Abstract
Mitochondria form filamentous networks that undergo continuous fission/fusion. In the pancreatic β-cells, mitochondria are essential for the transduction of signals linking nutrient metabolism to insulin granule exocytosis. Here we have studied mitochondrial networks in the insulinoma cell line INS-1E, primary rat and human β-cells. We have further investigated the impact of mitochondrial fission/fusion on metabolism-secretion coupling in INS-1E cells. Overexpression of hFis1 caused dramatic mitochondrial fragmentation, whereas Mfn1 evoked hyperfusion and the aggregation of mitochondria. Cells overexpressing hFis1 or Mfn1 showed reduced mitochondrial volume, lowered cellular ATP levels, and as a consequence, impaired glucose-stimulated insulin secretion. Decreased mitochondrial ATP generation was partially compensated for by enhanced glycolysis as indicated by increased lactate production in these cells. Dominant-negative Mfn1 elicited mitochondrial shortening and fragmentation of INS-1E cell mitochondria, similar to hFis1. However, the mitochondrial volume, cytosolic ATP levels, and glucose-stimulated insulin secretion were little affected. We conclude that mitochondrial fragmentation per se does not impair metabolism-secretion coupling. Through their impact on mitochondrial bioenergetics and distribution, hFis1 and Mfn1 activities influence mitochondrial signal generation thereby insulin exocytosis.
Mitochondria play a central role in energy metabolism, other biosynthetic processes, Ca2+ signaling, and the integration of diverse apoptotic stimuli. Mitochondria establish networks of filamentous structures that continuously move, divide, and fuse (1). This continuous remodeling is under the control of several fission and fusion proteins (2). The major components of the fission machinery are Fis1 (3) and Drp1 (dynamin-related protein 1) (4). Fis1 is a small (17-kDa) protein, anchored in the outer mitochondrial membrane (5). Drp1 has a dynamin-like central domain and a C-terminal GTPase domain, which couples GTP hydrolysis to mitochondrial membrane constriction during fission (6). In the mitochondrial fission process, Fis1 has been suggested to be a limiting factor. The number of expressed Fis1 molecules on the mitochondrial outer surface determines the frequency of mitochondrial fission (5).
Mitochondrial fusion is regulated by mitofusin 1 (Mfn1) and mitofusin 2 (Mfn2) as well as Opa1 (2). Mfn1 and Mfn2 are GTPases localized to the outer mitochondrial membrane that form homo- or hetero-oligomeric complexes during the fusion process. Mfn1 has higher GTPase activity and causes more efficient fusion than Mfn2 (7). Suppression of mitofusin function promotes fission, yielding highly fragmented mitochondria (8). A point mutation in the GTPase domain of Mfn1 (Mfn1T109A) allows the protein to act as a dominant-negative, inducing excessive fission when overexpressed (9).
The biological impact of the mitochondrial fission/fusion process is not restricted to structural changes. Fission and fusion allow mitochondria to exchange lipid membranes and intramitochondrial content including mtDNA (2). Thus, damage in the organelle can be restored by fusion with healthy mitochondria (10). Furthermore, dysfunctional parts of the reticulum can be separated by fission and selectively targeted to autophagy, which may protect cells from apoptosis by preventing the release of proapoptotic proteins (11, 12). Mitochondrial dynamics are also important during apoptosis as the organelle undergoes massive fragmentation during this process. For instance, overexpression of fission protein leads to apoptosis (3). Mitochondrial fission/fusion proteins also directly or indirectly influence mitochondrial energy metabolism. Knock-out or down-regulation of fission or fusion proteins reduces mitochondrial respiration and ATP generation (13–15).
Filamentous mitochondrial networks have also been observed in the pancreatic β-cell (16), and a recent electron microscope tomography study further described branched mitochondria in β-cells within the islet (17). In this cell type, mitochondria are of particular importance as they are both the center of energy production as well as the source of coupling factors that link nutrient metabolism to insulin exocytosis. Because of extremely low lactate dehydrogenase levels, more than 90% of glucose-derived carbons are funneled into the β-cell mitochondria (18, 19). mtDNA-depleted β-cell lines showed complete absence of glucose-stimulated insulin secretion, illustrating the importance of the organelle in signal generation (20). Moreover, patients having mtDNA mutations develop diabetes. The prevalence of mitochondrial diabetes varies between populations, accounting for up to 1% of diabetes cases (21, 22). In addition, postmortem islets from type 2 diabetes patients display ultrastructural changes in mitochondria and reduced glucose-stimulated insulin secretion resulting from reduced glucose oxidation and attenuated ATP generation (23–25).
In islets from the Zucker diabetic fatty rat, β-cell mitochondria are fragmented into smaller units when compared with those of control animals (16). These studies give a first insight into the possible importance of mitochondrial structure and dynamics for β-cell function, also raising the question whether mitochondrial structural changes contribute to the defects of the β-cell in the diabetic state. Here we have studied mitochondrial morphology in primary human and rat β-cells as well as INS-1E cells. Furthermore, we have manipulated the mitochondrial filamentous network and analyzed the impact of the resulting mitochondrial morphology changes on cell viability and metabolism-secretion coupling in INS-1E cells. The mitochondrial network was manipulated by overexpression of either hFis1 or dominant-negative Mfn1 (DN-Mfn1; Mfn1T109A). Overexpression of hFis1 deteriorated mitochondrial signal generation and insulin secretion, also causing a limited degree of apoptosis, whereas such changes were not observed in DN-Mfn14-overexpressing cells. We suggest that hFis1 induces a specific metabolic defect in the β-cell. This defect is not simply due to fragmentation of the mitochondria as massive fragmentation caused by DN-Mfn1 did not mimic any of these defects.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfection—Human pancreatic islets were isolated from two different donors obtained through the EURODIA European Union sixth framework programme consortium (25). Rat islets were isolated from male Wistar rats (250 g, Elevage Janvier, Le Genest-St-Isle, France) after collagenase digestion. Islets were dissociated with 0.02% trypsin and seeded on coverslips coated with A431 cell-derived extracellular matrix (26). Human islet cells were cultured in CMRL-1066 (Invitrogen) medium at 5.6 mm glucose supplemented with 10% heat-inactivated fetal calf serum, 100 units/ml penicillin, 100 μg/ml streptomycin, and 100 μg/ml gentamycin (Sigma). Rat islet cells were cultured in RPMI 1640 (11.1 mm glucose, Invitrogen) supplemented with 10% fetal calf serum and antibiotics. INS-1E cells were cultured in a humidified atmosphere (37 °C) containing 5% CO2 in complete medium composed of RPMI 1640 supplemented with 10% fetal calf serum, 1 mm sodium pyruvate, 50 μm 2-mercaptoethanol, 2 mm glutamine, 10 mm HEPES, 100 units/ml penicillin, and 100 μg/ml streptomycin. Transient transfection of INS-1E cells was performed using the Lipofectamine™ 2000 reagent (Invitrogen).
