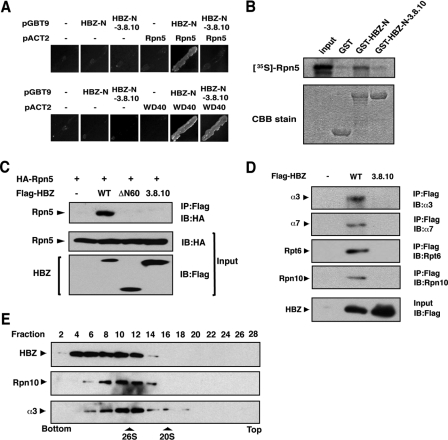
The N-terminal Region of HBZ Directly Interacts with the 26 S Proteasome—It is plausible that the N-terminal region of HBZ recruits cellular factors that regulate c-Jun degradation. We performed a yeast two-hybrid screen to identify these factors. We used a fusion protein that combined the GAL4 DNA-binding domain with the HBZ N-terminal 120 amino acids as bait, because the HBZ N-terminal 60 amino acids alone possessed intensive transactivation ability. Some positive clones were found to correspond to Rpn5 (also known as PSMD12 or p55), a 19 S proteasome subunit. As shown in Fig. 6A (upper panel), the HBZ N-terminal 120 amino acids (HBZ-N) interacted with Rpn5 in yeast, whereas the mutant HBZ-N-3.8.10, which lacked the ability to destabilize c-Jun (Fig. 5C), did not associate with Rpn5. On the other hand, WD40 protein, another target identified in the screen, interacted with both HBZ-N and HBZ-N-3.8.10 (Fig. 6A, lower panel).
FIGURE 6.
The N-terminal region of HBZ directly interacts with the 26 S proteasome. A, identification of Rpn5 as a target for the N-terminal region of HBZ using a yeast two-hybrid screen. The yeast strain Y190 was transformed as indicated. Transformants were grown in -Leu/-Trp/-His medium and selected for histidine prototrophy. B, HBZ associates with Rpn5 in vitro. GST, GST-HBZ-N, or GST-HBZ-N-3.8.10 fusion proteins were incubated with 35S-labeled Rpn5. Bound proteins were detected by autoradiography. The abundance of GST proteins is shown by CBB staining. C, HBZ associates with Rpn5 in cells. HEK-293T cells were transfected as indicated. Cell lysates were immunoprecipitated with an anti-FLAG antibody and then immunoblotted with an anti-HA antibody. D, HBZ associates with the 26 S proteasome. HEK-293T cells were transfected with pcDNA3-FLAG-HBZ or pcDNA3-FLAG-HBZ-3.8.10 and then treated with MG132 for 12 h. Cell lysates were immunoprecipitated with an anti-FLAG antibody, followed by immunoblotting with the antibodies indicated. E, cells transiently expressing HA-tagged HBZ were fractionated by 10-40% glycerol density gradient centrifugation. Each fraction was analyzed by immunoblotting with anti-HA, anti-Rpn10, or anti-α3 antibodies. The peaks corresponding to the 26 S and 20 S proteasomes are indicated.
We next confirmed the interaction between HBZ and Rpn5 using an in vitro GST pull-down assay. Recombinant HBZ-N and HBZ-N-3.8.10 fused to GST (GST-HBZ-N and GST-HBZ-N-3.8.10) were incubated with 35S-labeled Rpn5 that was translated in vitro. The 35S-labeled Rpn5 copurified with GST-HBZ-N but not with GST-HBZ-N-3.8.10 (Fig. 6B). To further confirm the interaction between HBZ and Rpn5, we performed a coimmunoprecipitation assay in HEK-293T cells transiently expressing HA-tagged Rpn5 and FLAG-tagged HBZ wild-type (-WT) or mutants (-ΔN60, -3.8.10). As shown in Fig. 6C, Rpn5 coimmunoprecipitated with HBZ-WT, but not with either the HBZ-ΔN60 or -3.8.10 mutants. These results indicate that the N-terminal region of HBZ associates with the proteasomal subunit Rpn5 and that this association correlates with the destabilization of c-Jun.
