Abstract
Epithelial-mesenchymal transition (EMT) is important during embryonic cell layer movement and tumor cell invasiveness. EMT converts adherent epithelial cells to motile mesenchymal cells, favoring metastasis in the context of cancer progression. Transforming growth factor-β (TGF-β) triggers EMT via intracellular Smad transducers and other signaling proteins. We previously reported that the high mobility group A2 (HMGA2) gene is required for TGF-β to elicit EMT in mammary epithelial cells. In the present study we investigated the molecular mechanisms by which HMGA2 induces EMT. We found that HMGA2 regulates expression of many important repressors of E-cadherin. Among these, we analyzed in detail the zinc-finger transcription factor SNAIL1, which plays key roles in tumor progression and EMT. We demonstrate that HMGA2 directly binds to the SNAIL1 promoter and acts as a transcriptional regulator of SNAIL1 expression. Furthermore, we observed that HMGA2 cooperates with the TGF-β/Smad pathway in regulating SNAIL1 gene expression. The mechanism behind this cooperation involves physical interaction between these factors, leading to an increased binding of Smads to the SNAIL1 promoter. SNAIL1 seems to play the role of a master effector downstream of HMGA2 for induction of EMT, as SNAIL1 knock-down partially reverts HMGA2-induced loss of epithelial differentiation. The data propose that HMGA2 acts in a gene-specific manner to orchestrate the transcriptional network necessary for the EMT program.
Epithelial-to-mesenchymal transition (EMT)3 is an important process during development but also during tumor progression (1). It converts adherent epithelial cells to motile mesenchymal cells. A key event in the process of EMT is the functional loss of E-cadherin (encoded by CDH1), a key protein maintaining epithelial cell-cell adhesion (2). Transcriptional repression of the CDH1 gene is one of the mechanisms for silencing of E-cadherin. Several transcriptional repressors, such as the zinc finger factors SNAIL1, SNAIL2 (also known as SLUG), ZEB1/δEF1, ZEB2/SIP1, and the basic helix-loop-helix (bHLH) factor E47, have been identified as strong repressors of E-cadherin expression and have been implicated in tumor progression (3). Another bHLH transcription factor, TWIST, has also been implicated with the onset of EMT in mammary epithelial cells and with tumor metastasis (4). Integrated to the function of the bHLH transcriptional repressors is the role of Id (inhibitor of differentiation/DNA-binding) proteins, such as Id2 and Id3, which form complexes with bHLH factors, such as E47, and inhibit the process of EMT (5, 6). Whether Id proteins act as negative regulators of EMT by interfering with the normal action of some of the more recently identified mediators of EMT, such as E2-2 or TWIST, in addition to E47 remains to be elucidated.
The TGF-β family of cytokines strongly promotes the EMT process not only during embryonic development but also during cancer progression (7). For example, TGF-β1 overexpression in keratinocytes of transgenic mice increases EMT and the development of aggressive spindle carcinoma upon exposure to chemical carcinogens (8). Moreover, TGF-β pathway inhibitors block EMT, carcinoma invasiveness, and metastasis in several mouse models of cancer progression (for review, see Ref. 9). Although TGF-β promotes tumorigenesis of late stage carcinomas via induction of EMT, suppression of anti-tumor immune responses, and stimulation of angiogenesis, the same factor acts as a tumor suppressor in normal epithelia and even in early stage adenomas through its ability to inhibit cell growth (9).
TGF-β signals via two distinct receptor serine/threonine kinases, the type I and type II receptors (10). After ligand binding, the type II receptor transphosphorylates the type I receptor, leading to phosphorylation of Smad2 and Smad3, which are known to constantly shuttle between the nucleus and the cytoplasm. Activated Smad2/3 then form complexes with Smad4 and accumulate in the nucleus, where they regulate gene expression in collaboration with specific transcription factors and coactivator/corepressor complexes (10, 11). The TGF-β receptors also activate alternative molecular pathways that provide further signaling specificity, as they operate in parallel or in direct coordination with the Smads (12). Signaling pathways downstream of TGF-β receptors that elicit EMT have recently been elucidated (13); in vitro studies and analyses of tumor growth in mouse models have established that Smads together with additional signaling proteins mediate the EMT response (7, 13).
We have previously described the high mobility group (HMG) protein A2 as a critical factor required for TGF-β-induced EMT in mouse mammary epithelial cells (14). Members of the HMGA family are non-histone chromatin binding factors containing three AT-hooks that enable these proteins to bind to A/T-rich sequences in the minor groove of DNA (15). HMGA proteins are involved in many fundamental cellular processes, such as regulation of gene expression, cell cycle, cellular senescence, differentiation, and viral integration (15). HMGA proteins are highly expressed during embryogenesis but poorly expressed in adult tissues. However, HMGA proteins are overexpressed or fused to other factors in various types of tumors primarily, but not exclusively, of mesenchymal origin. In our previous work we demonstrated that ectopically expressed HMGA2 induces a mesenchymal phenotype characterized by a strong down-regulation of E-cadherin (14). This observation prompted us to dissect the molecular mechanisms by which HMGA2 induces EMT. In the present study we demonstrate that HMGA2 regulates the expression of specific repressors of E-cadherin. We focused on SNAIL1 and demonstrated that HMGA2 cooperates with the TGF-β/Smad signaling pathway during regulation of SNAIL1 gene expression. The mechanism behind this cooperation involves the physical interaction between HMGA2 and the Smads, leading to increased binding of Smad proteins on the SNAIL1 promoter. We also found that SNAIL1 acts as a major effector downstream of HMGA2 for induction of EMT, as SNAIL1 knock-down partially reverses the effect of HMGA2 on mesenchymal differentiation.
MATERIALS AND METHODS
Cells and Reagents—Mouse mammary epithelial NMuMG cell clones overexpressing HA-tagged human HMGA2 (HMGA2-NMuMG) have been described (14). Human hepatocarcinoma HepG2, human embryonic kidney 293T, and green monkey kidney COS1 cells have been described (16). Recombinant mature TGF-β1 was from PeproTech EC Ltd, London, UK.
