Abstract
The microbial degradation of the plant cell wall is a pivotal biological process that is of increasing industrial significance. One of the major plant structural polysaccharides is mannan, a β-1,4-linked d-mannose polymer, which is hydrolyzed by endo- and exo-acting mannanases. The mechanisms by which the exo-acting enzymes target the chain ends of mannan and how galactose decorations influence activity are poorly understood. Here we report the crystal structure and biochemical properties of CjMan26C, a Cellvibrio japonicus GH26 mannanase. The exo-acting enzyme releases the disaccharide mannobiose from the nonreducing end of mannan and mannooligosaccharides, harnessing four mannose-binding subsites extending from -2 to +2. The structure of CjMan26C is very similar to that of the endo-acting C. japonicus mannanase CjMan26A. The exo-activity displayed by CjMan26C, however, reflects a subtle change in surface topography in which a four-residue extension of surface loop creates a steric block at the distal glycone -2 subsite. endo-Activity can be introduced into enzyme variants through truncation of an aspartate side chain, a component of a surface loop, or by removing both the aspartate and its flanking residues. The structure of catalytically competent CjMan26C, in complex with a decorated manno-oligosaccharide, reveals a predominantly unhydrolyzed substrate in an approximate 1S5 conformation. The complex structure helps to explain how the substrate “side chain” decorations greatly reduce the activity of the enzyme; the galactose side chain at the -1 subsite makes polar interactions with the aglycone mannose, possibly leading to suboptimal binding and impaired leaving group departure. This report reveals how subtle differences in the loops surrounding the active site of a glycoside hydrolase can lead to a change in the mode of action of the enzyme.
The plant cell wall represents the dominant source of organic carbon in the biosphere and as such supports many facets of terrestrial and marine life. Access to this valuable source of carbon is mediated by extensive microbial enzyme consortia, which generate sugars that are utilized by microbial ecosystems to the benefit of plants, mammalian herbivores, and insects. These degradative enzymes are increasingly deployed in industrial processes, and it is apparent that their use in producing second generation, lignocellulose-based, biofuels is of considerable environmental significance (1, 2). Among the major polysaccharides in softwoods and angiosperms are the mannans. These polysaccharides comprise a backbone of β-1,4-linked d-mannopyranose sugars (known as “mannan”) or a heterogeneous combination of β-1,4-d-mannose and β-1,4-d-glucose units (termed “glucomannan”), which can be decorated with α-1,6-galactose side chains to yield “galactomannan” and “galactoglucomannan,” respectively (3). For complete enzymatic degradation, the galactose decorations are removed by α-galactosidases (4, 5), whereas the internal glycosidic linkages in mannans are cleaved by endo-β-1,4 mannanases into mannooligosaccharides, which are subsequently hydrolyzed by β-mannosidases to release d-mannose from the nonreducing end of the oligosaccharides (6). Accessory enzymes, such as the α-galactosidases are required essentially because side chains appended to plant cell wall matrix polysaccharides restrict the capacity of the backbone-cleaving enzymes to cleave their target substrates. Although galactose decorations restrict the capacity of mannanases to hydrolyze the mannan backbone (e.g. see Refs. 7 and 8), the structural basis for this inhibition is unknown.
In the sequence-based classification of glycoside hydrolases (9) (recently reviewed in Ref. 10), endo-acting mannanases are located primarily in families GH5 and GH26, whereas GH4, GH27, GH36, and GH57 contain α-galactosidases, and β-mannosidases are predominantly located in families GH1 and GH2. All of these enzymes, except α-galactosidases, belong to clan GH-A, consistent with their (β/α)8 barrel fold, hydrolysis of the glycosidic bond by a general acid/base-catalyzed double displacement mechanism with retention of anomeric configuration, and the presentation of the two catalytic residues, the acid/base and the nucleophile at the C terminus of β-strands 4 and 7, respectively (11).
With the rapid expansion of genomic sequence data, one of the major challenges is how to harness this treasure trove of sequence information to identify the full repertoire of catalytic activities displayed by the different glycoside hydrolase families and thus to inform the application of these enzymes for improved applied purposes. Such analyses should provide a biological rationale for the repertoire of glycoside hydrolases produced by both prokaryotic and eukaryotic systems. Indeed, until recently, GH26 was thought to be exclusively an endo-β-1,4-mannanase family, but two members that display, respectively, β-1,3-xylanase (12) and β-1,4:β-1,3-glucanase activities (13, 14) have now been described. This diversity in function is further exemplified by the recent identification of a GH5 enzyme, CmMan5A, which, although displaying extensive sequence identity to endo-acting mannanases, is an exo-acting mannanase that releases mannose from the nonreducing end of polymeric substrates (7).
The saprophytic soil bacterium Cellvibrio japonicus exemplifies how the analysis of the biochemical properties of glycoside hydrolases, revealed by the recent completion of its genome sequence (15), will make a significant contribution to understanding the mechanism by which the organism can utilize the plant cell wall as a carbon and energy source. C. japonicus synthesizes one of the most extensive mannan-degrading systems known. The bacterium is reported to express two GH26 and three GH5 endo-acting mannanases (8). Analysis of the genome sequence of the bacterium revealed two additional genes encoding the mannanases CjMan5D, which is a homologue of CmMan5A, and an additional membrane-bound GH26 enzyme, CjMan26C (15). Clones encoding CjMan26C were not identified when genomic libraries of C. japonicus were previously subjected to extensive screening for endo-mannanase activity, suggesting that the enzyme displays unusual catalytic activities.
Here we report the biochemical analysis of this novel enzyme, CjMan26C, along with its high resolution crystal structures. We show that the enzyme is an exo-acting mannanase that releases the disaccharide mannobiose from the nonreducing end of oligo- and polysaccharides, an activity not previously described in mannan-degrading systems to our knowledge. The crystal structure shows that the substrate binding cleft adopts a blind canyon topography that presents a steric block at the -2 glycone binding site, explaining why the enzyme displays an exo-mode of action. Modification of the residues that present the steric block changes the mode of action of the enzyme from exo to endo. Additionally, the structure of the enzyme in “Michaelis complex” with decorated oligosaccharide suggests a possible means by which the galactoside appendages are accommodated but restrict the activity of the enzyme.
MATERIALS AND METHODS
Gene Cloning and Protein Expression—The region of the C. japonicus mannanase gene, man26C, encoding mature CjMan26C (residues 25-419 of the full-length enzyme), was amplified by PCR from genomic DNA using the thermostable DNA polymerase Pfu Turbo (Stratagene) and the primers 5′-GCGCATATGAGCGAGAAGCCTGCAGAATCGCGG-3′ and 5′-GCGCTCGAGGCGATACAAGGGCGGCAGCTC-3′ that contain NdeI and XhoI restriction sites, respectively, depicted in boldface type. The DNA product was cloned into the NdeI and XhoI sites of the Escherichia coli expression vector pET20b (Novagen) to generate pVT1. CjMan26C encoded by pVT1 contains a C-terminal His6 tag.
Protein Expression and Purification—E. coli BL21 cells harboring pVT1 were cultured in Luria-Bertani broth at 37 °C to midexponential phase (A600 = 0.6), and recombinant protein expression was induced by the addition of 1 mm isopropyl 1-thio-β-d-galactopyranoside and incubation for a further 5 h at 37 °C. CjMan26C was purified by immobilized metal ion affinity chromatography using Talon™ resin and elution in 20 mm Tris/HCl buffer, pH 8.0, containing 300 mm NaCl and 100 mm imidazole deploying a Bio-Rad fast protein liquid chromatography system. The eluted mannanase was then dialyzed against 50 mm sodium phosphate buffer, pH 7.0. The fractions containing the mannanase were concentrated using a 30,000 molecular weight cut-off Vivaspin 20 centrifugal concentrator. Purified CjMan26C was judged homogenous by SDS-PAGE.
