Abstract
We recently modeled transient receptor potential (TRP) channels using the Gestalt Domain Detection Algorithm-Basic Local Alignment Tool (GDDA-BLAST), which derives structural, functional, and evolutionary information from primary amino acid sequences using phylogenetic profiles (Ko, K. D., Hong, Y., Chang, G. S., Bhardwaj, G., van Rossum, D. B., and Patterson, R. L. (2008) Physics Arch. Quant. Methods arXiv:0806.2394v1). Herein we test our functional predictions for the TRP_2 domain of TRPC3; a domain of unknown function that is conserved in all TRPC channels. Our functional models of this domain identify both lipid binding and trafficking activities. In this study, we reveal: (i) a novel structural determinant of ion channel sensitivity to lipids, (ii) a molecular mechanism for the difference between diacylglycerol (DAG)-sensitive and DAG-insensitive TRPC subfamilies, and (iii) evidence that TRPC3 can comprise part of the vesicle fusion machinery. Indeed, the TRPC3 TRP_2 domain mediates channel trafficking to the plasma membrane and binds to plasma membrane lipids. Further, mutations in TRP_2, which alter lipid binding, also disrupt the DAG-mediated fusion of TRPC3-containing vesicles with the plasma membrane without disrupting SNARE interactions. Importantly, these data agree with the known role of DAG in membrane destabilization, which facilitates SNARE-dependent synaptic vesicle fusion (Villar, A. V., Goni, F. M., and Alonso, A. (2001) FEBS Lett. 494,117 -120 and Goni, F. M., and Alonso, A. (1999) Prog. Lipid Res. 38,1 -48). Taken together, functional models generated by GDDA-BLAST provide a computational platform for deriving domain functionality, which can have in vivo and mechanistic relevance.
Transient receptor potential (TRP)3 channels are a superfamily of diverse ion channels that are central to the regulation of physiological processes such as sensory perception (4), cell development and growth (5, 6), hearing (7, 8), taste (9), and fertility (10) as well as multiple diseases including mucolipodosis (11, 12) and polycystic kidney disease (11, 13, 14). TRP channels comprise a cellular Ca2+ entry pathway that responds to stimulation with Ca2+-mobilizing agonists (15). Their channel activation can be influenced by phosphorylation, nitrosylation, glycosylation, protein-protein interactions, protein-lipid interactions, mechanosensation, temperature, Ca2+ store-depletion, and exocytosis (14, 16). This polymodal activation scheme is complex and underscores the reason why it is difficult to study the role of TRP channels within biological processes. This is compounded by the fact that: (i) TRP channels can form multimers with other TRP isoforms, and (ii) there is, in general, a lack of domain annotation for TRP channels. Therefore, reconciling the results obtained with an exogenously expressed channel to those observed with endogenous channels is challenging as is predicting protein regions underlying specific functionalities.
The canonical TRP (TRPC) channels are so named because they are closest to the prototypical Drosophila TRP channel, which mediates photoreception in the fly eye (17). Importantly, TRPC channels have been directly linked to brain development (14, 18), smooth muscle contraction (14, 19), neuronal guidance and outgrowth (6, 20, 21), as well as cardiovascular disease and glomerulosclerosis (22, 23). Through a yet unknown mechanism, TRPC channels are activated in response to the receptor second messengers inositol 1,4,5-trisphosphate (IP3) and DAG (reviewed in Refs. 14, 16). Importantly, TRPC channels can be subdivided into two subfamilies: the DAG-sensitive TRPC channels (C3/6/7) and the DAG-insensitive TRPC channels (C1/4/5). Multiple binding and regulatory activities have been empirically identified for TRPC channels. For example, two lipid binding sites have been identified in the DAG-sensitive TRPCs (24, 25). Specifically, the extreme N-terminal lipid binding domain binds to phosphatidylinositol 4,5-bisphosphate (PIP2) and regulates TRPC3 plasma membrane surface expression. Similarly in TRPC6, a the C-terminal lipid binding domain exists that binds to phosphatidylinositol 3,4,5-trisphosphate (PIP3), mediating calmodulin binding (24). This domain is conserved in all DAG-sensitive TRPCs, suggesting that they too share this functional domain. Conversely, formal queries of TRPC protein sequences using popular domain detection algorithms (e.g. Pfam, SMART, NCBI-CDD, InterProScan), yield no information about the nature or location of lipid binding domains. In general, these programs predict that TRPC channels contain only one functional domain (ion channel), and two domains whose function is unknown (ankyrin, TRP_2), thus leaving the majority of the protein without functional annotation.
We present here a comparative and physiological analysis of the TRP_2 domain within TRPC3. Our in silico (1), in vitro, and cellular assays reveal that the TRP_2 domain in TRPC channels is a lipid binding/trafficking domain, which, when mutated, alters the fusion of DAG-sensitive TRPC3-containing vesicles with the plasma membrane. Further, these results suggest that like TRPM7 (26), TRPC channels also have a “SNARE-like” activity and are directly involved in vesicle fusion.
MATERIALS AND METHODS
Lipid Binding Assays
PIP-strip—These assays were performed per the manufacturer's instructions using 500-1000 ng of purified protein (25).
Lipid Binding Liposomes—These assays were performed as previously described (27). Briefly, lipid mixtures phosphatidylethanolamine (29.2%), phosphatidylcholine (29.2%), phosphatidylserine (29.2%), and phosphatidyl-inositol (12.5%) (all in CHCl3) were dried down to form a thin film in a 0.5-ml minifuge tube (Beckmann) and then bath-sonicated in 0.2 m sucrose, 20 mm KCl, 20 mm Hepes, pH 7.4, 0.01% azide to yield a 10× dense lipid stock. This was diluted 1:10 in dilution buffer (0.12 m NaCl, 1 mm EGTA, 0.2 mm CaCl2 (free Ca2+ concentration of ∼50 nm), 1.5 mm MgCl2, 1 mm dithiothreitol, 5 mm KCl, 20 mm Hepes, pH 7.4, 1 mg/ml bovine serum albumin) containing 500-1000 ng of recombinant protein. Protein complexes were allowed to form by incubation at 30 °C for 5 min prior to centrifugation (100,000 × g for 30 min). After spinning, supernatants were carefully removed, and the pellets retrieved by addition of an equal volume of 60 °C SDS sample buffer and subsequent bath sonication. Both PIP-strip and liposomal assays were visualized via Western analysis. Films were scanned and analyzed using the Bio-Rad Gel-dock system (p values from Student's t test).