Adenovirus Construction and Infection—Doxycycline-inducible adenoviruses encoding hFis1, WT-Mfn1, and DN-Mfn1 were constructed using the Adeno-X Tet-On expression system (Clontech Laboratories, Inc.). Adenoviruses encoding the transgene were applied together with the adenovirus expressing the reverse tetracycline-transactivator (Clontech laboratories, Inc.) to islet cells and INS-1E cells for 90–120 min 1 day after seeding (27).
Mitochondrial Morphology—Mitochondria of human and rat islet cells were visualized following infection with adenovirus encoding enhanced cyan fluorescence protein (mitoECFP). Human β-cells were identified by immunohistochemical staining of insulin as described previously (28). INS-1E cells were transfected with mitochondria-targeted red fluorescence protein (mitoRFP) plasmid followed by infection with adenoviruses. Mitochondrial morphology was observed 48 h after infection and analyzed using a confocal laser-scanning microscope (LSM510meta, Zeiss, Zurich, Switzerland) after fixation with 4% paraformaldehyde. Mitochondrial volume and skeleton length were calculated as described in supplemental Figs. S3 and S4.
Western Blot—Total protein extracts (50 μg) were resolved by 11% SDS-PAGE and transferred onto nitrocellulose membranes. The membranes were blocked in 20 mm Tris-HCl, pH 7.5, 500 mm NaCl, 0.1% Tween 20, 5% milk and then incubated with first antibodies: anti-glyceraldehyde-3-phosphate dehydrogenase (1:8000; Sigma), anti-hFis1 (1:1000) (29), or anti-Mfn1 (1:1000), a generous gift from Dr. D. C. Chan (California Institute of Technology, Pasadena, CA). Anti-chicken and anti-rabbit IgG-horseradish peroxidase conjugates (Sigma) were used as secondary antibodies.
Glucose-stimulated Insulin Secretion—INS-1E cells seeded on 24-well plates (3 × 105 cells/well) were infected with adenoviruses. After 48 h, infected cells were preincubated with glucose-free medium for 2 h subsequently with KRBH buffer (135 mm NaCl, 3.6 mm KCl, 2 mm NaHCO3, 0.5 mm NaH2PO4, 0.5 mm MgSO4, 1.5 mm CaCl2, 10 mm HEPES, pH 7.4, and 0.1% bovine serum albumin) containing 2.8 mm glucose for 30 min. The cells were then stimulated for 30 min in KRBH buffer at different glucose concentrations. Insulin was measured by rat insulin enzyme immunoassay kit (Spi-Bio, Montigny le Bretonneux, France).
Cellular ATP Content and Lactate Production—For the measurement of ATP content and lactate production, INS-1E cells were prepared as described for insulin secretion. Total cellular ATP was measured using a bioluminescence assay kit according to the manufacturer's instructions (HS II, Roche Diagnostics). Lactate in the cell supernatant was measured using a lactate assay kit (BioVision, Mountain View, CA).
Measurement of Mitochondrial Membrane Potential— INS-1E cells were seeded (5 × 104 cells/well) on black-walled 96-well plates (Greiner Bio-One GmbH, Frickenhausen, Germany) and infected with adenoviruses. Cells were cultured for 48 h and then loaded with 500 nm JC-1 (Invitrogen) in KRBH buffer (2.8 mm glucose) for 30 min at 37 °C. After washing twice, JC-1 fluorescence was measured ratiometrically using a Flex-Station (Molecular Devices) with excitation/emission wavelengths of 490/540 nm (green; monomer) and 540/590 nm (red; J-aggregates). Changes in fluorescence ratios from basal levels were normalized to ratio differences between resting and maximal depolarization induced by 10 μm FCCP, and expressed as the percentage of changes ((R - RFCCP)/(R0 - RFCCP) × 100) (30).
Measurement of Cytosolic and Mitochondrial Calcium Concentration—Cytosolic calcium ([Ca2+]i) was measured using “yellow cameleon” (YC3.6). Cells grown on glass coverslips were transiently co-transfected with mitochondrial fission genes, mitoRFP and YC3.6, and cultured for 72 h. The cells were placed on an inverted microscope (Axiovert 200 m, Zeiss) equipped with an array laser confocal spinning disk (QLC100; VisiTech, Sunderland, UK). Cells expressing YC3.6 were excited at 440 nm. The fluorescent images were acquired at 490 and 535 nm emission wavelengths using a cooled charge-coupled device camera (CoolSnap HQ; Roper Scientific, Trenton, NJ) and analyzed using Metamorph software (Molecular Devices). [Ca2+]i was calculated from the intensity ratio of two emission wavelengths (F535/F490) according to Nagai et al. (31). Mitochondrial calcium ([Ca2+]mito) was measured using mitochondrial-targeted aequorin as described (28).
Data Analysis—Data are presented as means ± S.E. Statistical significance was determined using Student's t test. p < 0.05 was considered significant.
RESULTS
Mitochondrial Morphology in Pancreatic β-Cells and INS-1E Cells—To visualize the mitochondria of primary pancreatic β-cells, we generated a virus encoding mito-ECFP. ECFP-labeled mitochondria formed tubular networks throughout the cytosol in the majority of human and rat β-cells (Fig. 1A; supplemental Fig. S1A). β-cells were identified by immunofluorescence labeling with an insulin antibody (Fig. 1A). The rat clonal β-cell line INS-1E displays a similar filamentous pattern of mitochondrial distribution as analyzed either with the mitoECFP virus or following transient transfection of mitoRFP (Fig. 2A and data not shown). To manipulate the mitochondrial filamentous network, INS-1E cells were co-transfected with genes of the fission/fusion machinery and mitoRFP. Overexpression of hFis1, DN-Mfn1, or Drp1 caused mitochondrial fragmentation in a large fraction of transfected cells (Fig. 2A, supplemental Fig. S2, A and B). In contrast, overexpression of WT-Mfn1 and WT-Mfn2 caused the aggregation of mitochondria (Fig. 2A and supplemental Fig. S2B).
FIGURE 1.