Although HBZ interacted with ectopically expressed Rpn5, it was not distinguishable whether HBZ interacts with free Rpn5 or complex form of proteasome. To investigate this point, we examined whether other subunits of the proteasome were coimmunoprecipitated with HBZ. As shown in Fig. 6D, all of the proteasomal subunits tested here coimmunoprecipitated with HBZ, but not with its mutant. α3 and α7 are α-subunits that form the outer rings of the 20 S proteasome, whereas Rpt6 and Rpn10 are subunits of the 19 S regulatory complex. The components of both the 19 S and 20 S proteasomes coprecipitated with HBZ, suggesting that HBZ interacts with the 26 S proteasome complex. This notion was supported by the cosedimentation of HBZ and the 26 S proteasome in a glycerol density gradient (Fig. 6E).
Our findings indicate that HBZ interacts with the 26 S proteasome and c-Jun via its N and C termini, respectively. This suggests that HBZ acts as a tethering factor between c-Jun and the 26 S proteasome, thereby facilitating delivery of c-Jun to the proteasome in a ubiquitin-independent manner.
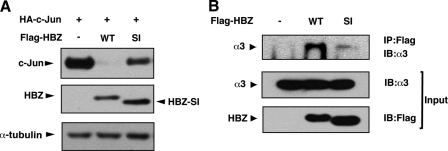
Recently, it has been reported that the HTLV-1 genome encodes an alternative splicing isoform of HBZ (35-37), termed HBZ-SI by Murata's group. We investigated whether the two HBZ isoforms differed in their ability to destabilize c-Jun. Interestingly, HBZ-SI promoted c-Jun degradation less efficiently than wild-type (Fig. 7A). As expected from the above results, HBZ-SI interacted with the proteasome less efficiently than wild-type (Fig. 7B). This correlation also supports the significance of the association between HBZ and the 26 S proteasome in c-Jun destabilization.
FIGURE 7.
HBZ splicing isoform has less ability to destabilize c-Jun than wild-type. A, HBZ-SI promotes degradation of c-Jun less effectively than wild-type. HEK-293T cells were transfected with 0.5 μg of pcDNA3-HA-c-Jun and 3 μg of either pcDNA3-FLAG-HBZ or pcDNA3-FLAG-HBZ-SI. Cell lysates were immunoblotted with anti-HA, anti-FLAG, or anti-α-tubulin antibodies. B, HBZ-SI associates with the proteasome less effectively than wild type. HEK-293T cells were transfected with 8 μg of pcDNA3-FLAG-HBZ or pcDNA3-FLAG-HBZ-SI and then treated with MG132. Cell lysates were immunoprecipitated with an anti-FLAG antibody and then immunoblotted with an anti-α3 antibody. The abundance of α3 and HBZ proteins in whole cell lysates are shown (middle panel and lower panel).
DISCUSSION
Viruses have evolved sophisticated strategies to utilize or manipulate the host ubiquitin-proteasome system for their propagation. For example, human papillomavirus type-16 E6 protein and adenovirus E1B55k-E4orf6 proteins act as part of E3 ubiquitin ligase complexes to promote ubiquitination and subsequent degradation of the tumor suppressor, p53 (14, 38). The human immunodeficiency virus type-1 Vif protein also acts as an E3 ubiquitin ligase that targets APOBEC3G, a potent cellular anti-viral factor, for proteasomal degradation in a ubiquitin-dependent manner (39). There are an increasing number of viral proteins that utilize cellular ubiquitination machinery to dysregulate cellular functions (15-17). On the other hand, the human cytomegalovirus (CMV) pp71 protein has been shown to promote proteasomal degradation of Rb protein without promoting its ubiquitination (12), the molecular mechanism of which, however, is unknown. Recently, a number of eukaryotic proteins, including ODC (8), p53 (9), p21waf1/cip1 (10, 11), and IκBα (40), have been shown to be degraded through a proteasome-dependent, ubiquitin-independent pathway, suggesting the significance of this alternative pathway in various cellular events (7). Here, we demonstrate that the HTLV-1 HBZ protein promotes degradation of c-Jun, a cellular transcription factor, using a proteasome-dependent but ubiquitin-independent mechanism. First, although HBZ-mediated degradation of c-Jun was prevented by proteasome inhibitor treatment, HBZ failed to promote c-Jun ubiquitination. Second, HBZ destabilized a c-Jun mutant lacking all lysine targets for ubiquitination. Third, a dominant-negative ubiquitin (K48R) mutant failed to abrogate HBZ-mediated c-Jun degradation. Fourth, c-Jun degradation mediated by HBZ was unaffected in ts20 cells at the restrictive temperature. Taken together, these results indicate that HBZ promotes c-Jun degradation in a ubiquitin-independent manner.
