Abstract
FBI-1 (also called Pokemon/ZBTB7A) is a BTB/POZ-domain Krüppel-like zinc-finger transcription factor. Recently, FBI-1 was characterized as a proto-oncogenic protein, which represses tumor suppressor ARF gene transcription. The expression of FBI-1 is increased in many cancer tissues. We found that FBI-1 potently represses transcription of the Rb gene, a tumor suppressor gene important in cell cycle arrest. FBI-1 binds to four GC-rich promoter elements (FREs) located at bp –308 to –188 of the Rb promoter region. The Rb promoter also contains two Sp1 binding sites: GC-box 1 (bp –65 to –56) and GC-box 2 (bp –18 to –9), the latter of which is also bound by FBI-1. We found that FRE3 (bp –244 to –236) is also a Sp1 binding element. FBI-1 represses transcription of the Rb gene not only by binding to the FREs, but also by competing with Sp1 at the GC-box 2 and the FRE3. By binding to the FREs and/or the GC-box, FBI-1 represses transcription of the Rb gene through its POZ-domain, which recruits a co-repressor-histone deacetylase complex and deacetylates histones H3 and H4 at the Rb gene promoter. FBI-1 inhibits C2C12 myoblast cell differentiation by repressing Rb gene expression.
The importance of the tumor suppressor retinoblastoma (Rb)3 protein in regulating key cellular events was first suggested by the identification of a tumor, retinoblastoma, in which the Rb locus was invariably deleted (1–3). Rb is implicated in the development of various cancers (4 and references therein). Rb suppresses tumorigenesis by inhibiting cell cycle progression at G1/S by preventing the transcription of several genes important in cell cycle control (5). Rb is phosphorylated in a cell cycle-dependent manner (6 and references therein). When Rb is hypophosphorylated, it forms complexes with E2F family proteins and inhibits transcription by recruiting proteins involved in transcriptional repression (7). Once phosphorylated, Rb can no longer form complexes with E2F proteins. E2F proteins, upon dimerization with their differentiation-regulated transcription factor partners, are capable of activating the expression of a number of genes that are likely to regulate or promote entry into S phase (6 and references therein).
Investigations on how transcription of the Rb gene is regulated are important in elucidating the cellular regulatory mechanism of Rb gene expression (8–12). For example, induction of Rb gene transcription by MyoD, via CREB, is a key event in muscle differentiation (9). Furthermore, transcriptional activation of the Rb gene by GABP and HCF-1 is also important in muscle differentiation (10, 11). In contrast, YY1 and MIZF repress transcription of Rb, and this repression is important for inhibiting myogenesis (10, 12). All these data suggest that multiple transcription factors act on the transcriptional regulation of Rb gene.
We have been investigating the biological functions of FBI-1 (also called Pokemon/ZBTB7A), which contains a BTB/POZ-domain at its N terminus and Krüppel-like zinc fingers at its C terminus (13, 14). Recently, there have been several reports on the function of FBI-1. FBI-1 stimulates the Tat activity of human immunodeficiency virus, type 1 long terminal repeat and represses human ADH5/FDH gene expression by interacting with Sp1 zinc fingers (14, 15). The mouse counterpart of FBI-1, LRF, co-immunoprecipitates and co-localizes with Bcl-6, and is involved in chondrogenesis and adipogenesis (16–18). The rat homolog of FBI-1, OCZF, is a transcriptional repressor and is involved in osteoclastogenesis (19). FBI-1 enhances NF-κB-mediated transcription through an interaction between the POZ-domain of FBI-1 and the RHD of NF-κB (20). Recently, FBI-1 was identified as a proto-oncogene (21). Serial analysis of gene expression analysis showed that the expression of FBI-1 is increased in cancer tissues. In transgenic mice overexpressing FBI-1, FBI-1 represses transcription of tumor suppressor gene, ARF, by binding to its promoter region. The product of ARF is a transcriptional activator of p53, another tumor suppressor. Thus, repression of ARF can eventually inhibit expression of p53, promoting oncogenesis in the thymus, liver, and spleen. In FBI-1 knockout mice, overexpression of ARF increases expression of p53, induces senescence, apoptosis, and eventually blocks cellular differentiation (21). FBI-1 is overexpressed in solid tumors, such as cancers of the colon and bladder, in which the normal function of the ARF/p53 pathway is frequently lost. It is likely that FBI-1 has multiple additional target genes by which it can exert oncogenic activity (22 and references therein).
We suspected that FBI-1 might be involved in the transcriptional regulation of the genes involved in differentiation, cell cycle control, and tumor suppression, such as Rb. We found that the Rb gene is the molecular target of proto-oncogene FBI-1, and we investigated the molecular mechanism of transcriptional regulation in detail.
EXPERIMENTAL PROCEDURES
Plasmids Construction—pGL3-Enhancer-Rb-Luc plasmid was kindly provided by Dr. Masayuki Sekimata (Fukushima Medical University, Japan) (12, 23). The pGL2-Rb-Luc fusion plasmid was prepared by cloning the Rb promoter (bp –370 to +106) into the pGL2-Basic plasmid (Promega, Madison, WI). Various mutant Rb-Luc plasmids were prepared using a site-directed mutagenesis kit (Stratagene). Expression plasmid vectors for the VP16-co-repressors, BCoR (aa 112–753), NCoR (aa 1709–2215), and SMRT (aa 194–657) fusion proteins (pKH135EF-BCoR, pKH73/110EF-NCoR, and pCMX-SMRT) were reported elsewhere (24). Construction of pcDNA3-FBI-1, pcDNA3-FBI-1ΔPOZ, and pG5-Luc were reported elsewhere (14). To prepare the recombinant GST-POZFBI-1 fusion protein expression vector, a cDNA fragment encoding the POZ-domain of FBI-1 was subcloned into pGEX4T3 (Amersham Biosciences) (14). The expression vectors for Sp1ZFDBD (zinc finger DNA binding domain) (aa 624–718) and FBI-1ZFDBD (aa 366–495) was prepared by cloning the PCR-amplified cDNA fragments into pGEX4T1 (24, 25). The oligonucleotide primers used for PCR amplification of FBI-1ZFDBD are as follows: forward, 5′-GATCGGATCCGCCTGGTCGCAGAAGGTGGAG-3′ and reverse, 5′-GATCGTCGACCGAGGGGACGCCGTTGCAGCCGTC-3′.
Mammalian expression vectors for the co-repressors used in GST-pulldown assays were prepared by subcloning the cDNA fragments encoding BCoR (aa 112–753), NCoR (aa 1709–2215), and SMRT (aa 194–657) into pcDNA3.0 (Invitrogen) as reported elsewhere (24). The FBI-1 expression vector in Drosophila SL2 cells was prepared by cloning FBI-1 cDNA into pPac-PL provided by Dr. Carl Thummel (University of Utah). pPac-Sp1 was provided by Dr. Robert Tjian (University of California, Berkeley, CA).
Cell Culture/Stable Cell Line—HeLa cells were cultured in Dulbecco's modified Eagle's medium (DMEM, Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen). Stable HeLa cells overexpressing FBI-1 were prepared by transfecting HeLa cells with a recombinant Lenti virus, LentiM1.4-FBI-1-FLAG. Control stable cells were prepared using recombinant LentiM1.4-LacZ virus. Drosophila SL2 cells were cultured in Schneider's medium (Invitrogen) supplemented with 10% fetal bovine serum.
Transcription Analysis of the Rb Gene Promoter—HeLa, HCT116, and HEK 293A cells were cultured with the medium recommended by the ATCC (Manassas, VA) on 6-well culture vessels and were transfected with 0.3 μg of pGL2-Rb-Luc Wt or mutants and 0.5 μg each of pcDNA3.0, pcDNA3.0-FBI-1, and pcDNA3.0-FBI-1ΔPOZ using Lipofectamine Plus reagent (Invitrogen). After 36 h of incubation, cells were harvested, lysed, and assayed for luciferase activities. Luciferase activity was normalized with the protein concentration of the lysate.
Electrophoretic Mobility Shift Assays—EMSAs were carried out as described previously (24). Binding reactions were carried out in 20 μl of binding buffer containing 10 mm HEPES, pH 7.9, 60 mm KCl, 5 μm ZnCl2, 1 mm dithiothreitol, 1% bovine serum albumin, 7% glycerol, 0.1 μg of recombinant FBI-1ZFDBD or Sp1ZFDBD, and 10,000 cpm of probe at room temperature for 30 min. Antibodies against GST-Tag, FBI-1, or Xpress were added to binding reactions for supershift of the probe-protein complex. The sequences of FREs and Sp1-binding GC-box oligonucleotides were as follows (only the top strand sequences are shown): FRE1, 5′-GATCGGATGAGGCCCACAGTCACC-3′; FRE2, 5′-GATCCCACAGTCACCCACCAGACT-3′; FRE3, 5′-GATCAGGGGGTGGTTCTGGGTAGA-3′; FRE4, 5′-GATCCGCCTGGACCCACGCCAGGT-3′; GC-box 1, 5′-GATCACGGGCGGAAGTGAC-3′; and GC-box 2, 5′-GATCAGTTGCCGGGCGGGGGAGGG-3′.
