Abstract
Unstable lipid-rich plaques in atherosclerosis are characterized by the accumulation of macrophage foam cells loaded with cholesterol ester (CE). Although hormone-sensitive lipase and cholesteryl ester hydrolase (CEH) have been proposed to mediate the hydrolysis of CE in macrophages, circumstantial evidence suggests the presence of other enzymes with neutral cholesterol ester hydrolase (nCEH) activity. Here we show that the murine orthologue of KIAA1363, designated as neutral cholesterol ester hydrolase (NCEH), is a microsomal nCEH with high expression in murine and human macrophages. The effect of various concentrations of NaCl on its nCEH activity resembles that on endogenous nCEH activity of macrophages. RNA silencing of NCEH decreases nCEH activity at least by 50%; conversely, its overexpression inhibits the CE formation in macrophages. Immunohistochemistry reveals that NCEH is expressed in macrophage foam cells in atherosclerotic lesions. These data indicate that NCEH is responsible for a major part of nCEH activity in macrophages and may be a potential therapeutic target for the prevention of atherosclerosis.
Atherosclerotic cardiovascular diseases are the leading causes of mortality in industrialized countries, despite advances in the management of coronary risk factors. Heart attacks arise from thrombotic occlusion of coronary arteries following the rupture of plaques. Lipid-rich plaques, which are characterized by a plethora of CE3-laden macrophage foam cells, are prone to rupture (1). Thus, it is important to clarify the mechanism that eliminates CE from macrophage-derived foam cells.
Foam cells are generated by the unlimited uptake of modified lipoproteins through scavenger receptors (2). Cholesterol in the lipoproteins is stored in lipid droplets as CE after re-esterification by acyl-CoA:cholesterol acyltransferase 1 (ACAT1) (3). Hydrolysis of CE is the initial step toward elimination of cholesterol from foam cells (4). Free cholesterol thus generated is re-esterified or is released from the cells primarily through ATP-binding cassette transporters (5). Thus, the balance between synthesis and hydrolysis of CE conceivably governs the level of CE in macrophages.
Hydrolysis of CE in macrophages has been known for over 40 years (6). However, its molecular mechanism has yet to be fully understood. Circumstantial evidence suggests that the hydrolysis of CE in foam cell macrophages is mediated by hormone-sensitive lipase (HSL), a multifunctional enzyme that catalyzes the hydrolysis of triacylglyerol (TG), diacylglycerol, CE, and retinyl ester in various organs such as adipose tissue, muscle, and testis (7, 8). This belief is supported by the following facts. First, it has been demonstrated that various lines of macrophages express HSL (9–12). Second, HSL expression is regulated coordinately with nCEH activity in murine macrophages (13). Third, we (14) and others (15) demonstrated that overexpression of HSL is associated with increased hydrolysis of CE stores in THP-1 and RAW264.7 macrophages.
However, recent studies by us (16) and others (17) have challenged this notion by finding that peritoneal macrophages isolated from HSL-deficient mice retained almost a normal level of nCEH activity, suggesting that enzyme(s) other than HSL are responsible for nCEH activity in macrophages. Moreover, HSL is expressed at a low level (11, 12) or is undetectable (17) in human macrophages. Recently, Ghosh et al. (18) reported cloning of cholesteryl ester hydrolase (CEH) which mediates the hydrolysis of CE in human monocyte-derived macrophages. They have further shown that macrophage-specific transgenic expression of CEH significantly reduced atherosclerosis in low density lipoprotein receptor knock-out mice (19). However, the CEH gene is identical to the human orthologue of TGH, which was originally reported as a microsomal TG lipase in rat liver by Lehner et al. (20). In our hands, both the mouse orthologue of TGH, designated as TGH-1, and its paralogue with 70.4% identity to TGH-1, designated as TGH-2, have negligible nCEH activity (21). These considerations have prompted us to employ a bioinformatic approach to identify the enzyme responsible for nCEH activity in macrophages.
EXPERIMENTAL PROCEDURES
Materials—Phorbol 12-myristate 13-acetate, triolein, lecithin, bovine serum albumin fraction V (BSA), p-nitrophenyl butyrate, and leupeptin were purchased from Sigma. Dexamethasone, 3-isobutyl-2-methylxanthine, fatty acid-free BSA, cholesterol esterase from Pseudomonas sp., cholesterol oxidase, horseradish peroxidase, p-hydroxyphenylacetic acid, and sodium taurocholate were purchased from Wako Pure Chemicals (Osaka, Japan). Pioglitazone was provided by Takeda Pharmaceutical (Osaka, Japan). Tri[3H]oleoylglycerol, cholesterol [1-14C]oleate, and [1-14C]oleic acid were purchased from GE Healthcare. LDL was isolated by ultracentrifugation and acetylated to prepare acetylated LDL (acLDL) as described previously (14).
Cells—HEK293 or RAW264.7 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) high glucose containing 10% (v/v) fetal bovine serum without or with 1 mm sodium pyruvate, respectively. THP-1 cells were cultured in RPMI 1640 medium containing 10% (v/v) fetal bovine serum and differentiated to THP-1 macrophages by treatment with 100 nm phorbol 12-myristate 13-acetate for 48 h. Murine peritoneal macrophages (MPM) were harvested as described previously (16). In brief, 1 ml of 5% thioglycolate broth was injected into the peritoneal cavities of mice. After 4 days, the peritoneal cavities were lavaged with 16 ml of ice-cold saline. The cells were washed three times with PBS and resuspended in DMEM to give a concentration of 106 cells per ml, and 10 ml per dish was plated in 10-cm dishes. After incubation at 37 °C for 2 h, the nonadherent cells were removed by washing three times with warmed PBS. Cells were incubated in DMEM containing 10% fetal calf serum for 24 h and harvested for the experiments. 3T3-L1 cells were cultured in medium A (DMEM containing 10% calf serum, supplemented with calcium pantothenate and biotin). To induce differentiation of 3T3-L1 cells into adipocytes, 2 day postconfluent preadipocytes (day 0) were incubated with 1 μm dexamethasone and 0.5 mm isobutylmethylxanthine, 5 μg/ml insulin, 1 μm pioglitazone for 48 h, following treatment with 5 μg/ml insulin and 1 μm pioglitazone for 48 h. After the incubation period, cells were switched to medium A, and the medium was renewed every other day. Mononuclear cells were isolated from a normal volunteer using Lymphoprep (NYCOMED, Roskilde, Denmark), and cells that attached to plastic dishes were cultured in RPMI 1640 medium containing 10% fetal calf serum and used as human monocyte-derived macrophages.
