Abstract
Pax-8 is a transcription factor belonging to the PAX genes superfamily and its crucial role has been proven both in embryo and in the adult organism. Pax-8 activity is regulated via a redoxbased mechanism centered on the glutathionylation of specific cysteines in the N-terminal region (Cys45 and Cys57). These residues belong to a highly evolutionary conserved DNA binding site: the Paired Box (Prd) domain. Crystallographic protein-DNA complexes of the homologues Pax-6 and Pax-5 showed a bipartite Prd domain consisting of two helix-turn-helix (HTH) motifs separated by an extended linker region. Here, by means of nuclear magnetic resonance, we show for the first time that the HTH motifs are largely defined in the unbound Pax-8 Prd domain. Our findings contrast with previous induced fit models, in which Pax-8 is supposed to largely fold upon DNA binding. Importantly, our data provide the structural basis for the enhanced chemical reactivity of residues Cys45 and Cys57 and explain clinical missense mutations that are not obviously related to the DNA binding interface of the paired box domain. Finally, sequence conservation suggests that our findings could be a general feature of the Pax family transcription factors.
Pax-8 is an important eukaryotic transcription factor that is responsible, during embryogenesis, for the differentiation of several organs such as kidney, thyroid, and neural tube. Pax-8 is also essential in the adult organism, where it activates thyroid hormone production. During development, Pax-8 functionality is regulated by alternative splicing, generating isoforms that exhibit different transactivation properties but maintain unaltered the DNA binding domain (1, 2). In mature thyroid follicular cells, the complete Pax-8 splicing isoform regulates the expression of thyroglobulin, thyroperoxidase (3, 4), sodium/iodide symporter, and thyrotropin receptor genes (5–7). Pax-8 activity is co-regulated by thyroid transcription factor-1, thyroid transcription factor-2, and Hex, indicating an active role in the early commitment and differentiation of thyrocytes (8, 9). The Pax family shares a bipartite functionality consisting of a N-terminal binding region and a C-terminal transactivation region.
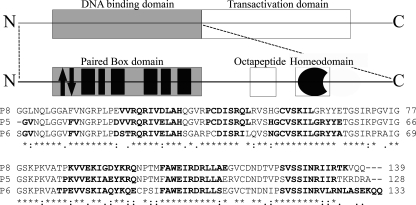
The N-terminal region is usually comprised of three domains, namely a Paired Box (Prd) domain, a conserved octapeptide, and a further homeodomain (see Fig. 1). Differences in conservation and functionality in these domains have been used as evolution markers for the Pax family (10), which has been subdivided into four groups according to the sequence homology in the N-terminal region. In particular, the N-terminal region of Pax-8 is composed by all three domains but presents an incomplete and inactive homeodomain. The Prd domain consists of a well conserved 128-residue-long region, formed by two distinct subdomains known in literature as PAI (N-terminal) and RED (C-terminal) (11).
FIGURE 1.
Schematic representation of Pax-8 protein organization. The upper frame enlightens the dualistic organization of this protein, with a N-terminal DNA binding region and a C-terminal transactivation region. The central frame is a magnification of the N-terminal region, showing the paired box domain, the evolutionary conserved octapeptide, and the non-functional homeodomain. The Paired domain black filled boxes represent α-helices, whereas the arrows indicate the β-hairpin. The lower part shows the primary structure of the Pax-8 Prd domain (P8) aligned with homologues Pax-5 (P5) and Pax-6 (P6) by means of ClustalW (44). Bold fonts highlight the residues found to be organized in a secondary structure. The investigated Pax-8 construct C-terminal linker plus the His6 tag are omitted for sake of clarity.
DNA binding studies have demonstrated that the two subdomains of the Pax-8 Prd domain bind DNA independently (12), are both required for proper promoter activation (13), and that binding is redox state dependent (14, 15). Although both subdomains contain cysteine residues, it was demonstrated both in vivo and in vitro that PAI subdomain DNA-binding activity depends on a reducing environment, whereas RED subdomain binding activity does not (16). Recently, the conserved PAI cysteines (Cys45 and Cys57), which are predicted to be in the DNA binding interface, were shown to be sensitive to oxidation by glutathionylation, whereas reduction by APE1/Ref-1 restores Pax-8 functionality.
The structures of three homologues of Pax-8 Prd domain have been characterized using x-ray diffraction techniques, in co-crystallization with their consensus DNA: the Drosophila (17), Pax-6 (18), and Pax-5 (19) Prd domains. For all the homologues, both subdomains are folded as helix-turn-helix (HTH)2 motifs connected by an extended linker region when bound to DNA. The N-terminal subdomain is preceded by an N-terminal β-hairpin that contacts DNA in the minor groove. With the exception of the Drosophila Prd domain, all the Prd homologues contact DNA both with the N- and C-terminal subdomains (12).
Several missense mutations involving Pax-8 Prd are associated with thyroid dysgenesis, which leads to congenital hypothyroidism (20–22). As a resulting phenotype, the thyroid is misplaced, severely reduced in size or totally absent. Most of these mutations are located in the N-terminal subdomain where a splicing variant has also been found (23), although only one mutation in the RED subdomain (20) has been characterized. Mutations in the Pax-8 Prd domain are also associated to Wilms' tumor (1). Interestingly, not all the affected residues are part of the predicted DNA binding surface (24–26), suggesting that these mutants could affect folding of the Prd domain.
In contrast to the DNA bound state of the Prd domains, the structure of the unbound state is still unknown. Early attempts using circular dichroism (CD) spectroscopy and classical homonuclear NMR approaches suggested that the free Prd domain is largely unstructured (27). Using CD spectroscopy, studies of the structure of the Pax-8 Prd domain under oxidizing and reducing conditions concluded that the free domain has a low α-helical content (≈19% at 277 K) that increases upon DNA binding (13). Interestingly, the unbound and isolated PAI and RED subdomains also maintain a basal, low α-helical content.