Site-directed Mutagenesis of the Rb Promoter—To investigate the role of FBI-1 binding sites on transcription, mutations were introduced into the promoter sequence using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). The core GGG of the binding sequences (5′-GDGGGYYYY-3′) of the FREs and the GC-boxes was replaced with AAA using the following mutagenesis oligonucleotides primers (only the top strands are shown): mFRE1, 5′-CCCGGGATAGGGATGAAATTTACAGTCACCCACCAGA-3′; mFRE2, 5′-ATGAGGCCCA; CAGTCATTTACCAGACTCTTTGTAT-3′; mFRE3, 5′-CACCCCGGCCTGGAGGAAAAAATTCTGGGTAGAAGCAC-3′; mFRE4, 5′-CTGGAAGGCGCCTGGATTTACGCCAGGTTTC; CCAG-3′; mGC-Box 1, 5′-ACGTGACGCCGCGGGCAAAAGTGACGTTTTCCCG-3′; and mGC-Box 2, 5′-CGCTCAGTTGCCGGGCAAGGGAGGGCGCGTCCGG-3′.
Mammalian Two-hybrid Assays—African green monkey kidney cells (CV-1) were grown in DMEM supplemented with 10% fetal bovine serum (Invitrogen). CV-1 cells were transiently transfected with pCMX-Gal4-POZFBI-1, pCMX-VP16-BCoR, pCMX-VP16-NCoR, pCMX-VP16-SMRT, and pG5-Luc using Lipofectamine Plus reagent (Invitrogen). Cells were cultured for 36 h. The cells were then harvested, lysed, and analyzed for luciferase activity on a luminometer (Microplate Luminometer LB 96V, EG&G Berthold, MD). Luciferase activity was normalized with the cotransfected β-galactosidase activity or protein concentration.
GST Fusion Protein Pulldown Assays—Escherichia coli BL21(DE3), transformed with either GST or GST fusion protein expression vector, was induced with 0.5 mm isopropyl-1-thio-β-d-galactopyranoside for 4 h at 37°C. After E. coli pellets were lysed and sonicated in lysis buffer (1× phosphate-buffered saline, 1 mm phenylmethysulfonyl fluoride, 2 mm EDTA, and 0.2 mg/ml lysozyme), recombinant proteins were purified by affinity chromatography using glutathione-agarose 4 beads (Peptron, Daejeon, Korea). Purified proteins were resolved on a 12% SDS-PAGE gel to quantitate them and assess their purity. The same volume of protein-agarose bead complex was used for all GST fusion protein pulldown assays.
Co-repressor proteins were prepared in vitro by incubating 1 μg of the pcDNA3.0 co-repressor expression vector with TnT Quick-coupled Transcription/Translation Extract (Promega), containing 40 μl of TnT Quick Master Mix and 2 μl of [35S]methionine (1175.0 Ci/mol, PerkinElmer Life Sciences) at 30 °C for 90 min.
Purified GST fusion proteins (5 μg) were incubated with GSH-agarose for 1 h at 4 °C in HEMG buffer (40 mm HEPES, pH 7.9, 100 mm KCl, 0.2 mm EDTA, 5 mm MgCl2, 0.1% Nonidet P-40, 10% glycerol, 1.5 mm dithiothreitol, protease inhibitor mixture (1 tablet/50 ml)). After agarose-GST protein complexes were washed, 10 μl of in vitro translated [35S]methionine-co-repressor was added, and this mixture was incubated in HEMG buffer for 4 h at 4°C. The reactions mixtures were centrifuged, and the pellets were washed thoroughly with cold binding buffer. Bound proteins were separated on a 12% SDS-PAGE gel, and the gels were exposed to x-ray film using an image-intensifying screen (Kodak).
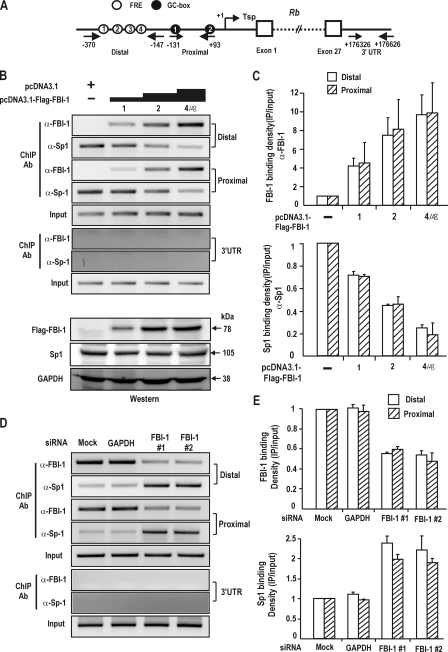
Chromatin Immunoprecipitation Assays—The binding of FBI-1 to the FRE elements on the Rb promoter in vivo was analyzed by ChIP assays. Sub-confluent HeLa cells on a 10-cm dish were transfected with 3μg of pcDNA3.0 or pcDNA3.0-FBI-1-FLAG using Lipofectamine Plus (Invitrogen) and grown for an additional 48 h. The HeLa cells were treated with formaldehyde (final 1%) to cross-link FBI-1totheRbpromoter. The remainder of the ChIP assay procedure was performed as reported previously (14). To amplify the proximal and distal promoter regions of the Rb gene, PCR reactions of the immunoprecipitated DNA were carried out using the following sets of oligonucleotide primers: distal FRE region (bp –370 to –147: forward primer, 5′-CACTAGCCAGATATTCCCTGCGGGG-3′ and reverse primer, 5′-TAAGTCATGAGGAATTAAACTGGGA-3′), proximal GC-box/FRE region (bp –131 to +93: forward primer, 5′-CACCGACCAGCGCCCCAGTTCCCCA-3′andreverseprimer, 5′-GGGAGGACGCGCGCGCACGTCG-3′) and downstream 3′ UTR region (bp +176326 to +176626: forward primer, 5′-GGATCTCAGGACCTTGGTGG-3′ and reverse primer, 5′-AGGGCCATTCTTACTATCCA-3′).
The binding competition between FBI-1 and Sp1 on the Rb promoter in vivo were analyzed by ChIP assays in the HEK293A cells transfected with pcDNA3.1-FBI-1 expression vector (1, 2, and 4 μg) or knockdown siRNA and also in Drosophila SL2 cells. Drosophila SL2 cells lacking mammalian transcription factors were transfected with pGL2-Rb-Luc, pPac-PL, pPac-Sp1 (2 μg), and pPac-FBI-1 (1, 2, 4 μg) using Cellfectin (Invitrogen) and grown for an additional 48 h. The cells were fixed with formaldehyde (final 1%) to cross-link FBI-1 and Sp1 protein onto the Rb promoter. The remaining ChIP procedure was performed as described previously (14). PCR was performed using the following cycling conditions: denaturation at 94 °C for 5 min, followed by 35 cycles of 94 °C for 30 s, 55 °C for 30 s, 72 °C for 30 s, and a final extension reaction at 72 °C for 5 min. We investigated whether co-repressor (BCoR) binding to the Rb promoter is dependent on FBI-1 using ChIP. HEK293A cells grown on 10-cm dishes were transfected with pcDNA3-FLAG-FBI-1 (3 μg) and/or pCMV-BCoR (3 μg). Chromatins were immunoprecipitated using anti-Myc or anti-FLAG antibody; the remaining ChIP procedures were performed as described above. Oligonucleotide primers used to amplify the distal FRE cluster region (bp –370 to –147) are described above.
We also investigated whether the acetylation status of histones H3 and H4 of nucleosomes at the proximal Rb gene promoter are modified by ectopic FBI-1 using antibodies specific to Ac-Histone H3 and Ac-Histone H4 (Upstate). The oligonucleotide primers used to amplify the Rb gene promoter region (bp –133 to +167) flanking the transcription start site were as follows: forward, 5′-CACCGACCAGCGCCCCAGTTCCCCA-3′; reverse, 5′-CCTGACGAGAGGCAGGTCCT-3′.
Knockdown of FBI-1 mRNA by siRNA—The siRNAs designed to knock down glyceraldehyde-3-phosphate dehydrogenase and FBI-1 were purchased from Ambion Inc. (Austin, TX). Their sequences were as follows: siFBI-1 #1, 5′-GCUGGACCUUGUAGAUCAAtt-3′ and 5′-UUGAUCUACAAGGUCCAGCtt-3′; siFBI-1 #2, 5′-AGUACCUCGAGUUCUUCCAtt-3 and 5′-UGGAAGAACUCGAGGUACUtt-3′. The siRNAs for FBI-1 (20 ng of each) were transfected into 6 × 106 HEK293A cells using Lipofectamine 2000 (Invitrogen). After culturing for 72 h, cells were harvested, and analyzed for protein and mRNA expression by Western blot and RT-PCR, respectively.
