Abstract
Brain-derived neurotrophic factor (BDNF) is a member of the neurotrophin family of neurotrophic factors. BDNF has long been recognized to have potential for the treatment of a variety of human neurodegenerative diseases. However, clinical trials with recombinant BDNF have yet to yield success, leading to the suggestion that alternative means of harnessing BDNF actions for therapeutic use may be required. Here we describe an approach to create low molecular weight peptides that, like BDNF, promote neuronal survival. The peptides were designed to mimic a cationic tripeptide sequence in loop 4 of BDNF shown in previous studies to contribute to the binding of BDNF to the common neurotrophin receptor p75NTR. The best of these peptides, the cyclic pentapeptide 2 (cyclo(-d-Pro-Ala-Lys-Arg-)), despite being of low molecular weight (Mr 580), was found to be an effective promoter of the survival of embryonic chick dorsal root ganglion sensory neurons in vitro (maximal survival, 68 ± 3% of neurons supported by BDNF). Pentapeptide 2 did not affect the phosphorylation of either TrkB (the receptor tyrosine kinase for BDNF) or the downstream signaling molecule MAPK, indicating that its mechanism of neuronal survival action is independent of TrkB. NMR studies reveal that pentapeptide 2 adopts a well defined backbone conformation in solution. Furthermore, pentapeptide 2 was found to be effectively resistant to proteolysis when incubated in a solution of rat plasma in vitro. These properties of pentapeptide 2 (low molecular weight, appropriate pharmacological actions, a well defined solution conformation, and proteolytic stability) render it worthy of further investigation, either as a template for the further design of neuronal survival promoting agents or as a lead compound with therapeutic potential in its own right.
BDNF5 is a member of the neurotrophin family of neurotrophic factors, along with nerve growth factor (NGF), neurotrophin (NT)-3 and NT-4/5 (1). These proteins play a key role in shaping the vertebrate nervous system during embryonic development by regulating naturally occurring neuronal death (2). They have also long been touted as having potential for the treatment for human neurodegenerative diseases, due to their neurotrophic effects on specific neuronal populations that are lost in these diseases. BDNF is particularly attractive in this respect, since it has been shown to promote the survival and/or prevent the degeneration of motor neurons (involved in amyotrophic lateral sclerosis) (3), populations of sensory neurons (sensory neuropathies) (4), basal forebrain cholinergic neurons (Alzheimer disease) (5), and dopaminergic neurons of the substantia nigra (Parkinson disease) (6). More recently, it has been demonstrated that BDNF probably plays a specific role in the etiology of Huntington disease, indicating an exciting potential for therapies aimed at replacing BDNF or otherwise mimicking its actions in this condition (7).
The effects of BDNF and the other neurotrophins are produced via two transmembrane receptors: members of the Trk family of receptor tyrosine kinases, and the glycoprotein p75NTR. The binding of a neurotrophin to the appropriate Trk member (NGF to TrkA, BDNF and NT-4 to TrkB, and NT-3 primarily to TrkC) causes stepwise homodimerization and subsequent autophosphorylation of the receptor, leading to initiation of multiple signaling cascades, including those for survival (8). p75NTR, on the other hand, binds all of the neurotrophins with a common low affinity (KD ≈ 10-9 m) but with differing kinetics (9). The precise biological function of p75NTR has yet to be fully elucidated, but there is strong evidence that it is involved in the signaling of apoptosis, as well as participating in interactions with other regulators of neuronal function, such as the Nogo receptor (10). It is also likely that p75NTR combines with Trk members to form the high affinity binding sites (KD ≈ 10-11 m) for neurotrophins found on responsive neurons (11). Thus, there is the opportunity for modulation and cross-talk between the signaling pathways of the Trk family members and p75NTR (12), with the ultimate response to a particular neurotrophin depending on the balance of signaling arising from the two receptors.
Neurotrophins exist as homodimers of ∼2 × 120 residues. Numerous x-ray crystal structures (e.g. see Refs. 13–15) demonstrate that the neurotrophins adopt a common three-dimensional fold, with each monomer consisting of seven longtitudinal β-strands connected by three solvent-exposed hairpin loops (loops 1, 2, and 4) and a longer loop (loop 3). The monomers are further characterized by three fully conserved disulfide bridges arranged in a cystine knot motif, characteristic of this superfamily of growth factors. Unlike most of the other members of the superfamily, however, the neurotrophin monomers in the homodimer are held together by noncovalent interactions only and are arranged in a parallel fashion. The latter feature places the six hairpin loops together at one end of the molecule.
A variety of studies of the interactions of the neurotrophins with their receptors have revealed that residues in the solvent-exposed loops play an important role in mediating contacts with Trk members and p75NTR. For example, replacement of the residues of loop 2 from NGF with the corresponding residues from loop 2 of BDNF gives a molecule able to bind to and activate TrkB (16). In support of this, we have shown that monomeric monocyclic peptides designed to mimic a single loop 2 of BDNF act as antagonists of BDNF-mediated neuronal survival in culture (17). The x-ray crystal structure of the complex of NGF with one of the Ig domains of TrkA shows that residues from loop 1 of NGF make direct contact with the receptor (18). In other site-directed mutagenesis studies, key interactions between neurotrophins and p75NTR have been demonstrated to be localized to three basic residues, distributed between loops 1 and 4 in NGF, NT-3, and NT-4/5, and as a contiguous cationic tripeptide (Lys-Lys-Arg) in loop 4 of BDNF (19, 20).
Despite promising preclinical data in a range of relevant animal models, neurotrophic factors generally have not lived up to their promise in the clinic. For example, recombinant BDNF failed to show demonstrable benefit to patients with amyotrophic lateral sclerosis in a large scale phase III trial (21). It is generally accepted that this failure is likely to have been caused, at least in part, by the poor pharmacokinetic behavior exhibited by recombinant BDNF, in particular a short plasma half-life (t½ of BDNF in rats is less than 1 min) (22). This outcome, along with similar results from trials with other neurotrophic factors, has led to the recognition that alternative means of harnessing the actions of BDNF and other neurotrophic factors for therapeutic use (i.e. other than using recombinant protein) will need to be found.
