Abstract
Human immunodeficiency virus Rev-binding protein (HRB), also called human Rev-interacting protein (hRIP) or Rev/Rex activation domain binding (RAB) is a partner of the tyrosine kinase substrate EPS15, and it has been recovered in the AP-2 interactome. EPS15 and AP-2 are involved in endocytosis, but the function of HRB in this process is still unknown. Here we identified HRB as a partner of the vesicular SNARE tetanus neurotoxin-insensitive vesicle-associated membrane protein (TI-VAMP, also called VAMP7) in yeast two-hybrid screens and using biochemical assays. In HeLa cells, HRB localized both in the nucleus and in the cytoplasm. In the cytoplasm, HRB colocalized with clathrin-, AP-2-, EPS15-, and transferrin receptor-containing vesicles. We did not see significant colocalization between HRB and TI-VAMP in HeLa cells, and we saw partial colocalization with green fluorescent protein-TI-VAMP in stably expressing Madin-Darby canine kidney cells. Nevertheless using a pHLuorin-tagged TI-VAMP construct, we found that HRB and TI-VAMP colocalize close to the plasma membrane after 5 min of anti-green fluorescent protein antibody uptake. These results suggest that TI-VAMP and HRB may interact only during the early stages of endocytosis. Furthermore uptake experiments followed by fluorescence-activated cell sorting showed that the endocytosis of fluorescent transferrin and pHLuorin-TI-VAMP is strongly reduced in HRB knockdown cells. Altogether these results suggest that HRB is involved in clathrin-dependent endocytosis and recruits TI-VAMP in this process.
A large body of evidence suggests that the formation of SNARE5 complexes between one vesicular SNARE (v-SNARE or VAMP) and a target SNARE (t-SNARE) complex mediates the fusion of intracellular membranes in eukaryotes (1). Target SNARE can be composed of two to three proteins related to syntaxin 1 or synaptosome-associated protein of 25 kDa (SNAP-25). Following membrane fusion, the vesicular SNARE protein localizes to its formerly target membrane in association with its target SNARE proteins. The ATPase N-ethylmaleimide-sensitive factor dissociates SNARE complexes in association with soluble N-ethylmaleimide-sensitive factor attachment proteins. Following this step, v-SNAREs are thought to be recycled, but the molecular mechanisms involved are only poorly understood (1-3).
Mammalian cells express only nine v-SNAREs (YKT6, SEC22, and VAMPs 1-5, 7, and 8), in fact even fewer in most cells because some VAMPs show a high cell/tissue-specific expression (4, 5). In this family of protein, TI-VAMP is a particularly intriguing member because of its original amino-terminal extension called the Longin domain (6). The presence of a Longin domain in v-SNAREs defines the Longin subfamily of VAMPs to which TI-VAMP, SEC22, and YKT6 belong (6, 7). Other v-SNAREs belong to the brevin subfamily or may correspond to intermediates in the case of VAMPs 4 and 8. The Longin domain of TI-VAMP plays important regulatory functions that are not fully understood but include the interaction with the molecular coat AP-3 (8, 9) and the inhibition of the capacity of TI-VAMP to associate with its t-SNARE partners (8, 10). The expression of the Longin domain alone inhibits neuritogenesis (10, 11), cell migration (12), exocytosis during phagocytosis (13), late endosome to lysosome transport (14), and lysosomal secretion (12) thus suggesting a dominant negative effect that may result from the titration of important endogenous regulators.
To identify the regulators of TI-VAMP, we carried out yeast two-hybrid screens using the Longin domain alone or the full cytoplasmic domain of TI-VAMP. Here we identify human immunodeficiency virus Rev-binding protein (HRB; also called Rev-interacting protein (RIP) or Rev/Rex activation domain binding (RAB)) as a partner of TI-VAMP in yeast two-hybrid screens and biochemical assays in vitro. HRB has been shown to be a cofactor of the retroviral protein Rev (15-17) and a partner of the tyrosine kinase substrate EPS15 (18, 19) involved in endocytosis. HRB is also implicated in the fusion of preacrosomal vesicle during the formation of the acrosome in spermatogenesis (20), and more recently, HRB has been found as an interactor of the clathrin adaptor AP-2 (21) involved in endocytosis. Here we show that HRB localized both to the nuclei and to AP-2+, EPS15+, clathrin+, and transferrin receptor (TfR)+ vesicles. Silencing the expression of HRB by RNA interference and shRNA led to the inhibition of transferrin (Tf) and TI-VAMP endocytosis in a dose-dependent manner. Thus our results suggest that HRB regulates clathrin-dependent endocytosis and may recruit TI-VAMP in this process.
EXPERIMENTAL PROCEDURES
Antibodies—The goat polyclonal antibody directed against a carboxyl-terminal peptide of human HRB (sc-1424) was from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Mouse monoclonal antibodies against the transferrin receptor (CD71), clathrin (X22), and AP-2 (clone AP6) were from Sigma, BD Transduction Laboratories, and Affinity Bioreagents (Golden, CO), respectively. Monoclonal antibody against EPS15 was kindly provided by A. Benmerah (Institut Cochin, Paris, France). The mouse monoclonal antibody (clone 158.2) and rabbit serum (TG18) directed against TI-VAMP/VAMP7 have been described previously (22, 23). Mouse monoclonal anti-GFP antibody was from Roche Diagnostics.
Affinity-purified Cy3- and Cy5-coupled anti-mouse, -rabbit, or -goat immunoglobulins were from Jackson Immuno-Research Laboratories (West Grove, PA). Alexa 488-coupled anti-goat immunoglobulins and Alexa 488-transferrin were from Molecular Probes (Carlsbad, CA). Bovine serum albumin and gelatin from cold water fish were from Sigma.
Constructs—The full-length cDNA sequence of human HRB was cleaved from the pMT-Rab cl 14 construct from Dr. P. P. Di Fiore (Fondazione Istituto FIRC di Oncologia Molecolare, Milan, Italy) by restriction digest using XbaI and SalI. The cleaved fragment was blunted using Klenow polymerase enzyme and was cloned into the pcDNA3.1 (Invitrogen) plasmid by using the EcoRV site. The new construct pcDNA-Hrb was checked by sequencing.
We previously described a pHLuorin-TI-VAMP construct (10). Here we improved the construction by adding a linker sequence between TI-VAMP and pHLuorin. Rat brain TI-VAMP cDNA bearing an AgeI site and a linker sequence (three Ser-Gly-Gly repeats) (24) in its 3′-end was obtained by PCR and cloned in the pcDNA3 vector. The superecliptic variant of pHLuorin was then cloned in the AgeI sites. The mutations (F64L and S65T) required to obtain the superecliptic variant (25) were obtained by site-directed mutagenesis.