Mutagenesis—Site-directed mutagenesis was carried out employing the PCR-based QuikChange mutagenesis kit (Stratagene) according to the manufacturer's instructions, using pVT1 as the template and primers listed in Table S1.
Enzyme Assays—Substrates used in the enzyme assays described below were purchased from Sigma except for konjac glucomannan, which was from Megazyme International. Degalatosylated carob mannan was generated from carob galactomannan as described previously (8). To evaluate enzyme activity, 4-ml reactions were set up comprising 50 mm sodium phosphate, buffer, pH 7.0, containing 1 mg/ml bovine serum albumin and soluble or insoluble substrate at 0.04-2% (w/v). The reaction, which was initiated by the addition of 200 μl of appropriately diluted enzyme, was incubated at 37 °C for up to 30 min, and at regular time intervals 500-μl aliquots were removed, and the quantity of reducing sugar was determined (16). The nature of the products was evaluated by HPLC5 (see below). The pH profile of the catalytic activity of the mannanases was determined using the following buffers: pH 3-5, 50 mm sodium acetate; pH 5-9, 50 mm sodium phosphate; pH 9-11, 50 mm CAPSO (Sigma). To determine the reaction products released from the mannose-containing substrates and to quantify the activity of the mannanases against manno-oligosaccharides, the enzymatic incubations were subjected to HPLC using enzyme reactions described above except that the 500-μl aliquots were treated with alkali to inactivate the enzyme, filtered, and then subjected to HPLC using an analytical CARBOPAC™ PA-100 anion exchange column (Dionex) equipped with a guard column. The elution conditions were as follows: 0-5 min, 66 mm NaOH; 5-25 min, 66 mm NaOH with a 0-75 mm sodium acetate gradient. Sugars were detected by pulsed amperometric detection. The catalytic efficiency of the mannanases against manno-oligosaccharides was determined using 20 pm to 5 μm enzyme and a substrate concentration of 30 μm, and the data were fitted to the equation, k = ln[S0]/[St], where k = (kcat/Km)[enzyme] × time, whereas [S0] and [St] represent substrate concentration prior to the start of the reaction and at a specified time during the reaction, respectively (17).
Crystallization, Data Collection, Structure Solution, and Refinement—Pure CjMan26C was washed into 5 mm Hepes-Na buffer, pH 7.0, by repeated dilution (40 volumes of water) and concentrated in a VIVASPIN 30-kDa concentrator. Crystals of wild type CjMan26C and the nucleophile mutant E338A were grown by vapor phase diffusion at 18 °C using the hanging drop method with an equal volume (1 μl) of protein (10 g/liter) and reservoir solution comprising 100 mm sodium citrate buffer, pH 5.6, containing 15% polyethylene glycol 3000. Crystals in the presence of various ligands were similarly grown with the exception that the protein solution contained 10 mm mannooligosaccharide. Crystals, which grew over a period of 1 week, were cryoprotected by the addition of 20% glycerol (v/v) to the crystallization mother liquor.
Diffraction data for four different complexes of Man26C (native, mannose complex, manno-oligosaccharide, and E338A-mannobiose) were collected on beamlines ID14-1 and ID23-1 of the European Synchrotron Radiation Facility, and data were processed in DENZO scaled with SCALEPACK from the HKL suite (18). All other computations used programs from within the CCP4 suite (19) unless stated otherwise. The structures of the four CjMan26Cs were solved by molecular replacement using the program MOLREP from the CCP4 suite with CjMan26A as the search model (Protein Data Bank code 1J9Y). Before refinement, 5% of the observations were chosen at random and set aside for cross-validation analysis and to monitor various refinement strategies, such as the weighting of geometrical and temperature factor restraints and the insertion of solvent water during maximum likelihood refinement using REFMAC5 (20). The structure of unliganded Man26C was observed in a different space group, and this structure was solved, with MOLREP, using the protein coordinates only of the mannose-CjMan26C structure. For all structures, cycles of maximum likelihood refinement were interspersed with manual corrections of the models using COOT (21). Ligands were incorporated toward the end of the refinement into CjMan26C. Data processing and refinement statistics are given in Table 1.
TABLE 1.
Data collection and refinement statistics for C. japonicus Man26C
| Apo-CjMan26C | CjMan26C-mannose | E338A CjMan26C-mannobiose | CjMan26C-6163-Gal2Man4 | |
|---|---|---|---|---|
| Data collection | ||||
| Space group | P212121 | P6122 | P6122 | P6122 |
| Cell dimensions (Å) | 52.3, 83.6, 84.3 | 84.7, 84.7, 244.7 | 84.3, 84.3, 243.3 | 84.5, 84.5, 244.3 |
| Resolution (Å) | 20-1.7 (1.76-1.70) | 50-1.47 (1.52-1.47) | 20-1.80 (1.86-1.80) | 20-1.57 (1.63-1.57) |
| Rmerge (%) | 8.3 (48.6) | 5.8 (28.7) | 8.7 (41.1) | 6.2 (24.6) |
| I/σI | 19.4 (3.7) | 47.4 (5.3) | 16.2 (2.9) | 38.4 (10.4) |
| Completeness (%) | 100 (100) | 99.1 (91.7) | 99.9 (100) | 99.4 (99.3) |
| Redundancy | 7.0 (7.0) | 9.1 (7.0) | 9.5 (8.8) | 11.3 (11.0) |
| Refinement | ||||
| Rwork/Rfree | 16/19 | 13/16 | 16/18 | 12/15 |
| Root mean square deviation bond lengths (Å) | 0.004 | 0.012 | 0.015 | 0.009 |
| Root mean square deviation bond angles (degrees) | 0.6 | 0.9 | 1.3 | 0.9 |
RESULTS
CjMan26C Defines a New Class of Mannobiohydrolase Activity—Recombinant CjMan26C, produced as described under “Materials and Methods,” was active against linear and decorated β-mannans but did not hydrolyze other plant cell wall polysaccharides, including xylans (derived from oat spelled, wheat, rye, or birchwood), pectins (homopolygalacturonic acid and rhamnogalacturonan), or β-glucans (cellulose, hydroxyethyl-cellulose, carboxymethyl-cellulose, laminarin, or lichenan). The CjMan26C-catalyzed hydrolysis of mannose-based polysaccharides was slow compared with classical endo-acting β-mannanases, such as GH5 and GH26 (8). For example, the activities of CjMan26C against insoluble β-mannan, carob galactomannan and konjac glucomannan were 1.6-, 9.3-, and 14.6-fold lower than its close endo-acting homolog CjMan26A (see below) (Table 2). CjMan26C, typical of the majority of GH26 enzymes, is a β-mannanase (mannanase). The pH and temperature optima of the enzyme, using galactomannan as substrate, were pH 8.0 and 50 °C (data not shown).
TABLE 2.
Catalytic activity of CjMan26C and CjMan26A mannanases
| Substratea | kcat/Km (CjMan26C)b | kcat/Km (CjMan26A)b |
|---|---|---|
| min−1m−1 | min−1m−1 | |
| Galactomannan | 1.4 × 104 ± 0.9 | 1.3 × 105 ± 0.8 |
| Glucomannan | 1.1 × 104 ± 0.8 | 1.6 × 105 ± 0.6 |
| β-Mannan | 2.2 × 104 ± 0.5 | 3.5 × 104 ± 1.0 |
| Mannohexaose | 2.7 × 109 ± 1.5 × 108 | NDc |
| Mannopentaose | 3.5 × 109 ± 1.2 × 108 | ND |
| Mannotetraose | 3.0 × 109 ± 3.3 × 108 | 1.4 × 107 ± 2.8 × 10−3 |
| Mannotriose | 2.9 × 106 ± 2.2 × 108 | 1.8 × 106 ± 1 × 10−2 |
The substrates galactomannan and glucomannan were from carob and konjac seeds, respectively.