Cell Culture and Transfection
HeLa cells and HEK293 cells were from American Tissue Culture Collection (Manassas, VA). Cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and 1% streptomycin (all from Atlanta Biologicals, Altanta, GA). HEK cells were transfected by electroporation, or, like HeLa cells, using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). PC12 cells were cultured as previously described (28).
Ca2+ Imaging
For standard fluorescence recording experiments, cells grown on coverslips were loaded with 2 μm Fura-2AM in the standard solution, that contained (in mm) 150 NaCl, 5 KCl, 1 CaCl2, 1 MgCl2, pH adjusted to 7.2 with 10 mm HEPES. Following 30 min of incubation at 37 °C and a wash, the coverslips were placed in the perfusion chamber that allows rapid exchange of bath solutions. The solutions were applied by bath perfusion with ten volumes of the perfusion chamber. Fura-2 was excited by alternating 340/380 nm of light, and the resulting 510 nm fluorescence was recorded through a CCD-cooled camera and analyzed using Metafluor software (Molecular Devices, Sunnyvale, CA). All data in supplemental Fig. S1, a and b are averages of ∼30 cells from three independent experiments. The quantification of the calcium imaging in Fig. 2b reflects the area under the curves for OAG and carbachol, respectively (% of WT, p values from Student's t test).
FIGURE 2.
Mutations in TRP_2 disrupt channel activity. a, Western analysis of HEK293 cell lysates transfected for 24 h with either YFP alone (con) or YFP + full-length WT, K202A/R204A (KR), S209R/S213F (SS), or S209R (S) TRPC3 mutants blotted with anti-Myc antibody. b, quantification of the area under the curve for Sr2+ entry in the presence of 100 μm OAG or 100 μm CCH stimulation in HEK293 cells transfected with either YFP alone (con) or YFP + full-length WT, K202A/R204A, S209R/S213F, or S209R TRPC3 mutants (S.E., n = 3, *, p value <0.01, Student's t test) (see supplemental Fig. S1, a and b for traces). c, whole cell electrophysiological recordings in HEK293 cells transfected with either full-length WT (red) or SS (black) TRPC3 and stimulated with 100 μm OAG or 100 μm carbachol.
Electrophysiology
For conventional whole cell recording, cells grown on coverslips were placed in a perfusion chamber that was secured on the stage of an inverted microscope equipped with a fluorescent illuminator and filters designed to identify GFP-expressing cells. Transmembrane currents were recorded using an HEKA EPC-10 amplifier (HEKA Instruments, Southboro, MA), stored in a PC and analyzed using Patchmaster and Origin software. Currents were digitized at 1 kHz and filtered through low-bandpass filter set at 5 kHz cutoff frequency. In whole cell mode, the pipette solution contained (in mm) 140 Cs-aspartate (to cancel endogenous K+ and Cl- conductances), 5 NaCl, 5 Mg-ATP, 10 HEPES, 2 EGTA, or 10 BAPTA, pH 7.2. The standard bath solution contained (in mm): 140 NaCl, 5 KCl, 1 CaCl2, 1 MgCl2, 10 HEPES, pH 7.5. In some experiments, NaCl was replaced with NMDG-glutamate and 0 or 10 mm divalent metals were included in the bath solutions. The experiments were performed at room temperature. The solutions were applied by bath perfusion with ten volumes of the perfusion chamber. Pipette resistance was in the 2-4 MOhm range, and seal resistance was always over 1 GOhm. Series resistance and capacitance were compensated using the proprietary protocol included with Patchmaster.
Confocal Immunofluorescence Analyses
For experiments with fixed cells, cells grown on glass coverslips were fixed and permeabilized by 10 min of incubation at -20 °C with 100% methanol, or were fixed by a 5-min incubation with 3.7% formaldehyde and permeabilized by incubation with 0.01% Triton X-100 at 4 °C for 5 min. After fixation, nonspecific sites were blocked by incubation in 5% goat serum. Subsequently, the cells were incubated with the primary antibodies in blocking solution. Following wash-out of the primary antibodies with phosphate-buffered saline, the cells were stained with fluorescent secondary antibodies and analyzed as above. The slides were placed on the stage of a Bio-Rad 1024 confocal microscope, and fluorescence was recorded and analyzed offline using ImageJ software.
Dextran Loading
Fibroblasts transfected with TRPC3 constructs were loaded with 5 mg/ml 10 kDa Texas Red-tagged dextran (Molecular Probes, Carlsbad, CA). Following a 3-h load, cells were fixed and processed for confocal immunocytochemistry as discussed above.
EM
HeLa cells were fixed by 30 min of incubation with a solution containing 2.5% glutaraldehyde in 0.1 m Na-cacodilate, washed with 0.1 m Na-cacodilate, and with freeze-thaw permeabilization of the membranes, cells were incubated overnight with α-Myc antibodies, rinsed, and biotinylated secondary antibodies were added. After an extensive wash, the samples were incubated with avidin-peroxidase and washed. Next, 3,3′-diaminobenzidine was added for 10 min and, after another wash, the samples were post-fixed with a solution containing 1% OsO4, washed with phosphate-buffered saline, and stained en block for 30 min with 2% uranyl acetate. Following dehydration by immersion in 30-100% ethanol, the samples were embedded in resin by immersion in 30-100% resin/propylene oxide mixtures. Fixed samples were mounted on grids and analyzed with a JEOL 100CX transmission electron microscope.
Stable Cell Lines
C-terminally tagged YFP-TRPC3 was engineered and cloned into the peYFP vector from Clontech (Mountain View, CA). These vectors were transfected into HEK-293 cells, using Lipofectamine2000 per the manufacturer's instructions. 24-h post-transfection, cells were passed at low density into 96-well plates and grown in 500 μg/ml G418. Wells that formed colonies were subcultured and assayed for stability by YFP-TRPC3 expression. These cells were expanded and frozen in liquid nitrogen. Stable cell lines were only used for 10 passages to ensure reproducibility.