Yeast Two-hybrid Cloning and Analysis—Yeast two-hybrid screens performed on human placenta cDNA library were described previously (8). For the screen performed on human fetal brain library, the same approach was used using only the TI-VAMP cytosolic domain (amino acids 1-188) as bait. The screens were carried out by Hybrigenics (Paris, France) using its standard procedures. Relevance of each identified interaction was noted on an A to E predicted biological score scale as follow. “A,” “B,” and “C” show “very high,” “high,” and “good” confidence, respectively, in the interaction.
Cell Culture—HeLa cells were cultured at 37 °C at 5% CO2 in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal calf serum, 100 units/ml penicillin, and 100 μg/ml streptomycin. Cells were regularly passaged by trypsin-EDTA to maintain exponential growth. All culture media were from Invitrogen.
Tet-Off MDCK cells overexpressing GFP and GFP-Longin were described previously (12). Briefly Tet-Off MDCK cells were cultured in DMEM with 7% fetal calf serum, 100 units/ml penicillin and streptomycin, 400 μg/ml hygromycin, 200 μg/ml G418, and 0.5 μg/ml doxycycline. Experiments were carried out for at least 5 days after the removal of doxycycline.
RNA Interference and Stable Cell Lines with shRNA—siRNA duplex corresponded to nucleotides 1593-1613 of the human HRB mRNA. Human HRB duplex sequences used are 5′-CAGCCCAAUGGUGCAGGUUTT-3′ and 5′-AACCUGCACCAUUGGGCUGTT-3′ (26). Oligofectamine (Invitrogen) was used for the delivery of siRNAs into HeLa cells according to the manufacturer's protocol. Control cells were transfected with luciferase siRNA (mock condition). Cells were incubated for 24 h with siRNA.
Four Sure Silencing™ shRNA plasmids targeting human HRB and one negative control shRNA targeting no known human gene and containing the neomycin resistance gene were purchased from SuperArray (Frederick, MD). HeLa cells were transfected by the different shRNAs against human HRB and by the negative control shRNA using FuGENE 6 (Roche Diagnostics) according to the manufacturer's protocol and were selected for neomycin resistance (400 μg/ml G418).
Western Blot—Cells were washed three times in cold PBS and lysed under continuous shaking with TSE (50 mm Tris-HCl, pH 8, 150 mm NaCl, 1 mm EDTA) buffer containing 1% Triton X-100 and protease inhibitors (Complete™, Roche Applied Science). Cells were scraped, and the supernatant resulting from centrifugation at 13,000 rpm for 30 min was used for Western blot analysis. Brains from embryonic day 16, P4, P14, P30, and adult rats were homogenized with a glass/Teflon homogenizer (10 strokes at 900 rpm) in homogenizing buffer (0.32 m sucrose, 150 mm NaCl, 10 mm HEPES, 1 mm EDTA) containing a protease inhibitor mixture (Complete, Roche Applied Science). After a 10-min centrifugation at 800 rpm at 4 °C, 1% Triton X-100 final concentration was added to the supernatant for 30 min, and finally the insoluble material was removed by centrifugation at 20,000 rpm at 4 °C for 30 min. Generally 25 μg of protein were boiled at 95 °C for 5 min in SDS buffer. SDS-PAGE analysis was performed using a 4-12% bis-Tris NuPAGE (Invitrogen) gradient gel and the manufacturer's buffers and then processed for Western blot. Primary antibodies were revealed with fluorescent secondary antibodies coupled to Alexa 680 or Alexa 800 and analyzed by Odyssey. Blots were quantified by using ImageJ (National Institutes of Health, Bethesda, MD).
Production of Recombinant Proteins—GST and GST-TI-VAMP constructs and production of recombinant proteins were described previously (8, 27).
In Vitro Interaction Assay and GST Pulldown—pcDNA3-Hrb construct was used for in vitro T7-driven transcription and translation using the TnT-coupled transcription/translation kit (Promega, Madison, WI). For binding experiments, 10 μl of the reaction or 500 μg of total brain extracts for GST pulldown were incubated overnight with GST, GST-TI-VAMP, or GST-Longin immobilized on glutathione-Sepharose beads at 1 μm final concentration in 200 or 500 μl of binding buffer (4 mm HEPES, pH 7.5, 100 mm NaCl, 3.5 mm CaCl2, 3.5 mm MgCl2, 1 mm EDTA, 0.1% Nonidet P-40 or Triton X-100 as indicated). The beads were collected by centrifugation and washed eight times with washing buffer (50 mm Tris-HCl, pH 8.0, 100 mm NaCl, 2.5 mm MgCl2, 0.1% Nonidet P-40 or Triton X-100 as indicated), and the final bead fraction was eluted with NuPAGE gel sample buffer and analyzed by Western blot.
Immunoprecipitation—Immunoprecipitation from rat brains was performed using a Triton X-100-soluble fraction prepared as described previously for Western blot. Immunoprecipitation experiments were done as described previously (28). Briefly P4 rat brain lysate was incubated with immunobeads overnight, and the beads were washed five times in homogenization buffer with 0.1% Triton X-100. Bound material was eluted with gel sample buffer, and the eluates were boiled for 5 min and run on 4-12% gradient NuPAGE gels (Invitrogen).
Immunocytochemistry—HeLa cells were fixed with 4% paraformaldehyde in PBS for 20 min; blocked 20 min in PBS, 50 mm NH4Cl; and permeabilized by treatment with 0.1% Triton X-100 in 0.125% gelatin in PBS for 4 min at room temperature. After treatment with 0.25% cold water fish skin gelatin in PBS for 30 min, cells were incubated overnight with the first antibodies in 0.125% gelatin in PBS at 4 °C. After several washings with PBS, cells were then incubated in the secondary antibodies in 0.125% gelatin in PBS for 1 h at room temperature, then washed several times in PBS, and mounted in Prolong medium (Invitrogen). Confocal laser-scanning microscopy was performed using an SP2 confocal microscope (Leica, Heidelberg, Germany). The images were assembled without modifications using Adobe Photoshop CS3 (Adobe Systems, San Jose, CA). For standard epifluorescence microscopy, images were acquired on a Leica DMRD microscope (objective 63×/1.32) using fluorescein isothiocyanate-, Cy3-, Cy5-, and 4′,6-diamidino-2-phenylindole-specific sets of filters and a high resolution camera (Coolsnap HQ) driven by the Metamorph Image Analysis System. Image files were merged for colocalization using Adobe Photoshop CS3.