One way to achieve this would be to develop small molecule mimetics of BDNF. We have previously described a structure-based design approach that has successfully yielded potent mimics of BDNF (23). Bicyclic dimeric peptides, consisting of a pair of disulfide-constrained mimics of loop 2 of BDNF tethered by either a disulfide or amide link, act as BDNF-like partial agonists in vitro, promoting the survival of chick sensory neurons in culture. Incorporation of an additional dimerizing linkage afforded a highly conformationally constrained tricyclic dimer with potent neuronal survival actions in vitro (EC50 = 11 pm). Further examinations with disulfide-constrained cyclic peptides based on loops 1 and 4 of BDNF revealed that generally, monomeric, monocyclic peptides as well as heterodimeric bicyclic peptides are BDNF inhibitors, whereas homodimeric, bicyclic peptides are BDNF-like agonists (24). However, although they are significantly reduced in size compared with BDNF itself, the molecular weight and complex nature of these dimeric peptides would pose considerable challenges for their development as clinically useful agents.
In an effort to obtain compounds with increased potential as drug leads, as well as to further probe the role of the solvent exposed loops of BDNF in mediating its neurotrophic actions, we describe here a series of small, monomeric circular peptides aimed at mimicking the putative p75NTR-binding cationic tripeptide sequence within loop 4 of BDNF. Cyclic peptides incorporating a variety of conformational constraints were designed, and selected compounds were synthesized and examined for (i) their ability to promote the survival of cultures of embryonic chick sensory neurons, (ii) their ability to affect signaling through the receptors for BDNF, and (iii) their susceptibility to proteolytic degradation in rat plasma in vitro. Two-dimensional NMR spectroscopy methods were used to determine the three-dimensional structure of one of these circular peptides in solution.
EXPERIMENTAL PROCEDURES
Molecular Modeling
Peptides were designed with the aid of the molecular modeling software packages Sybyl version 6.4 and Hyperchem version 4.0. The three sequential p75NTR-binding residues in loop 4 of BDNF (Lys94-Lys95-Arg96) were isolated from a model of the three-dimensional structure of BDNF derived via homology modeling techniques (17, 23). Using visual examination, a wide range of means of constraining this tripeptide were explored, from which a selection were built by adding appropriate amino acid(s) to the tripeptide and forming a single bond between the N and C termini. These residues/constraints explored included pairings of Gly, Ala, and Pro (including d-forms), β-amino acids, and other ω-alkyl amino acids of varying lengths. Each molecule thus obtained was then optimized to a local minimum energy conformation using the AMBER molecular mechanics force field implemented in HyperChem. Cyclic peptides that yielded low energy conformations following optimization were then subjected to a more rigorous conformational search using the Conformational Search command in HyperChem, in which backbone torsion angles were varied randomly and the resultant structure was minimized as described above. Unique low energy conformations were stored, whereas high energy or duplicate structures (i.e. root mean square (r.m.s.) deviation of nonhydrogen atoms <0.25 Å or backbone torsion angles within 5° of previously obtained conformations) were discarded. The similarity of the generated conformations to the native tripeptide in BDNF was determined by measuring the r.m.s. deviation of the α- and β-carbon atoms of the Lys-Lys-Arg sequence between the native and cyclic peptides.
Peptide Synthesis
Linear peptides were assembled on chlorotrityl resin from fluorenylmethoxycarbonyl-protected amino acids, using standard solid phase synthesis methods (25) on a PS3 peptide synthesizer (Peptide Technologies, Tucson Arizona). The side chain-protected linear peptides were liberated by treating the resin-bound peptide with acetic acid/trifluoroethanol/dichloromethane (1:1:8). The crude side chain-protected peptides were cyclized by dissolving them in dichloromethane (0.5 mg/ml) and adding O-(7-azabenzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate (2 eq) and diisopropyl-ethylamine (3 eq). The progress of the cyclization reaction was monitored by reverse phase HPLC. The final fully deprotected cyclic peptides (and linear peptides as appropriate) were obtained by treating the corresponding protected peptides in a mixture of trifluoroacetic acid/ethanedithiol/H2O (18:1:1) for 90 min), followed by precipitation in cold ether and purification by HPLC.
Peptide Purification and Characterization
Peptides were purified (and cyclization reactions were monitored) by reverse phase HPLC on analytical (53 × 7 mm C18 Rocket column; Alltech) and semipreparative (250 × 10 mm C18 (Alltech); 250 × 7 mm diphenyl (Vydac)) columns as appropriate. All final peptides eluted as single peaks after purification. The identity of all linear and protected intermediates and final peptides was confirmed by electrospray ionization mass spectrometry (performed either by Dr. Stuart Thomson (Victorian College of Pharmacy, Monash University) or Dr. Nicholas Williamson (ImmunoID Pty. Ltd.)). All peptides (intermediates and final products) gave molecular ions with a mass/charge ratio within 0.1% of calculated values.
Sensory Neuron Cultures
Peptides were examined for effects on neuronal survival in primary cultures of sensory neurons prepared from 8-day-old embryonic chicks as described previously (23). Peptides (1 × 10-8 to 1 × 10-5 m) were added to wells in triplicate 30 min after plating of neurons either alone (for concentration-response studies) or in combination with mouse recombinant BDNF (4 × 10-10 m). BDNF alone was used a positive control. Negative control wells contained neither peptide nor BDNF. After a 48-h incubation (37 °C, 5% CO2), the number of surviving neurons in each well was determined by counting phase bright cells at ×200 magnification in 80 randomly chosen fields (0.25 × 0.25 mm). Data are expressed as percentage of surviving neurons compared with BDNF-only positive controls (typically 30–45% of plated sensory neurons).
TrkB Autophosphorylation Studies
Cell Culture—293 cells were maintained in Dulbecco's modified Eagle's medium with 10% fetal bovine serum, 2 mm l-glutamine, 50 IU/ml penicillin, and 50 mg/ml streptomycin (MultiCel). In preparation for signaling studies, medium was aspirated, and cells were washed in one volume of sterile phosphate-buffered saline and then serum-starved for 4 h in Dulbecco's modified Eagle's medium, l-glutamine, penicillin, and streptomycin.
Generation of TrkB Stable Cell Lines—The trkB gene (a gift from Barbara Hempstead, Cornell University, Ithaca, NY) was subcloned in the pcDNA3 plasmid (Invitrogen) using standard molecular biology. 293 cells were transfected using the Gene-juice transfection reagent (Merck) and selected with 500 mg/ml G418 (A1720; Sigma). Surviving clones were expanded, and whole cell lysates were screened by Western blot for TrkB expression. Low expressing TrkB clones were identified and analyzed for BDNF responsiveness to identify cell lines that expressed nonconstitutively active TrkB receptors and responded to BDNF.