Mice—C57BL/6J and apoE knock-out mice were purchased from Clea (Tokyo, Japan) and The Jackson Laboratories (Bar Harbor, ME), respectively. Mice were maintained and cared for according to the regulations of the Animal Care Committee of the University of Tokyo. For the preparation of atherosclerotic aortas, mice were caged separately with 12-h light/dark cycles and given free access to standard chow diet containing 0.075% cholesterol (MF; Oriental Yeast Co., Ltd.; Osaka, Japan).
Data Base Search and Amino Acid Sequence Analysis—The search for a novel lipase containing lipase consensus motifs and α/β-hydrolase folds was performed in the GENES protein data base of Kyoto Encyclopedia of Genes and Genomes by using the MOTIF search program (22), followed by prediction of secondary structure by PSIPRED and the PREDICT PROTEIN PHD-sec program (23). The search for orthologues or paralogues of NCEH was performed by Sequence Similarity Database in the Kyoto Encyclopedia of Genes and Genomes Database (22). Alignment of the deduced protein sequence with other lipases, calculation of the percent identity, and generation of a phylogenetic tree were performed using the ClustalW program. Prediction of the transmembrane domain was performed using SOSUI and the PREDICT PROTEIN PHDhtm program (23).
cDNA Cloning for the Expression of Recombinant Proteins— Total RNA isolated from murine peritoneal macrophages was reverse-transcribed into cDNA using reverse transcriptase following the manufacturer's protocol (Thermoscript RNaseH-reverse transcriptase (Invitrogen)). The coding sequences of murine NCEH were amplified by PCR from macrophage cDNA using cDNA polymerase (AmpliTaq DNA polymerase (Roche Applied Science)). The primers used for PCR were as follows: murine NCEH forward 5′-CCGCCTGGGGACAATGAG-3′ and murine NCEH reverse 5′-TCTGAGGAGTTTCCTCCGTCA-3′.
The construction of cDNA for HSL was described previously. The products containing the complete open reading frame were ligated into compatible sites of pGEM T easy vector (Promega).
Recombinant Adenovirus for NCEH and HSL Expression (Ad-NCEH, Ad-HSL)—Murine NCEH or HSL cDNA fragment was amplified by PCR from mouse cDNA containing plasmids (described above) using forward and reverse primers. LacZ cDNA was amplified by PCR from SV40 β-galactosidase vector (Promega). The PCR products were ligated to pENTER4 vector (Invitrogen). Subsequently, murine NCEH or HSL cDNA fragment was subcloned into pAd/CMV/V5-DEST vector by Gateway Technology (Invitrogen). The adenoviral vectors were linearized using restriction enzyme and transfected into HEK293 using Superfect reagent (Promega). Large scale production of high titer recombinant Ad-NCEH, Ad-HSL, or Ad-LacZ was performed as described elsewhere (14). The purified viruses were stored in 10% (v/v) glycerol/phosphate-buffered saline (PBS) at -80 °C.
Recombinant Adenovirus Expressing Short Hairpin (sh) RNA Directed against Murine NCEH—Recombinant adenovirus expressing shRNA for NCEH (Ad-shNCEH) was produced by the Gateway Technology (Invitrogen). The sense and antisense templates of NCEH shRNA were produced from nucleotides 136 to 154 of the NCEH coding sequence.
The following oligonucleotides were used: shRNA for NCEH sense template, 5′-GT GAG TAA TCT GAT ACG TTA CGT GTG CTG TCC GTA ATG TAT CAG GTT ACT CAC-3′; shRNA for NCEH antisense template, 5′-GT GAG TAA CCT GAT ACA TTA CGG ACA GCA CAC GTA ACG TAT CAG ATT ACT CAC-3′. An adenovirus containing shRNA for β-galactosidase (Ad-shLacZ) was used as a control.
Murine peritoneal macrophages were seeded in 6-well plates at a cell density of 1.7 × 106 cells/well. After transduction with Ad-shNCEH or Ad-shLacZ for 48 h at 37 °C, cells were harvested, and whole cell lysates were assayed for enzymatic activities.
Generation of Rabbit Polyclonal Antibodies against Murine NCEH—To prepare polyclonal anti-murine NCEH antiserum, amino acid residues containing the catalytic domain of murine NCEH (amino acids 99–250) were expressed in bacteria as a glutathione S-transferase (GST) fusion protein, which was purified by glutathione affinity chromatography, and used for immunization of rabbits according to a standard protocol as described previously (24). Serum of rabbits before immunization was used as a control. From serum samples with high antibody titers, IgG fractions were isolated using a protein G column (GE Healthcare).