TI-VAMP Endocytosis Assay—Cells were transfected with pHLuorin-TI-VAMP construct using FuGENE 6 (Roche Diagnostics). After 24 h, cells were starved for 1 h in DMEM without serum and incubated in the same medium containing a monoclonal antibody against GFP (5 μg/ml) at 37 °C for 5, 15, or 30 min. Cells were then washed extensively in PBS, 1% BSA at 4 °C; fixed with 4% paraformaldehyde in PBS for 20 min; and processed for immunofluorescence as described below.
Transferrin Uptake Assay—Cells were starved for 1 h in DMEM without serum. Cells were collected in PBS-EDTA and incubated in DMEM, 15 mm HEPES, pH 7.5, 20 mm glucose, 1% BSA with fluorescent human transferrin for the indicated times at 37 °C; washed extensively with PBS, 1% BSA at 4 °C; and either incubated in acid buffer (50 mm glycine, 100 mm NaCl (i.e. 5.84 g/liter), pH 3.0) for 3 min at 4 °C to remove surface labeling before fixation or fixed immediately. Cells were then processed for immunofluorescence or for flow cytometry analysis. pHLuorin-TI-VAMP Uptake Assay—Cells were transfected with FuGENE 6 (Roche Diagnostics). 24 h later, cells were starved, collected in PBS-EDTA, and incubated in DMEM, 15 mm HEPES, pH 7.5, 20 mm glucose, 1% BSA with a monoclonal antibody against GFP at 4 °C for 30 min. Cells were then extensively washed with PBS, 1% BSA at 4 °C and then placed at 37 °C to allow endocytosis for 20 min prior to fixation. Cells were then incubated with an anti-mouse antibody coupled to Cy5. Cells were then processed for flow cytometry analysis.
Flow Cytometry—Analyses were performed with a CyAn ADP flow cytometer (DakoCytomation) equipped with two solid state lasers: 488 nm for Alexa 488 and phycoerythrin and 635 nm for Cy5. Alexa 488 was collected through a 530/40 nm band pass. Forward angle light scatter and right angle scatter were used to select cells; resultant fluorescence histograms were taken on 20,000 cells. Data analysis was performed with Summit v4.3.01. Median fluorescence intensity and mean fluorescence intensity values were determined for both positive and negative populations of cells. For statistical analysis, all experiments were repeated at least three times. Results are expressed as mean ± S.E. Comparisons of median values between mock and shRNA cells were performed using an unpaired Student's t test. Statistical significance was accepted at p < 0.05. Significant differences with the corresponding internal control values are indicated by asterisks (*, p < 0.05; **, p < 0.01; ***, p < 0.001 in Student's t test).
RESULTS
To identify the partners of TI-VAMP we carried out yeast two-hybrid screens using either the Longin domain of TI-VAMP alone or the full cytoplasmic domain of the human isoform as baits in human placenta (screen with both baits (8)) and fetal brain (screen with the full cytoplasmic domain only) cDNA libraries. These screens were validated by the identification of the cognate t-SNARE partners of TI-VAMP, syntaxin 3 and SNAP-23 in the human placenta library (8) and syntaxin 1 and SNAP-25 in the human fetal brain library.6 Syntaxin 3 and SNAP-23 represent the apical plasma membrane t-SNARE in epithelial cells (27), whereas syntaxin 1A and SNAP-25 constitute the neuronal synaptic t-SNARE (29), both of which are relevant to TI-VAMP exocytic function in epithelial and neuronal cells, respectively. Furthermore all screens identified the δ subunit of the molecular coat AP-3 that was previously functionally validated both in non-neuronal and neuronal cells (8, 9). In all these screens we also identified HRB. HRB appeared as an intriguing partner because it also binds EPS15 (18, 19), a tyrosine kinase substrate involved in endocytosis (30-32), and was recovered in the AP-2 interactome by GST pulldown experiments (21, 33). The phenotype of HRB-/- mice further reinforces the mysterious function of HRB because defects were seen only in sperm cells that lack an acrosome because of the inability of proacrosomal vesicles to fuse (20, 34). We decided to investigate the possible regulation of TI-VAMP function by HRB because of the evidence listed above suggesting a role of HRB in membrane trafficking.
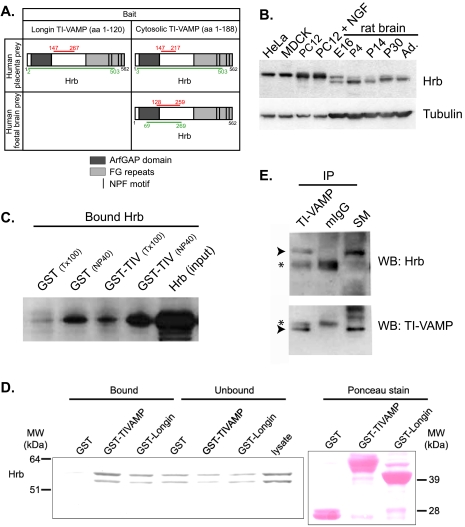
The intersection coverage of all prey clones identified in the yeast two-hybrid screens (Fig. 1A) spanned the region 147-267 in the screen with the Longin domain alone, the region 147-217 in the screen with the full cytoplasmic domain in the placenta library, and the region 128-259 in the screen with the same bait in the fetal brain library. Therefore, we can reasonably conclude that the minimal interaction domain with TI-VAMP is contained within the 147-217 region of HRB. This region of HRB is located downstream of an in silico predicted ADP-ribosylation factor GTPase-activating protein zinc finger domain motif and upstream of FG and NPF repeats that are thought to mediate other important interactions including that with EPS15 (18, 19, 35). The fact that HRB was identified in the screen using the Longin domain alone suggests that HRB interacts with this domain in TI-VAMP. We checked the expression profile of HRB by Western blotting extracts obtained from HeLa, MDCK, and PC12 cells and also from rat brain at different stages of development. We found expression in all cases with the notable presence of two close bands that could correspond to different isoforms of HRB expressed during brain development (Fig. 1B). The broad expression profile suggests that HRB may have a general function despite the sperm-restricted phenotype of HRB-/- mice (20).
FIGURE 1.