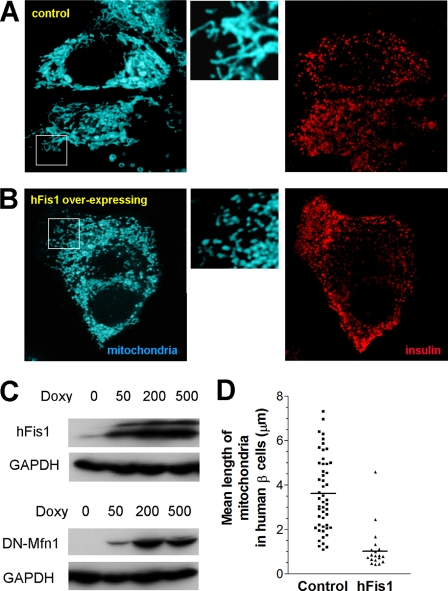
Mitochondrial morphology of human pancreatic β-cells. Dispersed human pancreatic islet cells were infected with adenoviruses expressing mitoECFP and reverse tetracycline-transactivator (A and B). The transgene was induced for 48 h in the presence of 500 ng/ml doxycycline. To identify β-cells, the cells were fixed and stained for insulin (right panels). MitoECFP revealed filamentous mitochondrial structures (A; left panel). Co-infection with adenovirus encoding hFis1 caused dramatic fragmentation of the mitochondria (B; left panel). The middle panels show larger magnifications to highlight the differences in mitochondrial morphology. C, doxycycline (Doxy)-dependent expression of hFis1 (upper panel) or DN-Mfn1 (lower panel) following virus infection of INS-1E cells. For control, endogenous glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was detected. D, quantification of mitochondrial length in human β-cells was performed as described in supplemental Fig. S3.
FIGURE 2.
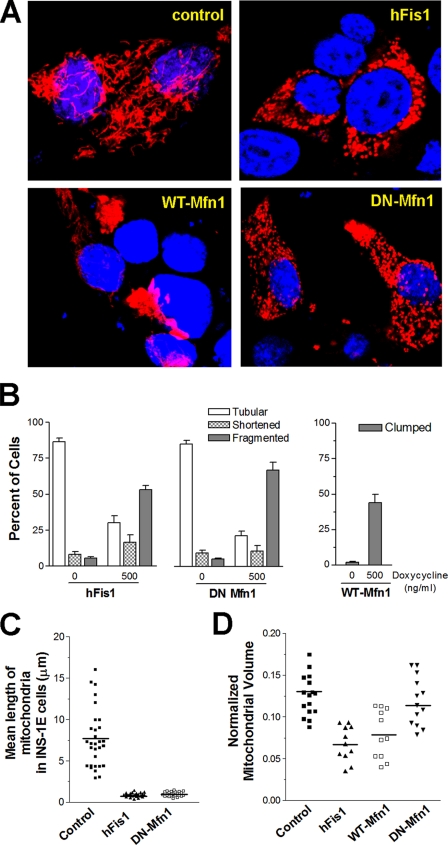
Mitochondrial morphology in INS-1E cells overexpressing fission/fusion genes. INS-1E cells were transiently transfected with mitoRFP to visualize the mitochondria. Following adenovirus infection, the fission/fusion proteins were induced by 500 ng/ml doxycycline for 48 h, and morphological changes were studied. A, representative confocal images show mitochondrial distribution and structure in cells after overexpression of hFis1, WT-Mfn1, and DN-Mfn1. B, the number of cells with tubular, shortened (rod shape), or fragmented (small round) mitochondria is expressed as the percentage of total mitoRFP-transfected cells (left and middle graph). Cells with clumped (aggregated) mitochondria were counted following induction of WT-Mfn1 (right graph). Data are presented as means ± S.E. (n = 6, for each experiment, more than 200 cells were counted). Mitochondrial volume normalized to cytosol was calculated from deconvoluted z-stack confocal images (see supplemental Fig. S4). Mean mitochondrial length (C) and mitochondrial volume (D) were compared between cells expressing different transgenes.
Among the fission/fusion genes tested, we decided to further focus on the role of hFis1 and DN-Mfn1 in insulin-secreting cells. First, we focused on them because hFis1 and Mfn1 mediate key, possibly rate-limiting, steps in fission and fusion, respectively. Second, they induced a pronounced effect on mitochondrial morphology in the majority of the expressing cells (supplemental Fig. S2, A and B). Third, we focused on them because hFis1 and DN-Mfn1 utilize different mechanisms to cause a very similar fragmentation phenotype. Overexpression of Mfn1 was further studied as it caused a mitochondrial morphology defect different from the other two transgenes. For this purpose, we generated adenoviruses for doxycycline-inducible expression of the transgenes in whole populations of INS-1E cells and primary β-cells. Doxycycline-dependent induction of hFis1 (17-kDa) and DN-Mfn1 (84-kDa) protein following infection of INS-1E cells is shown in Fig. 1C. Virus-mediated expression of the transgenes caused morphological changes of the mitochondria in INS-1E cells similar to those observed after transient transfection (Fig. 2B). Similarly, hFis1 overexpression led to the fragmentation of mitochondria in human (Fig. 1B) and rat β-cells (supplemental Fig. S1B). To get a quantitative read-out of the fragmentation phenotype in individual cells, we measured the average length of mitochondria. For this purpose, the mitochondria were “skeletonized” using Metamorph as illustrated in supplemental Fig. S3. This technique allows us to study relative length changes of mitochondria, albeit without taking into consideration mitochondrial distribution over several planes of the z axis. Mean mitochondrial skeleton length of human pancreatic β-cells and rat β-cells was markedly decreased by the overexpression of hFis1 (Fig. 1D and supplemental Fig. S1C). Adenovirus-driven overexpression of hFis1 or DN-Mfn1 elicited similar mitochondrial shortening and fragmentation in INS-1E cells (Fig. 2C). Despite the comparable fragmentation phenotype after induction of either of these two transgenes, we noticed that hFis1 overexpression reduced the mitochondrial area (mitoRFP staining) in the INS-1E cells, possibly reflecting a reduction in mitochondrial volume. To quantify this difference, we calculated the total mitochondrial area relative to the cell size from whole z-stack images of INS-1E cells. The area covered by the cell was measured with the plasma membrane targeted probe YC3.6 pm as described previously (32) and illustrated in supplemental Fig. S4, A–C. The filamentous mitochondria of control INS-1E cells reach the peripheral subplasmalemmal area of the cytosol (Fig. 2A and supplemental Fig. S4A). Overexpression of hFis1 caused clustering of mitochondria in the perinuclear area and therefore a reduction of the area occupied by the fragmented mitochondria (Fig. 2, A and D, and supplemental Fig. S4D). In contrast, the fragmented mitochondria of DN-Mfn1-overexpressing cells reached the cell periphery like in control cells (Fig. 2, A and D, and supplemental Fig. S4F). Mitochondrial aggregation, the consequences of WT-Mfn1 overexpression, also resulted in a relative decrease of area occupied by mitochondria in the cell (Fig. 2D and supplemental Fig. S4E).
hFis1 Induced Apoptotic Cell Death—Several fission/fusion genes have been found to modulate the apoptotic response (3, 33). To assess the effect of hFis1 and DN-Mfn1 on cell viability in the INS-1E cells, we used the colorimetric MTT assay (34). Increasing the virus titer or the doxycycline concentration caused a dose-dependent reduction of the MTT-derived signal, suggesting that the observed defect was proportional to the level of hFis1 expression (supplemental Fig. S5A and data not shown). After infection with the hFis1 virus (60 infection units/cell) and induction of the transgene with 500 ng/ml doxycycline for 48 h (the condition used throughout this study), we observed a decrease of the MTT signal to 66 ± 4% of control. In contrast, overexpression of DN-Mfn1 did not significantly alter cell viability even after infection with 120 infection units/cell of the adenovirus (supplemental Fig. S5A). The 34% reduction of the MTT signal after hFis1 overexpression does not, however, necessarily reflect cell death or loss as the MTT assay also measures the generation of reducing equivalents in this cell type (34). For instance, the observed reduction of mitochondrial volume as demonstrated in Fig. 2D could lower the amount of the mitochondrial enzyme succinate dehydrogenase, one of the main enzymes catalyzing the reduction of MTT. As an alternative measure of cell death or loss, we therefore also monitored the total amount of DNA after extraction. Total DNA content per well was lowered by 10% after hFis1 overexpression (8.18 ± 0.42 in non-induced versus 7.36 ± 0.39 μg/well in induced cells, n = 11, p < 0.01). The MTT assay may therefore overestimate the effect of hFis1 on cell death.