Cloning and Recombinant Protein Expression—The plasmid pcDNA3-HA-hHMGA2ΔC (amino acids 1–83) was cloned by PCR into the vector pcDNA3-HA C-terminally of the HA tag using the EcoRI and XhoI restriction sites. GST-hHMGA2 and GST-hHMGA2ΔC (1–83) were constructed by digesting pcDNA3-HA-hHMGA2 (14) and pcDNA3-HA-hHMGA2ΔC respectively, at EcoRI and HindIII sites and inserting the HMGA2 fragments into pGEX4T1 (GE Healthcare). The following hHMGA2 deletion mutants were constructed by PCR amplification of GST-hHMGA2 and subcloned into pGEX4T1 at EcoRI and HindIII sites: GST-hHMGA2 ΔAT3 (amino acids (aa) 1–73), GST-hHMGA2 N1 (aa 1–35), GST-hHMGA2 N2 (aa 1–25), GST-hHMGA2 ΔN2 (aa 26–109), GST-hHMGA2 ΔAT1 (aa 35–109), and GST-hHMGA2 C (aa 94–109). The hHMGA2 mutants were verified by DNA sequencing. The pcDNA3-FLAG-Smad2, -Smad3, and -Smad4 as well as pcDNA3–6xmyc-Smad3 full-length, MH1, linker, and MH2 mammalian expression vectors were described before (16). Restriction enzymes and DNA-modifying enzymes used were from New England Biolabs (Ipswich, MA) or Fermentas GmbH (St. Leon-Rot, Germany).
Cell Transfections—The empty vector (mock) and the vector containing the small hairpin RNA against Snail1 (shSnail1) described in Olmeda et al. (17) were transfected into HMGA2-NMuMG cells using Lipofectamine 2000 (Invitrogen). Two days post-transfection, cells were cultured in 1 mg/ml G418, and individual antibiotic-resistant clones were derived.
HepG2 and 293T cells were transiently transfected with calcium phosphate as described (16). When cell extracts were used for DNA affinity precipitation (DNAP) experiments, HepG2 and COS1 cells were transfected, respectively, with FuGENE HD (Roche Applied Science) and Lipofectamine 2000 (Invitrogen) as recommended by the manufacturers.
Reverse Transcription (RT)-PCR—Total DNA-free cellular RNA was extracted with the RNeasy kit (Qiagen AB, Solna, Sweden). RT-PCRs were performed as described (6) and analyzed using specific primers (Ref. 14 and Table 1). Primers for mouse glyceraldehyde-3′-phosphate dehydrogenase (Gapdh) were used as a reference. Lack of DNA contamination was verified by omitting reverse transcriptase (-RT). Quantitative real-time PCR reactions were as described (18). Gene expression levels were determined with the comparative Ct method using Gapdh as a reference. The control condition was set to 1 or 100%, and expression levels are presented as bar graphs of average values plus S.D.
TABLE 1.
Mouse oligonucleotide primers used for quantitative real-time RT-PCR
| Gene | Primer sequence | Strand | Product size | T | Accession number |
|---|---|---|---|---|---|
| bp | °C | ||||
| Snail1 | 5′–CCACTGCAACCGTGCTTTT–3′ | + | 66 | 60 | NM_011427 |
| 5′–CACATCCGAGTGGGTTTGG–3′ | – | ||||
| Snail2 | 5′–CTCACCTCGGGAGCATACAGC–3′ | + | 146 | 60 | NM_011415 |
| 5′–TGAAGTGTCAGAGGAAGGCGGG–3′ | – | ||||
| ZEB1 | 5′–ACAAGACACCGCCGTCATTT–3′ | + | 121 | 60 | NM_011546 |
| 5′–GCAGGTGAGCAACTGGGAAA–3′ | – | ||||
| ZEB2 | 5′–CACCCAGCTCGAGAGGCATA–3′ | + | 101 | 60 | NM_015753 |
| 5′–CACTCCGTGCACTTGAACTTG–3′ | – | ||||
| Twist | 5′–CGGGTCATGGCTAACGTG–3′ | + | 196 | 60 | NM_011658 |
| 5′–CAGCTTGCCATCTTGGAGTC–3′ | – | ||||
| E47 | 5′–CACAGAGACCTCCCGACTCCTA–3′ | + | 101 | 60 | NM_011548 |
| 5′–TGCGATTTCCTCATCCTCTTTC–3′ | – | ||||
| E2-2 | 5′–CTGCCTTAGGGACGGACAAA–3′ | + | 101 | 60 | NM_013685 |
| 5′–CGCCAAAGAAGTTGGTCCAT–3′ | – | ||||
| Gapdh | 5′–TGTGTCCGTCGTGGATCTGA–3′ | + | 76 | 60 | NM_001001303 |
| 5′–CCTGCTTCACCACCTTCTTGA–3′ | – |
Immunoblotting and Immunofluorescence Microscopy—Total protein extracts subjected to SDS-PAGE were analyzed by immunoblotting as described (6). Mouse monoclonal anti-α-tubulin and mouse monoclonal FLAG M5 were from Sigma-Aldrich; mouse monoclonal anti-HA (12CA5) was from Roche Applied Science; mouse monoclonal anti-ZO-1 was from Invitrogen/Zymed Laboratories Inc., Stockholm, Sweden; mouse monoclonal anti-phospho-Smad3 was from Cell Signaling Technology, Inc., Danvers, MA; rabbit monoclonal Smad3 was from Epitomics Inc., Burlingame, CA; mouse monoclonal anti-E-cadherin was from BD Biosciences. Mouse monoclonal anti-Snail1 and rabbit polyclonal anti-CAR were gifts from I. Virtanen (University of Helsinki, Finland) and R. Pettersson (Karolinska Institute, Sweden), respectively. Secondary anti-mouse-IgG and anti-rabbit-IgG coupled to horseradish peroxidase were from GE Healthcare. The enhanced chemiluminescence detection system was from Santa Cruz Biotechnology, Inc., Santa Cruz, CA.
For immunofluorescence cells were treated as indicated in the figure legends, fixed, and stained with TRITC-labeled phalloidin (Sigma-Aldrich) or with mouse anti-ZO-1, rabbit anti-CAR antibodies as primary antibodies, and TRITC-conjugated anti-mouse- or anti-rabbit-IgG antibodies as secondary antibodies (Jackson ImmunoResearch Europe Ltd, Suffolk, UK), as described (6). Photomicrographs were obtained by a Zeiss Axioplan 2 microscope with a Hammamatsu C4742–95 digital camera using the Zeiss Plan-neofluar 40×/0.75 objective lens. For phase-contrast microscopy, live cells growing on the culture dish were analyzed on a Zeiss Axiovert 25 microscope with a Kodak DC290 Zoom digital camera using the Zeiss Achrostigmat 20×/0.35 objective lens. All photography was at ambient temperature in the absence of immersion oil. Primary images were acquired with the camera's QED software. Image memory content was reduced and brightness-contrast was adjusted using Adobe Photoshop 6.0.