Interaction of HRB and TI-VAMP. A, HRB was identified as a new partner of TI-VAMP in three independent yeast two-hybrid screens. Results of yeast two-hybrid screens using either the Longin domain (amino acids 1-120) or the cytoplasmic domain of TI-VAMP (amino acids 1-188) and either a human placenta or a human fetal brain cDNA library are shown. HRB was found in the three different screens. Green lines highlight the total coverage of all prey clones identified in the screens, and red lines represent the intersection of the clones. Relevance of each identified interaction was noted as A on an A to E predicted biological score scale, showing a very high confidence on the interaction. The interaction with TI-VAMP is mediated by a minimal interaction domain of 70 amino acids (amino acids 147-217) of HRB. B, Western blot analysis of the expression of HRB in different cell lines and in rat brain. A 60-kDa band corresponding to HRB is observed in different cell types and in rat brain. Two bands of different apparent molecular mass that could correspond to different isoforms of the protein are observed in brain during development. C, in vitro interaction of HRB and TI-VAMP. Glutathione beads coated with GST or GST-TI-VAMP (1 μm) were incubated overnight with in vitro translated human HRB in buffer containing Triton X-100 (Tx100) or Nonidet P-40 (NP40) as detergent. The fractions bound to glutathione beads were eluted and separated by SDS-PAGE, and HRB was detected by Western blot. More in vitro translated HRB is bound to GST-TI-VAMP beads than to GST beads, indicating an interaction between HRB and TI-VAMP. D, in vitro interaction of HRB and the Longin domain of TI-VAMP. GST pulldown experiments were performed with recombinant GST, GST-TI-VAMP, and GST-Longin and with total rat brain extracts. HRB bound to the cytoplasmic domain and to the Longin domain of TI-VAMP. E, HRB interacts with TI-VAMP in the brain. Five P4 rat brains were homogenized, and the Triton X-100-soluble fraction was immunoprecipitated with a monoclonal antibody against TI-VAMP or with mouse IgG as a negative control. Co-immunoprecipitated endogenous HRB (arrowhead) and IgG (asterisk) were detected by Western blot. aa, amino acids; E16, embryonic day 16; NGF, nerve growth factor; TIV, TI-VAMP; mIgG, mouse IgG; WB, Western blot; IP, immunoprecipitation; Ad., adult; SM, starting material; Arf-GAP, ADP-ribosylation factor GTPase-activating protein.
Next we validated the interaction unraveled by the yeast two-hybrid screen by biochemical methods. To this purpose we produced GST-tagged cytoplasmic domain of TI-VAMP immobilized on glutathione-Sepharose beads and in vitro translated HRB and incubated these two together; GST alone was used as a negative control. We were able to identify a significant binding of HRB (bound to TI-VAMP rather than to GST) in the presence of Triton X-100 as well as Nonidet P-40 (Fig. 1C), but the recovery was rather limited. We also carried out GST pulldown experiments with recombinant GST, GST-TI-VAMP, and GST-Longin. We found an interaction of HRB with the full-length cytoplasmic and the Longin domains (Fig. 1D) in full agreement with the yeast two-hybrid results. Furthermore we were able to co-immunoprecipitate HRB with TI-VAMP from rat P4 brain extracts (Fig. 1E), thus demonstrating that the full-length native proteins are able to interact. All our attempts to co-immunoprecipitate HRB together with TI-VAMP or vice versa were negative when we used HeLa or MDCK cell lysates. We interpret these results as a possible indication that the interaction of TI-VAMP and HRB may be transient and/or have a rather low affinity.
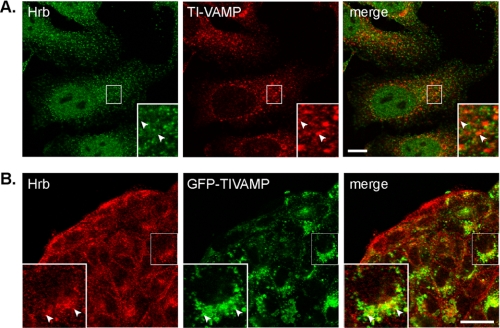
To gain further insights into HRB function, we then studied the subcellular localization of HRB by immunocytochemistry in HeLa cells (Figs. 2 and 3A) and GFP-TI-VAMP-expressing MDCK cells (Fig. 3B). HRB staining was found both in the nuclei and cytoplasm in HeLa cells. The specificity of the antibody directed against HRB was demonstrated by the loss of the cytoplasmic staining in HRB knockdown cells (see Fig. 5B). The nuclear staining may be due to cross-reaction with a nuclear antigen or to the fact that the residual expression of HRB in knockdown cells preferentially localizes in the nucleus (see below). There a significant colocalization was found with AP-2, EPS15, clathrin light chain, and TfR. These results suggest that HRB concentrates in AP-2+, EPS15+, clathrin+, and TfR+ membranes that very likely correspond to clathrin-coated structures. We did not see any significant colocalization between HRB and TI-VAMP in HeLa cells suggesting that these proteins may only interact transiently in a dynamic manner. In MDCK cells HRB concentrated in the perinuclear region where it partially colocalized with GFP-TI-VAMP (Fig. 3B).
FIGURE 2.
The subcellular localization of HRB is compatible with a role in endocytosis. HeLa cells were fixed and stained for endogenous HRB (in green) and the indicated markers of the clathrin-dependent endocytic pathway (in red). Arrowheads point to structures labeled by both HRB and the indicated marker (bar, 10 μm). Note the high degree of colocalization of HRB with AP-2 and EPS15. HRB also partially colocalizes with clathrin light chain and transferrin receptor, indicating that HRB likely localized in clathrin-coated structures. HRB was also present in a diffuse manner in the nucleus.
FIGURE 3.
Localization of TI-VAMP and HRB in HeLa and MDCK cells. A, HeLa cells were fixed and stained for endogenous HRB (in green) and TI-VAMP (in red). Arrowheads point to structures positive for only one of the proteins. No significant colocalization was observed at steady state, indicating that HRB and TI-VAMP could interact transiently (bar, 10 μm). B, MDCK cells expressing inducible GFP-TI-VAMP were fixed and stained for the endogenous HRB (in red). Arrowheads point to structures labeled by both HRB and GFP-TI-VAMP (bar, 10 μm).
FIGURE 5.