FLIP (Fluorescence Loss in Photobleaching)/FRAP (Fluorescence Recovery after Photobleaching)
Stably transfected WT or SS YFP-TRPC3 cells were plated onto poly-d-lysine-coated coverslips 24 h before use. Cells were treated with 10 μg/ml cyclohexamide for 1 h before use to block new protein synthesis. All experiments were performed on the Olympus FV1000 laser scanning confocal microscope (Olympus America Inc., Melville, NY). This was performed with an inverted Olympus IX-81 microscope using a blue argon laser anda60× oil immersion (1.42 NA) PlanApo lens. FV10-ASW software was used for the analysis. All experiments were performed at room temperature.
FRAP—Cells were bleached at 100% laser power for 5 s over a series of 3 different (oval, rectangle, square) standardized ROI sizes. Recovery from bleaching was then measured for 90 s at 3-s intervals.
FLIP—Cells were bleached at 100% laser power for 10 s and then given 20 s for recovery (12 total bleaches). Three standardized (total pixel volume) ROI's (oval, rectangle, square) were measured at the beginning of each bleach. Each ROI was further measured using the following method. Three standardized circles were placed either in the area under the membrane or on the plasma membrane. These circles were then quantified and averaged, and p values determined by Student's t test. This allowed us to separate cytosolic TRPC3 movement from TRPC3 movement at the plasma membrane.
TIRF
Total internal reflection fluorescence (TIRF) microscopy measurements were obtained using an enclosed Nikon 2000U epi-microscope maintained at 37 °C. The microscope is outfitted with a TIRF module, a Nikon 60× Apo 1.6N/A oil-immersion lens, Prior stage, and Roper512 CCD camera. Excitation was performed at 488 nm. Cells were incubated in Krebs buffer and allowed to equilibrate 5 min before measurements. Z-position was standardized using the Prior stage controls. Cells were imaged without stimulation for 180 cycles, followed by 180 cycles of measurement in the presence of 100 μm carbachol. Analysis was performed using MetaFluor software.
Protein Purification
TRPC3 fragments were cloned into Pet28c HIS-tagged vectors and transformed into BL21 bacteria. Protein production was induced by 100 μm isopropyl-1-thio-β-d-galactopyranoside for 30 min at 37 °C. Cells were lysed by sonication in lysis buffer (phosphate-buffered saline containing 100 mm EDTA, 1 mm phenylmethylsulfonyl fluoride, 5 mm dithiothreitol, and complete protease inhibitor mixture). After lysis, debris was pelleted by a 5-min 10,000 × g centrifugation. The supernatant was incubated on Talon beads for 30 min, washed 10 times with TBST, and eluted with 500 μl of 10 mm EDTA in TBS, pH 8. Proteins were then dialyzed 2× with 4 liters of TBS, pH 7.4 to remove detergent and EDTA.
Cell Surface Biotinylation
This was performed as previously described (25).
Antibodies and Reagents
HIS Pet28c vectors from Novagen (San Diego, CA). Lipids for liposomal assays were from Avanti (Alabaster, AL). PIP-strips were from Echelon (Salt Lake City, UT). Talon beads from Clontech (Mountain View, CA). Streptavidin beads, Fura 2AM, and Lipofectamine 2000 from Invitrogen (Carlsbad, CA). All other reagents were from Sigma. DIIKA: The dominant-negative dynamin 2 K44A mutant (D2KA) unable to hydrolyze GTP was a gift of Dr. Joseph P. Albanesi (Dept. of Pharmacology, UT Southwestern). Antibodies were used per the manufacturer's instructions.
RESULTS
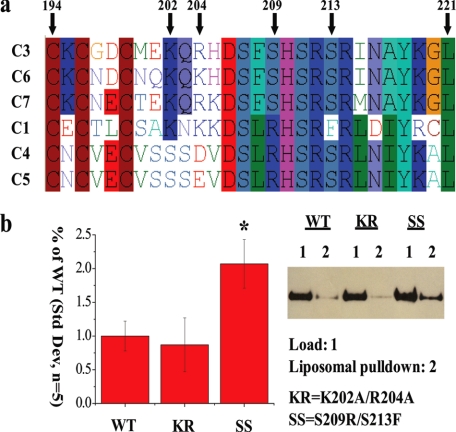
Peripheral Lipid Binding and Activity—TRPC3 has two known lipid binding domains that are not predicted by conventional domain detection algorithms (24,25). Using GDDA-BLAST, we observed multiple putative lipid binding regions in TRPC channels, including predictions for both regions of TRPC3 that have been experimentally validated to bind lipid (1). We also predicted lipid binding activity in the domain of unknown function, TRP_2. We observed that most of the conserved residues from GDDA-BLAST analysis occur between Gly-197 and Asp-233 in TRPC3 (1). We performed multiple sequence alignments between all of the human TRPC channels in this region and observed that DAG-sensitive TRPC3/6/7 have a non-synonymous substitution (Ser-209) when compared with DAG-insensitive TRPC1/4/5 (Fig. 1a). Additionally, Ser-213 in TRPC3 is a phenylalanine in TRPC1.
FIGURE 1.
TRP_2 domain in TRPC3 binds to plasma membrane lipids, in vitro. a, multiple sequence alignment of human TRPC channels within the TRP_2 domain. S209/S213 residues are noted on the alignment and are numbered with respect to TRPC3. The serine residues are conserved in DAG-sensitive TRPC3/6/7 channels, but are substituted in either one or both positions within DAG-insensitive TRPC1/4/5 channels. b, Western analysis of bacterially purified amino acids 161-280 WT, K202A/R204A (KR), or SS fragments cloned into His vector (1-μg load). These same fragments were tested for binding to liposomes containing phosphatidylcholine, phosphatidylethanolamine, phosphatidylserine, and phosphatidylinositol. The bar graph shows the quantification of the extent of liposomal pull-down (S.D., n = 5, *, p value <0.01, Student's t test).