Promoter-reporter Assays—HepG2 cells were transiently transfected with calcium phosphate (16). The full-length (-900) and deletion constructs of the mouse Snail1 promoter fused to luciferase have been previously described (19). The triple point mutant m1 (TAA to CGG, see Fig. 3A) mouse Snail1 promoter fused to luciferase was constructed using the -625 promoter as backbone and PCR-based site-specific mutagenesis. The reporter plasmid pCMV-β-Gal was transfected in each condition and used as a reference. The enhanced luciferase assay kit from BD Biosciences Pharmingen was used. Normalized promoter activity data are plotted in bar graphs representing average values from triplicate determinations with S.D. Each independent experiment was repeated at least twice.
FIGURE 3.
HMGA2 binds and increases binding of Smad3 to the Snail1 promoter. A, nucleotide sequences of wild type (wt) and mutant (m1–4) Snail1 promoter probes used for the DNAP experiments. Nucleotide numbers were assigned relative to the ATG codon. A line indicates that the sequence has not been altered. B, binding of HMGA2 (HA-HMGA2) and C-terminal-phosphorylated Smad3 (P-Smad3) to wild type Snail1 promoter probes described in panel A was assessed by DNAP experiments using extracts of transiently transfected HepG2 cells with FLAG-Smad3, FLAG-Smad4, and HA-HMGA2 expression constructs treated (+) or not (-) with 5 ng/ml TGF-β1 for 2 h. The probes used are indicated on the left. TCL, total cell lysates. WB, Western blot. C, binding of HMGA2 to wild type and mutant (m1, m2) Snail1 promoter probes spanning from -131 to -92 described in panel A was assessed by DNAP experiments using extracts of transiently transfected HepG2 cells with the HA-HMGA2 expression construct. D, luciferase reporter assays of wild type or mutant (m1) -625 Snail1 promoter constructs in HepG2 cells transiently transfected with HA-HMGA2 and/or FLAG-Smad3 and FLAG-Smad4 expression constructs as indicated. E, binding of endogenous Smad4 to the Snail1 promoter was assessed by chromatin immunoprecipitation (IP) of NMuMG cells treated (+) or not (-) with 5 ng/ml TGF-β1 for 2 h. ctrl, control. F, binding of phosphorylated Smad3 (P-Smad3) to wild type and mutant (m3, m4) probes spanning -230 to -178 of the Snail1 promoter described in panel A was assessed by DNAP experiments using extracts of HepG2 cells described in panel B.
Chromatin Immunoprecipitation Assays—Chromatin immunoprecipitation assays were performed as described (14). Briefly, the equivalent of 107 cells was used per chromatin immunoprecipitation reaction. Cells were cross-linked using 1% formaldehyde. Cells were lysed, and their DNA was sheared by sonication. After preclearing, the sonicated cell extracts were incubated with 5 μg of rabbit anti-Smad4 antibody (H-552; Santa Cruz Biotechnology Inc., Santa Cruz, CA) or preimmune rabbit antiserum as a negative control. Protein-DNA complexes were precipitated with protein A-Sepharose. Immunoprecipitated complexes were washed and then eluted with 1% SDS, 0.1 m NaHCO3. After purification, immunoprecipitated and input DNA were analyzed by PCR using TaqDNA polymerase (Invitrogen). The primers used were 5′-TCGAATCCTCTGTTTATTCTGTCTGT-3′ and 5′-GGAGCCAGAAAGTGCGATGA-3′ for the Snail1 promoter and 5′-TAATGCGCTTGCCTGAGCTA-3′ and 5′-GCTGTCAAATCGGGCATCA-3′ for the Hmga2 distal promoter.
DNAP Experiments—HepG2 or COS1 cells transiently transfected with the indicated constructs were lysed in buffer containing 0.5% Nonidet P-40, 100 mm EDTA, and 100 mm Tris-HCl, pH 8.0. After preclearing, protein extracts were incubated overnight with biotin-labeled probes described in Fig. 3 and in Table 2. The biotin-labeled probe Snail1 promoter -230/-92 was obtained by PCR amplification using the forward -230/-178 and the reverse -92/-131 probes described above. The biotin-labeled 4× CAGA DNA probe was synthesized in vitro. Streptavidin beads were then added for 1 h followed by 3 washes with lysis buffer and resuspended in SDS loading buffer. Bound proteins were subjected to SDS-PAGE.
TABLE 2.
Snail1 promoter oligonucleotide probes used for DNAP experiments
| Snail1 promoter | Primer sequence | Strand | |
|---|---|---|---|
| –131/–92 | wt | 5′–CCCAGCCCTGGTACTTAAAGGAATTTGCTGCTGCTAGGGG–3′ | + |
| 5′–CCCCTAGCAGCAGCAAATTCCTTTAAGTACCAGGGCTGGG–3′ | – | ||
| m1 | 5′–CCCAGCCCTGGTACTCGGAGGAATTTGCTGCTGCTAGGGG–3′ | + | |
| 5′–CCCCTAGCAGCAGCAAATTCCTCCGAGTACCAGGGCTGGG–3′ | – | ||
| m2 | 5′–CCCAGCCCTGGTACTCGGAGGAGCCTGCTGCTGCTAGGGG–3′ | + | |
| 5′–CCCCTAGCAGCAGCAGGCTCCTCCGAGTACCAGGGCTGGG–3′ | – | ||
| –177/–132 | wt | 5′–TGGTTCAGCCTTGACAAAGGGGCGTGACCAACAGTACGGTCACGCC–3′ | + |
| 5′–GGCGTGACCGTACTGTTGGTCACGCCCCTTTGTCAAGGCTGAACCA–3′ | |||
| –230/–178 | wt | 5′–GCCCAGGCGCACCTGCTCCGGTCTCAGTCTCCGGCCGCGCCGCCGCCAGCCAT–3′ | + |
| 5′–ATGGCTGGCGGCGGCGCGGCCGGAGACTGAGACCGGAGCAGGTGCGCCTGGGC–3′ | |||
| m3 | 5′–GCCCAGGCGCACCTGCTCCGATATCAATATCCGGC–3′ | + | |
| 5′–GCCGGATATTGATATCGGAGCAGGTGCGCCTGGGC–3′ | – | ||
| m4 | 5′–CTCCGATATCAATATCCGGCCGCGCCGCCGCCAGCCAT–3′ | + | |
| 5′–ATGGCTGGCGGCGGCGCGGCCGGATATTGATATCGGAG–3′ | – | ||
| –230/–92 | wt | Sequence identical to the combination of probes –230/–178, –177/–132, and –131/–92 | |
| 4× CAGA promoter | 5′–CAGACAGTCAGACAGTCAGACAGTCAGACAGT–3′ | + | |
| 5′–ACTGTCTGACTGTCTGACTGTCTGACTGTCTG–3′ | – |
GST Pulldown—The recombinant proteins were transformed into BL21 bacteria and purified on glutathione beads using standard techniques (GE Healthcare). The GST fusion proteins (10 μg) were mixed with FLAG-tagged protein expressing cell lysates from transfected 293T cells in lysis buffer with protease inhibitors (20 mm Tris, pH 7.5, 150 mm NaCl, 1% Triton X-100, 1 mm phenylmethylsulfonyl fluoride, and 10 μg/ml aprotinin) at 4 °C for 16 h. The GST beads were washed 4 times in a salt buffer (0.3 m NaCl, 20 mm Tris, pH 7.5, 1% Triton X-100) and once in phosphate-buffered saline. Bound proteins were resolved by SDS-PAGE and detected by immunoblotting. Input of the GST fusion proteins was visualized by Ponceau Red staining. GST pulldown assays were also done with in vitro transcribed-translated [35S]methionine-labeled proteins encoded by pcDNA3-FLAG-Smad3 and pcDNA3-FLAG-Smad4 using the TNT quick coupled in vitro transcription-translation system (Promega Biotech AB, Stockholm, Sweden). Input (2%) and bound proteins were resolved by SDS-PAGE, and the products were visualized by phosphorimaging using a FUJIFILM FLA-3000 unit and its associated software.