Northern Blot Analyses—Total RNA was isolated from various tissues or cultured cells using TRIzol reagent according to the manufacturer's protocol (Invitrogen). RNA (10 μg) was subjected to formaldehyde/agarose gel electrophoresis and blotted to a Hybond-N membrane (GE Healthcare). After cross-linking, mouse NCEH, HSL, and TGH-1 mRNA were detected using 32P-labeled probes. Probes for murine NCEH were originally prepared by reverse transcription-PCR from mouse peritoneal macrophage mRNA. Probes for HSL (exon 8 probe), TGH-1 were constructed from cDNA fragments amplified by reverse transcription-PCR using cDNA obtained from mouse adipose tissue as a template (14). After hybridization and washing, signals were visualized by exposure to a PhosphorImager Screen (FUJIFILM) and analyzed using BASTATION (FUJIFILM) software.
Western Blot Analysis—Cells were sonicated in buffer A and centrifuged at 40,000 or 100,000 × g for 45 min at 4 °C. The supernatant was used as S-40 or S-100 cytosolic fraction, and the precipitates were resuspended and used as the microsomal fraction (14). Ten micrograms of proteins of various cellular fractions were separated on a 10% SDS-PAGE and transferred to a nitrocellulose membrane. For detection of the proteins, the membranes were incubated with either anti-murine NCEH, anti-murine HSL antiserum, monoclonal anti-mouse ACAT1 antibody (25), or anti-F4/80 antibody (AbD serotec) at a dilution of 1:100–1,000. Specifically bound immunoglobulins were detected in a second reaction with a horseradish peroxidase-labeled IgG conjugate and visualized by enhanced chemiluminescence detection (ECL Plus, GE Healthcare) using a Kodak image system, and the intensity of immunoreactive bands were quantified by NIH-image software.
Assays for CE Hydrolase and TG Lipase Activities—Whole cell lysates were prepared from transfected HEK293, murine peritoneal macrophages, or THP1 cells and used for the enzyme activity assays. These cells were sonicated in buffer A (50 mmol/liter Tris-HCl, pH 7.0, 250 mmol/liter sucrose, 1 mmol/liter EDTA, 2 μg/ml leupeptin) and centrifuged at 100,000 × g for 45 min at 4 °C. The supernatant was used as S-100 cytosolic fraction, respectively. The precipitates at 100,000 × g were resuspended and used as the microsomal fraction (14). nCEH activity was measured essentially as described by Hajjar et al. (26), using a reaction mixture containing 6.14 μm cholesterol [1-14C]oleate (48.8 μCi/μmol; 1 μCi = 37 kBq). Triacylglycerol lipase activity was measured according to a modification of the method of Hajjar et al. (26). In brief, the samples were incubated at 37 °C for 30 min in a final volume of 200 μl of a reaction mixture containing 105 μm tri[3H]oleoylglycerol (99.4 μCi/μmol), 23.7 μm lecithin, 12.5 μm sodium taurocholate, 1 m NaCl, and 85 mm potassium phosphate, pH 7.0. The high concentration of NaCl was included to inactivate lipoprotein lipase.
In Vitro Adenovirus Experiments—Two days after THP-1 cells were treated with phorbol 12-myristate 13-acetate, THP-1 macrophages were incubated in RPMI 1640 medium containing 5 mg/ml BSA and transduced with recombinant adenovirus carrying NCEH, HSL, or LacZ as a control. After 24 h, acLDL was added to the medium at a final concentration of 100 μg/ml without or with 1% [14C]oleate-BSA complex (14), and cells were incubated for 48 h at 37 °C. Cellular lipids were extracted using hexane/isopropyl alcohol (3:2), and cholesterol content was determined by enzymatic fluorometric microassay according to the method of Heider and Boyett (27), with minor modifications (25). The radioactivity of intracellular CE formed from [14C]oleate was determined as described (14).
Immunohistochemical Localization—Cells were fixed with 3% paraformaldehyde after washing with PBS. Tissue samples were fixed in 10% formalin, pH 7.0, embedded in paraffin, cut into sections, and placed on glass slides. Tissue samples were deparaffinized and rehydrated before use. The samples were incubated in 3% H2O2 in PBS to quench endogenous peroxidase activity and washed with PBS, and nonspecific binding sites were blocked by incubation with 1% goat serum. The sections were then incubated with affinity-purified anti-NCEH polyclonal antibody. After washing with PBS, the sections were incubated with a biotinylated goat anti-rabbit antibody (1:200, Vector Laboratories, Burlingame, CA). The antibody-specific staining was visualized with a Vecstatin ABC reagent (Vector Laboratories) and diamine benzidine, resulting in a brown precipitate. Cell nuclei were counterstained with methyl green. Combined immunohistochemical staining was performed to identify the cell types expressing NCEH as described previously with minor modifications (28). NCEH was first localized by using the protocol above. Subsequently, the sections were treated with 0.1% pepsin (Roche Applied Science) in 0.1 n HCl for 30 min and incubated with rat monoclonal anti-murine F4/80 (1:10, serotec) for 30 min. The respective sections were incubated with a biotinylated rabbit anti-rat antibody (1:200, Vector Laboratories) for 30 min, followed by a 30-min incubation with an alkaline phosphatase-coupled ABC reagent (1:200, Vector Laboratories). Alkaline phosphatase activity was visualized by using the Alkaline Phosphatase Kit III (Vector Laboratories), resulting in a blue precipitate.
Statistical Analyses—Results are presented as means ± S.E. Student's t test was employed to compare the means. All calculations were performed with STAT view version 5.0 for Macintosh (SAS Institute Inc.).
RESULTS AND DISCUSSION
To search for enzymes with CE hydrolase activity, we screened a gene data base for murine and human proteins with homology to structures in known lipases, i.e. α/β-hydrolase folds (29), the GXSXG active serine motif for serine esterases, and the HG dipeptide motif that is present 70–100 amino acids N-terminal of the catalytic site serine in many lipases (29), yielding 53 candidates that included HSL, lipoprotein lipase, hepatic lipase, endothelial lipase, pancreatic lipase, carboxyl ester lipase, gastric lipase, lysosomal acid lipase, monoacylglycerol lipase, TGH-1, and TGH-2 (21). Thirty three genes had not been previously annotated. We expressed the candidate genes in HEK293 cells for measurement of nCEH activity. Northern blot analysis was used to verify the expression in murine peritoneal macrophages. Besides HSL, only one fulfilled these requirements.