Based on these data, we mutated TRPC3 to mirror TRPC1 (S209R/S213F, herein referred to as SS mutant). We tested the prediction that TRP_2 binds peripheral lipids in the physiologically relevant liposomal assay constructed to mimic plasma membrane lipid composition. We observed that SS TRPC3 significantly increased lipid binding versus WT or a K202A/R204A (KR) mutant (Fig. 1b). We were unable to test whether DAG is capable of binding to this peptide as DAG disrupts liposomal formation; however, we did not observe WT or SS binding to PIP-strips containing immobilized DAG (data not shown).
Next, we tested whether the SS mutation alters channel activity of overexpressed TRPC3 in response to the G-protein receptor agonist carbachol (which increases endogenous IP3 and DAG) or the cell permeant DAG analogue 1-oleoyl-2-acetylglycerol (OAG). In HEK293 cells transiently transfected with either Myc-tagged WT or SS TRPC3, the SS mutation reduces overall channel activity in response to either carbachol or OAG as measured by Fura-2 AM and whole cell electrophysiological recordings (Fig. 2, a-c and supplemental Fig. S1, a and b). We observed small but reproducible SS activity in a subpopulation of cells (data not shown).
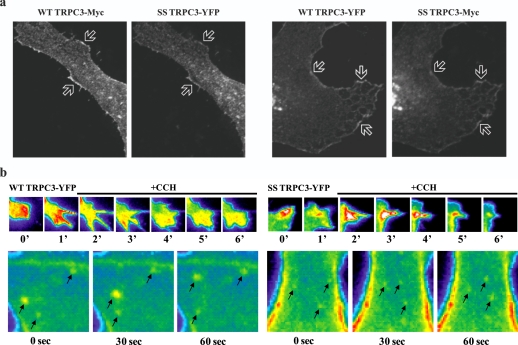
Localization—The above observations suggest that the SS mutation alters either channel activity and/or levels of TRPC3 in the plasma membrane (i.e. trafficking). This agrees with predictions derived by GDDA-BLAST for trafficking activity with the TRP_2 domain of TRPC3 (1). Therefore, we conducted a series of experiments to determine if the SS mutation alters TRPC3 trafficking. We performed confocal microscopy on HeLa cells transfected with full-length WT or SS TRPC3 (Fig. 3a and supplemental Fig. S1c). We observed no overt differences in localization of SS compared with WT expression. Further, co-expressed WT and SS TRPC3 colocalize within the same cellular regions (Fig. 3a). To address whether vesicle trafficking near the plasma membrane was altered, we performed TIRF on WT or SS TRPC3 stably expressed in HEK293 cells (Fig. 3b and supplemental Fig. S1d, and supplemental movies), which assays vesicle movement within ∼100 nm of the plasma membrane. We found that SS-positive vesicles are indistinguishable from WT in both size and movement. We next performed transmission electron microscopy on HeLa cells overexpressing Myc-tagged TRPC3 stained with anti-Myc antibodies (Fig. 4a). We do not observe any gross changes between WT and SS localization, although irregular blebbing can be observed near the plasma membrane in the SS preparations. These results show either there is no abnormality in SS mutant localization or that such abnormalities manifest at a stage immediately preceding the mutant interaction with the plasma membrane.
FIGURE 3.
Confocal localization of WT and SS TRPC3. a, confocal microscopy of HeLa cells co-transfected with Myc-tagged full-length WT and YFP-tagged SS TRPC3 or vice versa. Arrows depict areas of co-localization. b, TIRF of HEK293 cells stably transfected with either YFP-tagged full-length WT or SS TRPC3. These cells were stimulated with 100 μm CCH over a time course. Arrows demarcate TRPC3-containing vesicles and their movement over time. See supplemental movies for real-time TIRF experiments.
FIGURE 4.
Electron microscopy and quantification of vesicle mobility at the plasma membrane. a, transmission electron microscopy of HeLa cells transfected with either full-length Myc-tagged WT or SS TRPC3. Arrows depict TRPC3 immunoreactivity at the plasma membrane. Examples of blebbing are presented in the two right panels of SS TRPC3-Myc. b, graph demonstrating the rate of fluorescence recovery (FRAP) for HEK293 cells stably transfected with either YFP-tagged full-length WT (black) or SS (red) TRPC3. (S.D., n = 5, *, p value < 0.01 determined by Student's t test). c, graphs demonstrating the rate of fluorescence loss (FLIP) for cells transfected as in b. Left to right, total (cytosolic and membrane) ROI bleaching rate, cytosolic ROI bleaching rate, and membrane ROI bleaching rate. (S.D., n = 5, *, p value < 0.01 determined by Student's t test). For b and c, see“Materials and Methods,” and supplemental Fig. S1e.
Trafficking—As obtaining precise measurements of vesicle mobility using TIRF is challenging, we sought a more quantifiable method. Thus we utilized FRAP, and FLIP experiments to more accurately quantify the mobility of vesicles containing WT and mutant channels (Fig. 4, b and c and supplemental Fig. S1e). We used FRAP to test forward trafficking and FLIP to assay endocytosis and plasma membrane mobility (see “Materials and Methods”). In the FRAP experiments, we observed recovery of WT TRPC3 fluorescence, an effect which is significantly decreased in SS preparations (Fig. 4b). This result suggests that the sites occupied by SS TRPC3 are not subject to normal recycling. Thus, the plasma membrane cannot be repopulated with unbleached SS TRPC3. Our FLIP experiments demonstrated that WT TRPC3 is depleted from the unbleached area in a time-dependent manner (Fig. 4c). This represents vesicle endocytosis and diffusion of WT in the plasma membrane. Conversely, SS TRPC3 is rapidly depleted from areas subjacent to the plasma membrane as WT (Fig. 4c, Cytosolic ROI), yet has significantly increased retention time with the plasma membrane (Fig. 4c, Membrane ROI). With respect to the Membrane ROI, we observed a biphasic response whereby WT and SS are indistinguishable at early time points but differ significantly at later time points. The lack of effect in early time points could be due to multimerization of SS TRPC3 with endogenous TRPC channels or TRPC channels within the ROI, which have yet to dock. Nevertheless, the reduced mobility of SS at the plasma membrane is consistent with our finding that this mutant has increased affinity for plasma membrane lipids (see Fig. 1b).