Co-immunoprecipitation Assays—The FLAG-FBI-1 and Myc-BCoR expression vectors were co-transfected, and cells were washed, pelleted, and resuspended in lysis buffer supplemented with protease inhibitors (20 mm Tris-HCl, pH 7.5, 150 mm NaCl, 10% glycerol, 1% Triton X-100). Cell lysates were pre-cleared, and the supernatant was incubated with anti-Myc antibody on a rotating platform overnight at 4 °C, followed by incubation with protein A-agarose beads (Santa Cruz Biotechnology, Santa Cruz, CA). Beads were collected, washed, and resuspended in equal volume of 5× SDS loading buffer (10% SDS, 10 mm dithiothreitol, 20% glycerol, 0.2 m Tris-HCl, pH 6.8, 0.05% Bromphenol Blue). Immunoprecipitated proteins were separated by 12% SDS-PAGE. Western blot analysis was performed as described under “Western Blot Analysis.”
Reverse Transcription and PCR—Total RNA was isolated from the cells using TRIzol® reagent (Invitrogen). The DNAs were synthesized in a 20-μl reaction tube containing 5 μg of total RNA, 10 pmol of random hexamer, and 200 units of superscript reverse transcriptase II. PCR cycling conditions were as follows: denaturation at 94 °C for 5 min, 23 cycles of at 94 °C for 30 s, 62 °C for 30 s, and 72 °C for 40 s, and a final extension at 72 °C for 7 min. The PCR primers used for FBI-1 cDNA were 5′-GGCCTGCTGTGCGACGTGGT-3′ and 5′-CAGCAGGCGGGCGGCGCTGA-3′. The primers for Rb were 5′-AAAGAAAAAGGAACTGTGGG-3′ and 5′-AACTGCTGGGTTGTGTCAAA-3′. The primers for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were 5′-ACCACAGTCCATGCCATCAC-3′ and 5′-TCCACCACCCTGTTGCTGTA-3′.
Western Blot Analysis of Rb and FBI-1—Cells were harvested and lysed in TEN buffer (10 mm Tris-HCl, pH 8.0, 1 mm EDTA, and 0.1 m NaCl). Cell extracts (40 μg) were separated on a 10% SDS-PAGE gel, transferred to Immun-Blot™ polyvinylidene difluoride membrane (Bio-Rad), and blocked with 5% skim milk (BD Biosciences) in TBST (20 mm Tris-HCl, pH 7.5, 140 mm NaCl, and 0.001% Tween 20) for 10 min. Blotted membranes were incubated with Ab-FBI-1 overnight at 4 °C (Abcam), Ab-Rb (BD Biosciences), GAPDH (Calbiochem), and Ab-α-tubulin (Calbiochem). Membranes were further incubated with horseradish peroxidase-conjugated anti-mouse IgG (Vector) or anti-rabbit IgG (Vector) or anti-goat IgG (Santa Cruz Biotechnology), and developed with ECL reagents (PerkinElmer).
C2C12 Myoblast Cell Culture, Differentiation, and Western Blot Analysis—Mouse C2C12 myoblast cells were maintained in DMEM (Invitrogen) supplemented with 10% fetal bovine serum. Stable C2C12 cells overexpressing FBI-1 were prepared by transfecting C2C12 cells with a recombinant LentiM1.4-FBI-1-FLAG virus. To induce differentiation of the C2C12 cells into myotubes, medium was switched to DMEM with 2% horse serum as described elsewhere (12). For the Rb rescue experiment, recombinant adenovirus overexpression Rb was purchased from Vector Biolabs (Philadelphia, PA) and infected at 100 multiplicity of infection.
RESULTS
FBI-1 Represses Transcription of the Rb Gene—FBI-1 shows proto-oncogenic activity by repressing the expression of the tumor suppressor ARF gene (21). We suspected that ARF might not be the only target gene of transcription repression and that FBI-1 may have multiple additional target genes by which it can exert oncogenic activity (22 and references therein). We investigated whether other tumor suppressor genes, such as the Rb gene, which is critically involved in cell cycle control and tumor suppression, is the target of FBI-1. Using the recently characterized FBI-1 consensus sequence, we identified four potential FBI-1 binding sites (FRE) on the Rb promoter. The four potential FREs are GC-rich and have varying degrees of homology with the consensus FRE sequence with either perfect homology or with one or two mismatches (Fig. 1A) (21, 26).
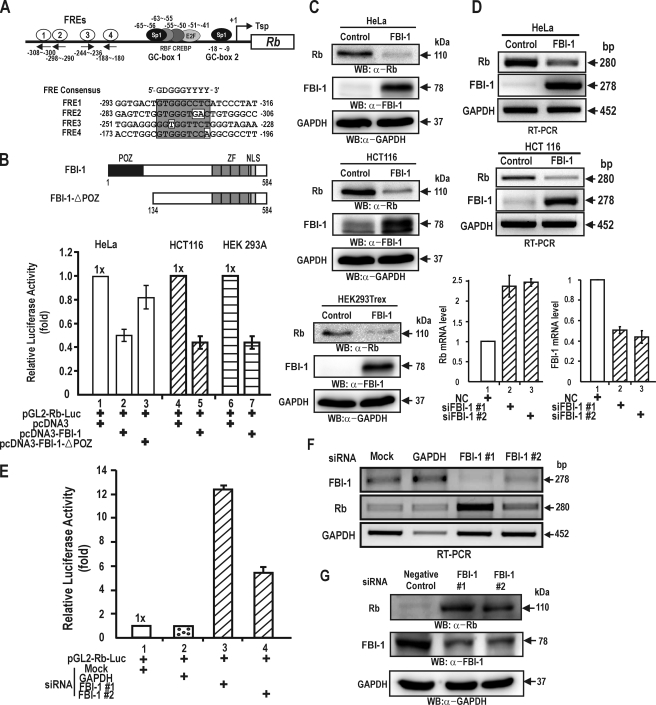
FIGURE 1.
FBI-1 represses transcription of the Rb gene. A, structure of the Rb gene promoter and sequence comparison of four potential FBI-1 binding sites (FRE). Transcription factor binding sites are indicated by circles. The FREs are located in the –180 to –308 bp region. FRE; FBI-1 binding site; Tsp (+1), transcription start site. B, structures of FBI-1 and the FBI-1 with a POZ-domain deletion. FBI-1 repressed transcription of the Rb gene by more than 50% in HeLa, HCT116, and HEK 293A cells. POZ, POZ-domain; ZF, Krüppel-like zinc fingers; NLS, nuclear localization sequence. C and D, RT-PCR and Western blot analysis of Rb gene repression by FBI-1 at mRNA and protein levels in stable HeLa and HCT116 cells overexpressing FBI-1 or β-galactosidase. GAPDH, control for RT-PCR and Western blot analysis. Histogram of the qRT-PCR analysis of FBI-1 and Rb mRNA is shown below. NC, negative control siRNA. E, transient transcription assays in HEK293A cells transfected with pGL2-Rb-Luc and siRNA designed to knock down endogenous FBI-1. Mock, negative control siRNA; GAPDH, positive control siRNA against GAPDH mRNA; FBI-1#1 and #2, siRNA designed to knock down endogenous FBI-1. F and G, RT-PCR and Western blot analysis of RNA interference of endogenous FBI-1 mRNA and protein expression. Knockdown of FBI-1 mRNA by siRNA increased Rb gene expression both at the mRNA and protein levels in HEK293A cells.
We transiently co-transfected HeLa cells with two FBI-1 expression plasmids (pcDNA3-FBI-1 and pcDNA3-FBI-1ΔPOZ) and the pGL2-Rb-Luc plasmid, and analyzed the luciferase activity of these transfected cells. FBI-1 repressed transcription of Rb by >50% relative to the control. FBI-1 lacking the POZ-domain had a much weaker effect, repressing the transcription of Rb by 18% relative to the control, suggesting that the POZ-domain is important in transcriptional repression of Rb gene (Fig. 1B). In HeLa cells, human papilloma virus E7 is known to influence the activity of Rb. One may argue that FBI-1 affects papilloma virus E7 and thereby affects Rb expression. Consequently, we carried out transcription assays in human colon cancer HCT116 cells and HEK293A cells, which do not have papilloma virus E7. In these two cell types, FBI-1 also repressed Rb gene transcription, suggesting that FBI-1 directly acts on the Rb promoter to repress transcription (Fig. 1B).
We also investigated whether FBI-1 represses Rb gene expression in stable HeLa and HCT116 cells overexpressing FBI-1 due to transfection with the recombinant control Lentivirus, LentiM1.4-FBI-1-FLAG. RT-PCR and Western blot analysis showed that FBI-1 decreased the mRNA and protein levels of the Rb gene, respectively, compared with those of the control cells transfected with the control Lentivirus-LacZ virus (Fig. 1, C and D, and supplement Fig. S1).