To analyze the effect of BDNF and cyclic pentapeptide 2 on TrkB phosphorylation and signaling, two distinct stable transfectants were analyzed. Briefly, ∼4 × 105 cells were subcultured onto 6-well plates and maintained in full serum overnight. The next day, cells were starved as indicated above and then treated with serum free medium alone or serum-free medium containing BDNF (100 ng/ml) (PeproTech) or pentapeptide 2 (10-5 m). Assays were performed over (i) differing time periods (1, 5, 15, 60, and 180 min), and (ii) differing concentrations of BDNF (1 pg/ml, 10 pg/ml, 100 pg/ml, 1 ng/ml, 10 ng/ml, and 100 ng/ml). Plates were then placed on ice, washed in cold phosphate-buffered saline, and lysed in TNE (Tris, NaCl, EDTA) buffer.
Western Blot Analysis—Cell lysates were incubated on ice for 15 min, centrifuged for 15 min at 4 °C, and cleared. Total protein content was determined using the Bradford method (Sigma). Equal amounts of total protein were reduced in standard Laemmli buffer and boiled, and proteins were separated by SDS-PAGE and transferred to polyvinylidene difluoride (Chemicon). Membranes were blotted for the presence of native proteins with anti-pan-Trk (C-14 sc-11; Santa Cruz Biotechnology), anti-phospho-TrkB (a gift from Moses Chao, New York University) (26), anti-MAPK(Erk1,2) (9102; Cell Signaling Technology), or anti-phospho-MAPK(Erk1,2) (9101; Cell Signaling Technology). Immunocomplexes were detected by the enhanced chemiluminescence technique (Amersham Biosciences) and imaged digitally (LAS-3000; Fujifilm).
NF-κB Luciferase Assay
The p75NTR-dependent NF-κB luciferase assay was recapitulated in 293 cells, following a previously published protocol (29). Briefly, 293 cells were cultured as above, plated at 2.5 × 106 cells/plate in a 100-mm plate, and transfected with an NF-κB reporter construct (catalog number 219078; Stratagene), constitutively active Renilla reporter (pRL-TK, E2241; Promega), Myc-RIP2 (a gift from Moses Chao; New York University), and p75NTR or an empty vector using the calcium phosphate method. After transfection, cells were incubated overnight and then collected and evenly distributed into a 12-well plate for 24 h. Cells were then serum-starved for 4 h and then treated with serum-free medium, serum-free medium containing 100 ng/ml NGF, and serum-free medium containing 100 ng/ml BDNF for 4 h. Cells were lysed and analyzed for luciferase and Renilla activity following the manufacturer's protocols (Dual Luciferase Reporter Assay, catalog number E1960; Promega), using a single tube dual injector luminometer (Lumat LB 9507; Berthold). Conditions were repeated in triplicate, and relative luciferase unit values were normalized to the constitutively active Renilla luminescence for the same sample. Data were normalized such that luciferase activity in the control condition = 1.
NMR Analysis and Structure Determination
NMR spectra of cyclic pentapeptide 2 were acquired on a Varian Inova 400-MHz NMR spectrometer. A lyophilized sample of peptide was dissolved in 550 μl of 10% D2O, and the pH was adjusted to 5.2. One-dimensional 1H spectra were obtained with a sweep width of 4000 Hz over 8192 points. Solvent suppression was achieved with selective low power saturation. Sample one-dimensional spectra were measured at a series of temperatures (30, 15, and 5 °C), and no significant temperature dependence was seen (data not shown). All subsequent spectra were recorded at 15 °C. Two-dimensional spectra were then acquired with a sweep width of 4000 Hz over 1024 points, with 800 t1 increments. TOCSY (28) and DQF-COSY (29) spectra were acquired for use in spin system assignments, whereas ROESY (30) spectra were acquired to generate distance constraints. Spectra were initially transformed using the Varian NMR software package to check for quality of the data. Subsequently, spectra were transformed using NMRpipe (B. A. Johnson; Merck) and analyzed using NMRview. Complete assignment of all nonexchangeable proton resonances relative to an internal TMS standard was made (see supplemental material). Dihedral constraints for the backbone φ angles were derived from 3JNH-CαH coupling constants measured from one-dimensional spectra. A total of 61 structurally important distance constraints and four backbone φ angles were determined.
Structure calculation was carried out using the software package DYANA (31). Cyclization of the peptide was achieved by introducing a set of special distance constraints to bring the N and C termini together and restrain the peptide bond angles to 180°. A modified version of the residue library DYANA containing a set of parameters defining a d-Pro residue was produced to allow calculations to include the d-Pro residue of peptide 2. A total of 100 structures were calculated on the basis of the NMR-derived constraint list using 10,000 steps of simulated annealing followed by 2000 steps of minimization of the DYANA target function. The 20 structures with the lowest target function were then selected as the final family representing the structure of pentapeptide 2.
Plasma Stability Studies
Peptides (circular pentapeptide 2, linear pentapeptide 1, and substance P (as a positive control); 1 mg/ml) were incubated (37 °C) in a solution of freshly prepared rat plasma (50% in phosphate-buffered saline; 200 μl) containing 4-isopropylbenzylacohol (0.05% v/v) as an internal standard ((32)). At various times, samples (10 μl) were removed, and l-lysine monohydrochloride (1000 equivalents in 10 μl) was added to displace peptide from plasma protein binding sites. Plasma proteins were then precipitated by the addition of acetonitrile (60 μl) and removed by centrifugation (4 °C, 10 min, 10,000 × g), and the supernatant (60 μl) was collected and diluted with water (100 μl). The remaining peptide was determined in aliquots (50 μl) using HPLC, by comparing the area of peptide peak and internal standard peaks. Where degradation was seen, one-phase exponential decay curves were fitted (GraphPad Prism software), and the half-life of the peptide was determined. Half-lives were determined in this manner from three independent experiments, which were then averaged and expressed as mean ± S.E. In instances where no degradation was observed, at the final time point, the peptide peak was collected and analyzed by mass spectrometry (i.e. to rule out the possibility of degradation having occurred without a noticeable shift in retention time).