To determine whether the reduced cell number is due to apoptosis, we employed the TUNEL assay. Doxycycline induction (500 ng/ml for 24 h) of hFis1 increased the percentage of TUNEL-positive cells 2.5-fold when compared with that of the non-induced control (supplemental Fig. S5, B and C). DN-Mfn1 overexpression did not increase the proportion of TUNEL-positive cells. Our results show that hFis1 only causes a modest decrease of cell number when overexpressed, which may be explained by the increased percentage of cells undergoing apoptosis.
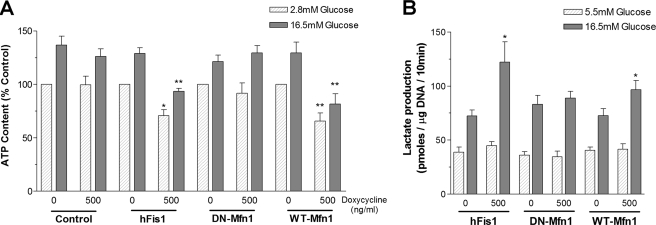
hFis1 Lowered Cellular ATP Levels and Augmented Lactate Production—Fission/fusion genes may directly affect mitochondrial metabolism as suggested by others (8, 14). We therefore tested whether hFis1 overexpression could have an impact on energy metabolism by measuring total cellular ATP content (Fig. 3A). ATP is expressed relative to the DNA content of the samples to correct for the moderate reduction of cell number after overexpression of hFis1. Incubation with stimulating glucose concentration (16.5 mm) for 10 min increased ATP content from 70.7 ± 15.4 to 94.1 ± 19.4 pmol/μg of DNA (p < 0.05) in control INS-1E cells. hFis1 overexpression lowered ATP content at basal (52.3 ± 9.2 pmol/μg of DNA, p < 0.05) and stimulating glucose concentrations (71.3 ± 12.6 pmol/μg of DNA, p < 0.01, Fig. 3A). On the other hand, DN-Mfn1 neither affected cellular ATP levels at low glucose nor affected cellular ATP levels at high glucose (67.7 ± 11.5 and 93.3 ± 10.9 pmol/μg of DNA, respectively). Overexpression of WT-Mfn1 reduced the ATP content at basal (49.8 ± 8.5 pmol/μg of DNA, p < 0.01) and stimulating glucose concentration (62.0 ± 10.3 pmol/μg of DNA, p < 0.01).
FIGURE 3.
hFis1 overexpression reduces cellular ATP content and increases lactate production. After infection with adenovirus coding for hFis1, DN-Mfn1, or WT-Mfn1, INS-1E cells were incubated for 48 h with or without doxycycline (500 ng/ml). A, ATP contents were measured 10 min after stimulation with 2.8 mm (basal) or 16.5 mm glucose. Cellular ATP was normalized to the DNA content and expressed as the percentage of control for each group (basal level in non-induced cells; n = 7). B, lactate production after a 10-min incubation with either 5.5 mm or 16.5 mm glucose was normalized to the DNA content (pmol of lactate/μg of DNA (n = 6)). Data are presented as means ± S.E. * and ** denote p < 0.05 and p < 0.01, respectively.
ATP is generated from glycolysis as well as mitochondrial oxidative phosphorylation. In cells with defects in mitochondrial oxidative phosphorylation such as mtDNA-depleted cells, accelerated glycolytic ATP production and lactate synthesis have been observed (35). To test whether a mitochondrial defect is responsible for reduced ATP production and whether INS-1E cells overexpressing hFis1 compensate by increasing glycolysis-derived ATP synthesis, we measured lactate production. Indeed, the increment of lactate production to high glucose was 2.3-fold higher in hFis1-overexpressing cells than in non-induced control cells (p < 0.05; Fig. 3B). WT-Mfn1 overexpression also augmented glucose-stimulated lactate production (1.7-fold, p < 0.05), whereas DN-Mfn1 did not (Fig. 3B). Thus, the hFis1- and Mfn1-dependent mitochondrial defect in ATP synthesis is partially compensated for by an increase in glycolytic flux.
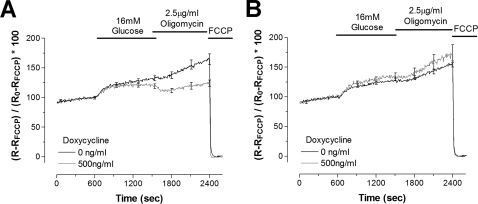
hFis1 Attenuated Hyperpolarization of ΔΨm by Glucose or Oligomycin—Glucose is known to hyperpolarize the mitochondrial membrane potential, which is essential for ATP generation (36). To estimate the impact of mitochondrial fragmentation on mitochondrial activation, we assessed mitochondrial membrane potential (ΔΨm) changes using the indicator dye JC-1, allowing ratiometric recording. Doxycycline induction of hFis1 attenuated hyperpolarization in response to glucose (Fig. 4A; p < 0.05). Conversely, expression of DN-Mfn1 did not decrease glucose-induced hyperpolarization when compared with the control (Fig. 4B).
FIGURE 4.
Effects of overexpression of fission genes on mitochondrial membrane potential. After infection with adenoviruses coding for hFis1 (A) or DN-Mfn1 (B), INS-1E cells were cultured and loaded with JC-1 (500 nm) as described under “Experimental Procedures.” The mitochondrial membrane potential (Ψm) was measured at 37 °C. Changes in a fluorescence ratio of red (excitation 540 nm/emission 590 nm) over green (490 nm/540 nm) were normalized to the ratio difference between resting (100%) and FCCP-induced depolarization (0%) and expressed as the percentage of change ((R - RFCCP)/(R0 - RFCCP) × 100). Averaged results (eight traces for each condition) from non-induced (black line) and cells overexpressing (gray line) hFis1 (A) and DN-Mfn1 (B) are shown including error bars every 300 s (S.E.; n = 8).