How, in the absence of polyubiquitination, does HBZ target c-Jun for proteasomal degradation? ODC was the first protein shown to be degraded in a ubiquitin-independent manner. Attachment of antizyme causes conformational changes in ODC, thereby exposing its C-terminal degradation signal for recognition by the 26 S proteasome (8, 41). Thus, antizyme promotes ODC degradation by enhancing the direct association of ODC with the 26 S proteasome. In contrast, degradation of p21waf1/cip1 and Rb proteins is shown to be mediated by the 20 S proteasome. These proteins are directly recognized by the α7 subunit of the 20 S proteasome (11, 13). Notably, the MDM2 oncoprotein has been shown to promote degradation of p21waf1/cip1 and Rb in a ubiquitin-independent manner (13, 42, 43). MDM2 directly associates with both these proteins and the 20 S proteasome and facilitates p21-20 S or Rb-20 S interactions. This tethering function of MDM2 is similar to HBZ-mediated c-Jun degradation. In this study, we have shown that HBZ interacts with the 26 S proteasome and c-Jun via its N and C termini, respectively. Moreover, we found that these associations are essential for c-Jun destabilization. These observations suggest that HBZ acts as a tethering factor between c-Jun and the 26 S proteasome, thereby facilitating the delivery of c-Jun to the proteasome without ubiquitination.
What is the physiological significance of HBZ-mediated c-Jun repression in the viral life cycle? In HTLV-1-infected cells, AP-1 is constitutively activated predominantly by Tax (44, 45). Although AP-1 contributes to T-cell transformation, aberrantly up-regulated AP-1 often accelerates cell death (46). HBZ may protect infected cells from cell death by preventing aberrant AP-1 activation. In addition, in this study we have shown that HBZ promotes protein degradation of various cellular bZIP proteins (e.g. ATF4, JunB, JunD, and c-Jun) that are involved in transcription from the HTLV-1 LTR. Because viral products (in particular Tax) are major targets of the host immune response (20), their overexpression is unfavorable for viral propagation. Therefore, as a negative regulator of viral expression, HBZ may play a key role in establishing and maintaining long, latent HTLV-1 infection.
In this study, we disclose a new viral strategy to utilize the host proteolytic apparatus for viral propagation. HBZ acts as a tethering factor between the 26 S proteasome and its substrate, thereby mimicking the targeting function of ubiquitination. Our findings also provide significant insight into the cellular proteasomal degradation pathway. It is possible that this direct tethering model between the proteasome and its substrate is a common cellular proteolytic event. We assume that in the future accumulating reports on ubiquitin-independent degradation will show the physiological significance of this alternative degradation pathway in various cellular events.
Acknowledgments
We thank Dr. Harvey Ozer for providing the mouse ts20 cells, and Dr. H. Miyoshi for providing the lentivirus-based transfection system.
This work was supported by grants-in-aid for cancer research and for the second-term comprehensive 10-year strategy for cancer control from the Ministry of Health, Labour and Welfare as well as by grants-in-aid for Scientific Research on Priority Areas “Integrative Research Toward the Conquest of Cancer” from the Ministry of Education, Culture, Sports, Science and Technology of Japan. In addition, this work was supported by research funds from the Takeda Science Foundation, the Suzuken Memorial Foundation, and the Japanese Leukemia Research Foundation. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: ODC, ornithine decarboxylase; Rb, retinoblastoma; GST, glutathione S-transferase; Ub, ubiquitin; Ni-NTA, nickel-nitrilotriacetic acid; CMV, cytomegalovirus; HA, hemagglutinin; CREB, cAMP-response element-binding protein; GADD34, growth arrest and DNA damage-inducible transcript 34; MCM5, minichromosome maintenance protein 5; AP-1, activating protein-1; HTLV-1, human T-cell leukemia virus type 1; HBZ, HTLV-1 bZIP factor.