RESULTS
HMGA2 Regulates Expression of a Specific Set of Repressors of the E-cadherin Gene—We previously reported that ectopic expression of HMGA2 in mouse mammary epithelial NMuMG cells induces a mesenchymal phenotype characterized by a strong down-regulation of E-cadherin expression (14). The observed E-cadherin down-regulation prompted us to analyze whether HMGA2 regulates key transcriptional repressors of the E-cadherin gene. We previously reported that two members of the mouse Snail family of zinc-finger transcription factors, Snail1, and Snail2 (also known as Slug) were induced in mammary epithelial cells stimulated by TGF-β1 or in the same cells after overexpression of HMGA2 (Fig. 1, A and B and Ref. 14). We extended our analysis to members of the zinc-finger/homeobox domain family, ZEB1 (δEF-1) and ZEB2 (SIP-1), and observed that their expression can be induced by TGF-β1 (Fig. 1C), whereas HMGA2 overexpression was also capable of inducing their levels (Fig. 1D), albeit to a lower degree compared with Snail1 and Snail2 induction (Fig. 1B). We then analyzed expression of the basic helix-loop-helix family of transcription factors (Twist, E47, and E2-2) and observed weak but statistically significant induction of these three genes by TGF-β1 especially at late time points (Fig. 1, E and F), whereas HMGA2 overexpression led to specific and dramatic up-regulation of Twist but no significant regulation of E47 or E2-2 expression levels (Fig. 1G). These experiments demonstrated that HMGA2 regulates the expression of a diverse set of transcription factor genes involved in regulation of E-cadherin transcription.
FIGURE 1.
HMGA2 regulates different E-cadherin transcriptional repressors. Expression levels of mRNA for Snail1 and Snail2 (A), ZEB1 and ZEB2 (C), Twist (E), and E47 and E2-2 (F) was assessed by quantitative RT-PCR in NMuMG cells treated or not with 5 ng/ml TGF-β1 for 2, 8, and 24 h. Expression levels of mRNAs for Snail1 and Snail2 (B), ZEB1 and ZEB2 (D), Twist, E47, and E2-2 (G) was assessed by quantitative RT-PCR in parental NMuMG or in a cell clone of NMuMG expressing constitutively human HMGA2 (HMGA2-NMuMG). Average values are plotted in the bar graphs with S.D. derived from triplicate determinations.
HMGA2 and TGF-β Cooperate to Activate the Snail1 Promoter—Among the seven transcriptional repressors of E-cadherin, Snail1 exhibited immediate-early responses to TGF-β1 and robust up-regulation by HMGA2 (Fig. 1). Moreover, using a specific siRNA against Hmga2, we previously described that HMGA2 was required for full induction of Snail1 expression by TGF-β in mammary epithelial NMuMG cells (Ref. 14 and reproduced in Fig. S1). Thus, we decided to analyze Snail1 gene expression regulation by TGF-β1 and HMGA2 in detail. We previously reported that HMGA2 activated a mouse Snail1 promoter-luciferase reporter containing 900 bp upstream of the ATG codon in human hepatocarcinoma HepG2 cells (Ref. 14 and reproduced in Fig. 2A). We also measured significant Snail1 promoter induction by TGF-β1 in HepG2 cells, which confirms additional previous reports from different cell systems (19, 20). When combined, TGF-β1 and HMGA2 activated the Snail1 promoter to considerably higher levels than each factor alone (Fig. 2A). The TGF-β1 effect on Snail1 promoter induction could be recapitulated by co-expression of Smad3 together with Smad4, which caused a strong induction of the Snail1 promoter (Fig. 2B); however, when expressed alone Smad3 or Smad4 did not activate the Snail1 promoter (Fig. 2B). HMGA2 co-expressed with Smad3 or with Smad4 led to the net induction observed by HMGA2 alone. However, when HMGA2 was co-expressed with both Smad3 and Smad4, a dramatic super-induction of the Snail1 promoter was measured (Fig. 2B). Moreover, the endogenous levels of SNAIL1 protein weakly increased by co-transfection of Smad3 and Smad4 or HMGA2 but were super-activated by the combination of Smad3, Smad4, and HMGA2 in the same human HepG2 cells (Fig. 2C). Activation of the Snail1 promoter by HMGA2 is selective, as HMGA2 was not able to activate the CAGA12-Luc reporter, a classic Smad3/Smad4-dependent reporter (Fig. S2A). However, co-expression of HMGA2 with Smad3 and Smad4 led to a stronger activation of the CAGA12 reporter by Smad3/Smad4 (Fig. S2A). This stronger activation could be explained by a more efficient binding of the Smad3/Smad4 complex to the CAGA12 promoter sequence when HMGA2 is present. Indeed, in DNAP experiments, Smad3 bound to a 4× CAGA probe more efficiently when HMGA2 was co-expressed with Smad3 and Smad4, although HMGA2 itself did not bind to this Smad-specific DNA sequence (Fig. S2B).
FIGURE 2.