For polysaccharide substrates activity is expressed as specific activity (mol of product/mol of enzyme/min) at a substrate concentration of 2 mg ml−1.
ND, not determined.
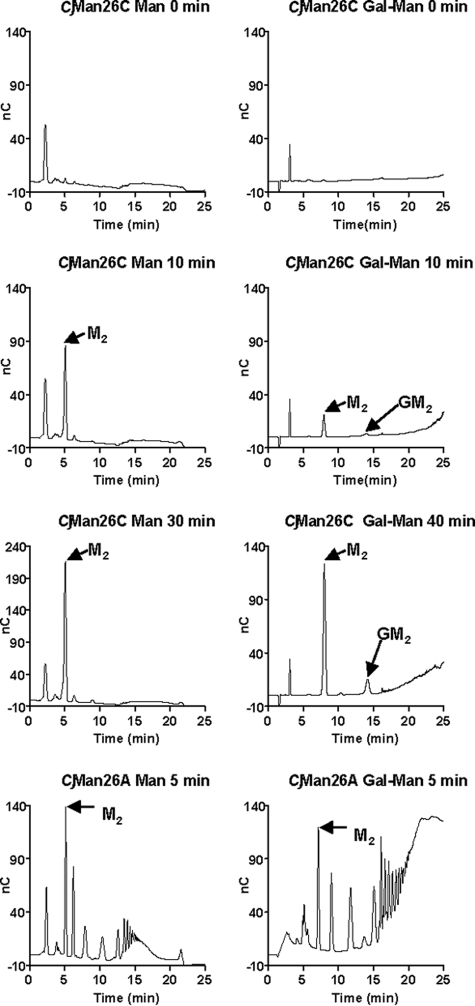
CjMan26C releases only mannobiose from β-mannan, whereas this disaccharide and, to a lesser extent, Gal-Man2, were generated from carob galactomannan, Fig. 1. This oligosaccharide fingerprint, comprising a single reaction product, is typical of an exo-acting enzyme; endo-acting glycoside hydrolases typically produce a range of reaction products that vary in size. These data all point to the novel catalytic activity of CjMan26C. In contrast to all of the other GH26 β-mannanases characterized to date (8, 22), which are endo-acting, CjMan26C is not only an exo-mannanase but is to our knowledge the first to be described that releases the disaccharide mannobiose and not mannose. By analogy to the terminology used to describe cellulose-degrading systems (see Refs. 23 and 24) for review), CjMan26C can be defined as a mannobiohydrolase. In light of the three-dimensional structure, described below, we can also say that the disaccharides are liberated at the nonreducing end of β-1,4-linked mannans and mannooligosaccharides; hence, the enzyme is a “-2 mannobiohydrolase” in the nomenclature advocated by Sinnott and co-workers (25).
FIGURE 1.
Hydrolysis of mannans by CjMan26C. CjMan26C (10 nm) and CjMan26A (79 nm) were incubated with 0.2% β-mannan (Man) and galactomannan (Gal-Man) in 50 mm sodium phosphate buffer, pH 7.0, containing 1 mg/ml bovine serum albumin. At regular time intervals, aliquots were removed and analyzed by HPLC. The peaks corresponding to mannobiose (M2) and 62-Gal-mannobiose (GM2) are indicated by arrows.
The activity of the mannobiohydrolase against mannooligosaccharides showed that the enzyme released mannose and mannobiose from mannotriose, exclusively mannobiose from mannotetraose, mannobiose and mannotriose from mannopentaose, and mannobiose and mannotetraose from mannohexaose, all of the expected patterns for a mannobiohydrolase. The enzyme displays no detectable activity against mannobiose, whereas the catalytic efficiency of CjMan26C against mannotriose, mannotetraose, mannopentaose, and mannohexaose was 2.9 × 106, 3.0 × 109, 3.5 × 109, and 2.9 × 109 min-1 m-1, respectively (Table 2). These data show that the enzyme contains four subsites that extend from -2 to +2, with bond cleavage occurring between the sugars located at -1 and +1, by definition (26). CjMan26C is an extremely efficient enzyme, with a kcat/Km against mannotetraose ∼20-fold higher than the catalytic efficiency of CjMan26A (27). Indeed, such a kcat/Km approaches catalytic perfection in which catalysis is limited by the diffusion rate (109 m-1 s-1) of molecules in aqueous environments (28). The enhanced activity of the mannobiohydrolase, compared with CjMan26A that also contains four subsites extending from -2 to +2, appears to be conferred by the +2 subsite, which contributes a productive binding energy of 4.0 kcal mol-1 to catalysis, determined from RTln(kcat/Km (mannotetraose)/kcat/Km (mannotriose)).
To investigate the capacity of CjMan26C to accommodate the galactosyl decorations present in galactomannan and also the glucosyl moieties in the backbone of glucomannan, the reaction products derived from Gal2Man5 and glucomannan were analyzed. Only mannobiose was released from glucomannan, demonstrating that glucose cannot bind to the glycone subsites, whereas the enzyme did not hydrolyze 6364-Gal2Man5, indicating that either the -1 or -2 subsites cannot accommodate mannosides decorated with galactose side chains (data not shown).
To explore further galactose recognition, 6364-Gal2Man5 was partially hydrolyzed by a GH27 α-galactosidase to generate, in equal proportions, 6364-Gal2Man5, 63-Gal-Man5, 64-Gal-Man5, and mannopentaose, and the hydrolysis of these oligosaccharides by CjMan26C was explored. The data show that CjMan26C liberates, albeit slowly, 61-Gal-Man2, demonstrating that galactose can be accommodated when bound to the mannoside at the -1 subsite (data not shown). The absence of 63-Gal-Man3 shows that the galactose side chain is not tolerated at the +1 subsite.
Three-dimensional Structure of CjMan26C—The crystal structure of CjMan26C was solved in various apo and ligand-bound forms (Table 1). The first to be solved, that co-crystallized with mannobiose (but subsequently shown to be a mannose complex), was determined by molecular replacement, using CjMan26A as the search model, to a resolution of ∼1.5 Å, from a P6122 crystal form with approximate cell dimensions a = b = 85, c = 245 Å and with one molecule in the asymmetric unit. The structure consists of 367 amino acids, equating to residues 53-419 of full-length (unprocessed) CjMan26C.
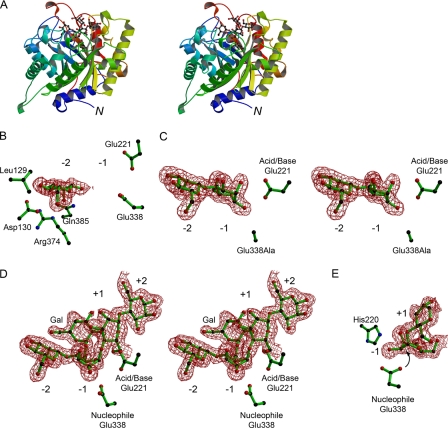
The structure of CjMan26C displays a canonical (β8/α8)-barrel topology, typical of GH26 (Fig. 2). The structure of CjMan26C (Fig. 2A) shows a high degree of similarity with GH26 endo-β1,4-mannanases, most notably with CjMan26A (27, 29). Overlaying the structures of the two Cellvibrio enzymes reveals a root mean square deviation of 1.46 Å for 311 equivalent Cα atoms using SSM (30). Structural comparison with GH26 and other clan GH-A enzymes renders it facile to identify the catalytic acid/base and nucleophile in the double displacement reaction as Glu-221 and Glu-338, respectively (described more fully below). Of particular note, with respect to exo/endo preference (described below) is the active center cleft that is restricted at one end by the presence of an extended 16-residue loop (defined henceforth as loop 3) connecting β-strand 2 and α-helix 3 (Fig. 3). In CjMan26A, the equivalent loop consists of only 12 amino acids, and as a result, the substrate binding cleft is open at both ends (discussed in more detail below).