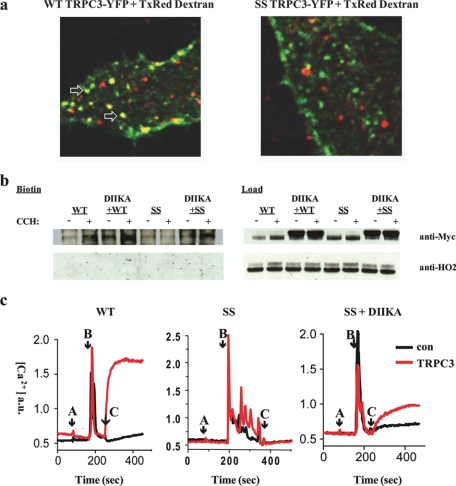
The decreased activity of the SS mutant could be due to altered vesicle insertion, endocytosis, and/or channel gating. To more rigorously evaluate whether endocytosis of the SS mutant is disrupted, we performed fluorescent dextran loading assays (Fig. 5a). WT TRPC3 (green) co-localizes with internalized dextran, while only a fraction of dextran-containing vesicles are positive for SS TRPC3, in accord with the results from our FLIP/FRAP experiments, indicating that endocytosis of the SS mutant is impaired. Next, we tested mutant channel activity in the presence of a dominant-negative form of dynamin (DIIKA), which blocks endocytosis (29) (Fig. 5, b and c). Under these conditions we observed increased SS TRPC3 channel cell surface expression, as well as activity via Fura-2AM using Ba2+ as the charge carrier. Combined with the dextran-loading assays, these data suggest that the SS defect(s) are not caused by decreased half-life in the plasma membrane or alterations in channel gating.
FIGURE 5.
Defects associated with the SS mutation are not caused by decreased half-life in the plasma membrane or alterations in channel gating. a, confocal microscopy of fixed HeLa cells transfected with either full-length WT or SS TRPC3 (green) and 10 kDa Texas Red-tagged dextran. Top, arrows show areas of co-localization. WT TRPC3 co-localizes with internalized dextran, while only a fraction of dextran-containing vesicles are positive for SS TRPC3. b, Western blot of biotinylated HEK293 cells (left) and loads (right) transfected with either Myc-tagged WT or SS TRPC3 alone, or coexpressed with a dominant-negative dynamin mutant (DIIKA), which blocks endocytosis. These preparations were treated with or without 100 μm CCH. Input lanes, 30 μg. Anti-HO2 blot serves as an intracellular negative control for biotinylation. c, Fura2AM measurements were made in HEK293 cells non-transfected (con, black) or transfected (red) with either WT or SS TRPC3 alone, or SS TRPC3 + dominant-negative DIIKA. Cells were acclimated first in nominally Ca2+-free medium (A, arrow), Ca2+ pools were released by 100 μm CCH in nominally Ca2+-free medium (B, arrow) followed by replacement with CCH and 1 mm Ba2+-containing media (C, arrow).
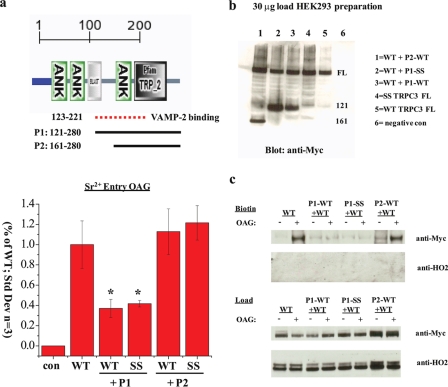
Although immunostaining, TIRF, and electron microscopy cannot distinguish between WT and SS channel localization at the plasma membrane (Figs. 3 and 4a), these data would not reveal alterations in SNARE activity. Indeed, amino acids 123-221 of TRPC3 have been shown to bind to VAMP-2 directly, while another uncharacterized region in the N terminus binds directly to the SNARE protein αSNAP (soluble N-ethylmaleimide-sensitive factor attachment protein) (30). These authors reported that the exocytotic insertion of TRPC3 channels is mediated by a VAMP-dependent, and calcium-independent process. Our in vitro lipid binding studies were performed with WT and SS TRPC3 preparations of amino acids 161-280 (see Fig. 1b), which includes a portion of the VAMP interaction site. Thus, to rule out possible effects directly on membrane fusion machinery, rather than lipid binding, we performed dominant-negative experiments using cloned fragments of TRPC3 (121-280 WT or SS, and 161-280 WT or SS), which were transfected into HEK293 cells (Fig. 6, a and b).
FIGURE 6.
Defects associated with the SS mutation are independent of SNARE interaction. a, top, domain architecture of TRPC3 amino acids 1-280 predicted by SMART (default settings). Amino acids 123-221 (dotted line) demarcate the region previously shown to be involved in binding VAMP-2, a SNARE protein (30). The two solid lines depict the boundaries of four experimental peptides P1 (WT or SS, amino acids 121-280), and P2 (WT or SS, amino acids 161-280). Bottom, quantification of the area under the curve for Sr2+ entry in the presence of 100 μm OAG stimulation in HEK293 cells transfected with either YFP alone (con), full-length WT or SS alone. TRPC3 WT was also coexpressed with cloned fragments P1 or P2. (S.D., n = 3, *, p value <0.01, Student's t test). b, Western analysis of HEK293 cell lysates (30 μg) transfected as above. All constructs are Myc-tagged and express at similar levels. c, Western blot of biotinylated HEK293 cells (top) and loads (bottom) transfected as in a. These preparations were treated with or without 100 μm OAG. Input lanes, 30 μg. Anti-HO2 blot serves as an intracellular negative control for biotinylation.