RESULTS
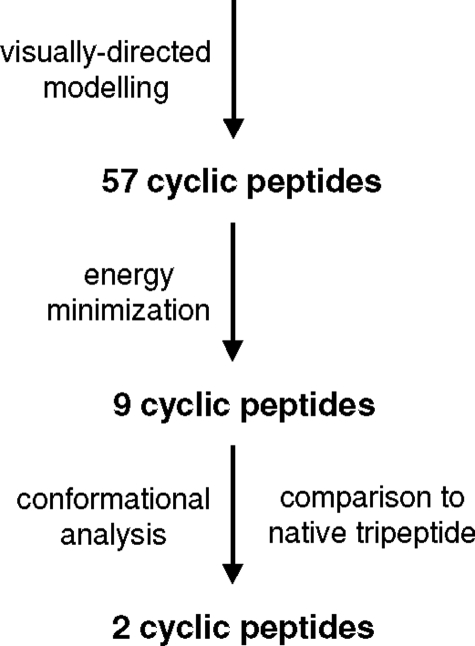
Design and Synthesis of Circular Peptide Mimetics of the p75NTR-binding Motif of BDNF—A computer-aided molecular design approach was used to determine possible means of constraining and presenting the p75NTR-binding tripeptide motif Lys94-Lys95-Arg96 in loop 4 of BDNF as a circular peptide (see Fig. 1). A wide range of constraints was considered, comprising both natural and unnatural amino acids. Using a visually directed approach, 57 peptides were selected and built in silico, and their energy was minimized. From this, we obtained nine compounds with little steric strain, which we thus considered likely to be readily synthetically accessible. Interestingly, seven of these compounds contained a d-Pro residue attached C-terminally to the Arg residue, whereas five contained flexible alkyl ω-amino acids. When subjected to a conformational search, two of these peptides (the d-Pro-containing pentapeptide 2 and the 6-aminohexanoyl-containing tetrapeptide 4) showed considerable similarity to the native tripeptide, in that 70% of their low energy conformations identified were deemed to be similar to the conformation of the native tripeptide (r.m.s. deviation of α and β carbon atoms of tripeptide sequence <0.4 Å).
FIGURE 1.
Schematic summary of molecular modeling approach used to design mimetics of Lys94-Lys95-Arg96 sequence in loop 4 of BDNF.
The circular peptides 2 and 4 were synthesized by standard solid phase peptide synthesis protocols, in which their linear side chain-protected precursors were liberated from acid-labile chlorotrityl resin, cyclized in solution, and deprotected. Analogues of circular pentapeptide 2 (peptides 5 and 6, to allow the importance of the nature of the cyclizing constraint to be examined, and peptides 7 and 8, to allow the role of the Lys residues in the tripeptide motif to be probed) were prepared using a similar strategy. The linear analogues of 2 and 4 (i.e. peptides 1 and 3) were obtained by omitting the cyclization step. All peptides synthesized were purified to homogeneity by HPLC and characterized by mass spectrometry (Table 1). It is interesting to note, however, that the synthesis of several other peptides was attempted (e.g. the Pro-Ala analogue of peptide 2), but these could not be induced to cyclize, despite attempts with a variety of cyclization reagents and conditions. This is probably due to the high degree of conformational rigidity exhibited by these peptides, exemplified by pentapeptide 2 (see below).
TABLE 1.
Sequence, observed and predicted mass spectral data, and maximal neuronal survival effects of peptides synthesized in this study
| Number | Structure | m/z ([M+H]+ predicted) | Maximum survival percentage (at concentration m) |
|---|---|---|---|
| 1 | H-d-Pro-Ala-Lys-Lys-Arg-OH | 598.0 (598.4) | NSa |
| 2 | Cyclo(d-Pro-Ala-Lys-Lys-Arg-) | 580.6 (580.4) | 38 ± 3 (10–6), 68 ± 3 (10–4) |
| 3 | H-Ahxb-Lys-Lys-Arg-OH | 544.0 (543.7) | NS |
| 4 | Cyclo(-Ahx-Lys-Lys-Arg-) | 526.2 (525.7) | 16 ± 3 (10–5) |
| 5 | Cyclo(-Ala-Pro-Lys-Lys-Arg-) | 581.7 (580.4) | NS |
| 6 | Cyclo(-Ala-d-Pro-Lys-Lys-Arg-) | 580.0 (580.4) | NS |
| 7 | Cyclo(-d-Pro-Ala-Ala-Lys-Arg-) | 523.6 (523.3) | 22 ± 0 (10–7) |
| 8 | Cyclo(-d-Pro-Ala-Lys-Ala-Arg-) | 524.4 (523.3) | 16 ± 3 (10–8) |
NS, no significant survival effect observed
Ahx, 6-aminohexanoyl
Effects of p75NTR-binding Motif Mimetics on Sensory Neuron Survival in Vitro—Peptides were examined for their ability to promote the survival of cultures of sensory neurons prepared from day 8 chick embryos. These well characterized cultures are known to contain a subpopulation of neurons that require exogenously added BDNF for their survival (17, 33). Of the compounds examined, the circular pentapeptide 2 produced a robust concentration-dependent survival effect in these cultures (Fig. 2A and Table 1). In multiple experiments with pentapeptide 2, we observed a biphasic response on neuronal survival, with a maximum of 38 ± 3% of the survival observed with BDNF at 10-6 m pentapeptide 2. Increasing the concentration to 10-4 m increased the maximum survival to 68 ± 3%. When added to cultures in combination with a fixed concentration (1 ng ml-1) of BDNF, pentapeptide 2 (at 10-7 and 10-6 m) was found to give a small increase in BDNF-mediated survival (Fig. 2B); however, pentapeptide 2 had no apparent effect on NGF-mediated neuronal survival (Fig. 2B).
FIGURE 2.
Effect of pentapeptide 2 (A) alone and (B) in combination with the neurotrophins BDNF (closed circles) or NGF (open circles) on survival of embryonic chick dorsal root ganglion sensory neurons in culture. BDNF and NGF were used at a concentration of 4 × 10-11 m. Surviving neurons were counted after 48 h in culture. Neuronal survival is expressed as mean ± S.E. from triplicate wells in 3–4 different preparations, after being normalized to survival in neurotrophin-only positive controls (100%) and negative controls (neither peptide nor neurotrophin; 0%).
All other peptides examined gave markedly reduced neuronal survival (Table 1). Substituting the rigid d-Pro-Ala cyclization constraint in pentapeptide 2 with a more flexible 6-aminohexanoyl constraint yielded the circular pentapeptide 4 with a markedly reduced maximal survival response. Peptides 1 and 3, the linear analogues of peptides 2 and 4, were devoid of a significant neuronal survival response, as were the circular peptides 5 and 6, containing other modifications to the cyclization constraint. Replacement of either Lys residue with Ala also yielded circular pentapeptides (7 and 8) with a reduced neuronal survival effect.