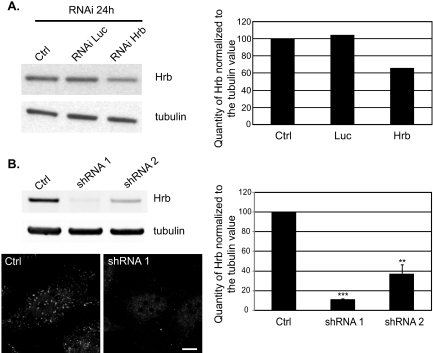
siRNA- and shRNA-mediated gene silencing of HRB in HeLa cells. A, cells were transfected by siRNA against HRB or luciferase (Luc) as a negative control, lysed, and analyzed by Western blot 24 h later. HRB protein is detected at 60 kDa. Extinction is about 35% with siRNA against HRB. B, stable HeLa cell lines were established using a control shRNA targeting no known human gene targets (Ctrl) or two different shRNAs directed against two different regions of the human HRB coding sequence (shRNA 1 and shRNA 2). Cells were lysed, and HRB was detected by Western blot. Extinction of HRB is around 90 and 50% for shRNA 1 and shRNA 2, respectively. Bottom panel, control (Ctrl) and HRB knockdown cells (shRNA 1) were fixed and stained for endogenous HRB (bar, 10 μm). Some HRB staining still remains in the nucleus but is virtually absent from the cytoplasm of shRNA 1 cells. RNAi, RNA interference. **, p < 0.01; ***, p < 0.001 in Student's t test. Error bars represent S.E. throughout.
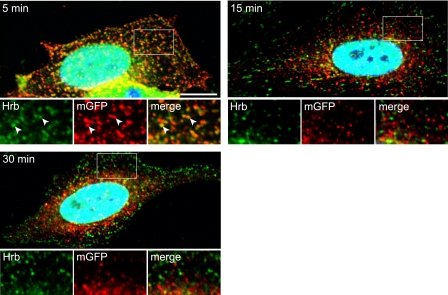
Because HRB localized in clathrin-coated structures and transiently interacted with TI-VAMP, we went on to investigate specifically the distribution of HRB during the endocytosis of TI-VAMP. To this end, we took advantage of a pHLuorin (pH-sensitive GFP) carboxyl-terminally tagged construct of TI-VAMP (10) that was here further refined by the introduction of a longer spacer between TI-VAMP and ecliptic pHLuorin sequences (see “Experimental Procedures”). We transfected HeLa cells and allowed them to bind and internalize anti-GFP antibodies for 5, 15, and 30 min prior to extensive washes at 4 °C, fixation, and immunocytochemical detection of both GFP-tagged TI-VAMP and HRB. We found a very significant degree of colocalization of GFP and HRB at 5 min but not at 15 or 30 min of endocytosis (Fig. 4). This result further supports the idea of a dynamic interaction between HRB and TI-VAMP and suggests that the interaction may be restricted to the early stages of clathrin-dependent endocytosis.
FIGURE 4.
HRB colocalizes with endocytosed TI-VAMP. HeLa cells were transfected with pHLuorin-TI-VAMP and then incubated with a GFP antibody at 37 °C to follow the endocytosis of pHLuorin-TI-VAMP. Cells were washed, fixed at the indicated time points, and stained for endogenous HRB (acquired in Cy5 channel, green) and endocytosed anti-GFP antibody (red). Arrowheads point to structure where HRB and pHLuorin-TI-VAMP are both present. Note the good degree of colocalization after 5 min of endocytosis suggesting that the interaction between both proteins may be restricted to the early stages of endocytosis (bar, 10 μm). mGFP, mouse monoclonal anti-GFP antibody.
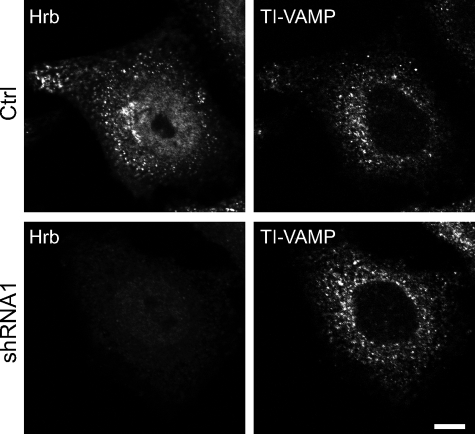
We then reasoned that our results and the presence of HRB in the AP-2 interactome (21) were likely to suggest a function of HRB in clathrin-dependent endocytosis. To test this hypothesis, we silenced the expression of HRB by siRNA and shRNA. In the first case, we found effective but not total silencing (∼35%) by transient transfection (Fig. 5A). We also selected stable HeLa cell lines expressing shRNA to analyze larger populations of cells. We cloned two cell lines: one with ∼10% (shRNA 1) and one with ∼40% (shRNA 2) remaining HRB (Fig. 5B). Immunocytochemistry demonstrated virtual absence of HRB in the cytoplasm but persistence of nuclear staining in shRNA 1 cells (Fig. 5B). Despite repeated efforts, we were not able to select HeLa cells with an expression level of HRB that would be undetectable by Western blotting. This suggests that HRB may not be dispensable in cultured cells like it is in the mouse (20). We did not see any significant change in the distribution of endogenous TI-VAMP; particularly we did not find significant accumulation at the plasma membrane (Fig. 6) in HRB-silenced cells. These results suggest that the interaction of TI-VAMP and HRB is not absolutely required for the proper localization of TI-VAMP but likely rather corresponds to a more subtle regulation.
FIGURE 6.
Inhibition of the expression of HRB has no effect on the localization of endogenous TI-VAMP. Control (Ctrl) and HRB knockdown cells (shRNA 1) cells were fixed and stained for endogenous HRB and TI-VAMP (bar, 10 μm). Note that endogenous TI-VAMP shows the same perinuclear pattern in both cases.