HMGA2 and Smad3/Smad4 cooperate to activate the Snail1 promoter. A, luciferase reporter assays of the Snail1 promoter construct in HepG2 cells transiently transfected (+) or not (-) with an HA-HMGA2 expression construct and treated (+) or not (-) with 1 ng/ml TGF-β1 for 24 h. B, luciferase reporter assays of the Snail1 promoter construct in HepG2 cells transiently transfected with FLAG-Smad3, FLAG-Smad4, and HA-HMGA2 expression constructs as indicated. Levels of expression of the transfected proteins were assessed by immunoblot using anti-FLAG and anti-HA antibodies. Stars indicate nonspecific bands. C, immunoblot analysis of endogenous SNAIL1 in HepG2 cells transiently transfected with FLAG-Smad3, FLAG-Smad4, and HA-HMGA2 expression constructs as indicated. α-Tubulin served as a loading control. D, luciferase reporter assays of the indicated deletion constructs of the Snail1 promoter in HepG2 cells transiently transfected with FLAG-Smad3, FLAG-Smad4, and HA-HMGA2 expression constructs as indicated. A schematic representation of potential binding elements for different transcription factors present in the Snail1 promoter and the breakpoints of the deletion constructs used are shown. In all panels normalized luciferase data are plotted in bar graphs as averages with S.D. derived from triplicate determinations.
We then used deletion constructs of the Snail1 promoter to identify the region of the promoter required for its activation by HMGA2 and Smad3/Smad4 (Fig. 2D) (19). Deletion of the promoter up to -170 bp relative to the ATG codon had no effect on its induction by HMGA2 or by Smad3/Smad4. This shorter promoter fragment also exhibited the observed super-induction by the combination of HMGA2 and Smad3/Smad4. In contrast, deletion of the region from -170 to -110 bp abolished its activation by HMGA2 and by Smad3/Smad4 (Fig. 2D). A similar region was identified to be required for induction of the Snail1 promoter by TGF-β1 (Fig. S2C). We, therefore, conclude that HMGA2 and Smad proteins mediate a transcriptional effect on the Snail1 promoter via a short region in its proximal part.
HMGA2 Binds to the Proximal Region of the Snail1 Promoter— We addressed HMGA2 binding to the Snail1 promoter by DNAP experiments using two biotinylated DNA probes encompassing the Snail1 promoter region previously identified to be required for its activation by HMGA2 and Smads (Fig. 3A, probes -131/-92 and -177/-132). HA-HMGA2 from transfected HepG2 cells bound to the probe covering the region between -131 and -92 (Fig. 3B). We identified two A/T-rich sequences as potential HMGA2-binding sites in this region of the Snail1 promoter. We mutated the first one (m1) or both of them (m2) (Fig. 3A) and assessed HMGA2 binding to these mutant probes in DNAP experiments. We observed a reduction of HMGA2 binding to the m1 mutant Snail1 promoter probe; this reduction was further enhanced by additional mutation of the second A/T-rich sequence in mutant probe m2 (Fig. 3C). Moreover, mutation of the first A/T-rich sequence in the context of the -625 Snail1 promoter-luciferase reporter caused a dramatic reduction in the activation by HMGA2 (Fig. 3D). The residual activation of the Snail1 promoter by HMGA2 might be due to a weak binding of HMGA2 to the second A/T-rich sequence. Intriguingly, mutation of the single A/T-rich motif (m1) led to a complete loss of the dose-dependent transactivation of the Snail1 promoter by HMGA2 (Fig. S3A). This strongly suggests that HMGA2 induces the Snail1 promoter activity by cooperative binding at the two A/T-rich motifs identified here. A second major effect of the m1 mutation was a strong reduction in Snail1 promoter activation by Smad3/Smad4 (Fig. 3D). These results demonstrate that HMGA2 binds the Snail1 promoter and that HMGA2-binding sites are required for proper activation of the promoter by Smads. HMGA2 and Smads, therefore, function in a closely interdependent manner on the Snail1 promoter.
Smads Bind Near the HMGA2-binding Sites on the Snail1 Promoter—We then asked whether Smads bind to the Snail1 promoter. Using chromatin immunoprecipitation in NMuMG cells, we demonstrated that Smad4 binds the Snail1 proximal promoter in a TGF-β1-dependent manner, whereas as previously reported by us (14), Smad4 did not bind to the Hmga2 distal promoter fragment used here as a negative control (Fig. 3E).
We then used DNAP experiments to further map the Smad-binding region on the Snail1 promoter. We co-transfected Smad3 and Smad4 in HepG2 cells and treated them with TGF-β1 for 2 h. Using the biotinylated oligonucleotides designed previously to identify the HMGA2-binding sites (Fig. 3A), we observed that phosphorylated Smad3 bound weakly to the -177/-132 probe and did not bind to the -131/-92 probe (Fig. 3B). The weak binding of Smad3 to the -177/-132 probe was slightly increased by co-transfection of HMGA2 and Smad3/Smad4 (Fig. 3B). When scrutinizing the Snail1 promoter sequence for putative Smad binding elements, we could identify a non-consensus Smad binding element and a G/C-rich sequence upstream of the -177/-132 probe (Fig. 3A). We explored whether Smad3 could bind to this sequence by designing an additional biotinylated probe from -230 to -178. Surprisingly, we observed that Smad3 had a greater affinity for this probe than for the -177/-132 probe (Fig. 3B). We designed two mutants (m3 and m4), each being mutated in the non-consensus Smad-binding sites (Fig. 3A). The 3′ G/C-rich sequence was deleted in probe m3, whereas the previously mapped E-box of the mouse Snail1 promoter (Fig. 2D) was deleted in probe m4. Smad3 was still able to bind to probe m4 but not to probe m3, demonstrating that Smad3 bound to the G/C-rich sequence and not to the non-consensus Smad binding element of the proximal Snail1 promoter (Fig. 3F). When HMGA2 was co-transfected with Smad3 and Smad4, once again we observed a significant and reproducible increase of Smad3 binding to the -230/-178 wt or to the m4 probe (Fig. 3, B and F). We also observed Smad3 and HMGA2 bound to a larger fragment of the Snail1 promoter encompassing both Smad- and HMGA2-binding sites identified above, Snail1 promoter -230/-92 (Fig. S3B). We conclude that the Smad3/Smad4 protein complex associates with a site residing about 50 bp from the A/T-rich motifs where HMGA2 binds, thus providing a proximity-based platform for transcriptional cooperation between these proteins on the Snail1 promoter.