FIGURE 2.
The three-dimensional structure of CjMan26C and its complexes. A, protein schematic diagram (divergent “wall-eyed” stereo) color ramped from N terminus (blue) to C terminus (red) with the Gal-Man4 ligand in a ball-and-stick representation. B, electron density (0.4 electrons/Å3) for mannose bound in the -2 subsite of wild-type Man26C with the acid/base Glu-221 and the nucleophile Glu-338 shown, as are residues implicated in the -2 exo activity. C, electron density (0.4 electrons/Å3) for the mannobiose complex of the E338A variant, in divergent (wall-eyed) stereo. D, electron density (0.4 electrons/Å3), in divergent (wall-eyed) stereo, for 6163-Gal2Man4 ligand in the -2 to +2 subsites of CjMan26C. There is very weak density, suggesting a +2 subsite 61-galactosyl moiety, but this could not be modeled appropriately. E, “End-on” view of the distorted sugar in the -1 subsite of CjMan26C, showing the resulting “in line” near attack geometry with the interaction of O2 with His-220 shown. This figure was drawn with BOBSCRIPT (47).
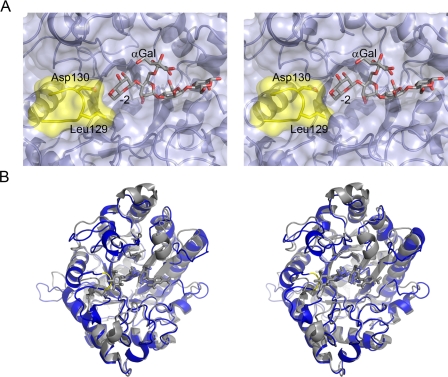
FIGURE 3.
Active center topography of a mannobiohydrolase. A, the solvent-accessible surface of the substrate-binding region of CjMan26C is shown (pale blue) in divergent (wall-eyed) stereo, with the 6163-Gal2Man4 ligand in licorice. Leu-129 and Asp-130, part of the loop which confers exo-specificity, are shaded yellow. B, overlap of the exo-enzyme CjMan26C (gray, the E338A complex with mannobiose for clarity) with the endo-enzyme CjMan26A (blue, with the “Michaelis complex” ligand, DNP-2F mannotrioside, in cyan). The CjMan26C “exo loop” is colored yellow. This figure was drawn with PyMol (DeLano Scientific LLC; available on the World wide Web).
Insights into Substrate Binding, Distortion, and Catalysis Revealed by Complex Structures of CjMan26C; Role of the -1 Subsite—The enzymes acting on mannosides, mannanases, and mannosidases are chemically interesting, since they catalyze nucleophilic substitution at the anomeric carbon of mannosides, itself a challenging reaction. Insights into the mechanism by which CjMan26C binds to its substrate and catalyzes glycosidic bond hydrolysis are revealed here by the resolution of the structure of the enzyme in complex with mannose, of a mutant with mannobiose, and, most insightfully, through the serendipitous trapping of a galactose-decorated oligosaccharide complex with what equates to 6163-Gal2-Man4 with the partially ordered galactoside bound to the -1 subsite mannoside. The +2 subsite shows extremely weak density for a 6-linked galactoside, but this cannot be modeled with any confidence. This complex was obtained through co-crystallization with commercial 61-Gal-Man2 (Megazyme) and probably represents either a slowly hydrolyzed contaminant or perhaps an unusual species generated by transglycosylation (as has often been observed with amylases (31, 32), for example). Indeed, 6163-Gal2-Man4 acts as a poor substrate but a potent inhibitor of mannohexaose hydrolysis with a Ki of 20 μm.
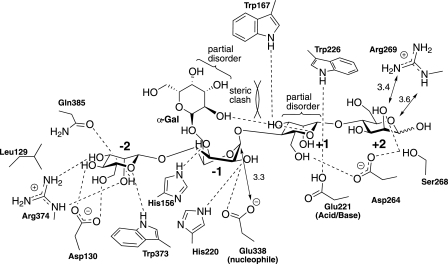
An intriguing feature of the CjMan26C-6163-Gal-Man4 complex, one that is rarely observed on glycosidases (a notable exception being the chitobiose complex of the Serratia marcescens GH20 chitobiase (33)) is that the substrate is, to a significant extent, uncleaved. So although density is stronger for the -2 and -1 subsite sugars, there is a substantial proportion of a “Michaelis” complex of unhydrolyzed substrate in which ligand binds from -2 to +2 and thus spans the critical -1 and +1 subsites. As a result, and consistent with current views on glycosidase chemistry and substrate distortion, the mannoside at the -1 subsite is distorted to a 1S5 conformation with the resulting axial orientation for the C1-O leaving group glycosidic bond. This is exactly the conformation observed previously on an acid/base mutant of a endo-mannnases with aryl 2F glycoside substrates (29), but here it is revealed on a wild-type active enzyme with a sugar leaving group and no fluorine substitution in the pyranose ring of the glycone sugar. The retention of uncleaved substrate in the proximal active site probably reflects a partial inhibitory effect of the galactose side chain at the -1 subsite. Thus, the C2-C3 edge of the pyranose ring clashes with O2 of the mannose at +1, which is likely to destabilize the proximal aglycone sugar, reducing its capacity to provide productive binding energy. The O2 of the galactose side chain also makes a hydrogen bond with the endocyclic oxygen with the +1 mannose. Thus, even when the initial glycosylation reaction occurs, leaving group departure is restricted, which probably promotes resynthesis of the glycosidic linkage through a transglycosylation reaction. Thus, the galactose side chain is not only reducing the rate of catalysis by making steric clashes but may also have an inhibitory effect by providing a bridge between sugars in the glycone and aglycone region of the active site. The structure of the CjMan26C-6163-Gal-Man4 complex, in common with recent studies, strongly implicates a B2,5 transition state during hydrolysis by retaining both β-mannanases (29) and β-mannosidases (7, 34, 35). Such a transition state would place the O2 hydroxyl group of the relevant mannoside pseudo-equatorial and avoid troublesome steric clashes at the anomeric centre. In contrast, some authors have advocated a 3H4 transition state for retaining β-mannosidase catalysis on theoretical grounds (36). Such a transition state, however, would require O4 to become pseudo axial and thus demand a major, unprecedented conformational change in the enzyme in order to prevent steric clashes of the -2 subsite sugar.
The complexes reveal the interactions between the substrate and the four subsites of the enzyme (Fig. 4). The catalytic apparatus, which is housed at the -1 subsite, comprises the catalytic acid/base, Glu-221, and nucleophile, Glu-338, which, in common with all clan GH-A enzymes, are located at the C terminus of β-strands 4 and 7, respectively. The identity of the catalytic residues is entirely consistent with the observation that the mutants E221A and E338A are essentially inactive (Table 3). The position of the catalytic nucleophile is stabilized through interactions with Arg-217 and Tyr-297. The side chain of the catalytic acid/base makes a hydrogen bond with the Nε1 of Trp-226, which is likely to contribute to both the position and ionization state of this critical amino acid. His-220, which is highly conserved in GH26 enzymes, and the catalytic nucleophile make a hydrogen bond with O2 of the sugar at the -1 subsite in its distorted conformation but not when the mannose adopts the relaxed 4C1 geometry in the E221A mutant mannobiose complex (Fig. 2). The only other polar interaction between the -1 subsite sugar and CjMan26C is at O3, which makes a polar contact with the imidazole ring of His-156. Unusually, the -1 sugar makes no further polar interactions with the enzyme. The substrate binding residues in the -1 subsite of CjMan26C are strictly conserved in GH26 mannanases. For example, the following residues in CjMan26C have structural equivalents in CjMan26A (the CjMan26A residues are in parentheses) His-156 (His-143), His-220 (His-211), Glu-221 (Glu-212), and Glu-338 (Glu-320).