Alternatively, we tried to show whether knockdown of endogenous FBI-1 mRNA expression could increase transcription of Rb by removing the transcription repression by FBI-1. Successful knockdown of endogenous FBI-1 mRNA by RNA interference using two independent siRNAs resulted in an increase in Rb gene transcription (Fig. 1E). RT-PCR and Western blots showed that siRNA treatment resulted in a significant decrease in endogenous FBI-1 mRNA and FBI-1 protein and a concomitant increase in Rb mRNA and protein expression, respectively (Fig. 1, E–G). These data suggested that FBI-1 is a transcriptional repressor of Rb gene expression.
FBI-1 Binds to the Rb Promoter Elements in Vivo and in Vitro—Having shown that FBI-1 can repress Rb gene transcription, we investigated whether FBI-1 could bind to the four potential FRE elements by EMSA and ChIP assays. Four [α-32P]dATP-labeled FRE oligonucleotide probes (bp –308 to –300, bp –298 to –290, bp –244 to –236, and bp –188 to –180) were allowed to interact with recombinant GST-FBI-1ZFDBD (zinc finger DNA binding domain) (aa 366–495). The FBI-1ZFDBD bound to the FRE 2, 3, and 4 probes, but bound the FRE 1 probe very weakly compared with the other FREs. Cold competitor oligonucleotides competed well, and the addition of GST-specific antibody caused the retarded band to disappear, as reported for Sp1ZFDBD, FBI-1ZFDBD, and NF-κB (Fig. 2A) (14, 27).
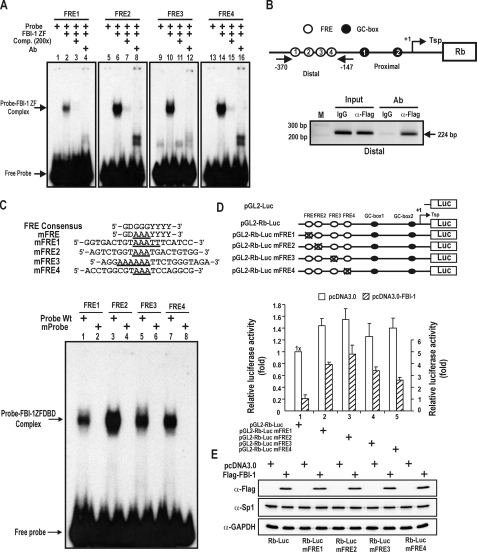
FIGURE 2.
FBI-1 binds to the distal FRE clusters of the endogenous Rb promoter. Characterization of the functional role of the FRE elements in transcription. A, EMSA. Four 32P-labeled FRE1–4 oligonucleotide probes (bp –308 to –300, –298 to –290, –244 to –236, and –188 to –180) were incubated with recombinant FBI-1ZFDBD (aa 382–490) and separated on a 4% non-denaturating polyacrylamide gel. B, ChIP assays of FBI-1 binding at the FRE cluster on the endogenous Rb gene. Structure of the Rb gene promoter and locations of PCR primer sets used for ChIP assays. pcDNA3-FLAG-FBI-1 expression plasmid was transfected into HeLa cells, and chromatin was immunoprecipitated with anti-FLAG antibody or control IgG. Arrows, primers used in ChIP; Tsp (+1), transcription start site. C, EMSA. Sequence comparisons of the FRE consensus sequence and mutant FRE probes. The core GGG sequence of the FREs was substituted with AAA in mutant probes. EMSA was carried out as in A. FBI-1ZFDBD was able to bind to the wild type, but not to any of the mutant probes. D, functional characterization of FBI-1 binding to FRE elements by transient transcription analysis in HeLa cells. Also shown are the structures of the pGL2-Rb-Luc Wt and mutant plasmids tested. Open circles, FBI-1 binding sites; black circles, Sp1-binding GC-boxes. X, mutation introduced. Mutations of the FREs increased transcription of Rb gene by 20–130% compared with the Wt promoter. E, Western blot analysis of Sp1 and FBI-1 in the HeLa cells transfected and described in D.
We further investigated whether FBI-1 binds to the endogenous Rb promoter in vivo using a ChIP assay. We transfected a FLAG-FBI-1 expression vector into HeLa cells. ChIP assays showed that the antibody against the FLAG-tag precipitated the distal four FRE cluster (bp –370 to –147) region. ChIP assays with IgG antibody as a control did not result in the precipitation of the distal region of the Rb gene promoter (Fig. 2B).
FBI-1 Binds to the FREs and This Binding Is Important for Transcription Repression—To investigate the functional importance of FBI-1 binding to the FRE elements in vivo, we mutated the FREs and analyzed the effect of these mutations on Rb gene transcription. First, we tested whether the mutations we introduced actually prevented FBI-1 from binding to the mutated FRE elements in vitro. We prepared mutant FRE oligonucleotides in which the core GGG sequence of the FREs was replaced with AAA (Fig. 2C). We analyzed the binding interactions between FBI-1 and FRE Wt or mutant FRE probes by EMSA. FBI-1 bound well to all the FRE Wt probes as in Fig. 2A, but FBI-1 did not bind to any of the mutant FREs, which indicates that the core GGG sequence is critical for FBI-1 binding (Fig. 2C).
Having shown that FBI-1ZFDBD does not bind to mutant probes, we introduced mutations into the promoter of the Rb gene by site-directed mutagenesis. We transiently transfected HeLa cells with Wt or mutant pGL2-Rb-Luc plasmids to investigate the in vivo function of the FRE1–4 elements. Mutations of all four FREs increased the transcriptional activity of the Rb promoter compared with the Wt promoter by 26–55%. And the transcriptional repression of the Rb promoter by FBI-1 was significantly decreased compared with the Wt promoter. In particular, mutation of the FRE2 sequence resulted in relatively potent derepression of Rb transcription, indicating that FRE2 may be the most important element in FBI-1-mediated transcription repression (Fig. 2D). In support of this, the FRE2 probe bound strongly to FBI-1ZFDBD (Fig. 2, A and C). These data suggested that FBI-1 acts as a transcriptional repressor of Rb by binding to all of the FREs of the Rb gene promoter in vivo. FBI-1 binds relatively strongly to the FRE2 element, and the FRE2 element appears more important for transcription repression of Rb transcription by FBI-1. Potentially the expression of Sp1 can be affected by the overexpressed FBI-1, which may affect Rb gene transcription. Western blot showed that ectopic FBI-1 does not affect Sp1 expression. Accordingly, the decrease in the transcription repression was likely caused by the decrease in FBI-1 binding on the FREs.
FBI-1 Binding to the Sp1-binding GC-box 2 Is Important for Transcriptional Repression—Having shown that FBI-1 is a GC-box-binding protein that could repress Rb gene transcription, we investigated whether FBI-1 could bind to the Sp1-binding GC-boxes of the proximal promoter of endogenous Rb gene in vivo, using a ChIP assay. We transfected a FLAG-FBI-1 expression plasmid into HeLa cells. ChIP assays showed that the antibody against the FLAG tag precipitated the proximal promoter region (bp –131 to +93). ChIP assays with IgG antibody as a control did not result in precipitation of the proximal region of the Rb gene promoter (Fig. 3A). To investigate the functional significance of FBI-1 binding to the GC-box element in vivo, we mutated GC-boxes 1 and 2 and analyzed the effect of this on Rb gene transcription and also possibly on the transcription repression by FBI-1 (Fig. 3B).
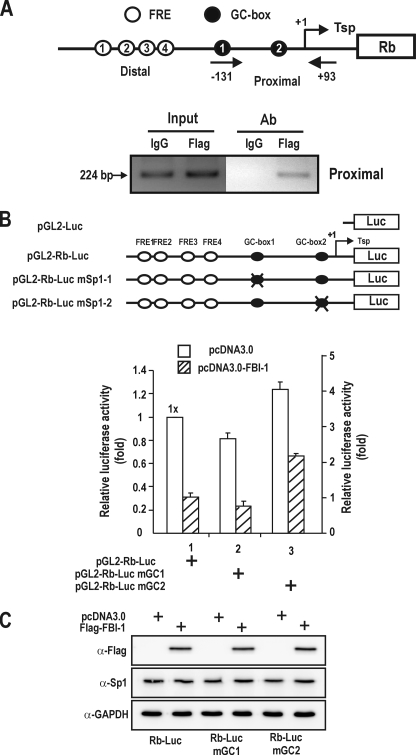
FIGURE 3.
FBI-1 binds to the proximal Rb promoter and may repress transcription via the GC-box 2 in vivo. A, ChIP assays of FBI-1 binding at the proximal promoter (bp –131 to +93) of the endogenous Rb gene. Structure of the Rb gene promoter and locations of PCR primer sets used for ChIP assays. pcDNA3-FLAG-FBI-1 expression plasmid was transfected into HeLa cells and chromatin was immunoprecipitated with anti-FLAG antibody or control IgG. Arrows, primers used in ChIP; Tsp (+1), transcription start site. B, structures of pGL2-Rb-Luc Wt and mutant plasmids and transcription analysis. Mutations were introduced into the proximal promoter. Open circles, FBI-1 binding sites; filled black circles, Sp1-binding GC-boxes; X, mutation introduced. Mutation of the GC-box 1 decreased transcription, but mutation of GC-box 2 increased Rb gene transcription compared with the Wt promoter, which suggests that FBI-1 may bind to GC-box 2 and repress transcription. C, Western blot analysis of Sp1 and FBI-1 in the HeLa cells transfected and described in B.