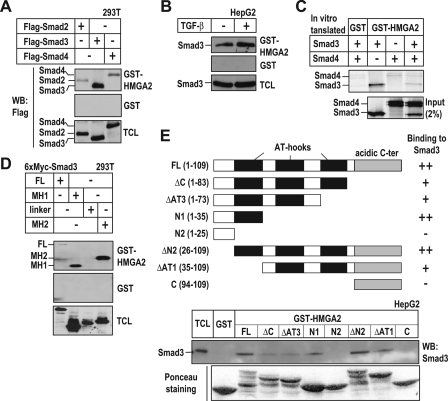
HMGA2 and Smads Form a Complex—The enhancement of Smad3 binding to the Snail1 promoter by HMGA2 and the relative proximity of the binding sites on the promoter DNA suggested a possible interaction between HMGA2 and Smads. To address this possibility, we designed GST pulldown experiments using GST-HMGA2 to affinity-select FLAG-Smad2, -Smad3, or -Smad4 from cell extracts of transiently transfected 293T cells. We observed an interaction between GST-HMGA2 and each of the Smads tested, whereas none of the Smads bound to GST alone (Fig. 4A). HMGA2 interacted stronger with Smad3 than with Smad2 or Smad4. We also observed an interaction between GST-HMGA2 and endogenous Smad3 from HepG2 cells (Fig. 4B) or with in vitro translated Smad3 and Smad4 (Fig. 4C). Using different domains of Smad3, we demonstrated that HMGA2 interacted with the MH1 and the MH2 domains of Smad3 but not with the linker domain (Fig. 4D). We then purified GST fusion proteins of HMGA2 with successive N- or C-terminal deletions (Fig. 4E) and tested their ability to interact with Smad3. We found that the AT-hook motif was important for the interaction of HMGA2 with Smad3 (Fig. 4E). These data support a model in which HMGA2 and Smads cooperate during transcriptional induction of Snail1 by forming ternary complexes and by binding on adjacent sites on the DNA.
FIGURE 4.
HMGA2 interacts with Smads. A, pulldown of 293T cell extracts transfected with FLAG-Smad2, -Smad3, or -Smad4 with purified GST or GST-HMGA2. The input transfected proteins are shown in an immunoblot of the total cell lysate (TCL). WB, Western blot. B, binding of endogenous Smad3 from HepG2 cell extracts treated (+) or not (-) with 2.5 ng/ml TGF-β1 for 2 h to GST or GST-HMGA2 was assessed. C, pulldown of in vitro translated Smad3 and Smad4 with purified GST or GST-HMGA2. Autoradiograms of the bound proteins and 2% of the input of in vitro translated proteins are shown. D, pulldown of 293T cell extracts transfected with domains of 6×Myc-Smad3 as indicated with purified GST or GST-HMGA2. FL, full-length. E, pulldown assay of HepG2 cell extracts with purified GST or deletion mutants of GST-HMGA2. HMGA2 deletion mutants are schematically depicted, and their ability to interact with Smad3 is summarized with +/-. Ponceau staining visualizes the input GST fusion proteins used in the pulldown.
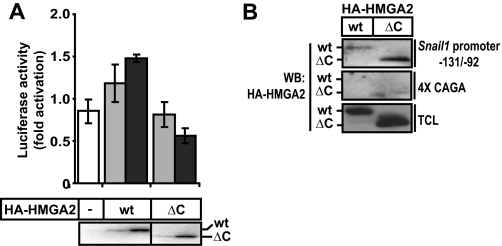
The Acidic C-terminal Domain of HMGA2 Is Required for Snail1 Promoter Activation—Although the previous data demonstrated that the region with the repeated AT-hook units of HMGA2 was required for interaction with Smad3, functional experiments using the same panel of deletion mutants of HMGA2 (Fig. 4E) drew our attention to the C-terminal tail of this protein. Snail1 promoter-reporter assays demonstrated that deletion of the last 27 amino acid residues of HMGA2 that encompass the entire C-terminal acidic domain led to a complete failure to transactivate the promoter (Fig. 5A). Even when the amount of transfected HMGA2 was increased, the Snail1 promoter activity remained insensitive to the C-terminal deletion mutant of HMGA2, whereas the promoter exhibited dose-dependent response to wild-type HMGA2. This result was not due to the lack of DNA binding of the mutant HMGA2 to the Snail1 promoter DNA, as proven by DNAP experiments (Fig. 5B), suggesting the implication of the C-terminal region of HMGA2 in transactivation. In fact the HMGA2 ΔC mutant bound very well to the Snail1 promoter sequence and in a specific manner, as it failed binding to the Smad-specific 4× CAGA promoter DNA (Fig. 5B). We conclude that HMGA2 binds to the Snail1 proximal promoter via its AT-hooks, uses at least one of the three AT-hooks to bind to the Smad3/Smad4 complex, which is also tethered to the same promoter nearby, and finally, that HMGA2 utilizes its C-terminal tail as a transactivation domain to mediate functional signals to the transcriptional machinery.
FIGURE 5.
The acidic C-terminal domain of HMGA2 is required for Snail1 promoter activation. A, luciferase reporter assays of the Snail1 promoter construct in COS1 cells transiently transfected with constructs for HA-HMGA2 wild type (wt) or HA-HMGA2 with its acidic C-terminal domain deleted (ΔC). Luciferase data are plotted as in Fig. 2, and immunoblot of the transfected HMGA2 proteins is shown below the graph. B, binding of wt or ΔC HMGA2 to the Snail1 promoter probe -131/-92 was assessed by DNAP experiments in COS1 cells. DNA binding specificity is shown using the Smad-specific 4× CAGA DNA probe. WB, Western blot; TCL, total cell lysates.
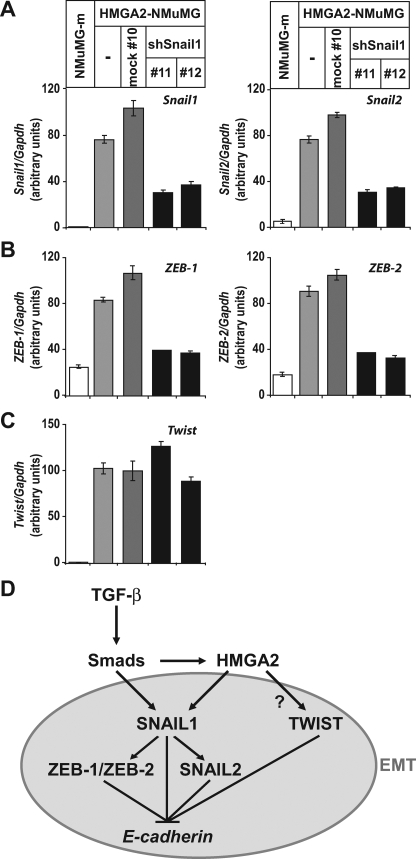
SNAIL1 Is a Major Effector of EMT Downstream of HMGA2— To examine whether the regulation of Snail1 gene expression by HMGA2 and Smads plays any role in the establishment of the mesenchymal phenotype induced by the ectopic expression of HMGA2, we performed RNA-mediated interference experiments. We stably expressed a small hairpin RNA construct against Snail1 (17) or an empty vector (mock#10) in the mesenchymal NMuMG cells stably transfected with HMGA2 (HMGA2-NMuMG cells). We obtained between a 40 and 60% decrease in Snail1 mRNA and protein levels in various stable cell clones (Fig. 6, A and B) (shSnail1#11 and shSnail1#12). Interestingly, shSnail1-expressing cell clones showed a more cuboidal phenotype compared with mock-transfected cell clones that appear as elongated, mesenchymal cells, similar to the parental HMGA2-overexpressing cells (Fig. 6C). In fact, we observed a considerable restoration of the tight junctions (Fig. 6D; ZO1 and CAR staining), a partial reorganization of actin stress fibers to a more cortical architecture of the cytoskeleton (Fig. 6D; phalloidin staining). However, E-cadherin was not relocalized at the cell-cell junctions nor was its expression restored (Fig. 6B). It is possible that another repressor of E-cadherin remained highly expressed and active in the HMGA2-NMuMG cell clones expressing shSnail1 or that the selected cell clones exhibit stable epigenetic silencing of E-cadherin. These results clearly establish that SNAIL1 is an important effector that elicits EMT downstream of HMGA2.