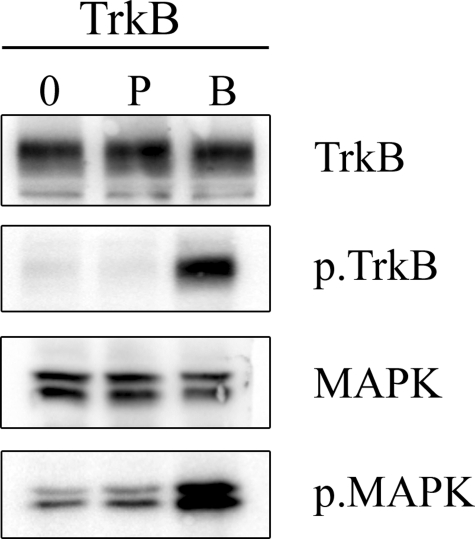
Effects of Circular Pentapeptide 2 on BDNF Receptor Signaling—To try to elucidate the mechanism of the sensory neuron survival of circular pentapeptide 2, we sought to examine its effects on signaling through the BDNF receptors. As expected, treatment of A293 cells stably transfected to express full-length TrkB with BDNF led to a robust increase in expression of the phosphorylated forms of both the BDNF receptor tyrosine kinase TrkB and the downstream signaling effector MAPK (Fig. 3). In contrast, pentapeptide 2 had no effect on the levels of either phosphorylated species. Levels of the unphosphorylated species were unchanged by either BDNF or peptide treatment (Fig. 3).
FIGURE 3.
Representative Western blot examining the effect of vehicle (0), pentapeptide 2 (P) and BDNF (B) on phosphorylation of TrkB and MAPK. Stably transfected 293-TrkB cells were treated with vehicle, pentapeptide 2 (10-5 m), or BDNF (4 × 10-9 m) for 5 min. Equal amounts of cell lysate were then analyzed by Western blot for TrkB (first panel), phosphorylated TrkB (p.TrkB; second panel), MAPK (third panel), and phosphorylated MAPK (p.MAPK; fourth panel).
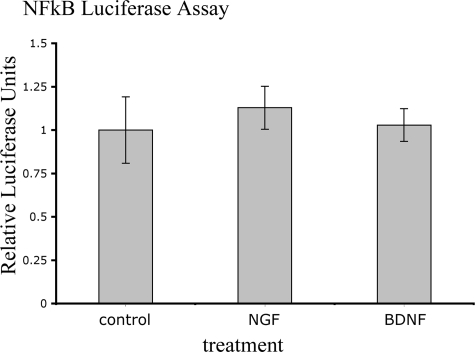
Previously published data have indicated that NGF signaling through p75NTR can activate NF-κB (27, 34–36). To identify whether pentapeptide 2 might be producing its neuronal survival effect by signaling through p75NTR, we attempted to repeat the NF-κB luciferase assay in 293 cells in vitro, as described by Khursigara et al. (27), which indicated that NGF stimulation of p75NTR results in NF-κB activation. However, despite numerous attempts, we were unable to replicate these positive control data. Indeed, our data indicate that neither NGF nor BDNF was able to activate NF-κB above control (vehicle)-treated cells (Fig. 4).
FIGURE 4.
Effect of NGF and BDNF on NF-κB activation in vitro. A293 cells (transiently transfected with an NF-κB luciferase reporter construct, constitutively active Renilla reporter construct, Myc-RIP-2 vector, and p75NTR vector) were treated with either NGF or BDNF (4 × 10-9 m) for 4 h, as previously described (27). The response is expressed as relative luciferase units, in which the luciferase readings have been standardized to the constitutively active Renilla luminescence for each sample and then normalized such that the standardized luciferase activity for the control (neither NGF nor BDNF treatment) is set to 1.
Structure of Circular Pentapeptide 2 in Solution—Complete resonance assignments of all the protons in the molecule were made on the basis of two-dimensional DQF-COSY, TOCSY, and ROESY spectra (see supplemental data). The presence of strong cross-peaks in the ROESY corresponding to interactions between Arg(CαH) and Pro(CδH2) indicates that the proline is in the trans conformation (37). Coupling constants (3JNH-αH) were obtained for all four amide resonances from one-dimensional spectra of the peptide. The measured values were as follows: Ala, 5.5 Hz; Lys-3, 9.8 Hz; Lys-4, ∼4 Hz; Arg, 9.8 Hz, corresponding to φ angles of approximately -110° for Lys3 and Arg and -60° for Ala and Lys4 (37) (see supplemental materials). The presence of well defined backbone angles indicates that there is very little, if any, conformational exchange on the NMR time scale for the backbone of the peptide.
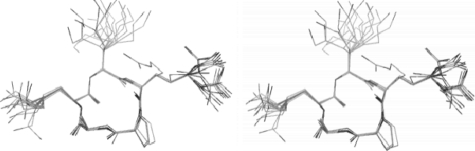
An ensemble of structures was calculated on the basis of distant constraints derived from the ROESY spectra and backbone dihedral constraints obtained from the 3JNH-CaH coupling constants. The resulting structures show a very well defined backbone core (backbone r.m.s. deviation of 0.32 Å) with structural variation restricted to atoms in the side chains of the peptide (Fig. 5). Overall, the physical constraints imposed on motion of the peptide by cyclization appear to have given rise to a peptide with limited conformational flexibility of its cyclic backbone. Even the side chains of the Arg5 and Lys3 residues exhibit restricted mobility out to the γ-carbon and beyond, perhaps as a result of charge-charge-repulsion between the side chain termini.
FIGURE 5.
Stereo image of the ensemble of 20 NMR-derived solution structures of pentapeptide 2.