In summary, these experiments suggested a lack of secondary and tertiary organization of the free domain in solution. However, the C-terminal subdomain of the Drosophila Prd domain is not bound to the DNA, but is nonetheless structured (18). This observation, together with the activity of Cys45 and Cys57 in the free protein, raises questions about the nature of the unbound state and justifies further research.
Here, we investigate the unbound state of the Pax-8 Prd domain using high resolution heteronuclear NMR, providing the first detailed characterization of a free Prd domain. Based on NOE and chemical shift data, we show that the two subdomains have a well defined secondary structure and a defined tertiary HTH-fold. Our data indicate that the two subdomains behave as independent “beads” on an otherwise flexible “string.”
Our data show that the conserved cysteines in the N-terminal PAI subdomain have reduced pKa values, whereas the RED subdomain cysteine is largely buried, thereby providing a structural basis for the difference in glutathionylation susceptibility of the PAI and RED subdomains. Moreover, our findings help to explain the deleterious effects of mutations involving residues that do not belong to the DNA binding interface.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification—The DNA sequence encoding residues 1–146 of the human PAX-8 gene (SwissProt ID Q06710) cloned into a pIVEX2.3 MCS vector (Roche Molecular Biochemicals) was a kind donation of Prof. Cao. This construct has been thoroughly described elsewhere (16) and encodes a 159-residue fusion protein containing a 13-residue C-terminal linker ending with a His6 affinity tag. The 13C-15N doubly labeled fusion protein was expressed and purified by ASLA Biotech ltd. (Riga, Latvia) using an optimized ad hoc protocol described in the supplemental data.
NMR Spectroscopy—NMR spectra were recorded on the in-house Bruker Avance 500 spectrometer and on a Bruker Avance II 750 spectrometer of the Large Scale NMR Facility, Utrecht, The Netherlands.
All NMR experiments were performed at 298 K using a sample containing 0.9 mm U-13C/15N Pax-8 Prd domain in 50 mm sodium phosphate buffer at pH 6.2 in 90%, 10% H2O/D2O, supplemented with 10–15 mm d10-dithiothreitol and 0.1% NaN3. Backbone resonance assignment was accomplished using standard sensitivity enhanced (28) decoupled 15N and 13C HSQC (29, 30) experiments, three-dimensional HNCA, HNCO (31), HN(CO)CA, HN(CA)CO (32), CBCANH (33, 34)–CBCACONH (35), H(CCO)NH (36), and residue selective MUSIC experiments specific for glycine, serine, and valine-isoleucine-alanine residues (37, 38).
Side chain resonances were assigned using 15N/13C-resolved HSQC-TOCSY (39), HCCH-TOCSY (39, 40), and 15N/13C-edited HSQC-NOESY (41, 42) experiments. Interproton distance restraints were obtained from 15Nor 13C resolved three-dimensional HSQC-NOESY spectra, both recorded with a mixing time of 100 ms. Further details are described in supplemental data.
Proton chemical shifts were referenced to 2,2-dimethyl-2-silapentane-5-sulfonic acid, whose resonance was set to 0.00 ppm whereas 13C and 15N chemical shifts were referenced indirectly to 2,2-dimethyl-2-silapentane-5-sulfonic acid, using the absolute frequency ratio (43). The program CARA3 was used for spectral analysis.
Collection of Conformational Restraints, Structure Calculation, and Refinement—Full backbone assignment of the Pax-8 Prd (residues 11–138 of the 159-amino acid protein construct) could be obtained with the exception of Pro44 and 85% of the nonexchangeable side chain resonances could be assigned.
For the full construct residues 1–2 and 153–159 could not be assigned due to unfavorable water exchange or overlap. Chemical shifts values and NOE data for the N- and C-terminal cloning artifacts indicate that they are unstructured and do not interact with the Prd domain. In the following, the discussion will focus only on the Prd domain.
NOE cross-peaks from the three-dimensional 15N- and 13C-resolved NOESY spectra were assigned and converted into distance restraints using a homemade script. Dihedral angle restraints for the ϕ and Ψ angles were derived from N, C′, Cα, Hα, and Cβ chemical shifts using TALOS (45). The collected restraints were analyzed to remove redundant information and resulted in 1794 distance values, which, together with 123 dihedral angle restraints, formed the experimental data set for the restrained modeling. A trans-conformation was assumed for Pro44, supported by our experimental NOE restraints for the surrounding region. After applying standard pseudoatom corrections (46), 300 structures were calculated using CYANA 2.1 (47) starting from random conformations, simulated annealing details are described in supplemental data. The 20 conformers with the lowest CYANA target function were subjected to refinement in implicit solvent (dielectric constant = 4 × r) using restrained energy minimization with the AMBER force field (48) in the program DISCOVER (Accelrys) to improve local structure quality and electrostatics (see details in supplemental data).
The structural quality was evaluated with Procheck-NMR (49) and WHATCHECK (50). The programs MOLMOL (51) and PyMol (52) (DeLano Scientific, Palo Alto, CA) were used to visualize and evaluate r.m.s. deviations between the structures. WHAT-IF (53) server has been used to evaluate salt bridges. A homemade script has been written to evaluate hydrophobic interactions, following the rules of Israelachvili and Pashley (55). Complete NMR restraints and structure calculation statistics are given in Table 1 for residues 11–138 of the fusion protein corresponding to the Prd and excluding the N- and C-terminal cloning artifacts.
TABLE 1.