The murine gene for NCEH (NCBI nucleotide entry AK045363) encodes a 408-amino acid protein (GenBank™ accession number NP_848887) with a calculated molecular mass of 45.7 kDa. KIAA1363, which is a human orthologue of murine brain chlorpyrifos oxon-binding protein (30) that was recently identified during our study, is the human orthologue of NCEH and encodes a 440-amino acid protein (GenBank™ accession number NP_065843) with 87.5% identity to the murine NCEH in the C-terminal 408 amino acids. The amino acid sequences of murine NCEH and its closely related proteins, murine, rat, and human HSL, are shown in Fig. 1. The structures of the predicted α-helix and β-sheet are well conserved between NCEH and HSL. NCEH contains domains that are present in HSL such as the catalytic core domain and lipid-binding domain (31, 32), and is thus apparently homologous to HSL in terms of domain structure. Based on the homology with HSL, it can be predicted that Ser-191, Asp-348, and His-378 of the murine NCEH protein form a putative catalytic triad, and His-113 and Gly-114 form an HG oxyanion motif. Despite the remarkably high structural homology between NCEH and HSL, NCEH possesses only 22.1% sequence identity to HSL. In addition to the absence of the regulatory domain of HSL, NCEH lacks the N-terminal region of HSL. The N-terminal region of NCEH instead contains a stretch of 23 hydrophobic amino acids, a putative transmembrane domain, which is absent in HSL, suggesting distinct subcellular localization of these two proteins.
FIGURE 1.
Structure of NCEH. Sequence alignment of murine NCEH with human, rat, and mouse HSL. The deduced amino acid sequence of murine NCEH protein (NCEH) is aligned and compared with murine, rat, and human HSL (murHSL, ratHSL, and humHSL). The first Met residue is numbered as 1. HSL has four functional domains as follows: 1) the N-terminal 300 residues domain, containing the ALBP-binding site (boldface); 2) catalytic core domain (line), containing the HG dipeptide oxyanion motif (shaded box) and GXSXG active serine motif (shaded box); 3) regulatory module domain, containing five serine residues, Ser-557, Ser-559, Ser-591, Ser-650, and Ser-65, which can be phosphorylated by cAMP-dependent protein kinase (solid circle) or other kinases such as AMP-activated protein kinase or extracellular signal-regulated kinase (ERK) (open circle); 4) lipid-binding domain (dashed line), containing a putative lipid-binding site and two of the catalytic triad, i.e. Asp-694 and His-724 (shaded box). The predicted secondary structure was shown as follows: α-helix is indicated by a line; β-sheet is indicated by an arrow; secondary structure of HSL and NCEH is shown in gray and black, respectively. The mature NCEH protein is highly conserved during evolution and has homology to HSL in these motifs and secondary structures, except for the regulatory domain and the N-terminal 300 residue domain. NCEH has an N-terminal transmembrane region (outlined letters). Identical amino acids are shown by asterisks. Amino acids considered as strongly conserved are as follows: STA, NEQK, NHQK, NDEQ, QHRK, MILV, MILF, HY, and FYW, and are indicated by colons. Amino acids considered as weakly conserved are as follows: CSA, ATV, SAG, STNK, STPA, SGND, SNDEQK, NDEQHK, NEQHRK, FVLIM, and HFY, and are indicated by dots.
Analysis using the ClustalW program supported the phylogenetic kinship between NCEH and HSL (supplemental Fig. S1). Neutral lipid hydrolases are clustered essentially into three large groups as follows: 1) an extracellular lipase superfamily comprising lipoprotein lipase, endothelial lipase, hepatic lipase, and pancreatic lipase; 2) a carboxylesterase family comprising carboxyl ester lipase, TGH-1, and TGH-2; and 3) a distinct “intracellular lipase” gene family comprising HSL and NCEH. Lysosomal acid lipase and adipose triglyceride lipase do not belong to any of these families. Arylacetamide deacetylase shows weak similarity (44.0%) to murine NCEH, and there is no other mouse protein with much sequence similarity to murine NCEH.
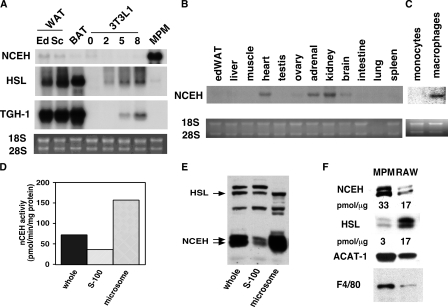
Northern blot analysis revealed that NCEH mRNA was expressed at high levels in peritoneal macrophages and kidney, and to a lesser degree in heart and adrenal tissue (Fig. 2, A and B). Compared with other enzymes with nCEH activity expressed in macrophages, such as HSL and TGH-1, a murine orthologue of CEH reported by Ghosh (18), the expression of NCEH was the most abundant and specific for macrophages (Fig. 2A). Although NCEH expression was barely detectable in freshly isolated human monocytes, it was robustly induced during the differentiation to mature macrophages (Fig. 2C).
FIGURE 2.