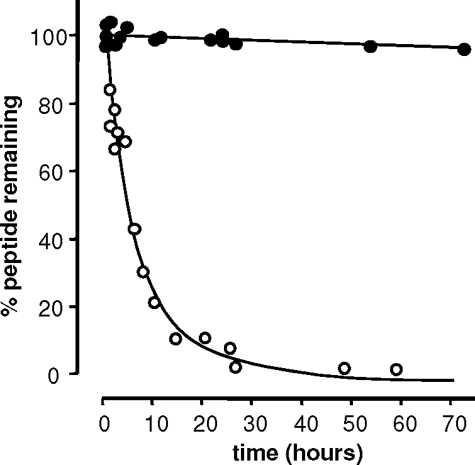
Plasma Proteolytic Stability of p75NTR-binding Motif Mimetics—The proteolytic stability of peptides was assessed by incubating them at 37 °C in a 50% solution of freshly prepared rat plasma in phosphate-buffered saline and assessing the amount of peptide remaining using HPLC (Fig. 6). Under these conditions, circular pentapeptide 2 appeared to be completely resistant to proteolytic degradation over the time period examined (72 h); indeed, it was not possible to determine the half-life of this process. Linear pentapeptide 1 (the acyclic analogue of peptide 2) gave a half-life of 5.4 ± 0.3 h, suggesting that the circular nature of pentapeptide 2 was responsible to a large degree for conferring proteolytic stability on pentapeptide 2. In contrast, the neuropeptide substance P, used as a positive control for plasma proteolytic activity, was rapidly degraded in the rat plasma solution, with a half-life of 22 ± 9 min. This result is consistent with literature data for substance P (t½ = 24 min (38)).
FIGURE 6.
Stability of cyclic pentapeptide 2 (closed circles) and linear pentapeptide 1 (open circles) in rat plasma in vitro. Peptides (1 mg/ml) were incubated at 37 °C in a solution of freshly prepared rat plasma. The percentage of peptide remaining in samples taken at various time points was determined by HPLC, using 4-isopropylbenzyl alcohol as an internal standard.
DISCUSSION
In this paper, we have used a structure-based design approach to develop small monomeric circular peptides aimed at mimicking the putative p75NTR-binding cationic tripeptide sequence within loop 4 of BDNF. Unlike their monomeric monocyclic loop 2 counterparts that are BDNF antagonists, we found that several of these circular pentapeptides act as BDNF-like agonists, promoting the survival of embryonic chick sensory neurons in vitro. The most potent of these compounds, the pentapeptide 2, was found by NMR spectroscopy to adopt a single, well defined backbone conformation in solution and to be proteolytically stable in a solution of rat plasma in vitro. Taken together, these data suggest that pentapeptide 2 offers a novel opportunity for further development either as a neurotrophic drug itself or as a template for the design of nonpeptidic BDNF mimetics.
The recognition that cyclic pentapeptides in general offer potential as privileged drug templates has led to experimental and theoretical studies of the structures of these compounds, including the cyclic pentapeptide cyclo(d-Pro-Ala-Ala-Ala-Ala) (39, 40) an analogue of pentapeptide 2. Our experimental determination of a highly constrained backbone conformation for pentapeptide 2 is in agreement with theoretical studies in DMSO of cyclo(-d-Pro-Ala4-) (35). Molecular dynamics simulations of this peptide sequence in explicit solvent showed that it switched between a major and a minor conformation, occupying the major conformation for up to 99% of the simulation period. In concurrence with this, the NMR spectra we obtained of pentapeptide 2 were consistent with a single, rigidly defined backbone conformation for the peptide. It is interesting to note that preliminary NMR analysis of several of the other peptides in this study yielded spectra indicative of poorly defined structures (data not shown). Comparison of the backbone angles for the ensemble of structures for pentapeptide 2 (see supplemental materials) with those derived from molecular dynamics simulation of DMSO-solvated cyclo(-d-Pro-Ala4-) (Table 1) (40) indicate that, although similar, the structures determined experimentally here in water are not identical to those modeled in DMSO. The differences are perhaps unsurprising, considering the impact of the different solvents and the steric and charge-related effects of the side chains present in pentapeptide 2 compared with the model peptide. Thus, although the backbones of cyclic pentapeptides are likely to be highly rigid (as exemplified by 2), it is likely that the actual conformation is determined at least to some extent by the nature of the side chains in the macrocycle.
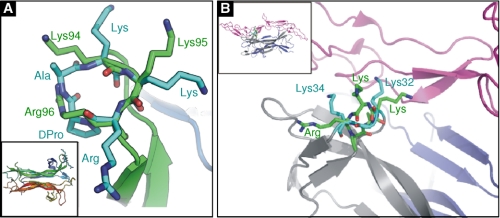
Although there are no high resolution structural data for the BDNF homodimer itself, the crystal structures of heterodimers of BDNF with NT-3 (1BND) (41) and NT-4 (1B8M) (15) have been determined. Loop 4 of BDNF in these structures is effectively identical with a backbone r.m.s. deviation of 0.1 Å, although the orientation of the loop with respect to the rest of the protein is somewhat different (overall backbone r.m.s. deviation between the BDNF monomers in the two structures is 0.71 Å). Comparison of the experimental pentapeptide 2 structure determined here with the conformation of the loop 4 residues Lys94-Lys95-Arg96 of BDNF (Fig. 7A) reveals that although the backbone of the equivalent cyclopeptide residues has the same shape as in BDNF, allowing superimposition of the three corresponding Cα atoms with an r.m.s. deviation of only 0.12 Å, the actual dihedral angles of the residues are quite different. Subsequently, although the two lysine side chains of pentapeptide 2 sit in roughly the same volume of space as Lys94 and Lys95 of BDNF, the Arg96 side chain of BDNF overlaps with the pentapeptide 2 backbone.
FIGURE 7.
A, superimposition of pentapeptide 2 (cyan) on the Lys94-Lys95-Arg96 tripeptide of loop 4 of BDNF (green). Inset, backbone trace of BDNF (green-blue)-NT-3 (red-orange) heterodimer, showing the positions of loop 4 tripeptide residues (sticks, top right). B, superimposition of pentapeptide 2 (green) on Lys32–Lys34 (cyan) of loop 1 of NGF (cyan). The remainder of the NGF backbone is shown in gray-blue; the backbone of the p75NTR structure is shown in pink. Inset, backbone trace of the complex of NGF (gray-blue) and p75NTR (pink), showing the position of the NGF loop 1 residues (sticks, green).