FIGURE 4.
Schematic diagram of the interactions of CjMan26C with manno-oligosaccharide 6163-Gal2Man4. The mannoside at -1 is distorted to an approximate 1S5 conformation with a “near attack” conformation for Glu-338, which is poised for in-line nucleophilic attack at the anomeric carbon with an Onucleophile -C1-Oleaving group angle of ∼155°. The α-1,6-linked galactoside bound to the -1 subsite sugar exhibits partial disorder (poor electron density around C3 and C4), reflecting unfavorable steric clashes with the +1 subsite mannose moiety. This clash is also reflected in multiple conformations for the +1 mannoside. Direct hydrogen bonds to protein/ligand, >3.2 Å, are shown with other relevant longer interactions indicated with arrows and the appropriate distance.
TABLE 3.
Catalytic activity of CjMan26C mutants
| CjMan26C varianta | kcat/Kmb | Activity relative to CjMan26Cc | Activity relative to CjMan26A |
|---|---|---|---|
| min−1M−1 | |||
| Wild type CjMan26C | 3.0 × 109 ± 3.3 × 108 | 1.0 | 2.1 × 102 |
| Wild type CjMan26A | 1.4 × 107 ± 2.8 × 105 | 4.6 × 10−3 | 1.0 |
| E221A | 7.5 × 102 ± 2.6 × 101 | 2.5 × 10−7 | 5.4 × 10−4 |
| E338A | 7.7 × 102 ± 1.3 × 101 | 2.6 × 10−7 | 5.5 × 10−4 |
| L129G | 4.0 × 106 ± 3.8 × 105 | 1.3 × 10−3 | 0.29 |
| L129A | 7.4 × 106 ± 1.5 × 105 | 2.5 × 10−3 | 0.53 |
| D130Gd | 2.7 × 105 ± 7.5 × 104 | 9.0 × 10−5 | 1.9 × 10−2 |
| D130Ad | 2.9 × 104 ± 3.1 × 103 | 9.7 × 10−6 | 2.1 × 10−3 |
| L129G/D130G | 8.2 × 107 ± 9.1 × 106 | 2.4 × 10−2 | 5.9 |
| L129A/D130Ad | 4.5 × 104 ± 4.5 × 103 | 1.5 × 10−5 | 3.2 × 10−3 |
| ΔL129/D130d | 2.7 × 105 ± 1.6 × 104 | 9.0 × 10−5 | 1.9 × 10−2 |
| ΔN128/L129/D130d | 2.7 × 104 ± 1.7 × 103 | 9.0 × 10−6 | 1.9 × 10−3 |
| ΔN128/L129/D130/A131d | 3.4 × 104 ± 6.6 × 102 | 1.1 × 10−5 | 2.4 × 10−3 |
The varants are all of CjMan26C.
The substrate used to measure catalytic efficiency was mannotetraose.
The activity is expressed as decrease in activity relative to wild type CjMan26C. For example, wild type CjMan26A is 210-fold less active than wild type CjMan26C.
Variants that display endo-activity (see Table 4).
Leaving Group Interactions at the +1 and +2 Subsites—In the CjMan26C-6163-Gal-Man4 complex, difference density indicates that the +1 and +2 sugars are partially disordered. Although the +2 mannose clearly adopts a relaxed chair conformation, it would appear that a disordered sugar, presumably galactose, is appended to O6. The +1 mannoside is also partially distorted, which may reflect unfavorable steric clashes with the -1′ subsite galactose moiety (described above).
The sugar at the +1 subsite makes limited polar interactions with the enzyme. O6 makes a polar interaction with the side chain of Asp-266, whereas O3 hydrogen bonds to Nε1 of Trp-167 (Fig. 4). In CjMan26A, the equivalent mannose at the +1 subsite will make similar polar contacts with the enzyme; O3 hydrogen bonds to the indole nitrogen of Trp-134, whereas O6 may make a polar contact with Asn-257, equivalent to Asp-266 in CjMan26C, although this will require a significant rotation of the β-carbon. CjMan26C contains a hydrophobic platform, which extends from the -1 subsite, where Trp-373 stacks against the sugar ring, whereas Trp-226 is in a planar orientation to the pyranose sugar at the +1 subsite (Fig. 4). These aromatic residues are highly conserved in GH26 enzymes exemplified by CjMan26A, where the structural equivalents to Trp-226 and Trp-373 are Trp-217 and Trp-360, respectively (27).
The +2 site differs substantially between CjMan26C and CjMan26A. In the mannobiohydrolase, Ser-268 interacts with O2 and O3, whereas Arg-269 may make long range polar contacts with O2 and the endocyclic oxygen. The equivalent loop in CjMan26A does not present any residues close to a putative +2, and it is not readily apparent why the enzyme is 10-fold more active against mannotetraose than mannotriose (8), although this equates to a binding energy of only 1.3 kcal mol-1. Indeed, since both enzymes exhibit similar activities against mannotriose, which occupies subsites -2 to +1, the higher catalytic efficiency displayed by CjMan26C reflects tighter binding at the +2 subsite, with a ΔΔG of 2.7 kcal mol-1 between the two mannanases at this subsite.
Structural Basis for exo-Mannobiohydrolase Activity Is Conferred by an Enclosed Topography at the -2 Subsite—The mannose at the -2 subsite of CjMan26C makes numerous interactions with the enzyme. O2 makes polar contacts with the side chains of Arg-374, Glu-385, and Trp-373, whereas O3 also interacts with Arg-374 and Gln-385; the endocylic oxygen and O6 also make hydrogen bonds with Trp-373, and the exocyclic oxygen also makes a polar contact with Asp-130 (Fig. 4). The interactions between the substrate at the -2 subsite of CjMan26C and CjMan26A are partially conserved. Thus, the CjMan26C residues Trp-373, Arg-374, and Gln-385 are, respectively, structurally equivalent to Trp-332, Arg-333, and His-349 in CjMan26A. The marked difference, however, is conferred by the CjMan26C loop 3, connecting β-strand 2 to α-helix 3, which contains Asp-158, which interacts with O4 and O6 of the bound sugar. It is this feature that creates the steric constraint, through the carbonyl of Leu-129 and the side chain of Asp-130 that prevents extension of the substrate beyond the -2 subsite (Fig. 3). In CjMan26A, the equivalent loop is four residues shorter and instead yields an open substrate binding groove. Indeed, the residue most equivalent to Asp-130 in CjMan26A, Glu-131, is displaced by 2 Å such that the glutamate interacts with O6 but not O4 of the mannose residue.
The enclosed -2 subsite unique to CjMan26C (among solved mannanase structures), in light of the various reaction product profiles, provides the rationale both for the exo-mannobiohydrolase activity and for the specificity toward heterogeneous substrates. Thus, in the -2 and +1 subsites, O6 of the mannose is pointing into the surface of the protein, explaining why a α-1,6-linked galactose cannot be accommodated at these sites. At subsites -2 and +2, the axial O2 of mannose is exploited as a specificity determinant, consistent with the action patterns on galactomannan and β-mannan. As discussed above and recently in the literature (29), mannose specificity at the -1 subsite is driven not by the relaxed 4C1 conformation of the sugar, in which the axial O2 does not interact with the enzyme, but by the equatorial and axial orientation of O2 and O3, respectively, at the transition state. The interactions at the -1 and -2 subsites explain why only mannobiose is released from glucomannan. Most notably, the enclosed -2 subsite explains the exo activity of CjMan26C in a manner than can be experimentally tested.