Dysfunctional and depolarized mitochondria are known to maintain ΔΨm by reversal of the F1F0-ATPase, resulting in ATP consumption instead of production (37). Blockage of the F1F0-ATPase by oligomycin causes hyperpolarization in normal mitochondria and depolarization in dysfunctional mitochondria. Here we added oligomycin (2.5 μg/ml) following glucose stimulation. Although control non-induced cells displayed further hyperpolarization by oligomycin, this response was strongly reduced when hFis1 was overexpressed (Fig. 4A, p < 0.01). Unlike hFis1, DN-Mfn1 induced hyperpolarization similar to the control. Moreover, oligomycin-induced hyperpolarization was even slightly increased (Fig. 4B). These results suggest that fragmentation by hFis1 impairs mitochondrial function, whereas defective fusion secondary to DN-Mfn1 overexpression does not.
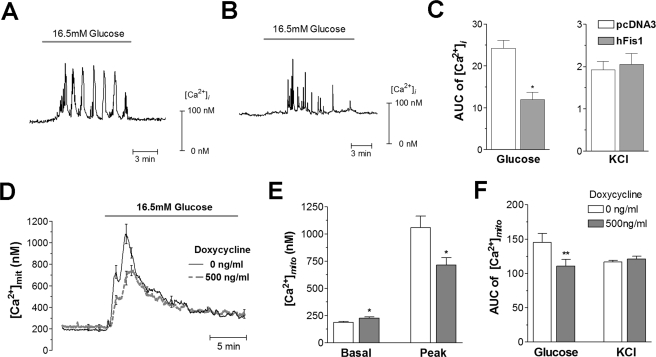
hFis1 Reduced Glucose-stimulated Cytosolic and Mitochondrial Calcium Responses—ATP acts as a central signal linking nutrient metabolism to plasma-membrane depolarization, calcium influx, and insulin exocytosis (38). Despite their defect in mitochondrial energy metabolism, hFis1-overexpressing cells still display a glucose-dependent increase in ATP generation. To test whether the observed reduction of ATP levels at low and high glucose concentrations affects downstream signals in metabolism-secretion coupling, we measured cytosolic and mitochondrial calcium. The cytosolic calcium concentration ([Ca2+]i) was analyzed following transient co-transfection of hFis1 with the yellow cameleon probe YC3.6 and mitoRFP (to analyze the fragmentation state of the mitochondria). In control cells, glucose evoked high frequency calcium transients or slow [Ca2+]i oscillations (Fig. 5A). Cells with fragmented mitochondria due to overexpression of hFis1 showed significantly lower amplitude of [Ca2+]i transients when compared with the control cells (Fig. 5B). To quantify this phenotype, we integrated the signal above basal during 10 min after glucose addition (in Fig. 5, AUC indicates area under the curve). The area under the curve in response to glucose was reduced by 52% in hFis1-overexpressing cells when compared with control cells (p < 0.05; Fig. 5C). DN-Mfn1-induced mitochondrial fragmentation did not reduce glucose-stimulated [Ca2+]i transients (data not shown). We also used KCl (30 mm), a stimulus that directly depolarizes the plasma membrane without requiring cellular metabolism (20). KCl caused a marked increase in [Ca2+]i, which was not significantly altered by hFis1 transfection (Fig. 5C). These results show that voltage-gated Ca2+ influx is unaffected, attributing the attenuated action of glucose to defective mitochondrial function.
FIGURE 5.
Reduced glucose-stimulated cytosolic ([Ca2+]i) and mitochondrial calcium ([Ca2+]mito) increases in hFis1-overexpressing INS-1E cells. A–C, changes in [Ca2+]i were measured using YC3.6. Representative traces from control (transfection with pcDNA3 empty vector: A, 29 cells from six experiments) and hFis1-overexpressing (B, 31 cells from nine experiments) cells are shown. Areas under the curve were calculated during the first 10 min after glucose application or the first 10 s after KCl addition (C). AUC, area under the curve. D–F, changes in [Ca2+]mito were measured using mitochondrial-targeted aequorin and compared between non-induced (n = 9) and doxycycline (500 ng/ml)-induced hFis1-overexpressing cells (n = 9; D). The basal and peak levels of [Ca2+]mito in response to glucose (E) and the areas under the curve following glucose or KCl stimulation (F) were compared between the two groups. Data are presented as means ± S.E. * and ** denote p < 0.05 and p < 0.01, respectively.
Mitochondrial calcium is an important signal in β-cell metabolism-secretion coupling with a potential feedback stimulatory effect on mitochondrial metabolism (28, 38). We therefore tested whether hFis1 overexpression alters the mitochondrial calcium response to glucose or KCl. In noninduced control cells, glucose rapidly increased the average mitochondrial calcium concentration ([Ca2+]mito) from 195 ± 8 nm to a peak amplitude of 1062 ± 75 nm, which thereafter gradually returned to resting levels (Fig. 5, D and E). Doxycycline-induced overexpression of hFis1 resulted in modest elevation of the resting [Ca2+]mito levels (226 ± 13 nm; p < 0.05) and in a lowering of the peak Ca2+ response (839 ± 55 nm; p < 0.05; Fig. 5E). The glucose-mediated area under the curve increase of [Ca2+]mito was reduced by 33% in hFis1-overexpressing cells (p < 0.01; Fig. 5F). Mitochondrial calcium responses to KCl were unchanged after hFis1 overexpression (Fig. 5F).
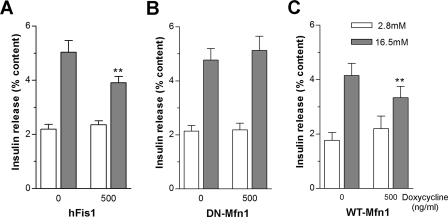
hFis1, but Not DN-Mfn1, Reduced Glucose-stimulated Insulin Secretion—Based on the reduced ATP levels in hFis1-overexpressing cells and the corresponding reduced cytosolic calcium responses, we tested whether glucose-stimulated insulin secretion is affected by hFis1-dependent mitochondrial fragmentation. Initially, experiments were performed by transiently co-transfecting plasmids encoding mitochondrial fission proteins and human growth hormone. This hormone was used as a marker for insulin in the subpopulation of transiently co-transfected cells since human growth hormone is co-stored with insulin in the secretory granules (39). hFis1 but not DN-Mfn1 overexpression in INS-1E cells decreased glucose-mediated release of human growth hormone containing granules both at intermediate and at high glucose concentrations (supplemental Fig. S6). Similar experiments were then performed using the doxycycline-inducible adenoviral vectors. The insulin content was reduced by 12% following induction of hFis1 (3.09 ± 0.24 in non-induced versus 2.72 ± 0.26 μg/well in induced cells, n = 7, p < 0.01). These results reflect the observed cell death caused by hFis1 and are consistent with the 10% reduction of DNA content as described earlier. Glucose-stimulated insulin release was therefore expressed as the percentage of cellular content to correct for hFis1-mediated cell loss (Fig. 6). In hFis1 infected non-induced control cells, insulin secretion was increased 2.3-fold when the glucose concentration was raised from 2.8 to 16.5 mm. After induction of hFis1, the glucose effect was reduced to 1.6-fold (p < 0.01; Fig. 6A). Shifting cells expressing DN-Mfn1 from 2.8 to 16.5 mm glucose resulted in 2.4-fold increase in insulin secretion (Fig. 6B), which did not differ from control. WT-Mfn1 overexpression attenuated the ratio of glucose-stimulated insulin release to 1.5-fold (p < 0.01; Fig. 6C). Our results demonstrate that hFis1 causes a specific defect in metabolism secretion coupling resulting in defective insulin secretion beyond and above its effect on cell viability. In contrast, the similar mitochondrial phenotype induced by DN-Mfn1 did not affect metabolism-secretion coupling.