The mechanism by which pentapeptide 2 produces neuronal survival in vitro remains to be fully elucidated. Given that pentapeptide 2 was designed to mimic a putative p75NTR binding region of loop 4 of BDNF, it was not surprising to find that it did not cause phosphorylation of either TrkB or the downstream signaling molecule MAPK. Nevertheless, these findings differ somewhat from the those of Saragovi and co-workers (42), who have showed that monomeric peptides derived from loop 4 of NGF can dimerize and activate TrkA, perhaps via an “inverse antagonism” mechanism, and may well reflect the different modes by which NGF and BDNF (and hence mimetic peptides derived from them) interact with p75NTR (for further discussion, see below). Unfortunately, our attempts to ascertain whether pentapeptide 2 could be acting directly via p75NTR did not prove fruitful, in that we were unable to recapitulate the experiments of others (e.g. see Ref. 27) showing that treatment of cells with neurotrophins could lead to activation of NF-κB via a p75NTR-dependent mechanism. Similarly, the use of 125I-BDNF, a notoriously difficult radioligand to work with, in competition binding experiments failed to give consistent specific binding of even the radioligand alone (data not shown). Nevertheless, the mechanism of action of pentapeptide 2 probably differs from that of loop 2-derived BDNF mimetic peptides previously reported by us; when added to sensory neurons in culture in combination with BDNF, pentapeptide 2 causes a slight promotion of the neuronal survival activity of BDNF. This is in clear contrast to the BDNF loop 2 peptides previously reported by us, in that monomeric loop 2 mimetics (a putative TrkB binding region of BDNF) behave as competitive antagonists, whereas the dimeric loop 2 mimetics show distinct partial agonist activity (i.e. they promote neuronal survival when used alone but act as competitive antagonists of BDNF-mediated survival). Furthermore, pentapeptide 2 does not show the very clear bell-shaped survival response curves seen for the loop 2-based dimeric BDNF mimetics.
Whatever the mechanism of its neuronal survival action, the structural requirements for the neuronal survival activity of pentapeptide 2 appear to be quite stringent. Removal of the d-Pro-Ala cyclization constraint (i.e. linear pentapeptide 1), the inclusion of a more flexible Ahx constraint (cyclic pentapeptide 4), or the introduction of other constraints that would probably induce different conformations on the Lys-Lys-Arg tripeptide (i.e. cyclic pentapeptides 5 and 6 with an Ala-Pro and Ala-d-Pro constraint, respectively) yielded peptides with markedly reduced or no neuronal survival activity. Replacement of either of the Lys residues of pentapeptide 2 with Ala also gave rise to peptides (7 and 8) with reduced neuronal survival activity. It is interesting in this context to note that in the structure of the NGF dimer complexed with p75NTR (43), NGF residues Gly94, Lys95, and Gln96 (which are equivalent to residues Lys94, Lys95, and Arg96 of BDNF) play no real role in the formation of the NGF-p75NTR complex. In contrast, loop 1 of NGF contains residues Lys32 and Lys34, which do contribute to p75NTR binding. (Indeed, it had previously been postulated from mutagenesis studies that Lys32 and Lys34 of NGF were probably the equivalents of Lys94 and Arg96 in BDNF (19).) Superimposition of the pentapeptide 2 structure on this loop in the NGF-p75NTR complex structure does allow Lys3 and Arg5 of the peptide to overlay closely with the positions of Lys32 and Lys34 of NGF (Fig. 7B). Although the significance of this superimposition is difficult to judge (the correspondence of such a short piece of sequence is possible through the limitations imposed by the structure of the peptide backbone alone), together the data suggest that all three charged side chains of pentapeptide 2 are not optimally positioned for maximal activity. Pentapeptide 2 may thus serve as a valid template from which to further explore analogues though a classical medicinal chemistry approach, in an attempt to maximize neuronal survival activity.
Pentapeptide 2 bears some structural resemblance to the αvβ3 integrin antagonist cilengitide (cyclo(-d-Phe-N(Me)Val-Arg-Gly-Asp-)), in that both are proteolytically stable cyclic pentapeptides containing a d-amino acid and are N-methylated at one site (cilengitide at its Val residue, pentapeptide 2 by way of its d-Pro residue). Cilengitide is currently in phase II clinical trials for the treatment of a variety of solid tumors (e.g. see Ref. 44). Whether pentapeptide 2 (or related cyclic pentapeptide analogues of it) might also possess such pharmaceutically acceptable properties that would render it suitable for further clinical development remains an area of further investigation.
Supplementary Material
Acknowledgments
We thank Alison Hunt-Sturman for excellent technical assistance.
The atomic coordinates and NMR data for pentapeptide 2 have been deposited with the Biological Magnetic Resonance Data Bank under accession number 20050.
This work was supported in part by the National Health and Medical Research Council of Australia, the Motor Neurone Disease Association of Australia, Calibre Biotechnology Pty. Ltd., and a publication grant from the University of Melbourne. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2.
Footnotes
The abbreviations used are: BDNF, brain-derived neurotrophic factor; NGF, nerve growth factor; NT, neurotrophin; HPLC, high pressure liquid chromatography; NTR, neurotrophin receptor; r.m.s., root mean square; MAPK, mitogen-activated protein kinase.