Tissue and subcellular distribution of NCEH expression. Total RNA (10 μg) from adipose tissues and 3T3-L1 adipocytes at various stages of differentiation and murine peritoneal macrophages (A), various murine tissues (B), and human monocytes/monocyte-derived macrophages (C) were subjected to Northern blot analysis. Specific mRNAs were detected with radiolabeled murine cDNAs for NCEH, HSL, and TGH-1. Ethidium bromide staining of the gels is shown. Abbreviations used are as follows: WAT, white adipose tissue; BAT, brown adipose tissue; MPM, murine peritoneal macrophages; ed, epididymal; sc, subcutaneous. Subcellular distribution of nCEH activity and NCEH and HSL proteins. MPM were sonicated and centrifuged at 100,000 × g and the supernatant (S-100) or microsomal (Ms) fractions along with whole cell lysate (whole) were subjected to the measurements of nCEH activity (D) and Western blot analysis (E) using anti-HSL and anti-NCEH antisera. F, quantification of NCEH or HSL proteins in macrophages. Ten micrograms of proteins of whole cell lysates from RAW264.7 and MPM were separated by SDS-PAGE on the same gel as the indicated amounts of GST fusion proteins (supplemental Fig. S2). Immunoblotting was performed, and the densities of bands from HSL or NCEH of RAW264.7 or MPM were quantified using NIH Image. Blots for ACAT1 and F4/80 are shown as controls. The moles of HSL or NCEH in 10 μg of protein from RAW264.7 or MPM were calculated from equations relating moles and densities of GST fusion proteins.
To determine the subcellular localization of endogenous nCEH activity and NCEH protein, we isolated S-100 and microsomal fractions from murine peritoneal macrophages, measured the nCEH activity (Fig. 2D), and performed Western blot analysis for either NCEH or HSL (Fig. 2E). The nCEH activity was distributed preferentially in the microsomal fraction. NCEH proteins, duplets with molecular mass of 45 and 50 kDa, were also distributed preferentially in the microsomal fraction. HSL was more equally distributed between the two fractions. Thus, the subcellular distribution pattern of nCEH activity is closer to that of NCEH protein than that of the HSL protein.
We compared the amounts of NCEH protein with those of HSL protein in murine peritoneal macrophages and RAW264.7 cells using recombinant proteins as standards for calculation (Fig. 2F and see supplemental Fig. S2). Here again, two bands were reacted with anti-NCEH antibody. Subsequent study showed that both bands are products of glycation of the 40-kDa protein.4 The ratio of NCEH protein to HSL protein was calculated to be 11 in murine peritoneal macrophages and 1 in RAW264.7 cells, respectively.
The preferential localization of HSL and NCEH in the cytosol and microsomal fractions, respectively, was also confirmed by measurement of the enzymatic activity of each fraction in cells transiently transfected with plasmids expressing HSL or NCEH. Although most of the nCEH activity of HSL-transfected cells was recovered in the S-40 fraction, nCEH activity was barely detectable in the S-40 fraction of NCEH-transfected cells as well as murine peritoneal macrophages (data not shown). This subcellular localization pattern is compatible with the microsomal distribution of NCEH and its preponderance in murine peritoneal macrophages. These results are apparently contrary to those reported by Khoo et al. (33), who showed that 60% of the CE hydrolase activity in the homogenate of J774 cells was recovered in the S-40 fraction; this difference could be attributed to the high level of expression of HSL in J774 cells (34) and the high level of expression of NCEH in murine peritoneal macrophages.
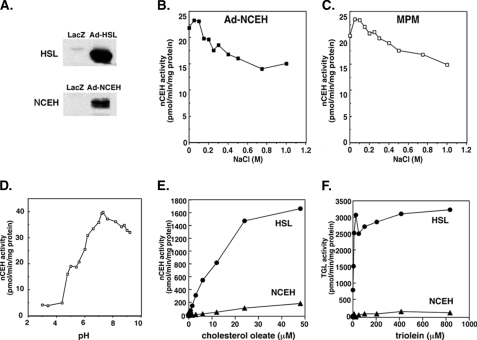
To further determine enzymological characteristics of NCEH, we overexpressed NCEH in HEK293 cells (Fig. 3A) and measured enzymatic activities of their whole cell lysate at various concentrations of NaCl (Fig. 3B), pH (Fig. 3D), and concentrations of cholesterol oleate (Fig. 3E) or triolein (Fig. 3F) as substrates.
FIGURE 3.
Enzymological characteristics of NCEH. HEK293 cells were infected
with Ad-LacZ, Ad-HSL, or Ad-NCEH and used for the experiments. A,
expression of NCEH or HSL was confirmed by Western blot analysis. Effects of
various concentrations of NaCl on the nCEH activity of whole cell lysate from
cells infected with Ad-NCEH (B) or of MPM (C). D,
effects of various pH on the nCEH activity of whole cell lysate from cells
infected with Ad-NCEH using 50 mm acetate buffer (pH  5), 50 mm phosphate buffer (5 < pH < 7.6), or 50
mm Tris-HCl buffer (pH
5), 50 mm phosphate buffer (5 < pH < 7.6), or 50
mm Tris-HCl buffer (pH  7.6). Shown are the effects of
various concentrations of cholesterol oleate (E) or triolein
(F) on the nCEH or triglyceride lipase activities, respectively, of
whole cell lysate from cells infected with Ad-NCEH or Ad-HSL. Values for
Ad-LacZ were subtracted from those for Ad-NCEH or Ad-HSL and plotted against
the concentrations of substrates. Km values were
calculated by fitting lines in Lineweaver-Burk plots.
7.6). Shown are the effects of
various concentrations of cholesterol oleate (E) or triolein
(F) on the nCEH or triglyceride lipase activities, respectively, of
whole cell lysate from cells infected with Ad-NCEH or Ad-HSL. Values for
Ad-LacZ were subtracted from those for Ad-NCEH or Ad-HSL and plotted against
the concentrations of substrates. Km values were
calculated by fitting lines in Lineweaver-Burk plots.