FIGURE 6.
Alteration in glucose-stimulated insulin release by adenoviral-driven overexpression of mitochondrial fission/fusion genes. After infection with adenovirus expressing hFis1 (A; n = 7), DN-Mfn1 (B; n = 7), or WT-Mfn1 (C; n = 6), INS-1E cells were incubated with or without doxycycline (500 ng/ml) for 48 h. Insulin release was measured at 2.8 and 16.5 mm glucose. Insulin release was normalized to the cellular insulin content. Data presented are means ± S.E., and ** denotes p < 0.01.
DISCUSSION
Here we investigated the role of the mitochondrial network in the physiological regulation of glucose-stimulated insulin secretion, a process depending on oxidative phosphorylation. Overexpression or suppression of fission/fusion proteins can shift the mitochondrial structure from an extensive network to fragmented and isolated mitochondria. We demonstrate that human β-cells display elongated and filamentous mitochondria distributed throughout the cytoplasm. To our knowledge, mitochondrial networks have not been described in human β-cells previously. Rat β-cells also show tubular mitochondrial structure, which is consistent with the observation by Bindokas et al. (16) in islets from lean rats stained with rhodamine 123. The extent of mitochondrial elongation varied between human β-cells. Overexpression of hFis1 invariably caused pronounced fragmentation of the organelle in both primary β-cells and INS-1E cells. The extent of mitochondrial shortening and mitochondrial fragmentation was similarly changed when fusion was inhibited after overexpression of DN-Mfn1 in INS-1E cells, consistent with earlier findings in HeLa cells (9).
hFis1 overexpression caused inhibition of glucose-induced insulin release, explained by attenuated nutrient-dependent hyperpolarization of the ΔΨm and reduced cytosolic and mitochondrial Ca2+ signals. The calcium signaling defects are the result of impaired mitochondrial metabolism as K+-induced cytosolic and mitochondrial Ca2+ increases were unaffected. In contrast, DN-Mfn1 expression preserved mitochondrial hyperpolarization and cellular ATP levels as well as a normal secretory response to glucose.
Oligomycin causes depolarization in dysfunctional mitochondria, which consume cytosolic ATP to maintain the ΔΨm by reversal of the F1F0-ATPase. Thus, evaluating the changes in ΔΨm by oligomycin could uncover latent mitochondrial defects (37, 40). Indeed, hyperpolarization in response to oligomycin was abolished in hFis1-overexpressing cells, consistent with previous observations in mouse embryonic fibroblasts (29). Fragmented mitochondria are known to be heterogeneous with regard to their ΔΨm (8). We therefore speculate that reduced hyperpolarization to oligomycin is due to an increased proportion of ATP-consuming mitochondria. In contrast, INS-1E cells expressing DN-Mfn1 retained oligomycin-induced hyperpolarization, which indicates that fragmentation of mitochondria per se does not alter control of the ΔΨm.
The lowered ATP levels in hFis1-overexpressing cells observed here are one of the consequences of mitochondrial dysfunction. Such cells actually compensated for their mitochondrial defect by accelerating glycolysis reflected by increased lactate production. This mechanism of compensation has also been observed in insulin-secreting βHC9 cells depleted of mtDNA (35). However, increased glycolysis cannot fully normalize cellular ATP levels in hFis1-overexpressing cells. In cells overexpressing DN-Mfn1, ATP levels were similar to control, and judging from the normal lactate response, mitochondrial activity was sufficient to maintain control ATP levels at both basal and stimulatory glucose concentrations. It is remarkable that despite similar mitochondrial fragmentation, the two transgenes had such strikingly different effects on mitochondrial energy metabolism.
Overexpression of wild type Mfn1 caused mitochondrial clumping, the result of excessive mitochondrial fusion in INS-1E cells as well as primary rat β-cells. To date, there is no information on the impact of mitochondrial hyperfusion by Mfn1 on mitochondrial energy metabolism. In this study, we show that Mfn1 overexpression reduced the ATP content, increased lactate production, and impaired glucose-stimulated insulin secretion. Consistent with our findings, several groups have reported that oxygen consumption and ATP generation are reduced in cells with highly fused mitochondria due to the down-regulation of fission proteins (14, 15).
It is interesting that INS-1E cells overexpressing hFis1 or WT-Mfn1 have a smaller total volume of mitochondria. Although we expected the reduced mitochondrial volume to be associated with decreased mtDNA copy number, direct quantitative PCR measurement of mtDNA did not substantiate this (data not shown). Reduced mitochondrial volume may, however, explain at least in part the observed reduced mitochondrial ATP production. In cells overexpressing DN-Mfn1, the mitochondrial volume was only little affected, and the fragmented organelles appeared to reach even peripheral areas of the cell. Taken together, there is a correlation between the mitochondrial volume and the ability of the cell to maintain cytosolic ATP levels on the one hand and to sense mitochondria-derived signals stimulating insulin secretion on the other.
In our study, hFis1 had a more marked impact on mitochondrial metabolism including reduction of MTT (reflecting oxidative metabolism in INS-1 cells (34)) than on cumulative cell loss, monitored by DNA and insulin contents. Even massive overexpression of hFis1 in INS-1E cells causes only moderate reduction in cell number by apoptosis. In other cell types, the consequences of hFis1 overexpression on apoptosis are more pronounced (41). On the other hand, DN-Mfn1 does not result in apoptosis, which is therefore not the consequence of mitochondrial fragmentation itself. In β-cells, Bcl-XL overexpression prevented apoptosis at the cost of impaired glucose-stimulated insulin secretion (42). In other cell types, it has been reported that overexpression of Bcl-XL attenuates apoptosis without affecting fragmentation triggered by hFis1 (3). Conversely, dominant-negative Drp1 prevented mitochondrial fragmentation but not apoptosis induced by hFis1 or apoptogenic stimuli (3, 43). Thus, converging evidence from a number of cell types suggests independent regulation of mitochondrial fission and apoptosis.