Our experimental predictions are as follows: (i) if the SS mutation interferes with VAMP-2/αSNAP binding, only the 121-280 WT fragment should act as a dominant-negative for WT TRPC3 activity/surface expression; (ii) if, however, the 121-280 SS fragment does not interfere with SNARE binding, both WT and SS peptides should function as a dominant-negatives; (iii) if the lipid binding TRP_2 region in TRPC3 (aa 161-280) binds to VAMP-2/αSNAP, the 161-280 WT fragment should act as a dominant-negative; (iv) if, however, the 161-280 WT and SS peptides do not act as dominant-negatives than this implies that the defects associated with the SS mutation are independent of SNARE interactions. Indeed, Fura2AM measurements within intact HEK293 cells transfected as above reveal that 121-280 WT and SS act as dominant-negatives and are equally effective at inhibiting TRPC3 activity, whereas 161-280 preparations do not (Fig. 6a). Furthermore, these data suggest that the VAMP-2 binding site can be more narrowly defined between amino acids 121 and 161. We extended these findings by assaying the surface expression of TRPC3 under the aforementioned preparations. Cell surface biotinylation experiments demonstrate that co-expression of either 121-280 WT or SS fragments inhibits the OAG-mediated surface expression of TRPC3, whereas the 161-280 WT fragment is similar to the control (Fig. 6c).
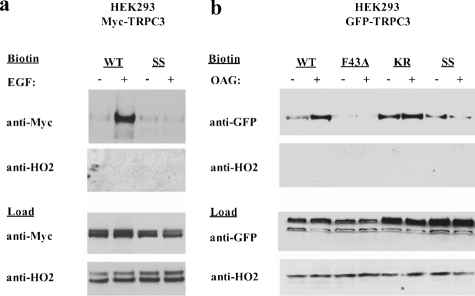
Fusion—The above results suggest that the SS mutation alters either the docking and/or fusion of channel-containing vesicles. To address this supposition, we assayed levels of overexpressed TRPC3 in the plasma membrane using cell surface biotinylation. Cells stimulated by tyrosine kinase receptor (epidermal growth factor (EGF)), OAG (Fig. 7, a and b), or G-protein-coupled receptor (carbachol, data not shown) manifest increased levels of WT TRPC3 in the plasma membrane, an effect that is inhibited by the SS mutation. Moreover, in non-transfected PC12 cells, which express endogenous TRPC3, treatment with OAG, RHC (a DAG-kinase inhibitor that increases endogenous levels of DAG), or carbachol elicits a substantial increase in endogenous TRPC3 surface expression within 1 min, while endogenous DAG-insensitive TRPC5 levels are unaltered (Fig. 8a).
FIGURE 7.
Mutations in TRP_2 disrupt the DAG-mediated surface expression of TRPC3. a, Western blot of biotinylated HEK293 cells (top) and loads (bottom) transfected with either Myc-tagged WT or SS TRPC3 alone and stimulated with or without 1 μm EGF. Input lanes, 30 μg. Anti-HO2 blot serves as an intracellular negative control for biotinylation. b, Western blot of biotinylated HEK293 cells transfected with either GFP-tagged full-length WT, F43A (PLC-γ binding mutant)(25), K202A/R204A (KR), or SS TRPC3 for 48 h, treated with or without 100 μm OAG. Input lanes, 20 μg. Anti-HO2 blot serves as an intracellular negative control for biotinylation.
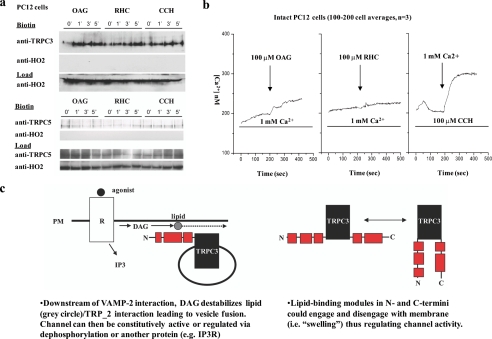
FIGURE 8.
DAG induces fusion of endogenous TRPC3-containing vesicles with the plasma membrane. a, top, Western blot of biotinylated non-transfected rat PC12 cells treated in a time course (0, 1, 3, and 5 min) with 100 μm OAG, 100 μm RHC (a DAG-kinase inhibitor which increases endogenous levels of DAG), or 100 μm CCH and blotted for endogenous TRPC3. Bottom, Western blot as above except blotted for endogenous TRPC5. b, free Ca2+ measurements were made in non-transfected rat PC12 cells. Cells were acclimated in normal 1 mm Ca2+ containing medium and then challenged with either left: 100 μm OAG (arrow) or middle: 100 μm RHC (arrow). Right:Ca2+ pools were released in cells by CCH (100 μm) (first bar), in nominally Ca2+-free medium followed by replacement with CCH and 1 mm Ca2+-containing media (arrow). These traces represent averages of 100-200 cells. c, oversimplified graphical representation of the proposed fusion mechanism of WT TRPC3.
OAG promotes TRPC3 activity, but the mechanism of this action has not been elucidated (2, 3, 31) perhaps due to inconsistencies between the endogenous and overexpressed channels (32, 33). This is further illustrated by our Ca2+ entry measurements in non-transfected PC12 cells (Fig. 8b). We do not observe significant Ca2+ activity following OAG or RHC stimulation despite the fact that TRPC3 cell surface levels increase with these treatments (Fig. 8a). The lack of channel activity in response to OAG and RHC treatment is likely due to phosphorylation of endogenous TRPC3 by protein kinase G or C, both of which have been shown to have an inhibitory action on OAG-mediated calcium flux (34, 35). These results suggest that DAG does not “activate” TRPC channels per se, rather it increases the fusion of TRPC3-containing vesicles with the plasma membrane (Fig. 5d) (15). DAG-induced fusion is well supported by the literature (2, 3) as this molecule is a known membrane destabilizer that promotes the fusion of neurotransmitter-containing vesicles both physiologically (e.g. via receptors) and patho-physiologically (e.g. via snake presynaptic phospholipase A2 neurotoxins) (36). In total, our results demonstrate that the TRP_2 domain in TRPC3 is a lipid binding domain that participates in membrane fusion.