Structural statistics and restraint information for the Pax-8 Prd domain
| Structural statistics and restraint informationa | |
|---|---|
| Restraint information | |
| Number of structures | 20 |
| Number of distance restraints | 1,794 |
| Intra/short/medium/long rangeb | 946/588/185/75 |
| Restraints per residue (overall) | 11.3 |
| Restraints per residue (N-terminal subdomain) | 16.3 |
| Restraints per residue (C-terminal subdomain) | 16.6 |
| TALOS derived dihedral angle restraints | 123 |
| Average r.m.s. deviation from experimental restraints | |
| Distance upper limits (Å) | 0.028 ± 0.050 |
| [Range] | [0.01, 0.57] |
| Distance lower limits | 0.035 ± 0.040 |
| [Range] | [–0.01, –0.57] |
| Dihedral angle restraints (°) | |
| Ramachandran quality parameters in selected subdomains (%) | |
| Residues in most favored regions | 90.20% |
| Residues in allowed regions | 8.20% |
| Residues in additionally allowed regions | 1.40% |
| Residues in disallowed regions | 0.10% |
| Consistency check over individual structures (mean value [ranges]) | |
| Abnormally short interatomic distances | 6 [2, 9] |
| Unsatisfied H bond acceptors (buried) | 1 [0, 2] |
| Unsatisfied H bond donors (buried) | 13 [6, 19] |
| Average G factors (53) | |
| Phi-psi | 0.140 |
| Chi1-chi2 | 0.360 |
| Chi1-only | 0.040 |
| Overall | 0.190 |
| Coordinate pairwise root mean square deviation (Å) | |
| Global backbone atoms | 11.1 ± 4.5 |
| Global side chain atoms | 11.6 ± 4.5 |
| N-terminal subdomainc | |
| Backbone | 1.61 ± 0.54 |
| Heavy atoms | 2.77 ± 0.56 |
| C-terminal subdomainc | |
| Backbone | 1.61 ± 0.32 |
| Heavy atoms | 2.75 ± 0.29 |
Statistics were calculated for residues 11–138 corresponding to the Prd domain using the 20 lowest energy structures
Distance restraints between hydrogens of residues i and j are classified as intra (intraresidue, i.e. i – j = 0), short (short-range, i.e. i – j = 1), medium (medium range, i.e. 1 < [i – j] <5), and long (long range, i.e. i – j ≥ 5)
The N-terminal subdomain encompasses fragment 29 – 62, whereas the C-terminal, fragment 87–135
Molecular Dynamics Simulations—Molecular dynamics simulation has been performed starting from the crystal structure of human Pax-6 Prd domain (Protein Data Bank code 6PAX) with the cognate DNA removed and chloride ions added to neutralize the overall charge (see details in supplemental data).
The system was simulated for 10 ns. In the simulation the temperature was kept constant through a simple velocity rescaling procedure, whereas the pressure was controlled through a Berendsen bath (57) using a relaxation time of 100 fs. The volume of the box was fluctuating about 540 nm3 with a standard deviation of less than 0.002 of its value. All structural analyses, in particular r.m.s. deviations, secondary structure, and angular order parameter analyses, have been performed using the program MOLMOL.
Cysteine pKa Calculation—pKa values for cysteines were calculated, based on the Poisson-Boltzmann equation, essentially according to the method of Antosiewicz et al. (58) with minor modifications (59). The starting structures were a model built by homology with Pax-6 (PDB code 6PAX) as template, and the 20 NMR structures. pKa calculations have been also performed on Pax-5 and Pax-6 structures.
Circular Dichroism—The purified Pax-8 Prd domain was used for CD spectroscopy at a concentration of 15 μm using a Jasco J-600 CD/ORD spectropolarimeter interfaced to a computer for data collection. Standard conditions were 50 mm Na2HPO4 (pH 6.2), 250 μm dithiothreitol, 277 K, 0.2-cm path-length cuvette. Spectra are presented in terms of mean residue molecular ellipticity ([θ]; deg cm2 dmol-1), based on a mean residue weight of 110.4 Da.
Data Deposition—Chemical shifts have been deposited at the BMRB data base, with access code 15693, whereas the protein coordinates were deposited in the protein data bank (PDB code 2K27).
RESULTS
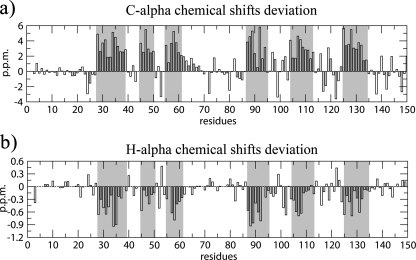
NMR Secondary Structure Identification—The 1H-15N HSQC spectrum is a sensitive indicator of the structural and dynamical properties of a protein. The spectrum of free Pax-8-Prd shows a chemical shift dispersion typical of an all α-helical folded protein (supplemental Fig. S1). Secondary structure assessment based on diagnostic sequential and medium-range NOEs and chemical shift deviation from random coil values (60, 61) revealed the presence of six α-helices, consistent with the Prd domain crystal structures described in literature (Fig. 2 and supplemental S2). The helices are named starting from the N-terminal by ordinal numbering.
FIGURE 2.
Residue by residue chemical shift deviation from random coil for the secondary structure diagnostic Cα and Hα resonances. Gray columns highlight the validated α-helices (60).
The total helix content of the free Pax-8 Prd amounts to ∼42%. This value is in agreement with control CD experiments (supplemental Fig. S3), which show an α-helical content that, depending on the analysis method adopted, varies from ∼28 to ∼39% (62, 63). CD experimental conditions were the same as those of the NMR with exception of protein and dithiothreitol concentrations. The reduced state of the Prd domain in solution is confirmed by α and β carbon chemical shifts of the conserved cysteines, which are in agreement with the values reported by Sharma and Rajarathnam (64).