We also suggest that hFis1-induced mitochondrial dysfunction is not causally related to structural changes. Recently, strong activation of autophagosome formation by hFis1 overexpression has been demonstrated in mouse fibroblasts, which correlated with mitochondrial dysfunction rather than with fragmentation (44). Fragmentation per se is not sufficient to trigger autophagy, which could explain why the mitochondrial volume was selectively decreased in hFis1-overexpressing cells. It is noteworthy that endogenous Fis1 protein is required for autophagy contributing to a quality control mechanism improving cellular function including mitochondrial respiration. Down-regulation of Fis1 impaired mitochondrial function and metabolism-secretion coupling in INS-1 cells due to reduced autophagy of defective mitochondria (12).
It is of interest that β-cells from Zucker diabetic fatty rats showed fragmented mitochondria and increased ROS production, preceding the onset of diabetes (16). Likewise, muscle mitochondria from obese hyperinsulinemic normoglycemic Zucker rats show fragmentation and reduced volume (1). In obese subjects and in type 2 diabetic patients, skeletal muscle mitochondria are also smaller than those from lean subjects (45). The altered mitochondrial morphology may be implicated in the development of type 2 diabetes or merely a consequence of the progression of the disease. It is noteworthy that mutations in Mfn2 have been linked to the Charcot Marie Tooth-2A (CMT-2A) disease, a peripheral neuropathy, which is associated with type 2 diabetes (46, 47). Here we report that the fission/fusion genes are able to induce dramatic alterations in mitochondrial structure that have a clear impact on metabolism-secretion coupling beyond a secondary effect on cell survival. Metabolism-secretion coupling may be affected by alterations of the bioenergetic properties of mitochondria when hFis1 or Mfn1 is overexpressed. Furthermore, reduced mitochondrial volume and displacement away from the cell periphery could alter metabolism secretion coupling by disconnecting mitochondrial signal generation from signal sensing at the plasma membrane. Our results also demonstrate that disruption of the mitochondrial network per se such as following overexpression of DN-Mfn1 does not impair metabolism-secretion coupling during glucose-induced insulin release. Moreover, the altered mitochondrial fragmentation alone does not incur cell death. Mitochondrial fragmentation following hFis1-mediated hyperfission has dramatically different effects on metabolism-secretion coupling than DN-Mfn1-mediated prevention of mitochondrial fusion.
Supplementary Material
Acknowledgments
We are grateful to Danielle Nappey, Olivier Dupont, Dr. Francoise Assimacopoulus-Jeannet, Dr. Oliver Hartley, and Dr. Sergei Startchik.
This work was supported by the Swiss National Foundation and EuroDia (Grant LSHM-CT-2006-518153), a European Community-funded project under framework program 6 (to C. B. W.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains six supplemental figures.
This article was selected as a Paper of the Week.
Footnotes
The abbreviations used are: DN, dominant-negative; WT, wild type; ECFP, mitochondrial targeted enhanced cyan fluorescent protein; mitoECFP, mitochondrial targeted ECFP; RFP, red fluorescent protein; mitoRFP, mitochondrial RFP; FCCP, carbonyl cyanide p-trifluoromethoxyphenylhydrazone; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; TUNEL, terminal deoxynucleotidyltransferase-mediated dUTP nick end-labeling.
References
- 1.Bach, D., Pich, S., Soriano, F. X., Vega, N., Baumgartner, B., Oriola, J., Daugaard, J. R., Lloberas, J., Camps, M., Zierath, J. R., Rabasa-Lhoret, R., Wallberg-Henriksson, H., Laville, M., Palacin, M., Vidal, H., Rivera, F., Brand, M., and Zorzano, A. (2003) J. Biol. Chem. 278 17190-17197 [DOI] [PubMed] [Google Scholar]
- 2.Chan, D. C. (2006) Annu. Rev. Cell Dev. Biol. 22 79-99 [DOI] [PubMed] [Google Scholar]
- 3.James, D. I., Parone, P. A., Mattenberger, Y., and Martinou, J. C. (2003) J. Biol. Chem. 278 36373-36379 [DOI] [PubMed] [Google Scholar]
- 4.Smirnova, E., Griparic, L., Shurland, D. L., and van der Bliek, A. M. (2001) Mol. Biol. Cell 12 2245-2256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoon, Y., Krueger, E. W., Oswald, B. J., and McNiven, M. A. (2003) Mol. Cell. Biol. 23 5409-5420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stojanovski, D., Koutsopoulos, O. S., Okamoto, K., and Ryan, M. T. (2004) J. Cell Sci. 117 1201-1210 [DOI] [PubMed] [Google Scholar]
- 7.Ishihara, N., Eura, Y., and Mihara, K. (2004) J. Cell Sci. 117 6535-6546 [DOI] [PubMed] [Google Scholar]
- 8.Chen, H., Chomyn, A., and Chan, D. C. (2005) J. Biol. Chem. 280 26185-26192 [DOI] [PubMed] [Google Scholar]
- 9.Santel, A., Frank, S., Gaume, B., Herrler, M., Youle, R. J., and Fuller, M. T. (2003) J. Cell Sci. 116 2763-2774 [DOI] [PubMed] [Google Scholar]
- 10.Detmer, S. A., and Chan, D. C. (2007) Nat. Rev. Mol. Cell. Biol. 8 870-879 [DOI] [PubMed] [Google Scholar]
- 11.Kim, I., Rodriguez-Enriquez, S., and Lemasters, J. J. (2007) Arch. Biochem. Biophys. 462 245-253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Twig, G., Elorza, A., Molina, A. J., Mohamed, H., Wikstrom, J. D., Walzer, G., Stiles, L., Haigh, S. E., Katz, S., Las, G., Alroy, J., Wu, M., Py, B. F., Yuan, J., Deeney, J. T., Corkey, B. E., and Shirihai, O. S. (2008) EMBO J. 27 433-446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen, H., Detmer, S. A., Ewald, A. J., Griffin, E. E., Fraser, S. E., and Chan, D. C. (2003) J. Cell Biol. 160 189-200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Benard, G., Bellance, N., James, D., Parrone, P., Fernandez, H., Letellier, T., and Rossignol, R. (2007) J. Cell Sci. 120 838-848 [DOI] [PubMed] [Google Scholar]