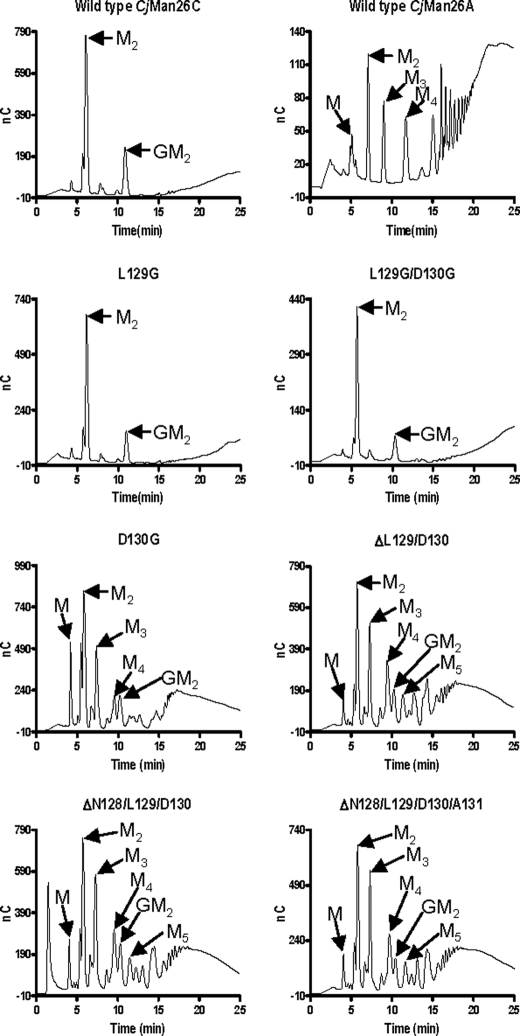
Engineering of Exo/endo-specificity—To dissect the structural basis for the exo-specificity of CjMan26C in more detail, several mutations were introduced into the loop that seals the -2 subsite. The D130A and D130G mutations substantially reduced catalytic activity (Table 3). This shows that the polar contacts between the carboxylate of the aspartate and O4 and O6 of the substrate play a key role in substrate recognition. Significantly, however, despite reduced activity, the mutations also alter the profile of the reaction products released from galactomannan and mannohexaose. Hydrolysis of the hexasaccharide via an exo-mode of action will generate initially mannobiose and mannotetraose, whereas the tetrasaccharide will subsequently be converted into the disaccharide. In contrast, an endo-enzyme will generate, in addition to mannobiose and mannotetraose, mannotriose, since the substrate can extend beyond both the distal glycone (-2) and aglycone (+2) subsites (Table 4). Rather than producing predominantly mannobiose, indicative of an exo-mode of action, the D130A mutant generated a range of oligosaccharides from galactomannan, now typical of an endo-acting enzyme (Fig. 5). Indeed, the proposed endo-mode of action displayed by D130A is consistent with the generation of mannotriose from mannohexaose (Table 4). These data indicate that the interaction between Asp-130 and the mannose at -2 locks the conformation adopted by loop 3, which presents a steric block preventing substrate extension distal to the -2 subsite. Similarly, deleting Asp-130 in combination with its flanking residues greatly reduced the activity of the enzyme while also altering its mode of action from exo to predominantly endo (Fig. 5). Reducing the side chain of Leu-129, which also presents a steric block at the -2 subsite, again resulted in a substantial decrease in the activity of the enzyme, although in this case, the exo-mode of action was retained (Fig. 5 and Table 4). The retention of exo activity in the L129A and L129G mutants is, maybe, not surprising, since the major clash with a sugar sitting in the putative -3 subsite would be through the invariant backbone carbonyl and amine groups. The low activity displayed by L129A and L129G suggests that the loss of the hydrophobic side chain has altered the conformation of loop 3 such that Asp-158 is no longer able to interact with the mannose at the -2 subsite, whereas the loss of the hydrophobic contact between the aliphatic side chain of the leucine and C6 of the sugar may also contribute to the decrease in activity. Intriguingly, the only mutation of Asp-130 that did not alter the mode of action of CjMan26C was L129G/D130G (Table 4 and Fig. 5). The double mutant retained significant catalytic activity and displayed an exo-mode of action, although the single mutation, D158G, greatly reduced activity of the enzyme, which displayed endo-activity. It is possible that the introduction of adjacent glycine residues introduces significant flexibility into the loop such that a polar residue can replace the function of Asp-130 and make hydrogen bonds with O4 and O6 of the -2 mannose, thereby maintaining the steric block that prevents substrate extension.
TABLE 4.
Ratio of reaction products generated from mannohexaose
| Enzyme | Ratio (M2 + M4)/M3a |
|---|---|
| Man26C wild type | 1:0 |
| Man26A wild type | 2.2:1 |
| Man26C L129G | 1:0 |
| Man26C L129A | 1:0 |
| Man26C D130G | 3.8:1 |
| Man26C D130A | 3.0:1 |
| Man26C L129G/D130G | 1:0 |
| Man26C L129A/D130A | 5.6:1 |
| Man26C ΔL129/D130 | 4.2:1 |
| Man26C ΔN128/L129/D130 | 2.1:1 |
| Man26C ΔN128/L129/D130/A131 | 2.3:1 |
The ratio of products was measured after 30% hydrolysis of 30 μM mannohexaose. A completely exo-acting enzyme will generate only mannobiose from mannohexaose; hence, a ratio of 1:0 is observed for wild type CjMan26C and three of the exo-acting mutants. A fully endo-acting mannanase, exemplified by CjMan26A, has a ratio of 2:1 ((mannobiose and mannotetraose)/mannotriose), similar to three of the mutants. A ratio higher ratio than 2:1, observed for four of the mutants, indicates an endo-activity, but substrate binding to the different subsites is not completely random.
FIGURE 5.
The hydrolysis of polysaccharides by mutants of CjMan26C. Galactomannan was treated with wild type CjMan26A, wild type CjMan26C, and mutants of CjMan26C, and the reaction profiles, after a 30-min incubation, were analyzed by HPLC. The peaks corresponding to mannose (M) mannobiose (M2), mannotriose (M3), mannotetraose (M4), mannopentaose (M5) and 62-Gal-mannobiose (GM2) are indicated by arrows.
There is a paucity of reports demonstrating the successful endo-exo conversion of glycoside hydrolases. Warren and co-workers (37) showed that the removal of the loop comprising the ceiling of the active site tunnel of the Cellulomonas fimi cellobiohydrolase Cel6B increased the endo character of the enzyme, reflected by increased viscosity of the hydrolyzed substrate, albeit at a significant catalytic cost. The modification of a key binding residue at the -5 subsite of a polygalacturonase is reported to convert its mode of action from endo-processive to endo-random (38). The conversion of an exo-acting C. japonicus GH43 arabinanase, CjAra43, into an endo-α-1,5-arabinanase (39) has some resonance with the modification of CjMan26C, reported here. In CjAra43, the sugar located at the -3 subsite makes hydrogen bonds with residues that create a steric barrier to any sugar appended to O5 of the arabinofuranose positioned at this distal glycone subsite. In contrast to CjMan26C, however, although the mutation of the polar residues released this steric block and thus converted the arabinanase into an endo-acting enzyme, the associated reduction in catalytic activity was modest, suggesting that the two hydrogen bonds do not make a substantial contribution to the overall binding energy of the -3 subsite (39). It is also possible that the distal glycone subsite in the inverting arabinanase makes a minor contribution to productive substrate binding compared with the glycone -2 subsite in CjMan26C, where, particularly in the deglycosylation step, its contribution to substrate binding is critical.