FIGURE 6.
Snai11 depletion partially reverts the mesenchymal phenotype associated with ectopic HMGA2 expression. A, analysis of Snail1 expression by quantitative RT-PCR in HMGA2-NMuMG cells non-transfected (-) and stably transfected with the empty vector (mock #10) or with a construct expressing a short hairpin RNA against Snail1 (shSnail1#11 and shSnail1#12, #11 and #12 being two independent clones). B, analysis of Snail1 and E-cadherin protein levels in cell clones described in panel A. α-Tubulin served as a loading control. NMuMG-m corresponds to a sub-clone of NMuMG stably transfected with the empty pMEP4 vector exhibiting a highly polarized epithelial phenotype. C, phase-contrast microscopy of cells stably transfected with the specific shRNA expressing vectors described in panel A. D, visualization of the epithelial tight junction markers ZO-1 and CAR and of the actin cytoskeleton by immunostaining of the cells stably transfected with the specific shRNA expressing vectors described in panel A. Bars represent 10 μm.
We then analyzed the effect of Snail1 depletion on expression of the other four E-cadherin repressors, whose levels become increased by HMGA2 overexpression, Snail2, ZEB1, ZEB2, and Twist in the same panel of cells (Fig. 7, A–C). Depletion of the induced Snail1 levels by the shRNA led to a concomitant and significant decrease in Snail2, ZEB1, and ZEB2 mRNA levels. In the case of ZEB1 and ZEB2, their expression was decreased almost down to the levels of the parental epithelial cells (NMuMG-m). In contrast, Twist levels remained unaltered by the knock-down of endogenous Snail1 (Fig. 7C). These results demonstrate that Snail1 regulates specifically the expression of Snail2, ZEB1, and ZEB2 but not that of Twist. The data suggest that HMGA2 causes EMT by inducing at least two primary transcriptional mediators of this process, Snail1 and Twist.
FIGURE 7.
Snail1 depletion decreases expression of specific repressors of E-cadherin induced by HMGA2. Expression levels of Snail1 and Snail2 (A), ZEB1 and ZEB2 (B), and Twist (C) were assessed by quantitative RT-PCR in cells described in Fig. 6. The data are plotted as described in Fig. 1. D, model of the molecular mechanism by which TGF-β via Smads induces HMGA2 that then, in collaboration with the Smads, regulates SNAIL1 expression, which in turn regulates additional repressors of E-cadherin to induce EMT. The connection between HMGA2 and TWIST remains unclear (question mark).
DISCUSSION
Intense research activity over the past few years led to a working model whereby a cohort of transcriptional repressors of the E-cadherin gene cooperate to elicit EMT (3). Although such transcriptional repressors that include Snail1, Snail2 (Slug), ZEB1, ZEB2, Twist, E47, and E2-2 all seem to negatively regulate E-cadherin expression, growing evidence suggests that their coordinate requirement during EMT must be due to the fact that these proteins regulate additional important regulators of the epithelial and mesenchymal differentiation programs.
Our study provides a new mechanistic angle to this exciting research field, as we explain how an additional transcription factor, HMGA2, already known to be linked with mesenchymal cell differentiation (15), provides a link between a major morphogenetic pathway, TGF-β, and the network of E-cadherin repressors. The data strongly suggest a hierarchical model of transcription factor mobilization during TGF-β-induced EMT (Fig. 7D). In mammary epithelial cells, TGF-β, via the Smad pathway, induces expression of HMGA2 (14). Then Smads and HMGA2 cooperatively bind to the Snail1 promoter and induce Snail1 expression, E-cadherin repression, and the overall EMT phenotype. However, Snail1 appears not to act in isolation and seems to be required for the induction of at least three more EMT mediators, Snail2, ZEB1, and ZEB2. Whether transcriptional induction of these three repressors requires the continuous input of HMGA2 and Smads remains an interesting open possibility. In parallel to Snail1 induction by Smads and HMGA2, the EMT mediator, Twist, is also induced by a molecular mechanism that is important to dissect in the future (Fig. 7D, question mark). The functional contribution of Snail2, ZEB1, ZEB2, and Twist downstream of HMGA2 may also be important but has not yet been addressed experimentally. We propose that regulation of Twist expression by HMGA2 might explain why E-cadherin levels remained repressed in otherwise epithelial-appearing cell clones with Snail1 knocked down. Alternatively, this could be due to the residual levels of Snail1, which although small, might be capable of exerting a significant degree of E-cadherin repression. Finally, existence of stable epigenetic silencing of the E-cadherin gene (i.e. promoter hypermethylation) in HMGA2-expressing cells cannot be fully disregarded at present.
The fact that Smad signaling induces HMGA2 levels in response to TGF-β and then HMGA2 binds to Smads to regulate Snail1 expression is another example of the feed-forward mechanism used by Smads for the regulation of many of their target genes (11) and which has been termed “self-enabling” mechanism by Massagué and Gomis (10). Our original observation that HMGA2 regulates many transcriptional regulators of the EMT process (Fig. 1) raised the possibility that this nuclear protein might confer some general chromatin reorganization that would target many genes, among which, the specific transcriptional repressors. However, the fact that HMGA2 does not have an impact on the bHLH factors E47 and E2-2 (Fig. 1F), in association with the detailed transcriptional mechanism that we have deciphered on the Snail1 promoter (Figs. 2 and 3), argues that HMGA2 acts as a specific regulator of Snail1 and possibly by extension of Twist and even Snail2, ZEB1, and ZEB2. Such specificity depends on direct DNA binding to specific A/T-rich promoter sequences but also depends on direct binding to transcriptional co-factors, such as the Smad proteins (Fig. 4). The specificity of the intermolecular interaction between Smads and HMGA2 relies on the AT-hook domains of HMGA2 and the two terminal domains of Smads, MH1 and MH2 (Fig. 4). In fact, the AT-hooks that we have mapped as the interaction interface with the Smads also serve as interface with the importins that carry HMGA2 to the nucleus, thus serving as a nuclear localization signal (21). The complexity of the interactions the AT-hook domain of HMGA2 can sustain is well exemplified by its previous analysis in the context of the enhanceosome on the interferon-β enhancer, where it was shown that one AT-hook domain in HMGA2 binds to DNA and to the p50 subunit of transcription factor NF-κB, which leads to transcriptional activation of the interferon-β gene (22). In agreement with this observation, we find that the AT-hook domains bind to Snail1 promoter DNA and also interact with the MH1 and MH2 domains of Smads, leading to transcriptional activation of the Snail1 gene (Figs. 3 and 4). Finally, experiments with the deletion mutant that lacks the C-terminal acidic sequence of HMGA2 (Fig. 5) demonstrated that this domain might play the role of a transactivation domain in the context of the Snail1 promoter. In the case of the Snail1 gene, our evidence (Fig. 5B) does not support a mechanism whereby the C-terminal acidic sequence affects the specificity or strength of DNA binding of HMGA2, as previously demonstrated for the interferon-β gene (22).