- 15.Lee, S., Jeong, S. Y., Lim, W. C., Kim, S., Park, Y. Y., Sun, X., Youle, R. J., and Cho, H. (2007) J. Biol. Chem. 282 22977-22983 [DOI] [PubMed] [Google Scholar]
- 16.Bindokas, V. P., Kuznetsov, A., Sreenan, S., Polonsky, K. S., Roe, M. W., and Philipson, L. H. (2003) J. Biol. Chem. 278 9796-9801 [DOI] [PubMed] [Google Scholar]
- 17.Noske, A. B., Costin, A. J., Morgan, G. P., and Marsh, B. J. (2008) J. Struct. Biol. 161 298-313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schuit, F., De Vos, A., Farfari, S., Moens, K., Pipeleers, D., Brun, T., and Prentki, M. (1997) J. Biol. Chem. 272 18572-18579 [DOI] [PubMed] [Google Scholar]
- 19.Wiederkehr, A., and Wollheim, C. B. (2006) Endocrinology 147 2643-2649 [DOI] [PubMed] [Google Scholar]
- 20.Kennedy, E. D., Maechler, P., and Wollheim, C. B. (1998) Diabetes 47 374-380 [DOI] [PubMed] [Google Scholar]
- 21.Maassen, J. A., Janssen, G. M., and 't Hart, L. M. (2005) Ann. Med. 37 213-221 [DOI] [PubMed] [Google Scholar]
- 22.Murphy, R., Turnbull, D. M., Walker, M., and Hattersley, A. T. (2008) Diabet. Med. 25 383-399 [DOI] [PubMed] [Google Scholar]
- 23.Deng, S., Vatamaniuk, M., Huang, X., Doliba, N., Lian, M. M., Frank, A., Velidedeoglu, E., Desai, N. M., Koeberlein, B., Wolf, B., Barker, C. F., Naji, A., Matschinsky, F. M., and Markmann, J. F. (2004) Diabetes 53 624-632 [DOI] [PubMed] [Google Scholar]
- 24.Anello, M., Lupi, R., Spampinato, D., Piro, S., Masini, M., Boggi, U., Del Prato, S., Rabuazzo, A. M., Purrello, F., and Marchetti, P. (2005) Diabetologia 48 282-289 [DOI] [PubMed] [Google Scholar]
- 25.Del Guerra, S., Lupi, R., Marselli, L., Masini, M., Bugliani, M., Sbrana, S., Torri, S., Pollera, M., Boggi, U., Mosca, F., Del Prato, S., and Marchetti, P. (2005) Diabetes 54 727-735 [DOI] [PubMed] [Google Scholar]
- 26.Maechler, P., Kennedy, E. D., Wang, H., and Wollheim, C. B. (1998) J. Biol. Chem. 273 20770-20778 [DOI] [PubMed] [Google Scholar]
- 27.Brun, T., Franklin, I., St-Onge, L., Biason-Lauber, A., Schoenle, E. J., Wollheim, C. B., and Gauthier, B. R. (2004) J. Cell Biol. 167 1123-1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ishihara, H., Maechler, P., Gjinovci, A., Herrera, P. L., and Wollheim, C. B. (2003) Nat. Cell Biol. 5 330-335 [DOI] [PubMed] [Google Scholar]
- 29.Alirol, E., James, D., Huber, D., Marchetto, A., Vergani, L., Martinou, J. C., and Scorrano, L. (2006) Mol. Biol. Cell 17 4593-4605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang, Y., Soboloff, J., Zhu, Z., and Berger, S. A. (2006) Mol. Pharmacol. 70 1424-1434 [DOI] [PubMed] [Google Scholar]
- 31.Nagai, T., Yamada, S., Tominaga, T., Ichikawa, M., and Miyawaki, A. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 10554-10559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frieden, M., Arnaudeau, S., Castelbou, C., and Demaurex, N. (2005) J. Biol. Chem. 280 43198-43208 [DOI] [PubMed] [Google Scholar]
- 33.Lee, Y. J., Jeong, S. Y., Karbowski, M., Smith, C. L., and Youle, R. J. (2004) Mol. Biol. Cell 15 5001-5011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Janjic, D., and Wollheim, C. B. (1992) Diabetologia 35 482-485 [DOI] [PubMed] [Google Scholar]
- 35.Noda, M., Yamashita, S., Takahashi, N., Eto, K., Shen, L. M., Izumi, K., Daniel, S., Tsubamoto, Y., Nemoto, T., Iino, M., Kasai, H., Sharp, G. W., and Kadowaki, T. (2002) J. Biol. Chem. 277 41817-41826 [DOI] [PubMed] [Google Scholar]
- 36.Maechler, P., and Wollheim, C. B. (2001) Nature 414 807-812 [DOI] [PubMed] [Google Scholar]
- 37.Irwin, W. A., Bergamin, N., Sabatelli, P., Reggiani, C., Megighian, A., Merlini, L., Braghetta, P., Columbaro, M., Volpin, D., Bressan, G. M., Bernardi, P., and Bonaldo, P. (2003) Nat. Genet. 35 367-371 [DOI] [PubMed] [Google Scholar]
- 38.Wiederkehr, A., and Wollheim, C. B. (2008) Cell Calcium 44 64-76 [DOI] [PubMed] [Google Scholar]
- 39.Iezzi, M., Kouri, G., Fukuda, M., and Wollheim, C. B. (2004) J. Cell Sci. 117 3119-3127 [DOI] [PubMed] [Google Scholar]
- 40.Wikstrom, J. D., Katzman, S. M., Mohamed, H., Twig, G., Graf, S. A., Heart, E., Molina, A. J., Corkey, B. E., de Vargas, L. M., Danial, N. N., Collins, S., and Shirihai, O. S. (2007) Diabetes 56 2569-2578 [DOI] [PubMed] [Google Scholar]
- 41.Barsoum, M. J., Yuan, H., Gerencser, A. A., Liot, G., Kushnareva, Y., Graber, S., Kovacs, I., Lee, W. D., Waggoner, J., Cui, J., White, A. D., Bossy, B., Martinou, J. C., Youle, R. J., Lipton, S. A., Ellisman, M. H., Perkins, G. A., and Bossy-Wetzel, E. (2006) EMBO J. 25 3900-3911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou, Y. P., Pena, J. C., Roe, M. W., Mittal, A., Levisetti, M., Baldwin, A. C., Pugh, W., Ostrega, D., Ahmed, N., Bindokas, V. P., Philipson, L. H., Hanahan, D., Thompson, C. B., and Polonsky, K. S. (2000) Am. J. Physiol. 278 E340-E351 [DOI] [PubMed] [Google Scholar]
- 43.Parone, P. A., James, D. I., Da Cruz, S., Mattenberger, Y., Donze, O., Barja, F., and Martinou, J. C. (2006) Mol. Cell. Biol. 26 7397-7408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gomes, L. C., and Scorrano, L. (2008) Biochim. Biophys. Acta 1777 860-866 [DOI] [PubMed] [Google Scholar]
- 45.Kelley, D. E., He, J., Menshikova, E. V., and Ritov, V. B. (2002) Diabetes 51 2944-2950 [DOI] [PubMed] [Google Scholar]
- 46.Celik, M., Forta, H., Parman, Y., Bissar-Tadmouri, N., Demirkirkan, K., and Battaloglu, E. (2001) Diabet. Med. 18 685-686 [PubMed] [Google Scholar]
- 47.Koc, F., Sarica, Y., Yerdelen, D., Baris, I., Battaloglu, E., and Sert, M. (2006) Int. J. Neurosci. 116 103-114 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.