DISCUSSION
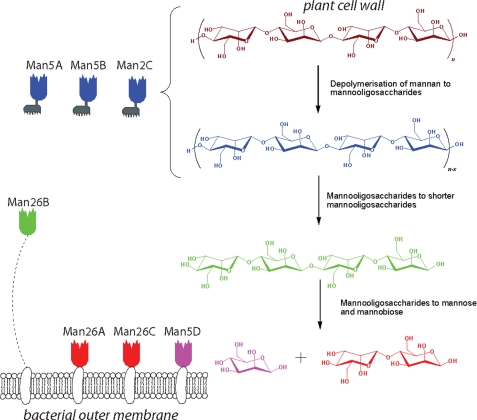
In light of the C. japonicus genome sequence (15), we believe that characterization of CjMan26C completes the analysis of the mannan-degrading system of C. japonicus. The bacterium expresses three GH26 and four GH5 mannanases. The molecular architecture and the cellular location of these enzymes provide insight into the hierarchy of the degradative process. The three endo-acting mannanases, CjMan5A, CjMan5B, and CjMan5C, all contain noncatalytic carbohydrate-binding modules (CBMs) that target crystalline cellulose (8). These enzymes are thus directed to the insoluble plant cell wall and are therefore predicted to catalyze the initial hydrolysis of mannans and glucomannans that are integral to these composite structures. The “solubilized” mannans are then hydrolyzed by the mannanases, CjMan5A, CjMan26A, CjMan26B, and CjMan26C, arrayed on the surface of the bacterium through their attachment to membrane lipids (their signal peptides are cleaved by type II signal peptidases, which append the mature enzyme to membrane lipid). These surface-bound mannanases lack CBMs, providing further evidence that these enzymes target soluble mannans. The catalytic activities of these enzymes provide some insight into their role within the hierarchy of mannan degradation (Fig. 6). There is little discrimination between the endo-acting mannanases CjGH26A, CjGH26B, CjGH5A, and CjGH5B against the various mannans (8, 22), indicating that the major determinants of substrate specificity are the cellular location of these enzymes and the presence or absence of cellulose-directed CBMs. In contrast, the catalytic activity displayed by the CBM-containing mannanase, CjMan5C, appears to be tailored to crystalline mannan, since it is the only enzyme to display activity against ivory nut mannan (8) (highly crystalline).
FIGURE 6.
The mannanase repertoire of C. japonicus. C. japonicus secretes three endomannanases, CjMan5A, -B, and -C, whose function is the depolymerization of plant cell wall mannans (magenta) into shorter mannooligosaccharides (blue). In this task, they are aided by their plant cell wall-targeting carbohydrate-binding modules (gray). Shorter manno-oligosaccharides are subsequently reduced in length by the, flexibly tethered, Man26B and then hydrolyzed to mannose and mannobiose by the outer membrane-bound Man26A, -26C, and -5D. Following disaccharide import, the disaccharide mannobiose is hydrolyzed to two molecules of mannose by intracellular β-mannosidase Man2 (not shown). The catalytic modules are colored with respect to their reaction products.
From a generic perspective, it would appear that GH26 mannanases are retained on the bacterial (outer) membrane, either within protein complexes (40, 41) or attached directly to the bacterial surface (8, 42), and do not contain CBMs that target crystalline cellulose. By contrast, endo-acting GH5 mannanases often contain cellulose-targeted CBMs (8, 43, 44), and thus the function of GH5 and GH26 C. japonicus mannanases proposed here is likely to have a generic biological resonance. Within this context, interesting variation between the activities of the surface mannanases was evident. CjMan26B displays canonical endo-mannanase activity and, intriguingly, is linked to the bacterial cell wall by a long ∼70-residue flexible linker sequence. This enzyme is thus able to explore a greater sequence space than the other surface mannanases, and its natural substrate is likely to be soluble decorated mannans (galactomannan or glucomannan). The catalytic activities of the other three membrane-bound mannanases are consistent with these enzymes targeting small oligosaccharides, since they all display activities against mannotriose and mannotetraose that are 3-4 orders of magnitude greater than the other Cellvibrio mannanases. It is not immediately obvious why the bacterium expresses both CjMan26A and CjMan26C, although they may display complementarity or endo-exo synergy in which (random) internal glycosidic bond cleavage by the endo-mannanase creates new reducing ends that are targeted by the exo-mannanase. The major oligosaccharides generated by these two enzymes, mannobiose and, to a lesser extent, mannotriose, will then be hydrolyzed by the exo-acting mannosidase, which, similar to its homologue in Cellvibrio mixtus (7), releases mannose from both the disaccharide and trisaccharide (data not shown). The likely biological rationale for the presentation of the mannanases that target mannooligosaccharides on the surface of the bacterium is that the mannose and mannobiose generated will be available for preferential uptake by C. japonicus rather than competing microorganisms within the milieu of the plant cell wall. Indeed, this hierarchical pattern of plant cell wall degradation is a common theme in this organism. Thus, xylan, the other major hemicellulosic polysaccharide, is initially degraded by enzymes containing cellulose-targeted CBMs, and the processing of the resultant xylooligosaccharides into their constituent sugars occurs on the surface or in the periplasm of the bacterium (45, 46). Indeed, this pattern of polymer degradation parallels the human intestine, where endo-acting enzymes generate oligopeptides and oligosaccharides that are further hydrolyzed on the surface of the epithelium into monomeric species.
In conclusion, CjMan26C provides an elegant example of how subtle modification to the loops surrounding the active site of an enzyme can result in dramatic topological changes that lead to an exo-activity that is tailored to the requirements of its host. Such knowledge will be invaluable when assigning the function of open reading frames revealed through the continued expansion in genome sequence data.
Supplementary Material
This work was supported by the Biotechnology and Biological Sciences Research Council (to H. J. G. and G. J. D.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1.
Footnotes
The abbreviations used are: HPLC, high pressure liquid chromatography; CAPSO, 3-(cyclohexamino)-2-hydroxy-1-propanesulfonic acid; CBM, carbohydrate-binding module.