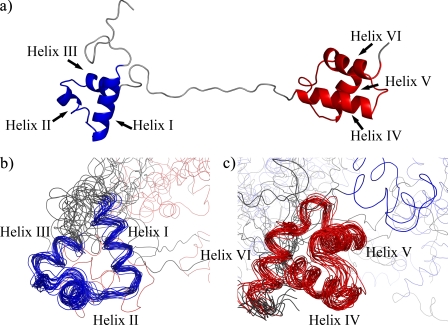
Pax-8 Prd Domain Conformation—Analysis of the NOESY cross-peaks resulted in the identification of 76 long range NOEs that reflect crucial interactions within each subdomain and are typical of a tertiary organization (supplemental Fig. S4). The backbone traces of the lowest energy structure and the ensemble of structures of the free Pax-8 Prd domain are shown in Fig. 3. Structural statistics are reported in Table 1.
FIGURE 3.
Structure of the free Pax-8 Prd domain. a, schematic representation of the lowest energy structure; b and c, backbone traces of the ensemble of structures superimposed on the PAI subdomain (in blue) (b) and the RED subdomain (in red) (c).
The tertiary organization of the free Pax-8 DNA binding domain shows the characteristics of a canonical Prd domain: two HTH motifs are connected by an unstructured linker region. NOE data and chemical shift differences from random coil values (60) support the random coil nature of the linker (Figs. 2 and supplemental S4). Furthermore, the backbone chemical shifts of the linker are consistent with a highly flexible backbone, according to the random coil index (65) (supplemental Fig. S5).
Additionally, the random coil index indicates a random coil-like structure and high backbone flexibility for the C- and N-terminal regions flanking the two HTH domains. The flexibility and lack of structure of the linker and the absence of NOEs between the two subdomains indicate that the two HTH subdomains are independent “beads on a string” and explain the high global coordinate r.m.s. deviation. The individual N- and C-terminal subdomains have considerably lower pairwise r.m.s. deviations values for both backbone and heavy atoms (∼1.6 and 2.7 Å, respectively) in comparison with the whole Prd domain; these values are consistent with a defined tertiary fold for both subdomains but are somewhat higher than typical for a well defined rigid structure.
Closer inspections reveals that, whereas the individual helices are well defined with pairwise r.m.s. deviation of ∼0.6 Å, the interhelical segments and the interhelical angles have considerable lower definition, resulting in an increased r.m.s. deviation. We suggest that the lack of a precise definition of the tertiary structure is a signature of protein dynamics. This is also reflected in the increased random coil index and predicted flexibility for the interhelical segments.
Thus, whereas the free domain has a well defined secondary structure and a defined tertiary fold, it is most likely a dynamic structure. Both the PAI and RED subdomains contain three helices consistent with the HTH motif (see Table 2 for an analysis).
TABLE 2.
Inter-helix tilt (degrees) comparisons in helix-turn-helix motifs
| Pax-8 | Pax-5 | Pax-6 | |
|---|---|---|---|
| Helix I–II | 114 ± 14 | 143 | 141 |
| Helix I–III | 108 ± 9 | 78 | 77 |
| Helix II–III | 103 ± 18 | 124 | 117 |
| Helix IV–V | 112 ± 7 | 120 | 122 |
| Helix IV–VI | 73 ± 8 | 75 | 77 |
| Helix V–VI | 121 ± 6 | 103 | 104 |
Helix I and II in the PAI subdomain pack against each other in an antiparallel arrangement and are almost perpendicular to helix III, which is the DNA recognition helix. Helices IV, V, and VI of the RED subdomain are packed analogously.
A striking difference between the two domains is the length of the DNA recognition helix. Although helix III spans on average only 1.5 helical turns, helix VI is 2.5 turns long. We next sought to identify the crucial side chain interactions responsible for the HTH-fold of the PAI and RED subdomains in the structural ensemble (supplemental Table S1).
Consistent with the NOE data, residue Ile47 in helix II is involved in hydrophobic interactions with residues Val42 in the loop between helices I and II, and Leu51 in helix II; Val58 interacts with Val53 and Leu62 in helix III interacts with Ile34 in most of the structures of the ensemble. These interactions stabilize the core scaffold of the whole PAI subdomain (supplemental Fig. S6a). Similarly, the scaffold of the RED domain is formed by the interactions between residue Val92 in helix IV, Ile107 in helix V, and Val122 in the loop between helixes V and VI (supplemental Fig. S6b). Additional hydrophobic interactions are found between residues in helix V and the turn connecting the latter to helix VI.
Comparison with the DNA-Pax Complex Structures—To assess the structural differences between the free and bound state of the Pax-8 Prd domain we compared the solution structures of Pax-8 with crystal structures of Pax homologues bound to DNA as the structure of DNA bound Pax-8 has not been determined. The DNA bound homologues Pax-5 and Pax-6 have 85 and 72% sequence identity with Pax-8, respectively (Fig. 1).
Superposition of the PAI and RED subdomains of these structures on the corresponding Pax-8 subdomains shows that the overall fold is highly similar. The backbone r.m.s. deviation between the PAI subdomain and the Pax-5 subdomain is 2.8 Å, averaged over the ensemble of structures. Analogously, the average backbone r.m.s. deviations for the RED subdomain is 1.8 Å. Similar values were obtained for the comparison with the Pax-6 subdomains. These somewhat high r.m.s. deviation values are in agreement with the structural definition of the ensemble itself.