DISCUSSION
In summary, GDDA-BLAST yielded accurate predictions of a new functional domain in TRPC channels (1). Our experimental evidence demonstrates that TRPC3 TRP_2 is a lipid/trafficking domain that contributes to DAG-sensitive vesicle fusion. The TRPC3 SS mutation within the TRP_2 domain has: (i) increased affinity for plasma membrane lipids, (ii) decreased mobility when associated with the plasma membrane, and (iii) deficits in DAG-induced plasma membrane insertion, upstream of SNARE interactions. We propose that the increased affinity for plasma membrane lipids inhibits the membrane destabilization of TRPC3-containing vesicles by DAG. In support, DAG is a well characterized cone-shaped lipid that destabilizes membranes at synapses, thus increasing vesicle fusion (2,3). DAG increases the negative curvature of membranes (i.e. propensity to form nonbilayer phases while keeping lamellar architecture), increases spacing between phospholipid headgroups (i.e. alters fluidity), and decreases membrane surface hydration, which may help the docking of proteins to membranes or micellar substrates.
While the N terminus of TRPC3-(121-280) can function as a dominant-negative of SNARE activity and vesicle fusion, this is independent of the lipid binding/trafficking activity of the TRP_2 domain (Fig. 7). Indeed, we and others (30, 37, 38) have previously suggested a secretion-like coupling model for Ca2+ entry mechanisms. For example, Ambudkar and co-workers (30) previously demonstrated that an N-terminal region of TRPC3, inclusive to the TRP_2 domain, can associate with synaptobrevin/VAMP and α-SNAP while co-localizing with many of the known SNARE machinery components. Importantly, VAMP and TRPC3 must exist in the same vesicles for fusion. Hence, TRPC3 likely comprises part of the SNARE machinery within vesicles it occupies, as has been proposed for TRPM7 (26). This idea is supported by our identification of SNARE domain “fingerprints” in TRPC3 channels with GDDA-BLAST (1). Furthermore, these data imply that cargo contained in SS TRPC3 vesicles also have defects in plasma membrane insertion.
Although many implications can be drawn from these data, many questions remain unanswered. How do the multiple lipid binding domains in TRPC channels work coordinately to regulate channel surface expression? Are these interactions regulated via phosphorylation? Nitrosylation? How is the formation of inter- and intramolecular domains regulated? And further, how does homo- and heteromultimerization alter any and/or all of these processes? These questions highlight the difficulty in studying the polymodal activity of TRP ion channels and demonstrate the need for computational methods capable of isolating the functional properties of protein domains. We propose that with further development and refinement, GDDA-BLAST will provide a computational modeling platform that translates directly to the laboratory benchtop, thereby speeding the scientific ability to answer difficult questions such as these.
Supplementary Material
Acknowledgments
We thank Drs. R. E. Rothe, Jim White, T. Gunn, Sascha C. Kendall, T. Chronos, and M. Murpa for creative dialogue. Some of the confocal microscopy was done at the Center for Quantitative Cell Analysis at the Huck Institutes of the Life Sciences, Penn State University. The Department of Health specifically disclaims responsibility for any analyses, interpretations, or conclusions.
This work was supported in part by the Searle Young Investigators Award and start-up funds from PSU (to R. L. P.), NCSA Grant TG-MCB070027N (to R. L. P. and D. V. R.), the T. O. Magnetti Fund (to R. L. P. and D. V. R.), a grant from the ML4 Foundation (to K. K.), Grant MH18501, and Research Scientist Award DA00074 (to S. H. S.). This project was also funded by a fellowship from the Eberly College of Sciences and the Huck Institutes of the Life Sciences (to D. V. R.) and a grant with the Pennsylvania Department of Health using Tobacco Settlement Funds (to D. V. R.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1 and movies.
Footnotes
The abbreviations used are: TRP, transient receptor potential; SS, S209R/S213F; CCH, carbachol; WT, wild type; TIRF, total internal reflection fluorescence microscopy; OAG, 1-oleoyl-2-acetylglycerol; DAG, diacylglycerol; SNARE, soluble NSF attachment protein receptors; FRAP, fluorescence recovery after photobleaching; FLIP, fluorescence loss in photobleaching; YFP, yellow fluorescent protein; GFP, green fluorescent protein.
References
- 1.Ko, K. D., Hong, Y., Chang, G. S., Bhardwaj, G., van Rossum, D. B., and Patterson, R. L. (2008) Physics Arch. Quant. Methods arXiv:0806.2394v1 [Google Scholar]
- 2.Villar, A. V., Goni, F. M., and Alonso, A. (2001) FEBS Lett. 494117 -120 [DOI] [PubMed] [Google Scholar]
- 3.Goni, F. M., and Alonso, A. (1999) Prog. Lipid Res. 381 -48 [DOI] [PubMed] [Google Scholar]
- 4.Clapham, D. E. (2003) Nature 426517 -524 [DOI] [PubMed] [Google Scholar]
- 5.Li, H. S., Xu, X. Z., and Montell, C. (1999) Neuron 24261 -273 [DOI] [PubMed] [Google Scholar]