The nCEH activity in the whole cell lysates from NCEH-transfected cells was slightly stimulated by low concentrations of sodium chloride (6.5% at 0.05 m), and inhibited only by 31% at higher concentrations (1 m) (Fig. 3B). This pattern closely resembled that of murine peritoneal macrophages (Fig. 3C). The CE hydrolase activity of NCEH-transfected cells was optimal at neutral pH (7.2) (Fig. 3D), which is close to the optimal pH for HSL, suggesting that both enzymes are not active or at least not fully functional in lysosomes.
The whole cell lysate of HSL-transfected cells exhibited increased hydrolase activities for both TG and CE, with apparent Km of 11.7 and 22.8 μm, respectively. On the other hand, whole cell lysate of NCEH-transfected cells hydrolyzed TG and CE with apparent Km of 30.3 and 324 μm, respectively.
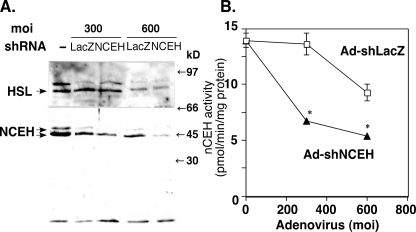
To determine more directly how much of the nCEH activity is accounted for by NCEH in murine peritoneal macrophages, we used RNA silencing technology (Fig. 4). Infection with increasing doses of Ad-shLacZ nonspecifically reduced the protein expression of both HSL and NCEH (Fig. 4A) as well as nCEH activities in the infected cells (Fig. 4B). Whereas infection with 300 and 600 m.o.i. of Ad-shNCEH did not specifically reduce the protein expression of HSL, it reduced the protein expression of NCEH by 51 and 41%, respectively, compared with Ad-shLacZ. In parallel with the inhibition of NCEH, nCEH activities were reduced by 49 and 42% by infection with 300 and 600 m.o.i. of Ad-shNCEH, respectively (Fig. 4B). These results indicate that at least half of the nCEH activity is mediated by NCEH in murine peritoneal macrophages.
FIGURE 4.
Effects of RNA interference of NCEH to inhibit protein expression and nCEH activity in murine peritoneal macrophages. Recombinant adenoviruses coding for shRNA against LacZ (Ad-shLacZ) or NCEH (Ad-shNCEH) were used to infect murine peritoneal macrophages at 300 and 600 m.o.i. Two days after infection, whole cell lysates were subjected to Western blotting (A) and the measurement of nCEH activity (B). This is a representative result of two independent experiments.
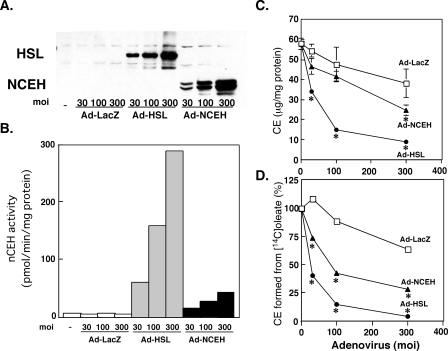
To further examine whether NCEH inhibits accumulation of CE in macrophages, we infected THP-1 cells, which had been induced to differentiate to mature macrophages by incubation with phorbol ester, with Ad-NCEH, Ad-HSL, or Ad-LacZ, and incubated the cells with acetyl-LDL, and compared the amounts of CE in the cells (Fig. 5C) and CE formation from [14C]oleate (Fig. 5D). The cells infected with Ad-HSL or Ad-NCEH expressed HSL or NCEH proteins, respectively, in a dose-dependent manner (Fig. 5A) and showed dose-dependent increases of nCEH activity in whole cell lysates (Fig. 5B). At 300 m.o.i., the nCEH activity in Ad-NCEH-infected cells was 14% that in Ad-HSL-infected cells (Fig. 5B). This increase in nCEH activity in cells overexpressing HSL or NCEH was associated with decreases in the intracellular CE content (Fig. 5C) as well as in the rate of CE formation from [14C]oleate (Fig. 5D). More pronounced inhibition of CE accumulation by Ad-HSL than by Ad-NCEH may be explained by the higher nCEH activity attained by Ad-HSL.
FIGURE 5.
Effects of overexpression of NCEH on cholesterol ester accumulation or cholesterol ester formation in THP-1 macrophages. Recombinant adenoviruses coding for LacZ (Ad-LacZ), HSL (Ad-HSL), or NCEH (Ad-NCEH) were used to infect THP-1 macrophages, and experiments were performed 3 days after infection. The cells were sonicated and used for Western blot analysis (A) and measurement of nCEH activity (B). Twenty four h after THP-1 macrophages were infected with recombinant adenovirus carrying NCEH, HSL, or LacZ as a control, the cells were incubated with 100 μg/ml acLDL. On day 3 after infection, intracellular content of CE (C) and CE formation from [14C]oleate (D) were measured. Data are presented as means ± S.E. of four measurements (C and D) (*, p < 0.001, Ad-HSL versus Ad-LacZ, or Ad-NCEH versus Ad-LacZ).
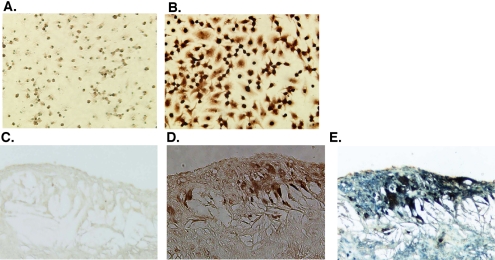
To determine whether NCEH protein is expressed in atherosclerotic lesions, we performed immunohistochemistry (Fig. 6). Cultured peritoneal macrophages were positively stained with anti-NCEH antibody (brown, Fig. 6B) but not with nonimmune IgG (Fig. 6A). Next, we stained tissue sections of aorta from apoE-/- mice (Fig. 6C), which contained advanced atherosclerotic lesions filled with cholesterol cleft. Double staining with F4/80, a macrophage-specific antibody (blue, Fig. 6E), showed prominent expression of NCEH protein (brown, Fig. 6, D and E) in macrophages surrounding the cholesterol cleft in the subintimal space of apoE-/- aorta.