We transiently transfected HeLa cells with Wt or mutant pGL2-Rb-Luc plasmids to investigate in vivo function of the Sp1-binding GC-boxes 1 and 2. Mutation of the Sp1-binding GC-box 1 (bp –65 and –56) reduced luciferase gene expression compared with the Wt Rb promoter. These data indicate that the first Sp1 binding site is important for transcriptional activation by Sp1, as reported previously (28). Unexpectedly, mutation of the second Sp1-binding GC-box 2 (bp –18 and –9) resulted in increased luciferase activity compared with the Wt Rb promoter (Fig. 3B). The data suggest that the second Sp1-binding site may be occupied by repressors such as FBI-1 and/or repressors of the Sp1-family. Mutation of the GC-box 2 increased the transcriptional of the Rb promoter by 2.2-fold compared with the Wt promoter in the presence of FBI-1 (Fig. 3B). These data indicate that GC-box 2 may be also the element in FBI-1-mediated transcription repression. In support of this, the GC-box 2 probe bound strongly to both Sp1 ZFDBD and FBI-1ZFDBD, compared with GC-box 1 (Fig. 4, C and D). As in Fig. 2, ectopic FBI-1 does not affect the expression of Sp1.
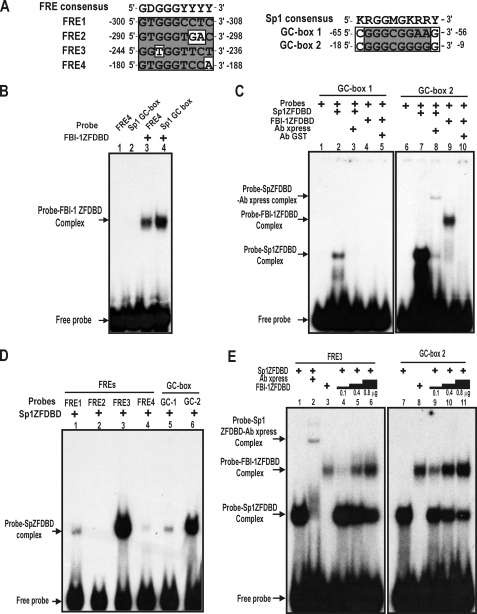
FIGURE 4.
FBI-1 binds to the proximal Sp1-binding GC-box 2, and Sp1 binds to the distal FRE3 element in vitro. FBI-1 and Sp1 compete with each other for binding to FRE3 and GC-box 2. A, comparisons of the nucleotide sequences of the FREs and Sp1-binding GC-boxes with their consensus sequences. B, EMSA. FBI-1ZFDBD was incubated with FRE4 or the typical Sp1 GC-box probe. FBI-1 binds to the Sp1-binding GC-box probe. FRE4, control FBI-1 binding probe. Sp1 GC-box, a typical Sp1-binding oligonucleotide probe with 5′-GGGCGGGG-3′ core sequence. C, EMSA. Recombinant Sp1ZFDBD or FBI-1ZFDBD was incubated with the GC-box probes. Sp1 binds both GC-box 1 and 2, but binds more strongly to GC-box 2 (lanes 2 and 7). FBI-1 binds to the Sp1 GC-box 2 probe only. D, EMSA and identification of a new Sp1 binding element, FRE3. Recombinant Sp1ZFDBD (0.1 μg) was incubated with the FRE probes or the GC-box probes. Sp1 significantly binds to FRE3 and GC-box 2 probes. E, EMSA. FBI-1 and Sp1 compete for binding to FRE3 and GC-box 2 in vitro. The probes were incubated with Sp1ZFDBD (0.1 μg) in the presence of increasing amounts of FBI-1ZFDBD (0.1, 0.4, and 0.8 μg). FBI-1 competes with Sp1 for binding to FRE3 and GC-box 2 in vitro.
Identification of a New Sp1 Binding Element (FRE3) in the Distal FRE Cluster and Proximal Binding of FBI-1 to GC-box 2—Although binding of FBI-1 to FRE1–4 and the resulting recruitment of the co-repressor-HDAC complex by FBI-1 may explain the observed transcriptional repression of Rb, binding competition between the transcriptional activator Sp1 and the transcriptional repressor FBI-1 could also result in transcriptional repression. Because the deletion of the POZ-domain of FBI-1 did not result in the complete derepression of transcription, transcription factor binding competition toward the same regulatory element might contribute to the observed transcriptional repression in HeLa cells (Fig. 1B).
Interestingly, both FBI-1 and Sp1 bind to GC-rich sequences, and some of the binding sites may be bound by both FBI-1 and Sp1. First, we investigated whether FBI-1 could bind to the typical Sp1-binding GC-box probe (5′-GATCATTCGATCGGGGCGGGGCGAGC-3′; underlined is the core Sp1 binding sequence) using EMSA (29). FBI-1ZFDBD was able to bind to the typical Sp1-binding GC-box probe (Fig. 4B). Furthermore, we carried out EMSA to determine whether the two GC-boxes of the Rb promoter can be bound by FBI-1. Although FBI-1ZFDBD was unable to bind to GC-box 1, it bound to GC-box 2 (Fig. 4C), indicating that FBI-1 can bind to some GC-boxes.
In turn, we also investigated whether Sp1 could bind to some of the FREs. We found that Sp1 bound to the FRE3 quite strongly (Fig. 4D), which is juxtaposed with FRE2 to which FBI-1 bound most strongly (Fig. 2C, lane 3). Sp1 binds weakly to FRE1 and FRE4. Sp1 binds to GC-box 2 more strongly than to GC-box 1 (Fig. 4, C and D).
FBI-1 and Sp1 Compete for the FRE3 and the GC-box 2 in Vitro and in Vivo—FBI-1 bound to the GC-box 2 and Sp1 bound to the FRE3 and GC-box 2. Thus, binding competition between Sp1 and FBI-1 on these elements may be important in the transcriptional repression of Rb gene. Therefore, we performed EMSA binding competition assays for the FRE3 and GC-box 2. Sp1ZFDBD (100 ng) was added to the probes in the presence of increasing amounts of FBI-1ZFDBD (100, 400, and 800 ng). Increasing the amount of FBI-1 decreased the FRE3-Sp1 or GC-box 2-Sp1 interaction, demonstrating molecular competition between FBI-1 and Sp1 (Fig. 4E). Interestingly, FBI-1 competed effectively for GC-box 2 compared with FRE3, again indicating the strong binding affinity of Sp1 for FRE3.
ChIP Assays Reveal that FBI-1 and Sp1 Compete for the FRE 3 Element and GC-box 2 of the Rb Gene Promoter in Vivo—We investigated the molecular competition between FBI-1 and Sp1 on the endogenous Rb promoter and 3′-untranslated regions in human kidney HEK293A cells. ChIP assays of the proximal promoter region containing GC-boxes 1 and 2, and also of the distal promoter region containing four FREs, showed that Sp1 binding to the Rb promoter regions is decreased and FBI-1 binding is increased, as an increasing amount of FBI-1 expression vector is co-transfected (Fig. 5B). Additionally, we knocked down the expression of endogenous FBI-1 mRNA and analyzed the promoter binding by FBI-1 and Sp1 on the endogenous Rb promoter by ChIP. Knockdown of FBI-1 expression decreased FBI-1 binding both on the proximal and distal FBI-1 binding regions, whereas Sp1 binding on the two regulatory regions was increased (Fig. 5D). In the control 3′-UTR region, no ChIP or binding competition was observed (Fig. 5, B and D). The data again support the idea that FBI-1 and Sp1 can compete with each other in binding to the FRE 3 element and GC-box 2 of the Rb promoter in vitro and also in vivo (Figs. 4E and 5, B–D).
FIGURE 5.
ChIP assays. FBI-1 and Sp1 compete with each other for binding to FRE3 and GC-box 2 in vivo. A, structure of human Rb gene. Arrows, primers used in ChIP; Tsp (+1), transcription start site. Circles with number, FBI-1 binding FREs; filled black circles with number, GC-box that binds Sp1; open boxes, exons; 3′-UTR, 3′-untranslated region with no known binding sites for either Sp1 or FBI-1. B, ChIP assays of binding competition between Sp1 and FBI-1 for the distal (bp –370 to –147), proximal promoter (bp, –131 to +93), and 3′-UTR elements (bp +176326 to +176626) of the endogenous Rb gene promoter in HEK293A cells. Human HEK293A cells were transfected with increasing amount of FLAG-FBI-1 expression vector and analyzed for Sp1 and FBI-1 binding using the antibodies indicated. Sp1 and FBI-1 compete with each other both on the distal and proximal promoter regions. C, histogram of the ChIP assay shown in B. D, ChIP assays on the endogenous Rb gene promoter after knockdown of endogenous FBI-1 expression in HEK293A cells. Knockdown of FBI-1 resulted in an increase in Sp1 binding to the endogenous Rb gene promoter, both on the proximal and distal promoters. E, histogram of the ChIP assay shown in C.