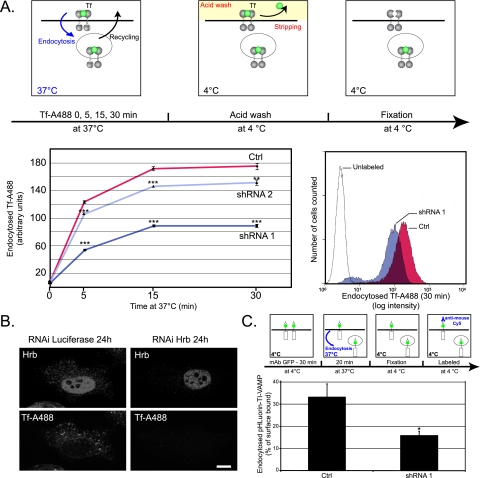
Next we directly assessed the role of HRB in clathrin-dependent endocytosis by measuring the kinetics of fluorescent Tf uptake in control and HRB-silenced HeLa cells by fluorescence-activated cell sorting. We choose this assay for clathrin-dependent endocytosis because of the localization of HRB in clathrin+ and TfR+ vesicles. Assaying the endocytosis of fluorescent Tf by flow cytometry over large populations of cells (up to 20,000 cells) proved to be extremely reliable and reproducible. We found a dose-dependent effect: shRNA 1 cells were able to take up only 50% of fluorescent Tf in comparison with control cells, whereas shRNA 2 endocytosed ∼90% of control (Fig. 7A). The fact that the initial rate of Tf endocytosis was affected in shRNA 1 cells strongly indicates that HRB has a direct function in clathrin-dependent endocytosis (Fig. 7A). We were able to confirm this result in siRNA transiently transfected HeLa cells by immunocytochemistry (Fig. 7B) thus demonstrating that the effect seen in stably silenced cells was not due to a cloning artifact. These results suggest that HRB is necessary to achieve efficient endocytosis of transferrin. Finally we tested the direct effect of silencing HRB on the endocytosis of TI-VAMP using the pHLuorin-TI-VAMP construct in control and HRB knockdown cells. We found that 33% of surface pHLuorin-TI-VAMP was endocytosed after 20 min in control cells, but only 16% was endocytosed in shRNA 1 cells (Fig. 7C). This decrease of ∼50% of TI-VAMP endocytosis in HRB-depleted cells demonstrates a role of HRB in the endocytosis of TI-VAMP.
FIGURE 7.
Inhibition of the expression of HRB inhibits transferrin and TI-VAMP endocytosis. A, control (Ctrl) or HRB knockdown cells (shRNA 1 and shRNA 2) were harvested and incubated with fluorescent Tf-Alexa 488 (A488) for 5, 15, or 30 min at 37 °C. Cells were then washed, and surface labeling was stripped with acid buffer. Endocytosed Tf was quantified by fluorescence-activated cell sorting. Each time point corresponds to 20,000 cells. The y axis represents arbitrary units of fluorescent Tf. HRB knockdown cells showed a significant inhibition of Tf endocytosis (Student's t test: control/shRNA 1, p = 1.8·10-6, 4·10-6, and 3.5·10-6 at 5, 15, and 30 min, respectively; control/shRNA 2, p = 2.8·10-4, 2.8·10-4, and 8.2·10-3 at 5, 15, and 30 min, respectively. Right panel, detailed analysis of one representative flow cytometry experiment measuring Tf endocytosis in control and HRB knockdown cells (shRNA 1) after 30 min of endocytosis. Control and HRB knockdown cells (shRNA 1) showed a significant increase in Tf median fluorescence compared with the unlabeled cells (corresponding to 0 min of endocytosis). HRB knockdown cells (shRNA 1) showed a significant decrease in Tf median fluorescence compared with the control cells (shown here by a left shift of the fluorescence intensity). B, HeLa cells were transfected with siRNA against HRB or luciferase. After 24 h, cells were starved and incubated at 37 °C with Tf-Alexa 488 for 30 min. Cells were then fixed and processed for immunofluorescence. HRB knockdown cells accumulate less fluorescent Tf than mock cells (treated with luciferase siRNA). C, control or HRB stably depleted cells (shRNA 1) were transfected with pHLuorin-TI-VAMP construct, harvested, and incubated with an anti-GFP antibody at 4 °C to stain surface TI-VAMP. Cells were then incubated at 37 °C for 20 min to allow endocytosis, extensively washed, fixed, and incubated with an anti-mouse antibody coupled to Cy5 to stain the pool of TI-VAMP that remains in the surface of the cells. This pool was quantified by fluorescence-activated cell sorting. Each measure corresponds to 20,000 cells. The y axis represents the percentage of surface-bound pHLuorin-TI-VAMP endocytosed in 20 min. HRB knockdown cells showed a significant decrease of ∼50% of TI-VAMP endocytosis (Student's t test: control/shRNA 1, p = 0.028). *, p < 0.05; ***, p < 0.001 in Student's t test. mAb, monoclonal antibody.
DISCUSSION
Here we identified HRB as a new partner of TI-VAMP and demonstrated that HRB is involved in clathrin-dependent endocytosis of TI-VAMP and transferrin receptor. The central role of SNARE proteins in membrane fusion makes the search for SNARE partners highly relevant to characterize the regulation of membrane trafficking. In the case of the v-SNARE TI-VAMP, we and others have identified several partners including t-SNAREs that are mainly located at the plasma membrane (syntaxins 1, 3, and 4; SNAP-23; and SNAP-P25 (8, 36)) both using yeast two-hybrid screens and biochemical approaches. We have also identified the δ subunit of the molecular coat AP-3 (8, 9), and we show here that HRB is a partner of TI-VAMP by using yeast two-hybrid screens and biochemical methods including in vitro interaction assays and immunoprecipitation. AP-3 is implicated in important sorting steps of TI-VAMP. Indeed we previously found that AP-3 is required for the targeting of TI-VAMP from early endosomes to late endosomes in epithelial cells (8). In the dentate gyrus, AP-3 is involved in the axonal targeting of TI-VAMP from the perinuclear VAMP4+ trans-Golgi network and recycling endosomal membranes in granule cells (9). We propose here that the interaction between HRB and TI-VAMP thus appears as another layer of regulation of the sorting of this v-SNARE. SNARE proteins are implicated in exocytosis and fusion events at the plasma membrane, but the molecular mechanisms of their recycling are still largely unknown. At the synapse, neurotransmitter are released after the fusion of synaptic vesicles with the presynaptic membrane, and clathrin-mediated endocytosis plays a major role in the recycling of the synaptic vesicle components (37-40), including SNARE proteins (41). In this way, clathrin assembly lymphoid-myeloid leukemia protein (CALM) was recently identified to be important for the endocytosis of synaptobrevin2/VAMP2 (42); thus it is likely that accessory proteins assist v-SNAREs for proper recycling as a more general mechanism. We demonstrate here that TI-VAMP interacts transiently with HRB, that this interaction occurs during early stages of endocytosis, and that HRB is required for optimal endocytosis of TI-VAMP indicating that HRB plays a role in the endocytosis of TI-VAMP. The fact that the localization of endogenous TI-VAMP was not apparently affected in HRB-silenced cells likely suggests that HRB is not absolutely required for the endocytosis of TI-VAMP and that other accessory proteins could play a redundant role in this process. The transient presence of TI-VAMP in HRB+ vesicles at early stages of the endocytosis of the v-SNARE suggests that TI-VAMP is endocytosed in a clathrin-dependent manner. The role of HRB in the regulation of TI-VAMP endocytosis further supports the concept that TI-VAMP mediates secretory processes as we have proposed based on previous work (5, 43). Furthermore we demonstrate here for the first time that HRB is implicated in clathrin-dependent endocytosis on the basis of its localization in clathrin+, AP-2+, EPS15+, and TfR+ vesicles and the inhibition of Tf endocytosis in cells in which the expression of HRB was silenced by siRNA or shRNA. However, at this point, we cannot define HRB as a critical factor of endocytosis because a significant amount (∼50%) of endocytosis could still occur in the virtual absence of HRB in the cytoplasm (our shRNA 1 cell line). Furthermore we did not see any significant decrease of epidermal growth factor endocytosis in HRB knockdown cells,6 thus suggesting that HRB has a limited function in this process. In addition, the lack of a strong phenotype of HRB-/- mice (20) makes it hard to think that HRB would be absolutely required for clathrin-dependent endocytosis. This is in contrast to the adaptor protein AP-2 because AP-2-deficient mice are not viable (44). Rather our data suggest that HRB regulates clathrin-dependent endocytosis. At the molecular level, the function of HRB may be to concentrate cargos including the v-SNARE TI-VAMP for internalization. In this model, HRB may function to internalize TI-VAMP-containing cis-SNARE complexes (45) or to concentrate TI-VAMP freed from SNARE complexes in clathrin-coated pits to limit its residence time at the cell surface and accelerate its endocytosis. This hypothesis will require further investigation.