FIGURE 6.
Expression of NCEH in murine peritoneal macrophages and in foamy macrophages in atherosclerotic plaques. Expression of NCEH protein in MPM (A and B) and the aorta of apoE-/- mice (C and D) was localized by immunohistochemistry using affinity-purified anti-NCEH rabbit IgG (visualized in brown) (B and D) or by combined immunohistochemical staining for NCEH (brown) and F4/80 (in blue, for macrophages) (E). Peroxidase activity was visualized using 3,3′-diaminobenzidine (brown) or Vector blue (blue), and sections were counterstained with 3% methyl green. As a control experiment, staining of MPM (A) and the aorta (C) with preimmune rabbit IgG as a primary antibody did not produce any horseradish peroxidase cross-reactivity. Objective magnifications are ×400 (A and B) and ×100 (C–E).
In summary, NCEH is substantially expressed in macrophages in atherosclerotic plaques and significantly contributes to nCEH activity of murine macrophages. This study is the first demonstration of the molecular identity of the dominant nCEH in macrophages, a long unidentified enzyme catalyzing the counter-reaction of acyl-CoA:cholesterol acyltransferase. Although our data do not exclude the possibility that HSL or possibly other enzymes account at least in part for the nCEH activity in macrophages, it is tempting to speculate that HSL evolves to have a more specialized role in adipocyte lipolysis and spermatogenesis (16), whereas NCEH serves in macrophages to hydrolyze CE or possibly other esters to maintain basic macrophage functions as scavengers. As mentioned above, the CEH gene is identical to the TGH gene in humans. Based on the negligible nCEH activity of overexpressed TGH-1 (21) and its marginal expression in murine peritoneal macrophages (Fig. 2A), it is unlikely that CEH plays a more significant role in CE hydrolysis than HSL or NCEH at least in mice. Further studies are needed to define the precise in vivo function of NCEH and its role in the development of atherosclerosis.
Given the relatively high expression of NCEH in human monocyte-derived macrophages (Fig. 2C), it is plausible that NCEH accounts for a major part of the CE hydrolysis in human macrophages, thereby contributing to the development of atherosclerosis. Although HSL (11) and CEH (18) have been reported to be expressed in human macrophages, there is no direct evidence for their relative contribution to the endogenous nCEH activity in human macrophages. It is necessary to clarify which enzymes are more relevant in human macrophages, before translating the current findings to clinical settings. Thorough characterization of these lipases in human macrophages, including relative expression and relative contribution to endogenous nCEH activity, deserves further study and is ongoing in our laboratory. Resolving this controversial issue would pave a way to the development of a new therapy targeted for the prevention and treatment of atherosclerosis.
Supplementary Material
Acknowledgments
We thank Drs. Joseph L. Goldstein, Michael S. Brown, David W. Russell, Hitoshi Shimano, and Nobuhiro Yamada for helpful discussions.
This work was supported by a grant-in-aid for scientific research from the Ministry of Education, Science, and Culture and the Program for Promotion of Fundamental Studies in Health Sciences of the National Institute of Biomedical Innovation and Takeda Science Foundation. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
Footnotes
The abbreviations used are: CE, cholesterol ester; HSL, hormone-sensitive lipase; CEH, cholesteryl ester hydrolase; nCEH, neutral cholesteryl ester hydrolase; Ad, adenovirus; BSA, bovine serum albumin; LDL, low density lipoprotein; acLDL, acetylated LDL; DMEM, Dulbecco's modified Eagle's medium; MPM, murine peritoneal macrophage; shRNA, short hairpin RNA; PBS, phosphate-buffered saline; TGH, triacylglycerol hydrolase; GST, glutathione S-transferase; m.o.i., multiplicity of infection; TG, triacylglycerol.