We also investigated the molecular competition between FBI-1 and Sp1 on the Rb promoter in Drosophila SL2 cells. pGL2-Rb-Luc and pPac-Sp1 expression vectors and increasing amounts of pPac-FBI-1 expression vector were co-transfected into Drosophila SL2 cells that do not express mammalian transcription factors. ChIP assays of the proximal promoter region and the distal promoter region showed that Sp1 binding to the Rb promoter regions is decreased as an increasing amount of FBI-1 expression vector is co-transfected (supplemental Fig. S2A).
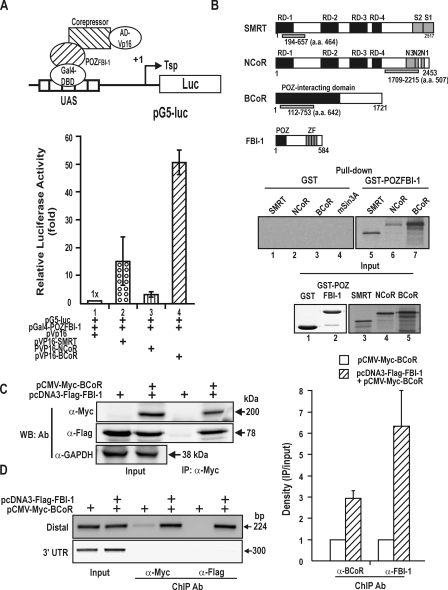
The POZ-domain of FBI-1 Interacts with the Co-repressors in Vitro and in Vivo—Because FBI-1 represses transcription of the Rb gene, we investigated whether the POZ-domain, which is important in transcriptional repression, interacted with the co-represssors using mammalian two-hybrid assays. We co-transfected CV-1 cells with pGal4-POZFBI-1, the reporter gene construct (pG5-Luc), and the pVP16AD-co-repressor (BCoR, NCoR, and SMRT) and analyzed for reporter luciferase activity. We found that the POZ-domain of FBI-1 was able to interact with BCoR, NCoR, and SMRT (Fig. 6A). To investigate whether the molecular interaction between the POZ-domain and the co-repressors was direct, we incubated recombinant GST-POZFBI-1or GST with in vitro synthesized [35S]methionine-labeled co-repressors and performed a pulldown assay. GST-POZFBI-1 interacted with BCoR, SMRT, and NCoR, suggesting that the molecular interaction between the POZ-domain and the co-repressors is direct (Fig. 6B).
FIGURE 6.
FBI-1 interacts with the co-repressors via its POZ-domain, and BCoR is recruited to the promoter by FBI-1. A, diagram of the mammalian two-hybrid assay and protein-protein interaction between the POZ-domain of FBI-1 and the co-repressors. Upstream activating sequence (UAS), binding site for Gal4DBD (B) GST fusion protein pulldown assays and structure of the co-repressors SMRT, NCoR, BCoR, and FBI-1. GST and GST-POZFBI-1 fusion proteins were incubated with in vitro synthesized [35S]methionine-labeled co-repressor polypeptides and pulled down. RD, repression domain; N3-1 and S1–2, domains involved in interaction with nuclear receptors; ZF, zinc finger domain; POZ, POZ-domain of FBI-1. Domains of co-repressors used for in vitro pulldown assays are indicated below by light gray filled bars. C, co-immunoprecipitation of FBI-1 and BCoR. Cell lysates prepared from HEK293A cells transfected with FLAG-FBI-1 and Myc-BCoR expression vectors were immunoprecipitated using anti-Myc antibody and analyzed by Western blotting using anti-FLAG antibody. D, ChIP assays of BCoR and FBI-1 binding at the distal FRE elements and 3′-UTR of Rb gene. HEK293A cells were transfected with FLAG-FBI-1 and/or Myc-BCoR expression vector and immunoprecipitated with the antibodies indicated. More BCoR is bound to the FRE cluster in the presence of FBI-1. A histogram of the ChIP assays is shown on the right.
Furthermore, we also examined whether FBI-1 can interact with a co-repressor by performing co-immunoprecipitation assays for BCoR. Cell extracts prepared from HEK293A cells transfected with FLAG-FBI-1 and/or Myc-BCoR expression vector were immunoprecipitated using anti-Myc antibody and analyzed by Western blot assay using antibody against Myc or FLAG. FBI-1 and BCoR interact with each other in vivo (Fig. 6C). Furthermore, we investigated whether the BCoR co-repressor is associated with the Rb gene promoter in an FBI-1-dependent manner. We transfected HEK293A cells with FLAG-FBI-1 or/and Myc-BCoR expression vectors. ChIP assays showed that BCoR binding to the FRE cluster (bp –370 to –147) was significantly increased in the presence of FBI-1. In contrast, in the absence of co-expressed FBI-1, BCoR binding was very weak. In the 3′-UTR control region, no ChIP of BCoR or FBI-1 was observed. These data suggested that the co-repressor BCoR was recruited by FBI-1 bound on the FRE cluster (Fig. 6D).
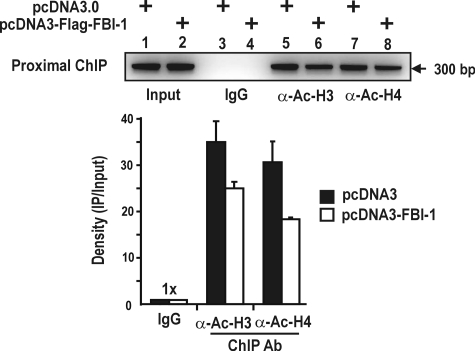
The FBI-1-co-repressor-HDAC Interaction Deacetylates Both Ac-H3 and Ac-H4 at the Proximal Promoter Region of the Rb Gene in Vivo—Because co-repressors repress transcription by recruiting HDACs, the molecular interaction between FBI-1 and co-repressors may be of importance in the transcriptional repression of the Rb gene by FBI-1. Accordingly, we investigated whether FBI-1 could deacetylate Ac-H3 and Ac-H4 histones bound at the proximal promoter region (bp –131 to +167). The ChIP assay showed that FBI-1 decreased acetylated histones 3 and 4 at the proximal promoter. The data suggest that histone deacetylation at the Rb proximal promoter is probably responsible for transcriptional repression by the complex containing FBI-1-co-repressor-HDAC (Fig. 7).
FIGURE 7.
FBI-1 deacetylates histones Ac-H3 and Ac-H4 at the proximal promoter of the endogenous Rb gene promoter. ChIP assays of H3 and H4 histone modification at the proximal promoter (bp –131 to +167) of the endogenous Rb gene using antibodies against Ac-H3 and Ac-H4. HEK293A cells were transfected with FLAG-FBI-1 expression vector, and immunoprecipitated with the antibodies indicated. A histogram of the ChIP assay is shown below.
Transcriptional Repression of Rb by FBI-1 Inhibits Myogenic Differentiation of C2C12 Myoblasts into Myotubes—Having shown that FBI-1 can repress Rb gene expression, we examined whether FBI-1 can affect the cellular processes controlled by Rb, such as myogenic differentiation. First, C2C12 cells were induced to differentiate by treatment with 2% horse serum. We analyzed the changes in the expression of Rb and FBI-1 by Western blot analysis. As C2C12 cells differentiated into myotubes, Rb gene expression increased and FBI-1 expression decreased (supplemental Fig. S3), suggesting a possible correlation between the two factors.
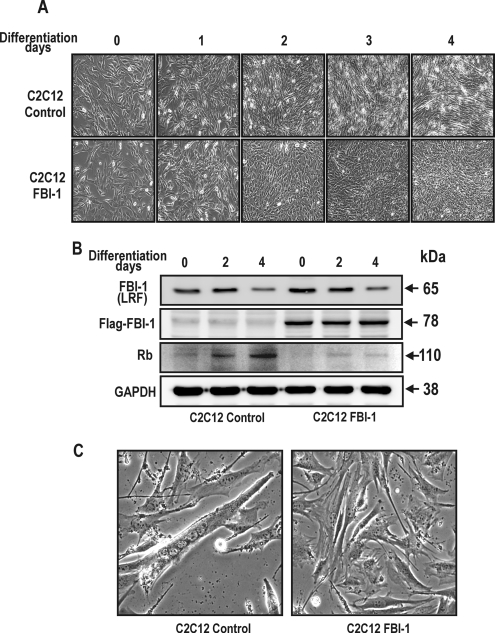
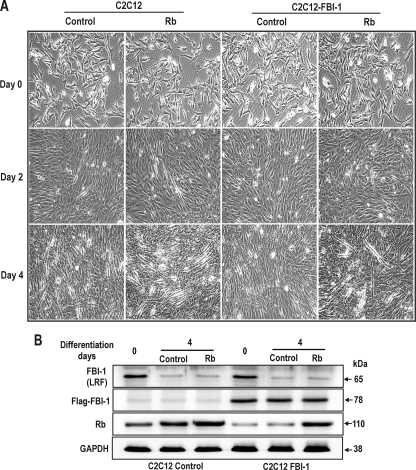
We examined the effect of ectopic FBI-1 expression on myoblast differentiation. We prepared stable cell lines by transfecting C2C12 cells with either recombinant Lenti-LacZ control virus or LentiM1.4-FLAG-FBI-1 virus. The stable control and FLAG-FBI-1 C2C12 myoblasts cells were treated with 2% horse serum to induce them to differentiate into myotubes. At day 0 of differentiation induction, there was no apparent change in the myoblast cell shape. However, after 3–4 days of culturing with differentiation medium, the cells showed marked differences in cell morphology. Abundant multinucleated myotubes were formed in the control cells, whereas very few myotubes were seen in C2C12 cells overexpressing FLAG-FBI-1. Western blot of the cell lysates at days 2–4 of differentiation showed that Rb protein was significantly decreased in the stable C2C12 cells (Fig. 8).