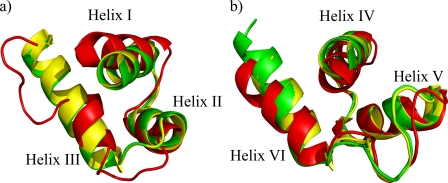
Although the RED subdomain closely resembles that of Pax-5 and Pax-6 in the DNA bound state, there are two interesting differences in the tertiary structure organization for the PAI domain (see Fig. 4). First, the N-terminal β-hairpin, which is characteristic of Paired box PAI subdomains and provides specific contacts at the DNA minor groove level (18), is absent. Second, the DNA recognition helix is structured only in its N-terminal part and for an average of 1.5 turns instead of the expected 3.5 turns. These observations are also in direct agreement with the observed chemical shifts. In addition, helix I is slightly tilted with respect to the crystallized DNA-bound Pax Prd domains, which could be a consequence of the lack of crucial DNA contacts. Finally, hydrophobic interactions and salt bridges that are conserved in Pax-5 and Pax-6 are also crucial for the HTH fold in Pax-8 as shown in supplemental Table S1.
FIGURE 4.
PAI (a) and RED subdomain (b) superimposition among the closest to mean structures of Pax-8 (red) and the crystal structures of Pax-6 (green) and Pax-5 (yellow).
Molecular Dynamics Simulation of Unbound Pax-6 Prd Domain—According to the currently accepted view, the Prd domains are mostly unfolded when free in solution. This means that the interaction with DNA should play a pivotal role in secondary and tertiary structuring of these domains. Following this hypothesis, intra-domain interactions should not be strong enough to maintain a rigid tertiary structure once DNA is removed. To test this hypothesis, we performed a molecular dynamics simulation starting from the crystal structure of the DNA bound conformation of Pax-6. Although Pax-6 Prd domain shows 72% sequence identity with Pax-8 Prd, i.e. less than Pax-5 (85% (10)), we preferred Pax-6 rather than Pax-5, because the latter was co-crystallized in a ternary complex with Ets-1.
The simulation ran for 10 ns in standard conditions. Snapshots were taken at 100-ps intervals to obtain a statistical ensemble for the system under study. The resulting trajectory revealed an early loss of the N-terminal β-hairpin (after about 1.5 ns) and confirmed the preservation of the two subdomains in the free state of Pax-6 that maintained a r.m.s. deviation of about 1 Å after 10 ns simulation. The mean r.m.s. deviation over the backbone indicates that the two subdomains do not loose the global scaffold in the absence of the DNA. Moreover, on the time scale probed by the simulation, no loss of structure is observed in the helical regions of the two subdomains.
Cysteine pKa Calculations—pKa calculations were performed on the Prd domains of Pax-5, Pax-6, homology modeled Pax-8, and the 20 NMR structures. We focus our attention on cysteine residues that have been found to be relevant for redox transcriptional regulation.
For all Pax-8 models a significant shift toward physiological pH has been found for the pKa of Cys45 (7.9 ± 0.2) and Cys57 (7.6 ± 0.3) thiol groups, whereas for Cys117 the pKa seems slightly shifted toward alkaline pH (8.9 ± 0.7). Structurally, the anion state of Cys45 and Cys57 seems to be stabilized by conserved (throughout the whole Pax family) positively charged residues in their vicinity, such as Arg43, Arg49, and Lys60. Similar pKa shifts have been found also for Pax-5 and Pax-6 homologue cysteines, showing that this feature is conserved. Furthermore, the N-terminal cysteines are accessible to the solvent (100% of the structures for Cys45 and 90% for Cys57), whereas Cys117 is more buried (accessible in 40% of the structures).
DISCUSSION
In this work we demonstrate, using NMR, CD, and molecular dynamics experiments, that the free Pax-8 Prd domain has a defined residual structure in solution. The solution structure of the Pax-8 Prd domain shows that the PAI and RED subdomains have a well defined secondary structure and a typical, albeit less precisely defined, HTH tertiary fold.
The secondary structure content is supported by CD measurements, whereas the presence of the HTH motif is supported by a 10-ns molecular dynamics simulation. Stabilization of the HTH fold can be explained by crucial hydrophobic interactions that are conserved in the DNA bound state of homologues Pax-5 and Pax-6. Finally, backbone chemical shifts indicate through the Random Coil Index that the HTH motifs of the two subdomains behave like beads on a string within an otherwise dynamic Prd domain.
Our findings provide the structural basis of the Prd domain activity regulation. Pax-8 activity has been found to depend on a redox reaction performed by APE1/Ref-1 (67) that results in the reduction/oxidation of two important and conserved cysteines located in helixes II and III, Cys45 and Cys57 (20). Specifically, glutathionylation of these N-terminal cysteines has been proposed to prevent DNA binding because they are part of the predicted interface (16).
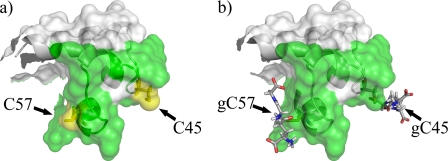
Importantly, such enzymatic modifications require accessible and reactive cysteine residues in the free state of the Prd domain. Indeed, Cys45 and Cys57 are highly accessible to the solvent in our structure. Furthermore, by pKa calculations we demonstrated that the local electrostatic environment of the thiol groups of these cysteines reduces their pKa, making them prone to ionization at physiological pH and therefore decreasing the redox potential (68). Finally, as these cysteines are solvent exposed, it is unlikely that glutathionylation will cause the unfolding of the Prd domain, rather it will mask a large part of the DNA binding interface (Fig. 5).
FIGURE 5.
Model of Pax8 glutathionylated at Cys45 and Cys57. Panel a shows the surface of the unbound Pax-8 Prd domain where the green surface represents the DNA binding interface. Panel b shows how glutathionylation of Cys45 and Cys57 hinders part of the DNA interaction surface. The model was built using PyMol (52).
Thus, the stability of the HTH fold in the unbound Prd domain is responsible for the reactivity of the cysteines 45 and 57. In opposition, Cys117 results to be inactive: it is mostly buried, with a thiol pKa shifted toward alkaline values. These features are conserved also in Pax-5 and Pax-6 Prd domains, thus suggesting an important functional role for the conserved PAI cysteines.