- 6.Shim, S., Goh, E. L., Ge, S., Sailor, K., Yuan, J. P., Roderick, H. L., Bootman, M. D., Worley, P. F., Song, H., and Ming, G. L. (2005) Nat. Neurosci. 8 730-735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corey, D. P., Garcia-Anoveros, J., Holt, J. R., Kwan, K. Y., Lin, S. Y., Vollrath, M. A., Amalfitano, A., Cheung, E. L., Derfler, B. H., Duggan, A., Geleoc, G. S., Gray, P. A., Hoffman, M. P., Rehm, H. L., Tamasauskas, D., and Zhang, D. S. (2004) Nature 432723 -730 [DOI] [PubMed] [Google Scholar]
- 8.Tabuchi, K., Suzuki, M., Mizuno, A., and Hara, A. (2005) Neurosci. Lett. 382304 -308 [DOI] [PubMed] [Google Scholar]
- 9.Zhang, Y., Hoon, M. A., Chandrashekar, J., Mueller, K. L., Cook, B., Wu, D., Zuker, C. S., and Ryba, N. J. (2003) Cell 112293 -301 [DOI] [PubMed] [Google Scholar]
- 10.Jungnickel, M. K., Marrero, H., Birnbaumer, L., Lemos, J. R., and Florman, H. M. (2001) Nat. Cell Biol. 3 499-502 [DOI] [PubMed] [Google Scholar]
- 11.Kiselyov, K., Soyombo, A., and Muallem, S. (2007) J. Physiol. 578641 -653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miedel, M. T., Rbaibi, Y., Guerriero, C. J., Colletti, G., Weixel, K. M., Weisz, O. A., and Kiselyov, K. (2008) J. Exp. Med. 2051477 -1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jordt, S. E., and Ehrlich, B. E. (2007) Subcell. Biochem. 45253 -271 [DOI] [PubMed] [Google Scholar]
- 14.Venkatachalam, K., and Montell, C. (2007) Annu. Rev. Biochem. 76387 -417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramsey, I. S., Delling, M., and Clapham, D. E. (2006) Annu. Rev. Physiol. 68619 -647 [DOI] [PubMed] [Google Scholar]
- 16.Soboloff, J., Spassova, M., Hewavitharana, T., He, L. P., Luncsford, P., Xu, W., Venkatachalam, K., van, R. D., Patterson, R. L., and Gill, D. L. (2007) Handb. Exp. Pharmacol. 575 -591 [DOI] [PubMed]
- 17.Montell, C., Jones, K., Hafen, E., and Rubin, G. (1985) Science 2301040 -1043 [DOI] [PubMed] [Google Scholar]
- 18.Chahl, L. A. (2007) Biochim. Biophys. Acta 1772968 -977 [DOI] [PubMed] [Google Scholar]
- 19.Firth, A. L., Remillard, C. V., and Yuan, J. X. (2007) Biochim. Biophys. Acta 1772895 -906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li, Y., Jia, Y. C., Cui, K., Li, N., Zheng, Z. Y., Wang, Y. Z., and Yuan, X. B. (2005) Nature 434894 -898 [DOI] [PubMed] [Google Scholar]
- 21.Wang, G. X., and Poo, M. M. (2005) Nature 434898 -904 [DOI] [PubMed] [Google Scholar]
- 22.Dietrich, A., Kalwa, H., Fuchs, B., Grimminger, F., Weissmann, N., and Gudermann, T. (2007) Cell Calcium 42 233-244 [DOI] [PubMed] [Google Scholar]
- 23.Winn, M. P., Conlon, P. J., Lynn, K. L., Farrington, M. K., Creazzo, T., Hawkins, A. F., Daskalakis, N., Kwan, S. Y., Ebersviller, S., Burchette, J. L., Pericak-Vance, M. A., Howell, D. N., Vance, J. M., and Rosenberg, P. B. (2005) Science 3081801 -1804 [DOI] [PubMed] [Google Scholar]
- 24.Kwon, Y., Hofmann, T., and Montell, C. (2007) Mol. Cell 25491 -503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Rossum, D. B., Patterson, R. L., Sharma, S., Barrow, R. K., Kornberg, M., Gill, D. L., and Snyder, S. H. (2005) Nature 43499 -104 [DOI] [PubMed] [Google Scholar]
- 26.Krapivinsky, G., Mochida, S., Krapivinsky, L., Cibulsky, S. M., and Clapham, D. E. (2006) Neuron 52 485-496 [DOI] [PubMed] [Google Scholar]
- 27.Cozier, G. E., Lockyer, P. J., Reynolds, J. S., Kupzig, S., Bottomley, J. R., Millard, T. H., Banting, G., and Cullen, P. J. (2000) J. Biol. Chem. 27528261 -28268 [DOI] [PubMed] [Google Scholar]
- 28.Patterson, R. L., van Rossum, D. B., Ford, D. L., Hurt, K. J., Bae, S. S., Suh, P. G., Kurosaki, T., Snyder, S. H., and Gill, D. L. (2002) Cell 111529 -541 [DOI] [PubMed] [Google Scholar]
- 29.Ekena, K., Vater, C. A., Raymond, C. K., and Stevens, T. H. (1993) Ciba Found. Symp. 176198 -211 [DOI] [PubMed] [Google Scholar]
- 30.Singh, B. B., Lockwich, T. P., Bandyopadhyay, B. C., Liu, X., Bollimuntha, S., Brazer, S. C., Combs, C., Das, S., Leenders, A. G., Sheng, Z. H., Knepper, M. A., Ambudkar, S. V., and Ambudkar, I. S. (2004) Mol. Cell 15635 -646 [DOI] [PubMed] [Google Scholar]
- 31.Hofmann, T., Obukhov, A. G., Schaefer, M., Harteneck, C., Gudermann, T., and Schultz, G. (1999) Nature 397259 -263 [DOI] [PubMed] [Google Scholar]
- 32.Ma, H.-T., Patterson, R. L., van Rossum, D. B., Birnbaumer, L., Mikoshiba, K., and Gill, D. L. (2000) Science 2871647 -1651 [DOI] [PubMed] [Google Scholar]
- 33.Kiselyov, K. I., Xu, X., Mohayeva, G., Kuo, T., Pessah, I. N., Mignery, G. A., Zhu, X., Birnbaumer, L., and Muallem, S. (1998) Nature 396478 -482 [DOI] [PubMed] [Google Scholar]
- 34.Kwan, H. Y., Huang, Y., and Yao, X. (2006) J. Cell Physiol. 207315 -321 [DOI] [PubMed] [Google Scholar]
- 35.Venkatachalam, K., Zheng, F., and Gill, D. L. (2003) J. Biol. Chem. 27829031 -29040 [DOI] [PubMed] [Google Scholar]
- 36.Rigoni, M., Caccin, P., Gschmeissner, S., Koster, G., Postle, A. D., Rossetto, O., Schiavo, G., and Montecucco, C. (2005) Science 3101678 -1680 [DOI] [PubMed] [Google Scholar]
- 37.Patterson, R. L., van Rossum, D. B., and Gill, D. L. (1999) Cell 98 487-499 [DOI] [PubMed] [Google Scholar]
- 38.Yao, Y., Ferrer-Montiel, A. V., Montal, M., and Tsien, R. Y. (1999) Cell 98 475-485 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.