FIGURE 8.
FBI-1 inhibits mouse C2C12 myoblast differentiation by inhibiting Rb gene expression. A, differentiation of control mouse C2C12 myoblasts and stable C2C12 myoblasts overexpressing FLAG-FBI-1, by horse serum treatment. Pictures of the cells were taken at days 0–4 after treatment with 2% horse serum differentiation medium. Differentiation into myotubes was observed only in control cells at days 3 and 4. Stable C2C12 myoblast cells overexpressing FBI-1 were prepared by transfection with either control lentri virus or recombinant Lenti-M1.4-FLAG-FBI-1 virus. B, Western blot analysis of the C2C12 cell lysates using the antibodies indicated. Rb expression was gradually increased in the control cells with concomitant decrease in FBI-1 (LRF). LRF is a mouse homologue of human FBI-1. In the stable cells, although endogenous LRF is decreased, FLAG FBI-1 is stably expressed and repressed Rb expression. Cell extracts (40 μg) were analyzed by Western blot analysis. GAPDH, control; Rb, retinoblastoma protein. C, myogenic differentiation is inhibited by FBI-1 in the stable C2C12-FLAG-FBI-1 cells. However, control cells underwent myogenic differentiation and showed multinuclear myotubes at days 3–4.
Alternatively, we also examined whether inhibition of myogenic differentiation by transcription repression of Rb gene by FBI-1 could be rescued by transfection with recombinant adenovirus overexpressing Rb in the stable C2C12-FBI-1 myoblasts cells. After 2–4 days of culturing with differentiation medium, the cells showed differences in cell morphology and many myotubes were formed in the Rb rescued stable C2C12-FBI-1 cells, whereas few myotubes were seen in C2C12-FBI-1 cells transfected with control adenovirus. Western blot of the cell lysates at days 0 and 4 of differentiation showed that Rb protein was significantly increased in the rescued stable C2C12-FBI-1 cells (Fig. 9). Our data suggest that FBI-1 represses Rb gene expression and thereby blocks myogenic differentiation of C2C12 myoblasts.
FIGURE 9.
Inhibition of mouse C2C12 myoblast differentiation by FBI-1 can be rescued by transfection with recombinant adenovirus overexpressing Rb gene. A, differentiation of control mouse C2C12 myoblasts and stable C2C12 myoblasts overexpressing FLAG-FBI-1, by horse serum treatment. Pictures of the cells were taken at days 0, 2, and 4 after treatment with 2% horse serum differentiation medium. Differentiation into myotubes was observed only in control cells at day 4. Stable C2C12 myoblast cells overexpressing FBI-1 were differentiation into myotubes only when transfected with recombinant adenovirus overexpressing Rb gene at day 4, but not in the cells rescued with control adenovirus. Ad, recombinant adenovirus. B, Western blot analysis of the C2C12 control or C2C12-FBI-1 cell lysates using the antibodies indicated. Rb expression was increased in the control cells with concomitant decrease in FBI-1 (LRF). LRF is a mouse homologue of human FBI-1. In the stable cells, although endogenous LRF is decreased, FLAG FBI-1 is stably expressed and repressed Rb expression. Transfection with recombinant adenovirus increases Rb expression both in control and stable cells. Cell extracts (40 μg) were analyzed by Western blot analysis. GAPDH, control; Rb, retinoblastoma protein.
DISCUSSION
We are interested in determining and characterizing the biological functions of FBI-1 (14, 20). Recently, FBI-1 was characterized as a proto-oncogene causing multiple cancers in the thymus, liver, and spleen. FBI-1 represses ARF gene expression, which in turns results in the repression of the tumor suppressor p53 (21). We investigated whether FBI-1 has additional target genes through which it can exert its oncogenic activity. Initially, we suspected that FBI-1 might regulate other genes that have critical roles in oncogenesis and differentiation, such as Rb, a tumor suppressor gene that regulates the G1 checkpoint of the cell cycle (6 and references therein). Most functional studies of Rb have been performed at the protein level. However, investigations into how transcription of the Rb gene is regulated are also important for understanding the cellular regulation of Rb functions (8–12).
We found that FBI-1 can potently repress Rb gene transcription, and its POZ-domain is important for this transcription repression. The POZ-domain of FBI-1 interacts with co-repressors such as BCoR, NCoR, and SMRT. Co-represssors in turn interact with HDACs, which suggests that FBI-1 may recruit HDACs through co-repressors to deacetylate histones H3 and H4 around the proximal promoter region of the Rb gene, thereby repressing Rb transcription.
We also found that the FBI-1 is a GC-box-binding protein and binds directly to FRE-1, -2, -3, and -4. Unexpectedly, we also found that FBI-1 binds to GC-box 2. Among the GC-rich FREs and two GC-boxes, FRE1–4, and GC-box 2 are important for transcriptional repression by FBI-1. FBI-1 bound to the FRE2 most strongly, and mutation of FRE2 resulted in potent derepression. Therefore, FBI-1 might repress transcription of the Rb gene primarily by binding to the FRE2 and by recruiting co-repressors.
The FBI-1 binding consensus sequence (5′-GDGGGYYYY-3′) and Sp1 consensus sequence (5′-KRGGMGKRRY-3′) are GC-rich (21, 26, 29). Sp1 binds to FRE3, and FBI-1 binds GC-box 2. Sp1 and FBI-1 share some regulatory elements that are rich in GC, and binding competition between the two factors for these elements may determine the level of transcription. EMSA and in vivo ChIP assay data suggested that transcription of the Rb gene can be repressed by competitive binding between Sp1 and FBI-1 for elements in the proximal and distal promoter regions.
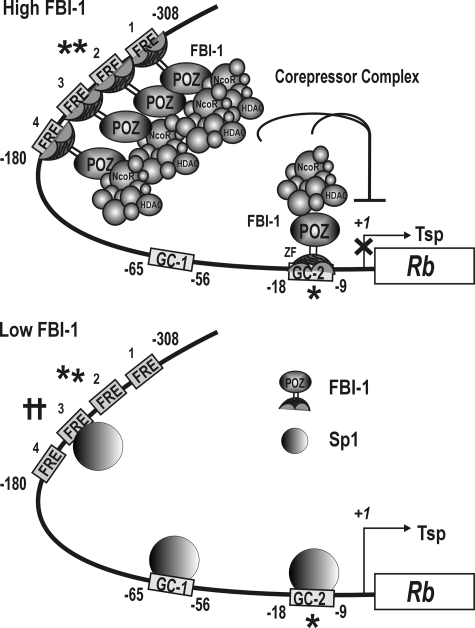
Rb gene transcription can be repressed by the combination of two molecular mechanisms: binding competition between FBI-1 and Sp1, and recruitment of co-repressors by the FBI-1 anchored onto FREs and/or GC-box 2. Based on our findings, we propose a hypothetical molecular mechanism of transcriptional regulation of the Rb gene by FBI-1 (Fig. 10). In cases where FBI-1 is highly expressed, such as in cancer cells, FBI-1 binds to FRE2 (and other FREs, but with slightly lower binding affinity) and GC-box 2 by competition with Sp1, thereby significantly decreasing Rb transcription; FBI-1 may also bind to FRE2, which does not involve binding competition with Sp1. Once anchored to FRE2, FRE3, FRE1, FRE4, and GC-box 2, the POZ-domain of FBI-1 can then recruit a co-repressor-HDAC complex, further repressing transcription through histone deacetylation.
FIGURE 10.
Hypothetical regulatory mechanism for the transcriptional regulation of Rb gene by FBI-1 and Sp1. When FBI-1 is highly expressed as in cancer cells, FBI-1 binds to the distal FREs and GC-box 2 on the Rb promoter by competing with Sp1. FBI-1 preferentially binds to FRE2 (⋆⋆). After FBI-1 binding, the co-repressor-HDAC complex is recruited by the POZ-domain of FBI-1 and represses transcription by deacetylating histones H3 and H4 of the nucleosomes around the proximal promoter. When FBI-1 is low, as in normal cells, Sp1 binds to FRE3 (††), a newly identified strong Sp1-binding element, GC-box 1, and GC-box 2 to activate transcription. FRE, FBI-1 binding element; GC-box, Sp1-binding GC-rich element; ⋆⋆, high affinity FBI-1 binding element; ⋆, high affinity FBI-1 binding GC-box2 element; ††, a newly identified high affinity Sp1-binding element; Tsp (+1), transcription start site; ⊥ transcription repression by histone modification at the proximal promoter.