The predefined nature of HTH motifs will also limit the total entropy costs of Pax-8 binding to its cognate DNA, as the subdomains are already right in overall conformation to dock to the DNA. Nevertheless, structural comparison with the DNA-bound homologues suggests crucial conformational changes within the PAI subdomain upon DNA binding. In particular, the conserved N-terminal β-hairpin and the C-terminal part of the DNA recognition helix (helix III) are unfolded in the free state. When bound to DNA, these two regions are stabilized by pivotal specific contacts to the DNA. The lower intrinsic stability of the N-terminal β-hairpin is also indicated in the molecular dynamics simulation of unbound Pax-6, in which this secondary structure element is lost shortly after release from the DNA. Overall, these local structural changes give some clues into the dissection of the thermodynamics of DNA binding by the Pax8 Prd domain.
Information gathered by CD spectra and random coil index, together with the NMR structure suggests the presence of unstructured regions in different parts of the Prd domain. This is in striking contrast with the conservation of the N-terminal subdomain region throughout eukaryotic PAX genes. Because the free protein is only partially structured and could thus easily accommodate mutations, the evolutionary conservation is most likely dictated by the properties of the protein when bound to DNA.
In keeping with this picture, most Mendelian inherited mutations, which are associated with diseases described in the OMIM (66) data base, are found in the N-terminal subdomain. Our structure suggests a molecular rationale for some dysfunctions associated with mutations in the Prd domain. In fact, based on homology modeling of Pax-8·DNA complex, the importance of DNA contacting residues such as Arg31, Ser54, and Cys57 is evident.
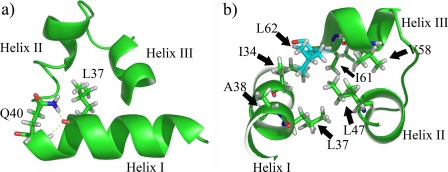
The presented data complete this view underlining the pivotal role of residues that do not obviously contact DNA, but contribute to the correct secondary and tertiary scaffold of the binding domain (as shown in Fig. 6). For instance, mutation Q40P (24) will destabilize helix I, due to the resulting absence of the hydrogen bonds with Leu37. Analogously, the mutation L62R (20) may reduce the stability of the whole PAI subdomain due to the crucial hydrophobic contacts with several other residues in the N-terminal HTH motif.
FIGURE 6.
Schematic representations of Pax-8 PAI subdomain. Panel a shows the hydrogen bonds found between the Leu37 carbonyl oxygen and Gln40 side chain and backbone amide nitrogens (Nε-2 and HN). Panel b depicts the hydrophobic residue side chains around Leu62 (in cyan), i.e. 5 Å apart. The cluster is an important part of the hydrophobic core of the PAI subdomain.
This interpretation could apply to the whole Pax family. For Pax-6 a very large number of mutations associated with diseases is present in the HGMD (56) data base. Interestingly, many of the mutations involve conserved residues that we highlighted as important for the structural arrangement of Pax-8, not only in the DNA bound conformation, but also in the unbound protein. Also in a study of the Pax-3 Prd domain (54), DNA binding impairment was observed upon mutation of residues that we address as fundamental in the tertiary scaffold.
In summary, in this work we show for the first time the unbound conformation of a Prd domain free in solution, demonstrating that Pax-8 adopts a tertiary structure even when not bound to DNA. The unbound structure provides the structural basis for the enhanced reactivity of Cys45 and Cys57 toward glutathionylation and is useful to explain the effect of some Prd pathologic mutations. This bears implications for the activity regulation and functionality of the whole Pax family.
Supplementary Material
Acknowledgments
We thank Prof. Cao for kindly providing the Pax-8 Prd expression vector and the assistance of Dr. A. Makek.
The atomic coordinates and structure factors (code 2K27) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
This work was supported by Italian Ministry for University and Research Grants COFIN 2006058958, FIRB RBNE03PX83, and FIRB RBNE03B8KK, European Union Grant LSHM-CT-2005-037525, and European Union project “EU-NMR-European Network of Research Infrastructures for Providing Access & Technological Advancement in Bio-NMR” FP-2005-RII3 Contract 026145. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6, Tables S1 and S2, additional text, and references.
Footnotes
The abbreviations used are: HTH, helix-turn-helix; Prd, paired box; CD, circular dichroism; r.m.s., root mean square; NOE, nuclear Overhauser effect.
Keller, R. (2004) The Computer Aided Resonance Tutorial, First Edition, ISBN 3-85600-112-3.