References
- 1.Hallbook, F. (1999) Curr. Opin. Neurobiol. 9 616-621 [DOI] [PubMed] [Google Scholar]
- 2.Oppenheim, R. W. (1991) Annu. Rev. Neurosci. 14 453-501 [DOI] [PubMed] [Google Scholar]
- 3.Askanas, V. (1995) Adv. Neurol. 68 241-244 [PubMed] [Google Scholar]
- 4.Lindsay, R. M. (1996) Philos. Trans. R. Soc. London 351 365-373 [DOI] [PubMed] [Google Scholar]
- 5.Siegel, G. J., and Chauhan, N. B. (2000) Brain Res. Brain Res. Rev. 33 199-227 [DOI] [PubMed] [Google Scholar]
- 6.Erickson, J. T., Brosenitsch, T. A., and Katz, D. M. (2001) J. Neurosci. 21 581-589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alberch, J., Perez-Navarro, E., and Canals, J. M. (2004) Prog. Brain Res. 146 195-229 [DOI] [PubMed] [Google Scholar]
- 8.Huang, E. J., and Reichardt, L. F. (2003) Annu. Rev. Biochem. 72 609-642 [DOI] [PubMed] [Google Scholar]
- 9.Rodriguez-Tebar, A., Dechant, G., and Barde, Y. A. (1990) Neuron 4 487-492 [DOI] [PubMed] [Google Scholar]
- 10.Wang, K. C., Kim, J. A., Sivasankaran, R., Segal, R., and He, Z. (2002) Nature 420 74-78 [DOI] [PubMed] [Google Scholar]
- 11.Hempstead, B. L., Martin-Zanca, D., Kaplan, D. R., Parada, L. F., and Chao, M. V. (1991) Nature 350 678-683 [DOI] [PubMed] [Google Scholar]
- 12.Teng, K. K., and Hempstead, B. L. (2004) Cell Mol. Life Sci. 61 35-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McDonald, N. Q., Lapatto, R., Murray-Rust, J., Gunning, J., Wlodawer, A., and Blundell, T. L. (1991) Nature 354 411-414 [DOI] [PubMed] [Google Scholar]
- 14.Butte, M. J., Hwang, P. K., Mobley, W. C., and Fletterick, R. J. (1998) Biochemistry 37 16846-16852 [DOI] [PubMed] [Google Scholar]
- 15.Robinson, R. C., Radziejewski, C., Spraggon, G., Greenwald, J., Kostura, M. R., Burtnick, L. D., Stuart, D. I., Choe, S., and Jones, E. Y. (1999) Protein Sci. 8 2589-2597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ibanez, C. F., Ilag, L. L., Murray-Rust, J., and Persson, H. (1993) EMBO J. 12 2281-2293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O'Leary, P. D., and Hughes, R. A. (1998) J. Neurochem. 70 1712-1721 [DOI] [PubMed] [Google Scholar]
- 18.Wiesmann, C., Ultsch, M. H., Bass, S. H., and de Vos, A. M. (1999) Nature 401 184-188 [DOI] [PubMed] [Google Scholar]
- 19.Ibanez, C. F., Ebendal, T., Barbany, G., Murray-Rust, J., Blundell, T. L., and Persson, H. (1992) Cell 69 329-341 [DOI] [PubMed] [Google Scholar]
- 20.Ryden, M., Murray-Rust, J., Glass, D., Ilag, L. L., Trupp, M., Yancopoulos, G. D., McDonald, N. Q., and Ibanez, C. F. (1995) EMBO J. 14 1979-1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.The BDNF Study Group (Phase III) (1999) Neurology 52 1427-1433 [DOI] [PubMed] [Google Scholar]
- 22.Poduslo, J. F., and Curran, G. L. (1996) Brain Res. Mol. Brain Res. 36 280-286 [DOI] [PubMed] [Google Scholar]
- 23.O'Leary, P. D., and Hughes, R. A. (2003) J. Biol. Chem. 278 25738-25744 [DOI] [PubMed] [Google Scholar]
- 24.Fletcher, J. M., and Hughes, R. A. (2006) J. Pept. Sci. 12 515-524 [DOI] [PubMed] [Google Scholar]
- 25.Chan, W. C., and White, P. D. (eds) (2000) Fmoc Solid Phase Peptide Synthesis: A Practical Approach, Oxford University Press, Oxford
- 26.Arevalo, J. C., Waite, J., Rajagopal, R., Beyna, M., Chen, Z. Y., Lee, F. S., and Chao, M. V. (2006) Neuron 50 549-559 [DOI] [PubMed] [Google Scholar]
- 27.Khursigara, G., Bertin, J., Yano, H., Moffett, H., DiStefano, P. S., and Chao, M. V. (2001) J. Neurosci. 21 5854-5863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Braunschweiler, L., and Ernst, R. R. (1983) J. Magn. Reson. 53 521-528 [Google Scholar]
- 29.Rance, M., Soerensen, O. W., Bodenhausen, G., Wagner, G., Ernst, R. R., and Wuethrich, K. (1983) Biochem. Biophys. Res. Commun. 117 479-485 [DOI] [PubMed] [Google Scholar]
- 30.Bax, A., and Davis, D. G. (1985) J. Magn. Reson. 63 207-213 [Google Scholar]
- 31.Guntert, P., Mumenthaler, C., and Wuthrich, K. (1997) J. Mol. Biol. 273 283-298 [DOI] [PubMed] [Google Scholar]
- 32.Bolin, D. R., Swain, A. L., Sarabu, R., Berthel, S. J., Gillespie, P., Huby, N. J., Makofske, R., Orzechowski, L., Perrotta, A., Toth, K., Cooper, J. P., Jiang, N., Falcioni, F., Campbell, R., Cox, D., Gaizband, D., Belunis, C. J., Vidovic, D., Ito, K., Crowther, R., Kammlott, U., Zhang, X., Palermo, R., Weber, D., Guenot, J., Nagy, Z., and Olson, G. L. (2000) J. Med. Chem. 43 2135-2148 [DOI] [PubMed] [Google Scholar]
- 33.Barde, Y. A., Edgar, D., and Thoenen, H. (1980) Proc. Natl. Acad. Sci. U. S. A. 77 1199-1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bhakar, A. L., Roux, P. P., Lachance, C., Kryl, D., Zeindler, C., and Barker, P. A. (1999) J. Biol. Chem. 274 21443-21449 [DOI] [PubMed] [Google Scholar]
- 35.Carter, B. D., Kaltschmidt, C., Kaltschmidt, B., Offenhauser, N., Bohm-Matthaei, R., Baeuerle, P. A., and Barde, Y. A. (1996) Science (N. Y.) 272 542-545 [DOI] [PubMed] [Google Scholar]
- 36.Kanning, K. C., Hudson, M., Amieux, P. S., Wiley, J. C., Bothwell, M., and Schecterson, L. C. (2003) J. Neurosci. 23 5425-5436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wuthrich, K. (1986) NMR of Proteins and Nucleic Acids, Wiley, New York
- 38.Berger, H., Fechner, K., Albrecht, E., and Niedrich, H. (1979) Biochem. Pharmacol. 28 3173-3180 [DOI] [PubMed] [Google Scholar]
- 39.Heller, M., Sukopp, M., Tsomaia, N., John, M., Mierke, D. F., Reif, B., and Kessler, H. (2006) J. Am. Chem. Soc. 128 13806-13814 [DOI] [PubMed] [Google Scholar]
- 40.Zhang, X., Nikiforovich, G. V., and Marshall, G. R. (2007) J. Med. Chem. 50 2921-2925 [DOI] [PubMed] [Google Scholar]
- 41.Robinson, R. C., Radziejewski, C., Stuart, D. I., and Jones, E. Y. (1995) Biochemistry 34 4139-4146 [DOI] [PubMed] [Google Scholar]
- 42.Maliartchouk, S., Debeir, T., Beglova, N., Cuello, A. C., Gehring, K., and Saragovi, H. U. (2000) J. Biol. Chem. 275 9946-9956 [DOI] [PubMed] [Google Scholar]
- 43.He, X. L., and Garcia, K. C. (2004) Science 304 870-875 [DOI] [PubMed] [Google Scholar]
- 44.Raguse, J. D., Gath, H. J., Bier, J., Riess, H., and Oettle, H. (2004) Oral Oncol. 40 228-230 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.