Addendum—While revising this article, Pryor et al. (45) also found that HRB interacts with TI-VAMP and regulates its endocytosis.
Acknowledgments
We are grateful to Pier Paolo Di Fiore (Fondazione Istituto FIRC di Oncologia Molecolare, Milan, Italy) for the HRB plasmid, Alexandre Benmerah (Institut Cochin, Paris, France) for the EPS15 antibody, Lucien Cabanié (Institut Curie, Paris, France) for the purification of Cl158.2, Etienne Formstecher (Hybrigenics, Paris, France) for help in the analysis of the yeast two-hybrid screen results, and Nicole Boggetto and Marie-Claude Gendron from the flow cytometry platform for technical assistance.
This work was supported in part by grants from INSERM (Avenir Program), the European Commission (“Signalling and Traffic” Specific Targeted Research Project Grant 503229), the Association pour la Recherche sur le Cancer, the Agence Nationale pour la Recherche (“PolarHiCell”), the Mairie de Paris Medical Research and Health Program, and the Fondation pour la Recherche Médicale (to T. G.). The CyAn ADP analyzer was bought with funds from the Ministry of Research and la Ligue Contre le Cancer (Paris committee). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: SNARE, soluble N-ethylmaleimide-sensitive factor attachment protein receptors; HRB, human immunodeficiency virus Rev-binding protein; TI-VAMP, tetanus neurotoxin-insensitive vesicle-associated membrane protein; VAMP, vesicle-associated membrane protein; GFP, green fluorescent protein; MDCK, Madin-Darby canine kidney; t-SNARE, target SNARE; v-SNARE, vesicular SNARE; SNAP, synaptosome-associated protein; Tf, transferrin; TfR, transferrin receptor; shRNA, short hairpin RNA; PBS, phosphate-buffered saline; DMEM, Dulbecco's modified Eagle's medium; siRNA, small interfering RNA; P, postnatal day; bis-Tris, 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol; GST, glutathione S-transferase; BSA, bovine serum albumin.
M. Chaineau, L. Danglot, V. Proux-Gillardeaux, and T. Galli, unpublished results.
References
- 1.Jahn, R., and Scheller, R. H. (2006) Nat. Rev. Mol. Cell Biol. 7 631-643 [DOI] [PubMed] [Google Scholar]
- 2.Hong, W. (2005) Biochim. Biophys. Acta 1744 493-517 [PubMed] [Google Scholar]
- 3.Galli, T., and Haucke, V. (2004) Sci. STKE 2004 re19. [DOI] [PubMed] [Google Scholar]
- 4.Bock, J. B., Matern, H. T., Peden, A. A., and Scheller, R. H. (2001) Nature 409 839-841 [DOI] [PubMed] [Google Scholar]
- 5.Proux-Gillardeaux, V., Rudge, R., and Galli, T. (2005) Traffic 6 366-373 [DOI] [PubMed] [Google Scholar]
- 6.Rossi, V., Banfield, D. K., Vacca, M., Dietrich, L. E., Ungermann, C., D'Esposito, M., Galli, T., and Filippini, F. (2004) Trends Biochem. Sci. 29 682-688 [DOI] [PubMed] [Google Scholar]
- 7.Filippini, F., Rossi, V., Galli, T., Budillon, A., D'Urso, M., and D'Esposito, M. (2001) Trends Biochem. Sci. 26 407-409 [DOI] [PubMed] [Google Scholar]
- 8.Martinez-Arca, S., Rudge, R., Vacca, M., Raposo, G., Camonis, J., Proux-Gillardeaux, V., Daviet, L., Formstecher, E., Hamburger, A., Filippini, F., D'Esposito, M., and Galli, T. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 9011-9016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scheuber, A., Rudge, R., Danglot, L., Raposo, G., Binz, T., Poncer, J. C., and Galli, T. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 16562-16567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martinez-Arca, S., Alberts, P., Zahraoui, A., Louvard, D., and Galli, T. (2000) J. Cell Biol. 149 889-899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martinez-Arca, S., Coco, S., Mainguy, G., Schenk, U., Alberts, P., Bouille, P., Mezzina, M., Prochiantz, A., Matteoli, M., Louvard, D., and Galli, T. (2001) J. Neurosci. 21 3830-3838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Proux-Gillardeaux, V., Raposo, G., Irinopoulou, T., and Galli, T. (2007) Biol. Cell 99 261-271 [DOI] [PubMed] [Google Scholar]
- 13.Braun, V., Fraisier, V., Raposo, G., Hurbain, I., Sibarita, J. B., Chavrier, P., Galli, T., and Niedergang, F. (2004) EMBO J. 23 4166-4176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pryor, P. R., Mullock, B. M., Bright, N. A., Lindsay, M. R., Gray, S. R., Richardson, S. C., Stewart, A., James, D. E., Piper, R. C., and Luzio, J. P. (2004) EMBO Rep. 5 590-595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bogerd, H. P., Fridell, R. A., Madore, S., and Cullen, B. R. (1995) Cell 82 485-494 [DOI] [PubMed] [Google Scholar]
- 16.Fritz, C. C., Zapp, M. L., and Green, M. R. (1995) Nature 376 530-533 [DOI] [PubMed] [Google Scholar]
- 17.Stutz, F., Neville, M., and Rosbash, M. (1995) Cell 82 495-506 [DOI] [PubMed] [Google Scholar]