M. Igarashi, manuscript in preparation.
References
- 1.Fuster, V., Moreno, P. R., Fayad, Z. A., Corti, R., and Badimon, J. J. (2005) J. Am. Coll. Cardiol. 46 937-954 [DOI] [PubMed] [Google Scholar]
- 2.Greaves, D. R., and Gordon, S. (2005) J. Lipid Res. 46 11-20 [DOI] [PubMed] [Google Scholar]
- 3.Chang, T. Y., Chang, C. C., Ohgami, N., and Yamauchi, Y. (2006) Annu. Rev. Cell Dev. Biol. 22 129-157 [DOI] [PubMed] [Google Scholar]
- 4.Brown, M. S., Ho, Y. K., and Goldstein, J. L. (1980) J. Biol. Chem. 255 9344-9352 [PubMed] [Google Scholar]
- 5.Oram, J. F., and Vaughan, A. M. (2006) Circ. Res. 99 1031-1043 [DOI] [PubMed] [Google Scholar]
- 6.Goodman, D. S. (1965) Physiol. Rev. 45 747-839 [DOI] [PubMed] [Google Scholar]
- 7.Holm, C., Kirchgessner, T. G., Svenson, K. L., Fredrikson, G., Nilsson, S., Miller, C. G., Shively, J. E., Heinzmann, C., Sparkes, R. S., Mohandas, T., et al. (1988) Science 241 1503-1506 [DOI] [PubMed] [Google Scholar]
- 8.Holm, C., Osterlund, T., Laurell, H., and Contreras, J. A. (2000) Annu. Rev. Nutr. 20 365-393 [DOI] [PubMed] [Google Scholar]
- 9.Khoo, J. C., Reue, K., Steinberg, D., and Schotz, M. C. (1993) J. Lipid Res. 34 1969-1974 [PubMed] [Google Scholar]
- 10.Small, C. A., Goodacre, J. A., and Yeaman, S. J. (1989) FEBS Lett. 247 205-208 [DOI] [PubMed] [Google Scholar]
- 11.Reue, K., Cohen, R. D., and Schotz, M. C. (1997) Arterioscler. Thromb. Vasc. Biol. 17 3428-3432 [DOI] [PubMed] [Google Scholar]
- 12.Harte, R. A., Hulten, L. M., Lindmark, H., Reue, K., Schotz, M. C., Khoo, J., and Rosenfeld, M. E. (2000) Atherosclerosis 149 343-350 [DOI] [PubMed] [Google Scholar]
- 13.Jepson, C. A., Harrison, J. A., Kraemer, F. B., and Yeaman, S. J. (1996) Biochem. J. 318 173-177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Okazaki, H., Osuga, J., Tsukamoto, K., Isoo, N., Kitamine, T., Tamura, Y., Tomita, S., Sekiya, M., Yahagi, N., Iizuka, Y., Ohashi, K., Harada, K., Gotoda, T., Shimano, H., Kimura, S., Nagai, R., Yamada, N., and Ishibashi, S. (2002) J. Biol. Chem. 277 31893-31899 [DOI] [PubMed] [Google Scholar]
- 15.Escary, J. L., Choy, H. A., Reue, K., and Schotz, M. C. (1998) Arterioscler. Thromb. Vasc. Biol. 18 991-998 [DOI] [PubMed] [Google Scholar]
- 16.Osuga, J., Ishibashi, S., Oka, T., Yagyu, H., Tozawa, R., Fujimoto, A., Shionoiri, F., Yahagi, N., Kraemer, F. B., Tsutsumi, O., and Yamada, N. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 787-792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Contreras, J. A. (2002) Biochem. Biophys. Res. Commun. 292 900-903 [DOI] [PubMed] [Google Scholar]
- 18.Ghosh, S. (2000) Physiol Genomics 2 1-8 [DOI] [PubMed] [Google Scholar]
- 19.Zhao, B., Song, J., Chow, W. N., St Clair, R. W., Rudel, L. L., and Ghosh, S. (2007) J. Clin. Investig. 117 2983-2992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lehner, R., and Vance, D. E. (1999) Biochem. J. 343 1-10 [PMC free article] [PubMed] [Google Scholar]
- 21.Okazaki, H., Igarashi, M., Nishi, M., Tajima, M., Sekiya, M., Okazaki, S., Yahagi, N., Ohashi, K., Tsukamoto, K., Amemiya-Kudo, M., Matsuzaka, T., Shimano, H., Yamada, N., Aoki, J., Morikawa, R., Takanezawa, Y., Arai, H., Nagai, R., Kadowaki, T., Osuga, J., and Ishibashi, S. (2006) Diabetes 55 2091-2097 [DOI] [PubMed] [Google Scholar]
- 22.Kanehisa, M., Goto, S., Kawashima, S., and Nakaya, A. (2002) Nucleic Acids Res. 30 42-46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rost, B., and Sander, C. (1993) J. Mol. Biol. 232 584-599 [DOI] [PubMed] [Google Scholar]
- 24.Okazaki, H., Osuga, J., Tamura, Y., Yahagi, N., Tomita, S., Shionoiri, F., Iizuka, Y., Ohashi, K., Harada, K., Kimura, S., Gotoda, T., Shimano, H., Yamada, N., and Ishibashi, S. (2002) Diabetes 51 3368-3375 [DOI] [PubMed] [Google Scholar]
- 25.Yagyu, H., Kitamine, T., Osuga, J., Tozawa, R., Chen, Z., Kaji, Y., Oka, T., Perrey, S., Tamura, Y., Ohashi, K., Okazaki, H., Yahagi, N., Shionoiri, F., Iizuka, Y., Harada, K., Shimano, H., Yamashita, H., Gotoda, T., Yamada, N., and Ishibashi, S. (2000) J. Biol. Chem. 275 21324-21330 [DOI] [PubMed] [Google Scholar]
- 26.Hajjar, D. P., Minick, C. R., and Fowler, S. (1983) J. Biol. Chem. 258 192-198 [PubMed] [Google Scholar]
- 27.Heider, J. G., and Boyett, R. L. (1978) J. Lipid Res. 19 514-518 [PubMed] [Google Scholar]
- 28.Faber, B. C., Cleutjens, K. B., Niessen, R. L., Aarts, P. L., Boon, W., Greenberg, A. S., Kitslaar, P. J., Tordoir, J. H., and Daemen, M. J. (2001) Circ. Res. 89 547-554 [DOI] [PubMed] [Google Scholar]
- 29.Ollis, D. L., Cheah, E., Cygler, M., Dijkstra, B., Frolow, F., Franken, S. M., Harel, M., Remington, S. I., Silman, I., Schrag, J., et al. (1992) Protein Eng. 5 197-211 [DOI] [PubMed] [Google Scholar]
- 30.Nomura, D. K., Leung, D., Chiang, K. P., Quistad, G. B., Cravatt, B. F., and Casida, J. E. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 6195-6200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kraemer, F. B., and Shen, W. J. (2002) J. Lipid Res. 43 1585-1594 [DOI] [PubMed] [Google Scholar]
- 32.Osterlund, T. (2001) Eur. J. Biochem. 268 1899-1907 [DOI] [PubMed] [Google Scholar]
- 33.Khoo, J. C., Mahoney, E. M., and Steinberg, D. (1981) J. Biol. Chem. 256 12659-12661 [PubMed] [Google Scholar]
- 34.Contreras, J. A., and Lasuncion, M. A. (1994) Arterioscler. Thromb. 14 443-452 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.