References
- 1.Kozmik, Z., Kurzbauer, R., Dörfler, P., and Busslinger, M. (1993) Mol. Cell. Biol. 13 6024-6035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Poleev, A., Fickenscher, H., Mundlos, S., Winterpacht, A., Zabel, B., Fidler, A., Gruss, P., and Plachov, D. (1992) Development 116 611-623 [DOI] [PubMed] [Google Scholar]
- 3.Zannini, M., Francis-Lang, H., Plachov, D., and Di Lauro, R. (1992) Mol. Cell. Biol. 12 4230-4241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Espinoza, C., R., Schmitt, T., L., and Loos, U. (2001) J. Mol. Endocrinol. 27 59-67 [DOI] [PubMed] [Google Scholar]
- 5.Ohno, M., Zannini, S., Levy, O., Carrasco, N., and Di Lauro, R. (1999) Mol. Cell. Biol. 19 2051-2060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Damante, G., and Di Lauro, R. (1994) Biochim. Biophys. Acta 1218 255-266 [DOI] [PubMed] [Google Scholar]
- 7.Santisteban, P., and Bernal, J. (2005) Rev. Endocr. Metab. Disord. 6 217-228 [DOI] [PubMed] [Google Scholar]
- 8.Pasca di Magliano, M., Di Lauro, R., and Zannini, M. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 13144-13149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lang, D., Powell, S. K., Plummer R. S., Young, K. P., and Ruggeri, B. A. (2007) Biochem. Pharmacol. 73 1-14 [DOI] [PubMed] [Google Scholar]
- 10.Balczarek, K., A., Lai, Z. C., and Kumar, S. (1997) Mol. Biol. Evol. 14 829-842 [DOI] [PubMed] [Google Scholar]
- 11.Jun, S., and Desplan, C. (1996) Development 122 2639-2650 [DOI] [PubMed] [Google Scholar]
- 12.Epstein, J. A., Glaser, T., Cai, J., Jepeal, L., Walton, D. S., and Maas, R. L. (1994) Genes Dev. 8 2022-2034 [DOI] [PubMed] [Google Scholar]
- 13.Pellizzari, L., Tell, G., and Damante, G. (1999) Biochem. J. 337 253-262 [PMC free article] [PubMed] [Google Scholar]
- 14.Kambe, F., Nomura, Y., Okamoto, T., and Seo, H. (1996) Mol. Endocrinol. 10 801-812 [DOI] [PubMed] [Google Scholar]
- 15.Tell, G., Scaloni, A., Pellizzari, L., Formisano, S., Pucillo, C., and Damante, G. (1998) J. Biol. Chem. 273 25062-25072 [DOI] [PubMed] [Google Scholar]
- 16.Cao, X., Kambe, F., Lu, X., Kobayashi, N., Ohmori, S., and Seo, H. (2005) J. Biol. Chem. 280 25901-25906 [DOI] [PubMed] [Google Scholar]
- 17.Xu, W., Rould, M. A., Jun, S., Desplan, C., and Pabo, C. O. (1995) Cell 80 639-650 [DOI] [PubMed] [Google Scholar]
- 18.Xu, H. E., Rould, M. A., Xu, W., Epstein J. A., Maas, R. L., and Pabo, C. O. (1999) Genes De. 13 1263-1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garvie, C. W., Hagman J., and Wolberger, C. (2001) Mol. Cell 8 1267-1276 [DOI] [PubMed] [Google Scholar]
- 20.Macchia, P. E., Lapi, P., Krude, H., Pirro, M. T., Missero, C., Chiovato, L., Souabni, A., Baserga, M., Tassi, V., Pinchera, A., Fenzi, G., Gruters, A., Busslinger, M., and Di Lauro, R. (1998) Nat. Genet. 19 83-86 [DOI] [PubMed] [Google Scholar]
- 21.Tell, G., Pellizzari, L., Esposito, G., Pucillo, C., Macchia, P. E., Di Lauro, R., and Damante, G. (1999) Biochem. J. 341 89-93 [PMC free article] [PubMed] [Google Scholar]
- 22.Meeus, L., Gilbert, B., Rydlewski, C., Parma J., Roussie, A. L., Abramowicz, M., Vilain, C., Christophe, D., Costagliola, S., and Vassart, G. (2004) J. Clin. Endocrinol. Metab. 89 4285-4291 [DOI] [PubMed] [Google Scholar]
- 23.Kozmik, Z., Czerny, T., and Busslinger, M. (1997) EMBO J. 16 6793-6803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Congdon, T., Nguyen, L. Q., Nogueira, C. R., Habiby, R. L., Medeiros-Neto, G., and Kopp, P. (2001) J. Clin. Endocr. Metab. 86 3962-3967 [DOI] [PubMed] [Google Scholar]
- 25.Vilain, C., Rydlewski, C., Duprez, L., Heinrichs, C., Abramowicz, M., Malvaux, P., Renneboog, B., Parma J., Costagliola, S., and Vassart, G. (2001) J. Clin. Endocrinol. Metab. 86 234-238 [DOI] [PubMed] [Google Scholar]
- 26.Grasberger, H., Ringkananont, U., LeFrancois, P., Abramowicz, M., Vassart, G., and Refetoff, S. (2005) Mol. Endocrinol. 19 1779-1791 [DOI] [PubMed] [Google Scholar]
- 27.Epstein, J., Cai, J., Glaser, T., Jepeal, L., and Maas, R. (1994) J. Biol. Chem. 269 8355-8361 [PubMed] [Google Scholar]
- 28.Kay, L. E., Keifer, P., and Saarinen, T. (1992) J. Am. Chem. Soc. 114 10663-10665 [Google Scholar]
- 29.Bax, A., Clore, G. M., and Gronenborn, A. M. (1990) J. Magn. Reson. 88 425-431 [Google Scholar]
- 30.Vuister, G. W., and Bax, A. (1992) J. Magn. Reson. 98 428-435 [Google Scholar]
- 31.Kay, L. E., Ikura, M., Tschudin, R., and Bax, A. (1990) J. Magn. Reson. 89 496-514 [DOI] [PubMed] [Google Scholar]