References
- 1.Davies, K. J. (2001) Biochimie (Paris) 83 301-310 [DOI] [PubMed] [Google Scholar]
- 2.DeMartino, G. N., and Slaughter, C. A. (1999) J. Biol. Chem. 274 22123-22126 [DOI] [PubMed] [Google Scholar]
- 3.Glickman, M. H., and Ciechanover, A. (2002) Physiol. Rev. 82 373-428 [DOI] [PubMed] [Google Scholar]
- 4.Goldberg, A. L. (2003) Nature 426 895-899 [DOI] [PubMed] [Google Scholar]
- 5.Prakash, S., Tian, L., Ratliff, K. S., Lehotzky, R. E., and Matouschek, A. (2004) Nat. Struct. Mol. Biol. 11 830-837 [DOI] [PubMed] [Google Scholar]
- 6.Takeuchi, J., Chen, H., and Coffino, P. (2007) EMBO J. 26 123-131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Orlowski, M., and Wilk, S. (2003) Arch. Biochem. Biophys. 415 1-5 [DOI] [PubMed] [Google Scholar]
- 8.Murakami, Y., Matsufuji, S., Kameji, T., Hayashi, S., Igarashi, K., Tamura, T., Tanaka, K., and Ichihara, A. (1992) Nature 360 597-599 [DOI] [PubMed] [Google Scholar]
- 9.Asher, G., Lotem, J., Sachs, L., Kahana, C., and Shaul, Y. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 13125-13130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sheaff, R. J., Singer, J. D., Swanger, J., Smitherman, M., Roberts, J. M., and Clurman, B. E. (2000) Mol. Cell 5 403-410 [DOI] [PubMed] [Google Scholar]
- 11.Touitou, R., Richardson, J., Bose, S., Nakanishi, M., Rivett, J., and Allday, M. J. (2001) EMBO J. 20 2367-2375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kalejta, R. F., and Shenk, T. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 3263-3268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sdek, P., Ying, H., Chang, D. L., Qiu, W., Zheng, H., Touitou, R., Allday, M. J., and Xiao, Z. X. (2005) Mol. Cell 20 699-708 [DOI] [PubMed] [Google Scholar]
- 14.Scheffner, M., Huibregtse, J. M., Vierstra, R. D., and Howley, P. M. (1993) Cell 75 495-505 [DOI] [PubMed] [Google Scholar]
- 15.Knight, J. S., Sharma, N., and Robertson, E. S. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 18562-18566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao, G., and Luo, H. (2006) Can. J. Physiol. Pharmacol. 84 5-14 [DOI] [PubMed] [Google Scholar]
- 17.Shackelford, J., and Pagano, J. S. (2005) Essays Biochem. 41 139-156 [DOI] [PubMed] [Google Scholar]
- 18.Takatsuki, K. (2005) Retrovirology 2 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoshida, M. (2001) Annu. Rev. Immunol. 19 475-496 [DOI] [PubMed] [Google Scholar]
- 20.Kannagi, M., Harada, S., Maruyama, I., Inoko, H., Igarashi, H., Kuwashima, G., Sato, S., Morita, M., Kidokoro, M., Sugimoto, M., Funahashi, S., Osame, M., and Shida, H. (1991) Int. Immunol. 3 761-767 [DOI] [PubMed] [Google Scholar]
- 21.Inoue, J., Yoshida, M., and Seiki, M. (1987) Proc. Natl. Acad. Sci. U. S. A. 84 3653-3657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nicot, C., Dundr, M., Johnson, J. M., Fullen, J. R., Alonzo, N., Fukumoto, R., Princler, G. L., Derse, D., Misteli, T., and Franchini, G. (2004) Nat. Med. 10 197-201 [DOI] [PubMed] [Google Scholar]
- 23.Gaudray, G., Gachon, F., Basbous, J., Biard-Piechaczyk, M., Devaux, C., and Mesnard, J. M. (2002) J. Virol. 76 12813-12822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Basbous, J., Arpin, C., Gaudray, G., Piechaczyk, M., Devaux, C., and Mesnard, J. M. (2003) J. Biol. Chem. 278 43620-43627 [DOI] [PubMed] [Google Scholar]