- 18.Doria, M., Salcini, A. E., Colombo, E., Parslow, T. G., Pelicci, P. G., and Di Fiore, P. P. (1999) J. Cell Biol. 147 1379-1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salcini, A. E., Confalonieri, S., Doria, M., Santolini, E., Tassi, E., Minenkova, O., Cesareni, G., Pelicci, P. G., and Di Fiore, P. P. (1997) Genes Dev. 11 2239-2249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kang-Decker, N., Mantchev, G. T., Juneja, S. C., McNiven, M. A., and van Deursen, J. M. (2001) Science 294 1531-1533 [DOI] [PubMed] [Google Scholar]
- 21.Schmid, E. M., Ford, M. G., Burtey, A., Praefcke, G. J., Peak-Chew, S. Y., Mills, I. G., Benmerah, A., and McMahon, H. T. (2006) PLoS Biol. 4 e262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paumet, F., Le Mao, J., Martin, S., Galli, T., David, B., Blank, U., and Roa, M. (2000) J. Immunol. 164 5850-5857 [DOI] [PubMed] [Google Scholar]
- 23.Muzerelle, A., Alberts, P., Martinez-Arca, S., Jeannequin, O., Lafaye, P., Mazie, J.-C., Galli, T., and Gaspar, P. (2003) Neuroscience 122 59-75 [DOI] [PubMed] [Google Scholar]
- 24.Miesenbock, G. (2000) in Imaging Neurons: A Laboratory Manual (Yuste, R., Lanni, F., and Konnerth, A., eds) pp. 59.1-59.12, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- 25.Sankaranarayanan, S., DeAngelis, D., Rothman, J. E., and Ryan, T. A. (2000) Biophys. J. 79 2199-2208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanchez-Velar, N., Udofia, E. B., Yu, Z., and Zapp, M. L. (2004) Genes Dev. 18 23-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Galli, T., Zahraoui, A., Vaidyanathan, V. V., Raposo, G., Tian, J. M., Karin, M., Niemann, H., and Louvard, D. (1998) Mol. Biol. Cell 9 1437-1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alberts, P., Rudge, R., Hinners, I., Muzerelle, A., MartinezArca, S., Irinopoulou, T., Marthiens, V., Tooze, S., Rathjen, F., Gaspar, P., and Galli, T. (2003) Mol. Biol. Cell 14 4207-4220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Söllner, T., Whiteheart, S. W., Brunner, M., Erdjument-Bromage, H., Geromanos, S., Tempst, P., and Rothman, J. E. (1993) Nature 362 318-324 [DOI] [PubMed] [Google Scholar]
- 30.Benmerah, A., Bayrou, M., Cerf-Bensussan, N., and Dautry-Varsat, A. (1999) J. Cell Sci. 112 1303-1311 [DOI] [PubMed] [Google Scholar]
- 31.Benmerah, A., Lamaze, C., Begue, B., Schmid, S. L., Dautry-Varsat, A., and Cerf-Bensussan, N. (1998) J. Cell Biol. 140 1055-1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Torrisi, M. R., Lotti, L. V., Belleudi, F., Gradini, R., Salcini, A. E., Confalonieri, S., Pelicci, P. G., and Di Fiore, P. P. (1999) Mol. Biol. Cell 10 417-434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Praefcke, G. J., Ford, M. G., Schmid, E. M., Olesen, L. E., Gallop, J. L., Peak-Chew, S. Y., Vallis, Y., Babu, M. M., Mills, I. G., and McMahon, H. T. (2004) EMBO J. 23 4371-4383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Juneja, S. C., and van Deursen, J. M. (2005) Hum. Reprod. (Oxf.) 20 881-893 [DOI] [PubMed] [Google Scholar]
- 35.de Beer, T., Hoofnagle, A. N., Enmon, J. L., Bowers, R. C., Yamabhai, M., Kay, B. K., and Overduin, M. (2000) Nat. Struct. Biol. 7 1018-1022 [DOI] [PubMed] [Google Scholar]
- 36.Rao, S. K., Huynh, C., Proux-Gillardeaux, V., Galli, T., and Andrews, N. W. (2004) J. Biol. Chem. 279 20471-20479 [DOI] [PubMed] [Google Scholar]
- 37.Morgan, J. R., Augustine, G. J., and Lafer, E. M. (2002) Neuromol. Med. 2 101-114 [DOI] [PubMed] [Google Scholar]
- 38.Morgan, J. R., Prasad, K., Jin, S., Augustine, G. J., and Lafer, E. M. (2003) J. Biol. Chem. 278 33583-33592 [DOI] [PubMed] [Google Scholar]
- 39.Schweizer, F. E., and Ryan, T. A. (2006) Curr. Opin. Neurobiol. 16 298-304 [DOI] [PubMed] [Google Scholar]
- 40.Takei, K., and Haucke, V. (2001) Trends in Cell Biol. 11 385-391 [DOI] [PubMed] [Google Scholar]
- 41.Blondeau, F., Ritter, B., Allaire, P. D., Wasiak, S., Girard, M., Hussain, N. K., Angers, A., Legendre-Guillemin, V., Roy, L., Boismenu, D., Kearney, R. E., Bell, A. W., Bergeron, J. J., and McPherson, P. S. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 3833-3838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harel, A., Wu, F., Mattson, M. P., Morris, C. M., and Yao, P. J. (2008) Traffic 9 417-429 [DOI] [PubMed] [Google Scholar]
- 43.Alberts, P., and Galli, T. (2003) Biol. Cell 95 419-424 [DOI] [PubMed] [Google Scholar]
- 44.Mitsunari, T., Nakatsu, F., Shioda, N., Love, P. E., Grinberg, A., Bonifacino, J. S., and Ohno, H. (2005) Mol. Cell. Biol. 25 9318-9323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pryor, P. R., Jackson, L., Gray, S. R., Edeling, M. A., Thompson, A., Sanderson, C. M., Evans, P. R., Owen, D. J., and Luzio, J. P. (2008) Cell 134 817-827 [DOI] [PMC free article] [PubMed] [Google Scholar]