When FBI-1 expression levels are low as in normal cells, FBI-1 cannot compete with Sp1 for FRE3 and GC-box 2, and these sites are open for Sp1 binding. The binding of Sp1 to FRE3, GC-box 1, and GC-box 2, will likely activate transcription of the Rb gene, and a certain cellular level of Rb gene transcription corresponding to the Sp1 level will be maintained. Also, transcription activators such as Sp1, ATF2, CREB1, and E2F1 bound at the –65 to –41 bp region can transcriptionally activate Rb resulting in cellular levels of Rb gene transcription (2–6).
Cellular regulators that regulate Rb gene expression can significantly affect cellular functions controlled by Rb. For example, MyoD via CREB induces Rb gene promoter transcription, which is a key event for muscle cell differentiation (9). Conversely, MIZF and YY1 repress transcription of Rb and inhibit muscle formation (10, 12, 23). In this report, we have shown that FBI-1 interferes with myoblastic C2C12 cell differentiation by inhibiting Rb gene transcription. We expect that FBI-1 could regulate other cellular functions such as the cell cycle, oncogenesis, and cell differentiation, by repressing Rb gene expression.
Supplementary Material
This work was supported by National Research Laboratory Grant ROA-2003-000-10318-0 (to M. W. H.) and Medical Research Center Grant R13-2002-054-05002-0 from Korea Science and Engineering Foundation of the Korea Ministry of Science and Technology. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
Footnotes
The abbreviations used are: Rb, retinoblastoma; ARF, alternative reading frame gene; Bcl-6, B-cell lymphoma-6; BCoR, BCL-6 interacting co-repressor; BTB/POZ, bric-a-brac tramtrack broad complex/poxvirus and zinc finger; ChIP, chromatin immunoprecipitation; CV-1, African green monkey kidney cell; DBD, DNA binding domain; DMEM, Dulbecco's modified eagle medium; FBI-1, factor that binds to the inducer of short transcripts of human immunodeficiency virus-1; FRE, FBI-1 response element; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GST, glutathione S-transferase; LacZ, β-galactosidase gene; Luc, luciferase gene; LRF, leukemia/lymphoma-related factor; NCoR, nuclear receptor co-repressor; Pokemon, POK erythroid myeloid ontogenic factor; SMRT, silencing mediator for retinoid and thyroid receptors; Sp1, specificity protein 1; RT, reverse transcription; Wt, wild type; ZF, zinc finger; ZFDBD, zinc finger DNA binding domain; aa, amino acid(s); siRNA, small interference RNA; CMV, cytomegalovirus; CREB, cAMP-response element-binding protein; HDAC, histone deacetylase.
References
- 1.Friend, S. H., Bernards, R., Rogelj, S., Weinberg, R. A., Rapaport, J. M., Albert, D. M., and Dryja, T. P. (1986) Nature 323 643–646 [DOI] [PubMed] [Google Scholar]
- 2.Lee, W. H., Bookstein, R., Hong, F., Young, L. J., Shew, J. Y., and Lee, E. Y. (1987) Science 235 1394–1399 [DOI] [PubMed] [Google Scholar]
- 3.Huang, H. J., Yee, J. K., Shew, J. Y., Chen, P. L., Bookstein, R., Friedmann, T., Lee, E. Y., and Lee, W. H. (1988) Science 242 1563–1566 [DOI] [PubMed] [Google Scholar]
- 4.Scambia, G., Lovergine, S., and Masciullo, V. (2006) Oncogene 25 5302–5308, review [DOI] [PubMed] [Google Scholar]
- 5.Goodrich, D. W., Wang, N. P., Qian, Y. W., Lee, E. Y., and Lee, W. H. (1991) Cell 67 293–302 [DOI] [PubMed] [Google Scholar]
- 6.Khidr, L., and Chen, P. L. (2006) Oncogene 25 5210–5219, review [DOI] [PubMed] [Google Scholar]
- 7.Nevins, J. R. (1992) Science 258 424–429 [DOI] [PubMed] [Google Scholar]
- 8.Gill, R. M., Hamel, P. A., Zhe, J., Zacksenhaus, E., Gallie, B. L., and Phillips, R. A. (1994) Cell Growth & Differ. 5 467–474 [PubMed] [Google Scholar]
- 9.Magenta, A., Cenciarelli, C., De Santa, F., Fuschi, P., Martelli, F., Caruso, M., and Felsani, A. (2003) Mol. Cell. Biol. 23 2893–2906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deléhouzée, S., Yoshikawa, T., Sawa, C., Sawada, J., Ito, T., Omori, M., Wada, T., Yamaguchi, Y., Kabe, Y., and Handa, H. (2005) Genes Cells 10 717–731 [DOI] [PubMed] [Google Scholar]
- 11.Sowa, Y., Shiio, Y., Fujita, T., Matsumoto, T., Okuyama, Y., Kato, D., Inoue, J., Sawada, J., Goto, M., Watanabe, H., Handa, H., and Sakai, T. (1997) Cancer Res. 57 3145–3148 [PubMed] [Google Scholar]
- 12.Sekimata, M., and Homma, Y. (2004) Biochem. Biophys. Res. Commun. 325 653–659 [DOI] [PubMed] [Google Scholar]
- 13.Morrison, D. J., Pendergrast, P. S., Stavropoulos, P., Colmenares, S. U., Kobayashi, R., and Hernandez, N. (1999) Nucleic Acids Res. 27 1251–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee, D. K., Suh, D., Edenberg, H. J., and Hur, M. W. (2002) J. Biol. Chem. 277 26761–26768 [DOI] [PubMed] [Google Scholar]
- 15.Pendergrast, P. S., Wang, C., Hernandez, N., and Huang, S. (2002) Mol. Biol. Cell 13 915–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davies, J. M., Hawe, N., Kabarowski, J., Huang, Q. H., Zhu, J., Brand, N. J., Leprince, D., Dhordain, P., Cook, M., Morriss-Kay, G., and Zelent, A. (1999) Oncogene 18 365–375 [DOI] [PubMed] [Google Scholar]
- 17.Liu, C. J., Prazak, L., Fajardo, M., Yu, S., Tyagi, N., Di, and Cesare, P. E. (2004) J. Biol. Chem. 279 47081–47091 [DOI] [PubMed] [Google Scholar]
- 18.Laudes, M., Christodoulides, C., Sewter, C., Rochford, J. J., Considine, R. V., Sethi, J. K., Vidal-Puig, A., and O'Rahilly, S. (2004) J. Biol. Chem. 279 11711–11718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kukita, A., Kukita, T., Ouchida, M., Maeda, H., Yatsuki, H., and Kohashi, O. (1999) Blood 94 1987–1997 [PubMed] [Google Scholar]
- 20.Lee, D. K., Kang, J. E., Park, H. J., Kim, M. H., Yim, T. H., Kim, J. M., Heo, M. K., Kim, K. Y., Kwon, H. J., and Hur, M. W. (2005) J. Biol. Chem. 280 27783–27791 [DOI] [PubMed] [Google Scholar]
- 21.Maeda, T., Hobbs, R. M., Merghoub, T., Guernah, I., Zelent, A., Cordon-Cardo, C., Teruya-Feldstein, J., and Pandolfi, P. P. (2005) Nature 433 278–285 [DOI] [PubMed] [Google Scholar]
- 22.Maeda, T., and Hobbs, R. M. (2005) Cancer Res. 65 8575–8578 [DOI] [PubMed] [Google Scholar]
- 23.Sekimata, M., and Homma, Y. (2004) Nucleic Acids Res. 32 590–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee, J. A., Suh, D. C., Kang, J. E., Kim, M. H., Park, H., Lee, M. N., Kim, J. M., Jeon, B. N., Roh, H. E., Yu, M. Y., Choi, K. Y., Kim, K. Y., and Hur, M. W. (2005) J. Biol. Chem. 280 28061–28071 [DOI] [PubMed] [Google Scholar]
- 25.Kadonaga, J. T., Carner, K. R., Masiarz, F. R., and Tjian, R. (1987) Cell 51 1079–1090 [DOI] [PubMed] [Google Scholar]
- 26.Pessler, F., and Hernandez, N. (2003) J. Biol. Chem. 278 29327–29335 [DOI] [PubMed] [Google Scholar]
- 27.Kwon, H. S., Kim, M. S., Edenberg, H. J., and Hur, M. W. (1999) J. Biol. Chem. 274 20–28 [DOI] [PubMed] [Google Scholar]
- 28.Sakai, T., Ohtani, N., McGee, T. L., Robbins, P. D., and Dryja, T. P. (1991) Nature 353 83–86 [DOI] [PubMed] [Google Scholar]
- 29.Suske, G. (1999) Gene 238 291–300, review [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.