- 25.Thebault, S., Basbous, J., Hivin, P., Devaux, C., and Mesnard, J. M. (2004) FEBS Lett. 562 165-170 [DOI] [PubMed] [Google Scholar]
- 26.Matsumoto, J., Ohshima, T., Isono, O., and Shimotohno, K. (2005) Oncogene 24 1001-1010 [DOI] [PubMed] [Google Scholar]
- 27.Gietz, D., St Jean, A., Woods, R. A., and Schiestl, R. H. (1992) Nucleic Acids Res. 20 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ciechanover, A., Orian, A., and Schwartz, A. L. (2000) BioEssays 22 442-451 [DOI] [PubMed] [Google Scholar]
- 29.Wertz, I. E., O'Rourke, K. M., Zhang, Z., Dornan, D., Arnott, D., Deshaies, R. J., and Dixit, V. M. (2004) Science 303 1371-1374 [DOI] [PubMed] [Google Scholar]
- 30.Nateri, A. S., Riera-Sans, L., Da Costa, C., and Behrens, A. (2004) Science 303 1374-1378 [DOI] [PubMed] [Google Scholar]
- 31.Ward, C. L., Omura, S., and Kopito, R. R. (1995) Cell 83 121-127 [DOI] [PubMed] [Google Scholar]
- 32.Treier, M., Staszewski, L. M., and Bohmann, D. (1994) Cell 78 787-798 [DOI] [PubMed] [Google Scholar]
- 33.Chowdary, D. R., Dermody, J. J., Jha, K. K., and Ozer, H. L. (1994) Mol. Cell. Biol. 14 1997-2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lemasson, I., Lewis, M. R., Polakowski, N., Hivin, P., Cavanagh, M. H., Thebault, S., Barbeau, B., Nyborg, J. K., and Mesnard, J. M. (2007) J. Virol. 81 1543-1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Satou, Y., Yasunaga, J., Yoshida, M., and Matsuoka, M. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 720-725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murata, K., Hayashibara, T., Sugahara, K., Uemura, A., Yamaguchi, T., Harasawa, H., Hasegawa, H., Tsuruda, K., Okazaki, T., Koji, T., Miyanishi, T., Yamada, Y., and Kamihira, S. (2006) J. Virol. 80 2495-2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cavanagh, M. H., Landry, S., Audet, B., Arpin-Andre, C., Hivin, P., Pare, M. E., Thete, J., Wattel, E., Marriott, S. J., Mesnard, J. M., and Barbeau, B. (2006) Retrovirology 3 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Querido, E., Blanchette, P., Yan, Q., Kamura, T., Morrison, M., Boivin, D., Kaelin, W. G., Conaway, R. C., Conaway, J. W., and Branton, P. E. (2001) Genes Dev. 15 3104-3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu, X., Yu, Y., Liu, B., Luo, K., Kong, W., Mao, P., and Yu, X. F. (2003) Science 302 1056-1060 [DOI] [PubMed] [Google Scholar]
- 40.Krappmann, D., Wulczyn, F. G., and Scheidereit, C. (1996) EMBO J. 15 6716-6726 [PMC free article] [PubMed] [Google Scholar]
- 41.Li, X., and Coffino, P. (1993) Mol. Cell. Biol. 13 2377-2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jin, Y., Lee, H., Zeng, S. X., Dai, M. S., and Lu, H. (2003) EMBO J. 22 6365-6377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang, Z., Wang, H., Li, M., Agrawal, S., Chen, X., and Zhang, R. (2004) J. Biol. Chem. 279 16000-16006 [DOI] [PubMed] [Google Scholar]
- 44.Fujii, M., Niki, T., Mori, T., Matsuda, T., Matsui, M., Nomura, N., and Seiki, M. (1991) Oncogene 6 1023-1029 [PubMed] [Google Scholar]
- 45.Mori, N., Fujii, M., Iwai, K., Ikeda, S., Yamasaki, Y., Hata, T., Yamada, Y., Tanaka, Y., Tomonaga, M., and Yamamoto, N. (2000) Blood 95 3915-3921 [PubMed] [Google Scholar]
- 46.Shaulian, E., and Karin, M. (2002) Nat. Cell Biol. 4 E131. [DOI] [PubMed] [Google Scholar]