References
- 1.Lynd, L. R., Laser, M. S., Bransby, D., Dale, B. E., Davison, B., Hamilton, R., Himmel, M., Keller, M., McMillan, J. D., Sheehan, J., and Wyman, C. E. (2008) Nat. Biotechnol. 26 169-172 [DOI] [PubMed] [Google Scholar]
- 2.Ragauskas, A. J., Williams, C. K., Davison, B. H., Britovsek, G., Cairney, J., Eckert, C. A., Frederick, W. J., Jr., Hallett, J. P., Leak, D. J., Liotta, C. L., Mielenz, J. R., Murphy, R., Templer, R., and Tschaplinski, T. (2006) Science 311 484-489 [DOI] [PubMed] [Google Scholar]
- 3.Brett, C. T., and Waldren, K. (1996) Physiology and Biochemistry of Plant Cell Walls: Topics in Plant Functional Biology, Chapman and Hall, London
- 4.Brumer, H., III, Sims, P. F., and Sinnott, M. L. (1999) Biochem. J. 339 43-53 [PMC free article] [PubMed] [Google Scholar]
- 5.Centeno, M. S., Guerreiro, C. I., Dias, F. M., Morland, C., Tailford, L. E., Goyal, A., Prates, J. A., Ferreira, L. M., Caldeira, R. M., Mongodin, E. F., Nelson, K. E., Gilbert, H. J., and Fontes, C. M. (2006) FEMS Microbiol. Lett. 261 123-132 [DOI] [PubMed] [Google Scholar]
- 6.McCleary, B. V. (1988) Methods Enzymol. 160 523-527 [Google Scholar]
- 7.Dias, F. M., Vincent, F., Pell, G., Prates, J. A., Centeno, M. S., Tailford, L. E., Ferreira, L. M., Fontes, C. M., Davies, G. J., and Gilbert, H. J. (2004) J. Biol. Chem. 279 25517-25526 [DOI] [PubMed] [Google Scholar]
- 8.Hogg, D., Pell, G., Dupree, P., Goubet, F., Martin-Orue, S. M., Armand, S., and Gilbert, H. J. (2003) Biochem. J. 371 1027-1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henrissat, B. (1991) Biochem. J. 280 309-316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davies, G. J., and Sinnott, M. L. (2008) Biochem. J., in press
- 11.Henrissat, B., Callebaut, I., Fabrega, S., Lehn, P., Mornon, J. P., and Davies, G. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 7090-7094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kiyohara, M., Sakaguchi, K., Yamaguchi, K., Araki, T., Nakamura, T., and Ito, M. (2005) Biochem. J. 388 949-957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taylor, E. J., Goyal, A., Guerreiro, C. I., Prates, J. A., Money, V. A., Ferry, N., Morland, C., Planas, A., Macdonald, J. A., Stick, R. V., Gilbert, H. J., Fontes, C. M., and Davies, G. J. (2005) J. Biol. Chem. 280 32761-32767 [DOI] [PubMed] [Google Scholar]
- 14.Money, V. A., Smith, N. L., Scaffidi, A., Stick, R. V., Gilbert, H. J., and Davies, G. J. (2006) Angew. Chem. Int. Ed. 45 5136-5140 [DOI] [PubMed] [Google Scholar]
- 15.DeBoy, R. T., Mongodin, E. F., Fouts, D. E., Tailford, L. E., Khouri, H., Emerson, J. B., Mohamoud, Y., Watkins, K., Henrissat, B., Gilbert, H. J., and Nelson, K. E. (2008) J. Bacteriol. 190 5455-5463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller, G. L. (1959) Anal. Chem. 31 426-428 [Google Scholar]
- 17.Matsui, I., Ishikawa, K., Matsui, E., Miyairi, S., Fukui, S., and Honda, K. (1991) J. Biochem. (Tokyo) 109 566-569 [DOI] [PubMed] [Google Scholar]
- 18.Otwinowski, Z., and Minor, W. (1997) Methods Enzymol. 276 307-326 [DOI] [PubMed] [Google Scholar]
- 19.Collaborative Computational Project 4 (1994) Acta Crystallogr. Sect. D 50 760-76315299374 [Google Scholar]
- 20.Murshudov, G. N., Vagin, A. A., and Dodson, E. J. (1997) Acta Crystallogr. Sect. D 53 240-255 [DOI] [PubMed] [Google Scholar]
- 21.Emsley, P., and Cowtan, K. (2004) Acta Crystallogr. Sect. D 60 2126-2132 [DOI] [PubMed] [Google Scholar]
- 22.Braithwaite, K. L., Black, G. W., Hazlewood, G. P., Ali, B. R., and Gilbert, H. J. (1995) Biochem. J. 305 1005-1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tomme, P., Warren, R. A., and Gilkes, N. R. (1995) Adv. Microb. Physiol. 37 1-81 [DOI] [PubMed] [Google Scholar]
- 24.Warren, R. A. (1996) Annu. Rev. Microbiol. 50 183-212 [DOI] [PubMed] [Google Scholar]
- 25.Davies, G. J., Sinnott, M. L., and Withers, S. G. (1997) in Comprehensive Biological Catalysis (Sinnott, M. L., ed) Academic Press, Inc., London
- 26.Davies, G. J., Wilson, K. S., and Henrissat, B. (1997) Biochem. J. 321 557-559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hogg, D., Woo, E. J., Bolam, D. N., McKie, V. A., Gilbert, H. J., and Pickersgill, R. W. (2001) J. Biol. Chem. 276 31186-31192 [DOI] [PubMed] [Google Scholar]
- 28.Samson, R., and Deutch, J. M. (1978) J. Chem. Phys. 68 285-290 [Google Scholar]
- 29.Ducros, V. M., Zechel, D. L., Murshudov, G. N., Gilbert, H. J., Szabo, L., Stoll, D., Withers, S. G., and Davies, G. J. (2002) Angew. Chem. Int. Ed. Engl. 41 2824-2827 [DOI] [PubMed] [Google Scholar]
- 30.Krissinel, E., and Henrick, K. (2004) Acta Crystallogr. Sect. D 60 2256-2268 [DOI] [PubMed] [Google Scholar]
- 31.Brzozowski, A. M., Lawson, D. M., Turkenburg, J. P., Bisgaard-Frantzen, H., Svendsen, A., Borchert, T. V., Dauter, Z., Wilson, K. S., and Davies, G. J. (2000) Biochemistry 39 9099-9107 [DOI] [PubMed] [Google Scholar]
- 32.Dauter, Z., Dauter, M., Brzozowski, A. M., Christensen, S., Borchert, T. V., Beier, L., Wilson, K. S., and Davies, G. J. (1999) Biochemistry 38 8385-8392 [DOI] [PubMed] [Google Scholar]
- 33.Tews, I., Perrakis, A., Oppenheim, A., Dauter, Z., Wilson, K. S., and Vorgias, C. E. (1996) Nat. Struct. Biol. 3 638-648 [DOI] [PubMed] [Google Scholar]
- 34.Vincent, F., Gloster, T. M., Macdonald, J., Morland, C., Stick, R. V., Dias, F. M. V., Prates, J. A. M., Fontes, C. M. G. A., Gilbert, H. J., and Davies, G. J. (2004) ChemBioChem. 5 1596-1599 [DOI] [PubMed] [Google Scholar]
- 35.Tailford, L. N., Offen, W. A., Smith, N. L., Dumon, C., Morland, C., Gratien, J., Heck, M.-P., Stick, R. V., Blériot, Y., Vasella, A., Gilbert, H. J., and Davies, G. J. (2008) Nat. Chem. Biol. 4 306-312 [DOI] [PubMed] [Google Scholar]
- 36.Nerinckx, W., Desmet, T., and Claeyssens, M. (2006) Arkivoc. 13 90-116 [Google Scholar]
- 37.Meinke, A., Damude, H. G., Tomme, P., Kwan, E., Kilburn, D. G., Miller, R. C., Jr., Warren, R. A., and Gilkes, N. R. (1995) J. Biol. Chem. 270 4383-4386 [DOI] [PubMed] [Google Scholar]
- 38.Pages, S., Kester, H. C., Visser, J., and Benen, J. A. (2001) J. Biol. Chem. 276 33652-33656 [DOI] [PubMed] [Google Scholar]
- 39.Proctor, M. R., Taylor, E. J., Nurizzo, D., Turkenburg, J. P., Lloyd, R. M., Vardakou, M., Davies, G. J., and Gilbert, H. J. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 2697-2702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Halstead, J. R., Vercoe, P. E., Gilbert, H. J., Davidson, K., and Hazlewood, G. P. (1999) Microbiology 145 3101-3108 [DOI] [PubMed] [Google Scholar]
- 41.Kurokawa, J., Hemjinda, E., Arai, T., Karita, S., Kimura, T., Sakka, K., and Ohmiya, K. (2001) Biosci. Biotechnol. Biochem. 65 548-554 [DOI] [PubMed] [Google Scholar]
- 42.Stoll, D., Stalbrand, H., and Warren, R. A. (1999) Appl. Environ. Microbiol. 65 2598-2605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hagglund, P., Eriksson, T., Collen, A., Nerinckx, W., Claeyssens, M., and Stalbrand, H. (2003) J. Biotechnol. 101 37-48 [DOI] [PubMed] [Google Scholar]
- 44.Puchart, V., Vrsanska, M., Svoboda, P., Pohl, J., Ogel, Z. B., and Biely, P. (2004) Biochim. Biophys. Acta 1674 239-250 [DOI] [PubMed] [Google Scholar]
- 45.Emami, K., Nagy, T., Fontes, C. M., Ferreira, L. M., and Gilbert, H. J. (2002) J. Bacteriol. 184 4124-4133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pell, G., Szabo, L., Charnock, S. J., Xie, H., Gloster, T. M., Davies, G. J., and Gilbert, H. J. (2004) J. Biol. Chem. 279 11777-11788 [DOI] [PubMed] [Google Scholar]
- 47.Esnouf, R. M. (1997) J. Mol. Graph. Model. 15 132-134 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.