In summary, we start deciphering a complex transcriptional program that orchestrates the EMT phenotypic transition downstream of the TGF-β pathway and provide interesting leads for an interconnected network of six embryonic transcription factors, whose re-emergence during cancer progression critically determines the evolution of carcinoma cells toward a mesenchymal phenotype. This study highlights how the Smads induce expression of several transcription factors, interact with the latter, and thus, regulate additional genes in a hierarchical manner to finally establish a complete change in cell differentiation.
Supplementary Material
Acknowledgments
We thank G. Manfioletti for useful discussions during the course of this work and I. Virtanen and R. Pettersson for providing important reagents.
This work was supported by Ludwig Institute for Cancer Research and the Swedish Cancer Society Projects 4855-B03–01XAC and CAN 2006/1078 (to A. M.) and Spanish Ministry of Education and Science Grants SAF2007-63051 and Consolider-Ingenio 2010-26102 (to A. C.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
Footnotes
The abbreviations used are: EMT, epithelial-to-mesenchymal transition; bHLH, basic helix-loop-helix; DNAP, DNA affinity precipitation; GST, glutathione S-transferase; HMG, high mobility group; MH, Mad homology; shRNA, short hairpin RNA; TGF-β, transforming growth factor β; RT, reverse transcription; TRITC, tetramethylrhodamine isothiocyanate; HA, hemagglutinin; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
References
- 1.Thiery, J.-P., and Sleeman, J. P. (2006) Nat. Rev. Mol. Cell Biol. 7 131-142 [DOI] [PubMed] [Google Scholar]
- 2.Peinado, H., Portillo, F., and Cano, A. (2004) Int. J. Dev. Biol. 48 365-375 [DOI] [PubMed] [Google Scholar]
- 3.Peinado, H., Olmeda, D., and Cano, A. (2007) Nat. Rev. Cancer 7 415-428 [DOI] [PubMed] [Google Scholar]
- 4.Yang, J., Mani, S. A., Donaher, J. L., Ramaswamy, S., Itzykson, R. A., Come, C., Savagner, P., Gitelman, I., Richardson, A., and Weinberg, R. A. (2004) Cell 117 927-939 [DOI] [PubMed] [Google Scholar]
- 5.Kondo, M., Cubillo, E., Tobiume, K., Shirakihara, T., Fukuda, N., Suzuki, H., Shimizu, K., Takehara, K., Cano, A., Saitoh, M., and Miyazono, K. (2004) Cell Death Differ. 11 1092-1101 [DOI] [PubMed] [Google Scholar]
- 6.Kowanetz, M., Valcourt, U., Bergström, R., Heldin, C.-H., and Moustakas, A. (2004) Mol. Cell. Biol. 24 4241-4254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zavadil, J., and Böttinger, E. P. (2005) Oncogene 24 5764-5774 [DOI] [PubMed] [Google Scholar]
- 8.Cui, W., Fowlis, D. J., Bryson, S., Duffie, E., Ireland, H., Balmain, A., and Akhurst, R. J. (1996) Cell 86 531-542 [DOI] [PubMed] [Google Scholar]
- 9.Pardali, K., and Moustakas, A. (2007) Biochim. Biophys. Acta 1775 21-62 [DOI] [PubMed] [Google Scholar]
- 10.Massagué, J., and Gomis, R. R. (2006) FEBS Lett. 580 2811-2820 [DOI] [PubMed] [Google Scholar]
- 11.Ross, S., and Hill, C. S. (2007) Int. J. Biochem. Cell Biol. 40 383-408 [DOI] [PubMed] [Google Scholar]
- 12.Moustakas, A., and Heldin, C.-H. (2005) J. Cell Sci. 118 3573-3584 [DOI] [PubMed] [Google Scholar]
- 13.Moustakas, A., and Heldin, C.-H. (2007) Cancer Sci. 98 1512-1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thuault, S., Valcourt, U., Petersen, M., Manfioletti, G., Heldin, C.-H., and Moustakas, A. (2006) J. Cell Biol. 174 175-183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fusco, A., and Fedele, M. (2007) Nat. Rev. Cancer 7 899-910 [DOI] [PubMed] [Google Scholar]
- 16.Morén, A., Hellman, U., Inada, Y., Imamura, T., Heldin, C.-H., and Moustakas, A. (2003) J. Biol. Chem. 278 33571-33582 [DOI] [PubMed] [Google Scholar]
- 17.Olmeda, D., Jordá, M., Peinado, H., Fabra, A., and Cano, A. (2007) Oncogene 26 1862-1874 [DOI] [PubMed] [Google Scholar]
- 18.Valcourt, U., Kowanetz, M., Niimi, H., Heldin, C.-H., and Moustakas, A. (2005) Mol. Biol. Cell 16 1987-2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peinado, H., Quintanilla, M., and Cano, A. (2003) J. Biol. Chem. 278 21113-21123 [DOI] [PubMed] [Google Scholar]
- 20.Cho, H. J., Baek, K. E., Saika, S., Jeong, M. J., and Yoo, J. (2007) Biochem. Biophys. Res. Commun. 353 337-343 [DOI] [PubMed] [Google Scholar]
- 21.Cattaruzzi, G., Altamura, S., Tessari, M. A., Rustighi, A., Giancotti, V., Pucillo, C., and Manfioletti, G. (2007) Nucleic Acids Res. 35 1751-1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Noro, B., Licheri, B., Sgarra, R., Rustighi, A., Tessari, M. A., Chau, K. Y., Ono, S. J., Giancotti, V., and Manfioletti, G. (2003) Biochemistry 42 4569-4577 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.