- 32.Ikura, M., and Bax, A. (1991) J. Biomol. NMR 1 99-104 [DOI] [PubMed] [Google Scholar]
- 33.Wittekind, M., and Mueller, L. (1993) J. Magn. Reson. Ser. B 101 201-205 [Google Scholar]
- 34.Grzesiek, S., and Bax, A. (1992) J. Magn. Reson. 99 201-207 [Google Scholar]
- 35.Grzesiek, S., and Bax, A. (1992) J. Magn. Reson. 96 432-440 [Google Scholar]
- 36.Grzesiek, S., Anglister J., and Bax, A. (1993) J. Magn. Reson. Ser. B 101 114-119 [Google Scholar]
- 37.Schubert, M., Smalla, M., Schmieder, P., and Oschkinat, H. (1999) J. Magn. Reson. 141 34-43 [DOI] [PubMed] [Google Scholar]
- 38.Schubert, M., Oschkinat, H., and Schmieder, P. (2001) J. Magn. Reson. 148 61-72 [DOI] [PubMed] [Google Scholar]
- 39.Davis, A. L., Keeler, J. E., Laue, D., and Moskau, D. (1992) J. Magn. Reson. 98 207-216 [Google Scholar]
- 40.Kay, L. E., Xu, G. Y., Singer, A. U., Muhandiram, D. R., and Formankay, J. D. (1993) J. Magn. Reson. Ser. B 101 333-337 [Google Scholar]
- 41.Kay, M. D., Sparks, L. E., Torchia, S. W., and Bax, A. (1988) J. Am Chem. Soc. 111 1515-1517 [Google Scholar]
- 42.Pascal, S. M., Muhandiram, D. R., Yamazaki, T., Formankay, J. D., and Kay, L. E. (1994) J. Magn. Reson. Ser B 103 197-201 [Google Scholar]
- 43.Wishart, D. S., Bigam, C. G., Yao, J. Abildgaard, F., Dyson, H. J., Oldfield, E., Markley, J. L., and Sykes, B. D. (1995) J. Biomol. NMR 6 135-140 [DOI] [PubMed] [Google Scholar]
- 44.Higgins, D., Thompson J., Gibson, T., Thompson J. D., Higgins, D. G., and Gibson, T. J. (1994) Nucleic Acids Res. 22 4673-4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cornilescu, G., Delaglio, F., and Bax, A. (1999) J. Biomol. NMR 13 289-302 [DOI] [PubMed] [Google Scholar]
- 46.Wuthrich, K., Billeter, M., and Braun, W. (1983) J. Mol. Biol. 169 949-961 [DOI] [PubMed] [Google Scholar]
- 47.Güntert, P. (2004) Methods Mol. Biol. 278 353-378 [DOI] [PubMed] [Google Scholar]
- 48.Weiner, S. J., Kollman, P. A., Case, D. A., Singh, U. C., Ghio, C., Alagona, G., Profeta, J. S., and Weiner, P. (1984) J. Am. Chem. Soc. 106 765-784 [Google Scholar]
- 49.Laskowski, R. A., Rullmann, J. A., MacArthur, M. W., Kaptein, R., and Thornton, J. M. (1996) J. Biomol. NMR 8 477-486 [DOI] [PubMed] [Google Scholar]
- 50.Hooft, R. W. W., Vriend, G., Sander, C., and Abola, E. E. (1996) Nature 381 272. [DOI] [PubMed] [Google Scholar]
- 51.Koradi, R., Billeter, M., and Wüthrich, K. (1996) J. Mol. Graph. 14 51-55 [DOI] [PubMed] [Google Scholar]
- 52.DeLano, W. L. The PyMOL Molecular Graphics System (2002) DeLano Scientific, Palo Alto, CA
- 53.Vriend, G. (1990) J. Mol. Graph. 8 52-56 [DOI] [PubMed] [Google Scholar]
- 54.Apuzzo, S., and Gros, P. (2006) Biochemistry 45 7154-7161 [DOI] [PubMed] [Google Scholar]
- 55.Israelachvili, J., and Pashley, R. (1982) Nature 300 341-342 [DOI] [PubMed] [Google Scholar]
- 56.Stenson, P. D., Ball, E., Howells, K., Phillips, A., Mort, M., and Cooper, D. N. (2008) J. Med. Genet. 45 124-126 [DOI] [PubMed] [Google Scholar]
- 57.Berendsen, H. J. C., Postma, J. P. M., van Gunsteren, W. F., Di Nola, A., and Haak, J. R. (1984) J. Chem. Phys. 81 3684-3690 [Google Scholar]
- 58.Antosiewicz, J., McCammon, J. A., and Gilson, M. K. (1994) J. Mol. Biol. 238 415-436 [DOI] [PubMed] [Google Scholar]
- 59.Fogolari, F., Ragona, L., Licciardi, S., Romagnoli, S., Michelutti, R., Ugolini, R., and Molinari, H. (2000) Struct. Funct. Genet. 39 317-330 [DOI] [PubMed] [Google Scholar]
- 60.Wang, Y., and Jardetzky, O. (2002) J. Am. Chem. Soc. 124 14075-14084 [DOI] [PubMed] [Google Scholar]
- 61.Wishart, D. S., and Sykes, B. D. (1994) Methods Enzymol. 239 363-392 [DOI] [PubMed] [Google Scholar]
- 62.Sreerama, N., and Woody, R. W. (1993) Anal. Biochem. 209 32-44 [DOI] [PubMed] [Google Scholar]
- 63.Chen, Y. H., Yang, J. T., and Chau, K. H. (1974) Biochemistry 13 3350-3359 [DOI] [PubMed] [Google Scholar]
- 64.Sharma, D., and Rajarathnam, K. (2000) J. Biomol. NMR. 18 165-171 [DOI] [PubMed] [Google Scholar]
- 65.Berjanskii, M. V., and Wishart, D. S. (2005) J. Am. Chem. Soc. 127 14970-14971 [DOI] [PubMed] [Google Scholar]
- 66.Hamosh, A., Scott, A. F., Amberger, J., Bocchini, C., Valle, D., and McKusick, V. A. (2002) Nucleic Acids Res. 30 52-55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tell, G., Pellizzari, L., Cimarosti, D., Pucillo, C., and Damante, G. (1998) Biochem. Biophys. Res. Commun. 252 178-183 [DOI] [PubMed] [Google Scholar]
- 68.Chivers, P. T., Prehoda, K. E., and Raines, R. T. (1997) Biochemistry 36 4061-4066 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.